Abstract
High consumption rates and a multitude of brands make multistate foodborne outbreaks of Salmonella infections associated with chicken challenging to investigate, but whole genome sequencing is a powerful tool that can be used to assist investigators. Whole genome sequencing of pathogens isolated from clinical, environmental, and food samples is increasingly being used in multistate foodborne outbreak investigations to determine with unprecedented resolution how closely related these isolates are to one another genetically. In 2014, federal and state health officials investigated an outbreak of 146 Salmonella Heidelberg infections in 24 states. A follow-up analysis was conducted after the conclusion of the investigation in which 27 clinical and 24 food isolates from the outbreak underwent whole genome sequencing. These isolates formed seven clades, the largest of which contained clinical isolates from a subcluster of case patients who attended a catered party. One isolate from a chicken processed by a large producer was closely related genetically (zero to three single-nucleotide polymorphism differences) to the clinical isolates from these subcluster case patients. Chicken from this large producer was also present in the kitchen of the caterer on the day before the event, thus providing additional evidence that the chicken from this producer was the outbreak source. This investigation highlights how whole genome sequencing can be used with epidemiologic and traceback evidence to identify chicken sources of foodborne outbreaks.
Keywords: Chicken, Foodborne outbreak, Salmonella, Whole genome sequencing
Multistate foodborne outbreaks associated with chicken can be difficult to track to the source for several reasons. Approximately 65% of Americans eat chicken at home during a given week (5), and many others also report eating chicken outside the home. With consumption rates this high, identifying chicken as the likely source of a multistate foodborne outbreak is challenging because traditional epidemiological methods (e.g., comparing foods eaten by ill people with foods eaten by people who were not ill) often fail to identify an association. When clusters of illness caused by Salmonella are identified, it can be difficult to attribute them to chicken because Salmonella is a contaminant of many foods, including chicken (4, 8, 12, 14, 16). Even when epidemiologists suspect chicken, it can be difficult to identify the specific source company because of the large number of chicken brands available, poor recall of the particular chicken brand (or lack of brand loyalty) by persons who were ill, and the fact that many different chicken brands may be produced by the same company. Each of these challenges arose during investigation of a multistate outbreak of Salmonella enterica serovar Heidelberg infections in 2014 in the United States.
Investigators were able to overcome these challenges in large part because of research that continued after the initial investigation. The Centers for Disease Control and Prevention (CDC) Enteric Diseases Laboratory Branch and the U.S. Department of Agriculture Food Safety and Inspection Service (FSIS) performed whole genome sequencing (WGS) on a subset of clinical and food isolates from the outbreak to determine their genetic relatedness (7, 13, 17, 20, 25). FSIS then provided select metadata regarding companies that operated the slaughter and processing facilities where the food samples were collected. This follow-up analysis provided key insights into the probable source of the outbreak.
The present article contains a description of the outbreak investigation and findings from the follow-up WGS and slaughter facility analysis. It also includes a discussion of the implications of these findings for this Salmonella Heidelberg infection outbreak and for chicken-associated Salmonella infection outbreaks more generally.
Materials and Methods
Outbreak investigation
Most foodborne outbreaks are detected and investigated at the local level. Other illness clusters, especially those involving individuals in multiple states, rely on PulseNet, a network of over 80 public health and food regulatory laboratories located throughout the United States, to identify cases and to help investigate potential sources of the infection. PulseNet laboratories use a standardized method, pulsed-field gel electro-phoresis (PFGE), to determine the DNA fingerprint of bacterial foodborne pathogens (6, 19). PulseNet database managers group isolates with indistinguishable PFGE patterns into clusters and notify CDC epidemiologists when the frequency of illnesses with any PFGE pattern exceeds a seasonally adjusted baseline. Among food isolates collected by FSIS during inspection activities, those that are indistinguishable from the clinical isolates by PFGE also are included in the cluster investigation. When a cluster is identified, CDC epidemiologists create a case definition and work with local and state health departments to identify and interview case patients. In this 2014 investigation, a case was defined as infection with the Salmonella Heidelberg strain with the PFGE XbaI pattern JF6X01.0051 isolated on or after 15 May 2014. This pattern accounts for only 4% of Salmonella Heidelberg clinical isolates uploaded to PulseNet each year.
During the hypothesis generation phase of this outbreak investigation, local and state public health officials interviewed case patients about foods and environmental exposures occurring in the 7 days before illness onset. Based on the results of preliminary interviews, a focused questionnaire was developed that included detailed questions about consumption of poultry, egg, tomato, pepper, onion, and fresh herbs, how these foods were packaged, and where and when they were purchased. The proportion of case patients reporting exposure to specific foods was compared with the proportion of healthy persons reporting consumption of the same foods in interviews from the Foodborne Diseases Active Surveillance Network (FoodNet) population survey of healthy adults in the United States conducted during 2006 and 2007 (5). A binomial probability distribution was used to determine which food exposures reported by case patients were significantly higher than those reported by FoodNet population survey respondents.
To provide additional clues regarding the source of this outbreak, health departments also investigated an illness subcluster by conducting a cohort study of persons who were exposed to foods served at a birthday party. Illness subclusters consist of two or more unrelated ill persons who ate or shopped at the same venue around the same time.
When suspected foods were identified during the course of the investigation, state government officials and FSIS conducted traceback investigations into the source of the foods. Product information, such as type of food, brand, and date and location of purchase, was collected. During this investigation, local health department staff also inspected a restaurant implicated in the illness subcluster to (i) identify restaurant practices that may have resulted in cross-contamination or undercooking of food and (ii) identify sources of Salmonella in the facility or its food.
Follow-up analyses
After the conclusion of the initial investigation, the CDC and FSIS technicians used WGS to determine how closely related a subset of the isolates—both clinical and food or animal—were to one another. Twenty-seven clinical isolates were chosen based on state of residence and isolation date. The aim was to select isolates from case patients who lived in different parts of the country and who were infected at different points during the outbreak. Four of these isolates were from case patients identified in the illness subcluster. Eight states were represented: California, Idaho, Minnesota, Nebraska, New York, Pennsylvania, Texas, and Utah. Twenty-four food or animal isolates were selected from 10 companies (13 establishments) that slaughter and/or process chicken. Nineteen of these isolates were collected during the outbreak investigation period, and five were collected in the months before the outbreak was identified.
Genomic DNA was extracted using QIAGEN DNeasy Blood and Tissue Kit (Qiagen, Aarhus, Denmark). The DNA libraries were prepared using a Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA), and the DNA sequencing was performed on a MiSeq Sequencing System (Illumina) using the 2 × 250 bp sequencing chemistry. High-quality single nucleotide polymorphism (hqSNP) analysis was performed using the LYVE-SET pipeline (1). The closed genome of Salmonella Heidelberg SL476 (10) was used as the reference, with prophages removed from the analysis. Read mapping was performed using SMALT, and SNPs were called using VarScan at >20×coverage and .95% read support, and clustered SNPs that were <5 bp apart were filtered out.
A phylogenetic tree was constructed using RAxML (21). Clades containing genetically related isolates were defined based on the SNP counts (<20 SNPs) and bootstrap values (>75). Isolates within a clade were considered genetically highly related when the SNP difference was <5. Each of the main clades in the tree was given a numeric identifier to simplify description and discussion. Once the phylogenetic tree was produced, FSIS provided metadata for the food isolates that were sequenced, including presumed corporate ownership information, which was deidentified and coded alphabetically in the phylogenetic tree.
Results
Outbreak investigation
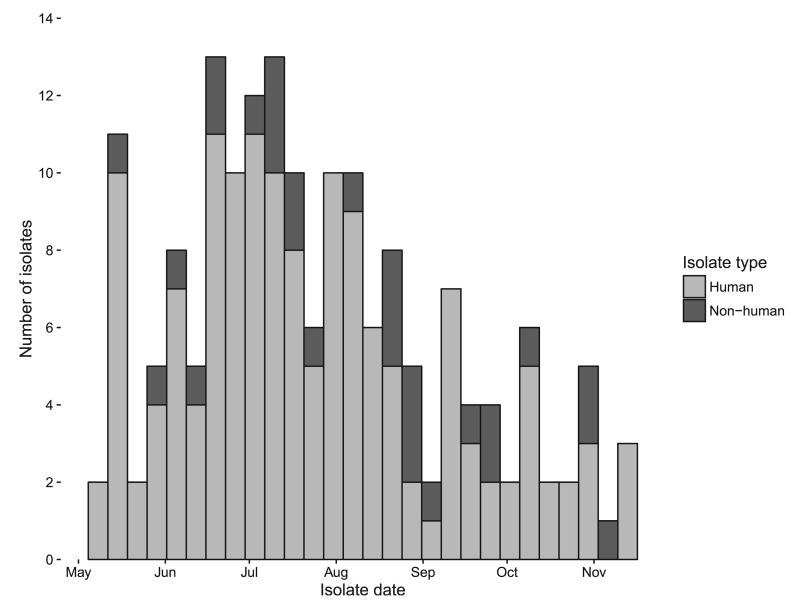
From 15 May through 15 December 2014, 146 salmonellosis cases were identified by PulseNet in 24 states (Figs. 1 and 2). Forty-six percent (63 of 138) of case patients were female, and their median age was 26 years (range, <1 to 92 years). Forty-two (47%) of 89 case patients were hospitalized, and no deaths were reported. The majority of isolates were from stool (110 of 145, 76%), with the remainder from blood (25 of 145, 17%) and from urine (10 of 145, 7%) (Table 1).
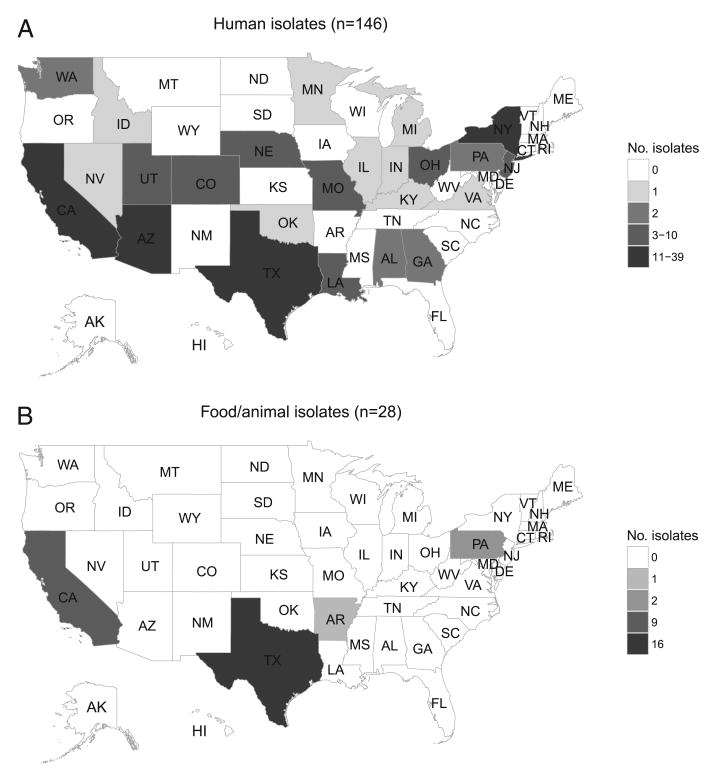
Figure 1.
Human (A) and food or animal (B) isolates by state of origin, Salmonella Heidelberg outbreak, United States, 2014.
Figure 2.
Epidemic curve by week, Salmonella Heidelberg outbreak, United States, 2014.
Table 1. Patient demographics and isolate type and source, Salmonella Heidelberg outbreak, United States, 2014.
| Variable | n | % |
|---|---|---|
| Sex (n = 146) | ||
| Female | 63 | 43 |
| Male | 75 | 51 |
| Unknown | 8 | 5 |
| Age (yr) (n = 146) | ||
| <1–10 | 45 | 31 |
| 11–20 | 16 | 11 |
| 21–40 | 31 | 21 |
| 41–60 | 25 | 17 |
| 61–80 | 17 | 12 |
| 81–100 | 4 | 3 |
| Unknown | 8 | 5 |
| Hospitalized (n = 89) | 42 | 47 |
| Died (n = 93) | 0 | 0 |
| Isolate type (n = 174) | ||
| Human | 146 | 84 |
| Food or animal | 28 | 16 |
| Isolate source | ||
| Human (n = 146) | ||
| Blood | 25 | 17 |
| Stool | 110 | 75 |
| Urine | 10 | 7 |
| Unknown | 1 | <1 |
| Food or animal (n = 28) | ||
| Beef | 1 | 4 |
| Chicken | 26 | 93 |
| Cecum (chicken) | 1 | 4 |
| No. of states involved | ||
| Human isolates | 24 | |
| Food isolates | 4 |
During the outbreak period, PulseNet also identified 27 isolates from food samples and 1 isolate from a chicken cecal sample with PFGE patterns indistinguishable from that of the outbreak strain (Fig. 2 and Table 1). Nearly all of the food isolates (26 of 27, 96%) were from slaughtered chicken, but one was from a beef sample from a facility that processed both beef and chicken. These isolates were collected during routine sampling in FSIS-regulated establishments (Fig. 1).
When local and state epidemiologists collected preliminary exposure information from case patients, they identified chicken and tomatoes as common food exposures. Sixty percent (39 of 65) of case patients reported eating chicken outside of the home (e.g., at a restaurant) in the 7 days preceding illness, a percentage significantly higher (P = 0.02) than expected (47%) according to the FoodNet population survey. Case patients also frequently consumed fresh tomatoes (44 of 70, 63%) and chicken (47 of 69, 68%) at home, but these proportions were not significantly higher than expected (Table 2). Thirty-two case patients were interviewed using a focused questionnaire that included more detailed questions on chicken exposure, but none of the exposures were significantly higher than expected, including those related to chicken consumption (Table 2).
Table 2. Case patient food exposures by investigation questionnaire and Foodborne Diseases Active Surveillance Network population survey, Salmonella Heidelberg outbreak, United States, 2014a.
| Question | Responses | Binomial P value | |||
|---|---|---|---|---|---|
|
| |||||
| Total | No. “yes” | % “yes” | % “yes” from FoodNet data | ||
| Preliminary questionnaire | |||||
| Chicken prepared and/or eaten at home | 69 | 47 | 68 | 65 | 0.34 |
| Chicken prepared and/or eaten away from home | 65 | 39 | 60 | 47 | 0.02 |
| Fresh tomatoes | 70 | 44 | 63 | 60 | 0.36 |
| Focused questionnaire | |||||
| Whole chicken prepared at home | 29 | 8 | 28 | 26 | 0.49 |
| Chicken parts or pieces prepared and/or eaten at home | 30 | 15 | 50 | 52 | |
| Chicken prepared and/or eaten away from home | 26 | 11 | 42 | 47 | |
| Egg or egg-containing dishes | 28 | 18 | 64 | 75 | |
| Ground beef prepared and/or eaten at home | 26 | 7 | 27 | 40 | |
| Fresh tomatoes | 26 | 14 | 54 | 60 | |
The Foodborne Diseases Active Surveillance Network (FoodNet) population survey collected information from healthy adults in the United States during 2006 and 2007 and included questions about food consumed during the 7 days prior to the interview.
An illness subcluster of 36 people was identified in Kern County, California, which included six laboratory-confirmed cases. Thirty-four of the case patients had eaten at a catered birthday party or consumed leftovers from that party. Two additional case patients with isolates with indistinguishable PFGE patterns had consumed food prepared by the same restaurant that had catered the birthday party on or around the same day. The Kern County Public Health Services Department conducted a cohort study of 52 persons who were exposed to food prepared by the restaurant. Chicken prepared by the restaurant was determined to be the most likely food vehicle: 74% (34 of 46) of those who ate the chicken were ill, and 33% (2 of 6) of those who did not eat the chicken were ill. The county staff collected samples of leftover food, including chicken, from two ill party attendees, but Salmonella was not isolated from these samples. The Kern County Environmental Health Department inspected the restaurant and noted multiple instances of potential cross-contamination, including uncovered cooked chicken stored on top of an open container of raw chicken, raw chicken in direct physical contact with cooked chicken on the grill, and a severely deteriorated cutting board used to cut multiple types of meats.
Follow-up analyses
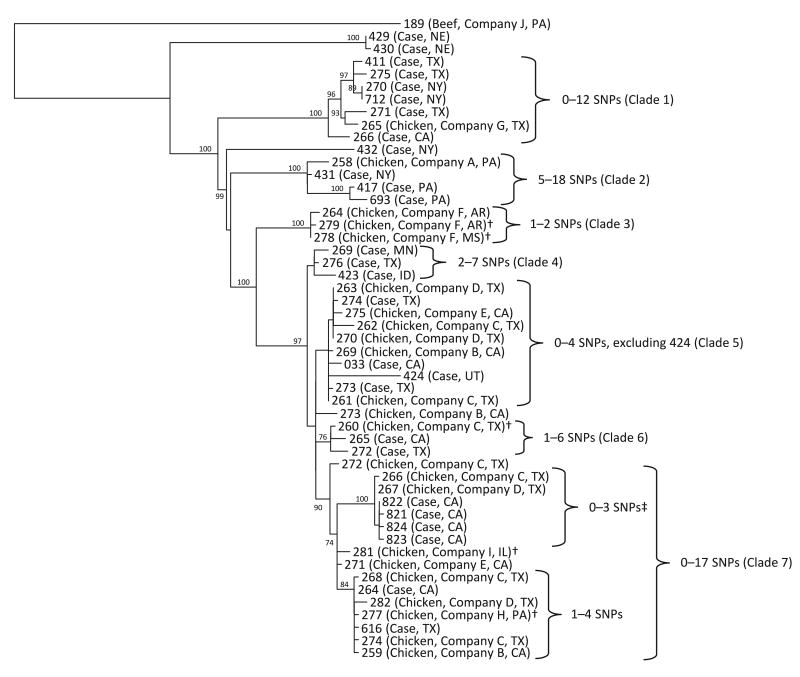
The CDC Enteric Diseases Laboratory Branch, FSIS Outbreaks Section of the Eastern Laboratory, and state health departments performed WGS on clinical and food isolates associated with the outbreak investigation (Fig. 3). The isolates formed seven main clades in the phylogenetic tree. These clades were genetically distinct from one another, differing by dozens of SNPs in some instances, and nearly all of the clades were composed of isolates that were genetically highly related to one another.
Figure 3.
Phylogenetic tree of select human (n = 27) and food (n = 24) isolates, Salmonella Heidelberg outbreak, United States, 2014. Reads were trimmed using run_assembly_trimClean.pl (2) with reads less than 50 bp in length removed. LYVE-SET version 1.1.4e was used with mapping by SMALT. SNPs were called using VarScan at >20× coverage, >95 % read support, and <5 bp apart. Reference Salmonella Heidelberg strain SL476 CP001120 with prophages was detected by PHAST and masked at coordinates 376790 to 417006, 1008480 to 1038446, 1102544 to 1148741, 2000469 to 2022881, 2870372 to 2902633, 3368422 to 3401394, and 4413389 to 4434285. † Food isolates selected from before the beginning of the outbreak; ‡ includes Kern County illness subcluster isolates.
Clade 7, the largest clade on the phylogenetic tree, contained all four clinical isolates from the Kern County illness subcluster that were sequenced and thus included the case patients with the strongest epidemiological link to a source in the outbreak. Five of the six case patients with isolates in clade 7 reported eating chicken; the sixth attended the Kern County birthday party.
Two chicken isolates in clade 7 were closely related to the isolates in the Kern County cases (0 to 3 SNP differences) and were from facilities owned by companies C and D. The restaurant implicated in the Kern County illness subcluster had purchased company C brand chicken 1 day before the catered birthday party and earlier in the week. Although company C chicken was not known to have been served at the catered birthday party, the chicken served may have been cross-contaminated with company C chicken on the restaurant's grill. No isolates from samples from this company were available for sequencing. Company D receives chicken from company C. Isolates from chicken from four other companies also grouped in clade 7, but it is not known whether there were any relationships between these companies (B, E, H, and I) and the other isolates in the clade.
Isolates from companies C and D also were present in two additional clades in the phylogenetic tree (Fig. 3). Clade 5 consisted of 10 isolates: 4 clinical isolates and 6 food isolates from companies C and D and other companies (B and E). Clade 6 contained one isolate from company C and two closely related clinical isolates.
Several of the food and clinical isolates were not closely related genetically to isolates from company C or company D and were found in clades 1 through 4. Chicken isolates from company F came from three different slaughter-processing establishments and were closely related genetically to one another (one or two SNP differences, clade 3), which indicates that the facilities probably shared a common source of chicken contaminated with Salmonella. Although comingling of chicken from different farms occurs at slaughter, multiple companies within a single corporate structure also may share source material during in-plant processing.
Discussion
Use of WGS following this investigation allowed a large cluster of isolates with the same PFGE pattern to be broken down into smaller groups of isolates that were more likely to be connected to a common source. The presence of isolates from company C chicken and its subsidiary, company D, in clades 5 through 7 of the phylogenetic tree suggests that their chicken probably played a role in some of the illnesses in this Salmonella Heidelberg infection outbreak, particularly in those individuals whose isolates were closely related to the company C or company D chicken isolates. The results of the epidemiologic and traceback investigations from the Kern County illness subcluster support this hypothesis. However, other isolates formed clades 1 through 4. These isolates might be unrelated to the isolates in clades 5 through 7, and those that were genetically highly related to one another in clades 1 through 4 could be from different outbreaks.
Chicken samples from four additional companies—B, E, H, and I—also contained isolates that were closely related to isolates from case patients and to company C and company D chicken isolates in clades 5 through 7, but whether or how these four companies were connected to company C and its subsidiary, company D, was not assessed via a traceback investigation. Company C is a large national poultry producer, and companies B, E, H, and I are small producers. At least two of these smaller companies make processed chicken foods in addition to producing chicken. One explanation for the high level of genetic similarity between chicken isolates from these companies and chicken isolates from companies C and D is that these larger companies sell raw chicken products to the smaller companies for use in processed food items containing chicken.
An alternative explanation for the close genetic relationship between isolates from these six poultry companies is that theses companies obtained live birds for slaughter that share a common contamination source further up the supply chain. The structure of the supply of broiler birds in the United States is pyramidal, with pedigree or primary breeders at the top. This genetic stock of birds is propagated though broiler breeders, which produce large quantities of chickens for consumption (3, 18, 22). Many poultry companies, particularly larger producers such as company C, are vertically integrated, meaning that they may control multiplier or broiler flocks that supply their own slaughter operations (24). Nevertheless, most large poultry companies initially receive their live birds from hatching breeder flocks controlled by a small number of companies. Because Salmonella Heidelberg can be transmitted vertically from hen to chick (9, 11), sustained intestinal colonization of multiple generations of live birds in seemingly separate supply chains is a biologically plausible explanation for the phylogenetic relationships observed in this investigation.
Although the evidence linking chicken from company C to at least a portion of illnesses in this outbreak is compelling, only 18% of the clinical isolates and 86% of the food isolates were included in this hqSNP-based phylogenetic analysis. Other isolates might have different genetic characteristics and if included could have produced a phylogenetic tree with a different shape and clade structure.
This analysis demonstrates how WGS, combined with epidemiologic, traceback, and routine product sampling and testing data, can be used to help identify the source of outbreaks in situations in which the source is difficult to determine, such as in outbreaks caused by Salmonella Heidelberg and chicken. This method also might be useful for investigating chicken-associated outbreaks caused by other Salmonella serotypes such as Salmonella Enteritidis and Salmonella Typhimurium, which can be transmitted vertically from hen to chick (15). WGS should be used as soon as evidence collected during case patient interviews identifies chicken as a suspected food vehicle. The greater discriminatory power of WGS allows heterogeneous clusters of isolates grouped by PFGE patterns to be broken into smaller groups of illnesses that are more likely to share a common source. Doing so makes it possible to exclude cases not likely to be a part of the outbreak and to determine whether one large outbreak is, in fact, a few smaller outbreaks, as might be the case with the outbreak described here. WGS also provides more confidence in the links between isolates from humans and those from foods. Routine product testing at slaughter and processing facilities provides a ready source of chicken isolates from a variety of companies that can be added to the phylogenetic tree. These isolates are becoming more accessible with the new FSIS information sharing protocols (23). When a food isolate is determined to be closely related genetically to an isolate from a case patient or a group of isolates from case patients, traceback can be quickly focused on a specific company, and outbreak investigators can simultaneously seek additional epidemiologic links between the case patients and the suspected foods. These tools used together can help promote timely and accurate outbreak investigations.
Acknowledgments
The conclusions, findings, and opinions expressed by the authors do not necessarily reflect the official position of the U.S. Department of Health and Human Services or the CDC.
References
- 1.Anonymous. LYVE-SET, a method of using hqSNPs to create a phylogeny, especially for outbreak investigations. [Accessed 28 October 2016];2016 Available at: https://github.com/lskatz/lyve-SET.
- 2.Anonymous. Genome assembly/prediction/annotation pipeline for the Linus command line. [Accessed 28 October 2016];2016 Available at: https://github.com/lskatz/CG-Pipeline.
- 3.Anthony NB. A review of genetic practices in poultry: efforts to improve meat quality. J Muscle Food. 1998;9:25–33. [Google Scholar]
- 4.Barrow PA, Simpson JM, Lovell MA. Intestinal colonisation in the chicken by food-poisoning Salmonella serotypes; microbial characteristics associated with faecal excretion. Avian Pathol. 1988;17:571–588. doi: 10.1080/03079458808436478. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Foodborne Diseases Active Surveillance Network (FoodNet) population survey atlas of exposures, 2006–2007. [Accessed 28 October 2016]; n.d. Available at: http://www.cdc.gov/foodnet/surveys/foodnetexposureatlas0607_508.pdf.
- 6.Centers for Disease Control and Prevention. [Accessed 28 October 2016];PulseNet. 2015 Available at: http://www.cdc.gov/pulsenet/
- 7.Chen Y, Burall LS, Luo Y, Timme R, Melka D, Muruvanda T, Payne J, Wang C, Kastanis G, Maounounen-Laasri A, De Jesus AJ, Curry PE, Stones R, K'Aluoch O, Liu E, Salter M, Hammack TS, Evans PS, Parish M, Allard MW, Datta A, Strain EA, Brown EW. Isolation, enumeration and whole genome sequencing of Listeria monocytogenes in stone fruits linked to a multistate outbreak. Appl Environ Microbiol. 2016 doi: 10.1128/AEM.01486-16. doi:10.1128/AEM 01486-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Gazzar FE, Marth EH. Salmonellae, salmonellosis, and dairy foods: a review. J Dairy Sci. 1992;75:2327–2343. doi: 10.3168/jds.S0022-0302(92)77993-4. [DOI] [PubMed] [Google Scholar]
- 9.Foley SL, Nayak R, Hanning IB, Johnson TJ, Han J, Ricke SC. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Environ Microbiol. 2011;77:4273–4279. doi: 10.1128/AEM.00598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fricke WF, Mammel MK, McDermott PF, Tartera C, White DG, Leclerc JE, Ravel J, Cebula TA. Salmonella enterica subsp. enterica serovar Heidelberg str. SL476, complete genome. [Accessed 28 October 2016];2014 :CP001120. Available at: http://www.ncbi.nlm.nih.gov/nuccore/
- 11.Gast RK, Guard-Bouldin J, Holt PS. Colonization of reproductive organs and internal contamination of eggs after experimental infection of laying hens with Salmonella Heidelberg and Salmonella Enteritidis. Avian Dis. 2004;48:863–869. doi: 10.1637/7204-05050R. [DOI] [PubMed] [Google Scholar]
- 12.Hanning IB, Nutt JD, Ricke SC. Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne Pathog Dis. 2009;6:635–648. doi: 10.1089/fpd.2008.0232. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann M, Luo Y, Monday SR, Gonzalez-Escalona N, Ottesen AR, Muruvanda T, Wang C, Kastanis G, Keys C, Janies D, Senturk IF, Catalyurek UV, Wang H, Hammack TS, Wolfgang WJ, Schoonmaker-Bopp D, Chu A, Myers R, Haendiges J, Evans PS, Meng J, Strain EA, Allard MW, Brown EW. Tracing origins of the Salmonella Bareilly strain causing a food-borne outbreak in the United States. J Infect Dis. 2016;213:502–508. doi: 10.1093/infdis/jiv297. [DOI] [PubMed] [Google Scholar]
- 14.Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerg Infect Dis. 2013;19:1239–1244. doi: 10.3201/eid1908.121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liljebjelke KA, Hofacre CL, Liu T, White DG, Ayers S, Young S, Maurer JJ. Vertical and horizontal transmission of Salmonella within integrated broiler production system. Foodborne Pathog Dis. 2005;2:90–102. doi: 10.1089/fpd.2005.2.90. [DOI] [PubMed] [Google Scholar]
- 16.Olsen SJ, Bishop R, Brenner FW, Roels TH, Bean N, Tauxe RV, Slutsker L. The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987–1997. J Infect Dis. 2001;183:753–761. doi: 10.1086/318832. [DOI] [PubMed] [Google Scholar]
- 17.Osterholm MT. Editorial commentary: the detection of and response to a foodborne disease outbreak: a cautionary tale. Clin Infect Dis. 2015;61:910–911. doi: 10.1093/cid/civ434. [DOI] [PubMed] [Google Scholar]
- 18.Paxton H, Anthony NB, Corr SA, Hutchinson JR. The effects of selective breeding on the architectural properties of the pelvic limb in broiler chickens: a comparative study across modern and ancestral populations. J Anat. 2010;217:153–166. doi: 10.1111/j.1469-7580.2010.01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 20.Rusconi B, Sanjar F, Koenig SS, Mammel MK, Tarr PI, Eppinger M. Whole genome sequencing for genomics-guided investigations of Escherichia coli O157:H7 outbreaks. Front Microbiol. 2016;7:985. doi: 10.3389/fmicb.2016.00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.U.S. Department of Agriculture. Poultry 2010: reference of health and management practices on breeder chicken farms in the United States, 2010. [Accessed 28 October 2016];2010 Available at: https://www.aphis.usda.gov/animal_health/nahms/poultry/downloads/poultry10/Poultry10_dr_Breeder.pdf.
- 23.U.S. Department of Agriculture, Food Safety and Inspection Service. FSIS notice: sharing information with state or local agencies, foreign government officials, and international organizations. [Accessed 28 October 2016];2016 Available at: http://www.fsis.usda.gov/wps/wcm/connect/9968da35-84c2-463b-813e-7f60682f21d9/45-16.pdf?MOD=AJPERES&CONVERT_TO=url&CACHEID=9968da35-84c2-463b-813e-7f60682f21d9.
- 24.Vukina T. Vertical integration and contracting in the U.S. poultry sector. J Food Distrib Res. 2001;32:29–38. [Google Scholar]
- 25.Wilson MR, Brown E, Keys C, Strain E, Luo Y, Muruvanda T, Grim C, Beaubrun JJG, Jarvis K, Ewing L, Gopinath G, Hanes D, Allard MW, Musser S. Whole genome DNA sequence analysis of Salmonella subspecies enterica serotype Tennessee obtained from related peanut butter foodborne outbreaks. PLoS ONE. 2016;11:e0146929. doi: 10.1371/journal.pone.0146929. [DOI] [PMC free article] [PubMed] [Google Scholar]