Abstract
Fungi are the causal agents of many of the world's most serious plant diseases causing disastrous consequences for large-scale agricultural production. Pathogenicity genomic basis is complex in fungi as multicellular eukaryotic pathogens. Here, we report the genome sequence of C. sojina, and comparative genome analysis with plant pathogen members of the genus Mycosphaerella (Zymoseptoria. tritici (synonyms M. graminicola), M. pini, M. populorum and M. fijiensis - pathogens of wheat, pine, poplar and banana, respectively). Synteny or collinearity was limited between genomes of major Mycosphaerella pathogens. Comparative analysis with these related pathogen genomes indicated distinct genome-wide repeat organization features. It suggests repetitive elements might be responsible for considerable evolutionary genomic changes. These results reveal the background of genomic differences and similarities between Dothideomycete species. Wide diversity as well as conservation on genome features forms the potential genomic basis of the pathogen specialization, such as pathogenicity to woody vs. herbaceous hosts. Through comparative genome analysis among five Dothideomycete species, our results have shed light on the genome features of these related fungi species. It provides insight for understanding the genomic basis of fungal pathogenicity and disease resistance in the crop hosts.
Keywords: Phytopathogenic fungi, Mycosphaerella pathogens, Genome sequence, Comparative genomics
1. Introduction
A number of genome sequences of plant pathogenic fungi in the genus Mycosphaerella that cause economically important disease of major crop hosts have been released [1], [2], [3], [4]. In addition, the fungus Cercosopora sojina is a plant pathogen that threatens global soybean supplies. The teleomorphs of Cercospora species with identified sexual stages are in the genus Mycosphaerella [5]. Recently, we sequenced and released the genome sequence of C. sojina, which would greatly expand the range for comparative analysis of the closely related members in the genus Mycosphaerella, and may provide new insight into the genomic basis of phytopathogenicity biology. It is essential for designing strategies to manage destructive disease in different major crop hosts effectively.
The genus Mycosphaerella and its associated anamorphs comprise one of the largest groups of plant-pathogenic fungi. Many Mycosphaerella species are important pathogens causing leaf spotting diseases in a wide variety of economically important crops including cereals, banana, woody plants, citrus, eucalypts, soft fruits and horticultural crops. Two of the most important pathogens of wheat and banana are Z. tritici (formerly known as synonyms M. graminicola) and M. fijensis, which cause Septoria leaf blotch and black Sigatoka leaf spot, respectively [6], [7]. These diseases occur in most wheat- and banana-producing areas throughout the world every year. Mycosphaerella pini and Mycosphaerella populorum are foliar pathogens of many pine and poplars species respectively, causing serious economic losses on forests and ecological deterioration world-wide. Pines account for the majority of commercial forest products and important members of native forests in many countries. And poplars, as the model organism for forest tree research, are valued as a future source for biofuel. Because of the undisputed economic and ecological importance, understanding these foliar pathogens at the genome level is the basis for developing new methods to manage the disease. These pathogens together represent the Mycosphaerella branch of the fungal evolutionary tree. Phylogenetically, species of Mycosphaerella are close relatives of the soybean frogeye leaf spot pathogen, C. sojina [8]. No sexual (teleomorphic) stage of C. sojina has been identified, but the teleomorphs of Cercospora species with identified sexual stages are in the genus Mycosphaerella. However, the host diversity of these pathogens suggests pathogen specialization besides the common pathogenicity mechanisms in fungi. Therefore, the availability of genome data from C. sojina as well as the closely related species provides the opportunity for comparative genome analysis. It could be valuable in identifying the genome features that can be exploited for better control of disease epidemics.
Through high-throughput DNA sequencing and large-scale comparative genomics of phytopathogenic fungi in this report, we present a genome profile of plant pathogenic fungi in the genus Mycosphaerella, and we investigate the genome sequence background of related pathogens to reveal the differences on genome features involved in pathogenicity to different hosts such as woody vs. herbaceous plants, small crop vs. large trees, tropical and temperate zones crops.
2. Material and methods
2.1. C. sojina whole-genome sequencing and assembly
Genome of C. sojina strain S9, from a soybean field in Georgia, was sequenced using the Illumina GA IIx next generation technology by paired-end sequencing method to a depth of 239 × at Keck Center at University of Illinois Urbana-Champaign. The produced sequences had read length of 124 base pairs (bp). A total of 29,619,123 reads from each end were produced for a total of 59,238,246 reads from one lane. C. sojina genomes were assembled using Velvet algorithm to obtain optimized results with high quality assembly.
2.2. C. sojina genome annotation
The C. sojina genes were predicted with ab initio gene finders (FGENESH, FGENESH +, and GENEWISE). We referred the gene models from Z. tritici (M. graminicola) as the closest species with C. sojina to train the gene finding programs. BlastX against publicly available non-redundant protein and BlastN against ESTs databases are used to validate and curate predicted complete coding regions of the gene models. The entire DNA sequence was also compared against the nonredundant protein databases in all six reading frames, using BlastX with threshold E < 1e-5 to identify any possible coding sequences previously missed by using ARTEMIS to collate data and facilitate annotation. Finally, a non-redundant set of gene models is produced, in which a single best gene model per locus is selected, preferring the candidate annotation with supporting evidence of homolog protein/EST sequence in public database and complete coding sequence region.
2.3. Genomics synteny mapping, genome wide orthologous genes annotation and evolutionary relationships across multiple species
Comparison of the genome of C. sojina with Joint Genome Institute (JGI) released other four related fungal genomes (M. graminicola v2.0, M. pini v1.0, M. populorum v1.0, M. fijiensis v2.0) was performed using Synteny Mapping and Analysis Program package (SyMAP v3.4) for detecting and displaying syntenic relationships between sequenced genome [9], [10] in compliance with instruction from the package. Genome wide comparison and annotation of orthologous genes across multiple species was performed using OrthoVenn [11] with the default settings. Basic cladogram for evolutionary relationships analysis was carried out with the software Mauve with the default settings from whole-genome ortholgous gene sequence data [12].
2.4. Genomics repeat structure profile and mapping
For the genome repeat sequences features of C. sojina and other four related fungal, genome repeat sequences structure were detected and repeat organization map were generated by Pygram pipeline [13], as an efficient genome repeat analysis tool, which provide an representation of the organization of repeated structures including frequency visualization in multi-genomes for discovering new structure features and specific repeat properties.
2.5. Genomics functional annotation
All predicted genes are annotated for function and physiology pathway using Blast2Go function annotation system [14], [15], according to Gene Ontology (GO), eukaryotic orthologous groups (KOGs), and KEGG metabolic pathways. For annotated C. sojina genes, where possible, assigned predicted protein functions using a combination of sequence comparison with BlastP and domains/motif identification with interProScan [14] and PFAM [16].
2.6. Inter-species genome-wide genes annotation comparison
Large-scale genome-wide genes GO functional comparison in all these related fungi were explored and plotted by program WEGO [17]. Comparison histogram were displayed with all items at different GO level separately, including the default second level and the third level, as well as level limited to only items with significant relationship for the genome dataset compared base on Pearson Chi-Square test (Significance level is below the 0.05, expected item counts are greater than 5).
3. Results
3.1. Overall genome comparison
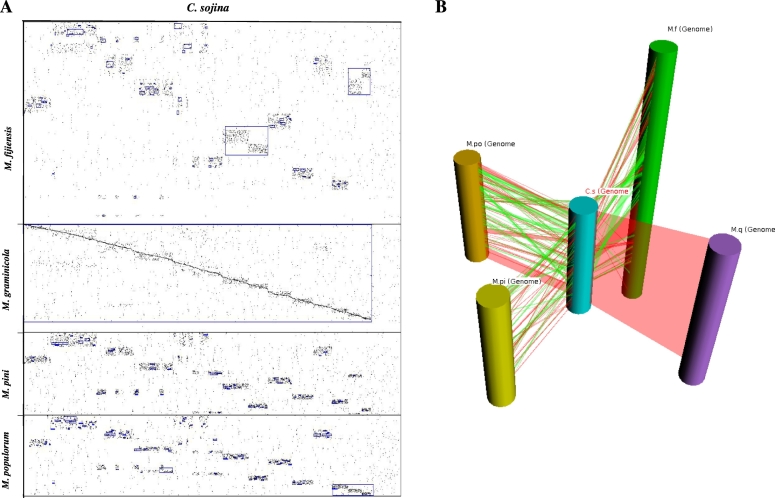
The C. sojina genome was compared with the four fungal genomes of close relatives in the same genus. Limited similarity was present among these phylogenetically close fungal genomes with the exception of the C. sojina and Z. tritici (M. graminicola) genomes, as shown in dot plot alignment mapping and 3D schematic view (Fig. 1). The homologous genome blocks between C. sojina and each of the other four species displayed 81.0% to 85.9% nuclear acid identity with an average of only 82.9%, for they are from close relatives (Additional file 1 Table S1). However, if counting the non-homologous regions at whole genome level, the sequences similarity decreases dramatically.
Fig. 1.
Dot plot alignment mapping and 3D view of C. sojina genome comparison with four close pathogen relatives.
(A) Genome dot plot alignment between C. sojina and Z. tritici (M. graminicola), M. pini, M. populorum, M. fijiensis. Dots represent anchors (also referred to as “hits”). A blue box indicates a Synteny Block determined by the SyMAP synteny-finding algorithm. (B) 3D schematic view of genome alignment between C. sojina and Z. tritici (M. graminicola), M. pini, M. populorum, M. fijiensis. The synteny blocks are shown as colored ribbons, with direct synteny blocks colored red, and inverted blocks colored green. The five species also differed considerably in genome size (Table 1). The largest, M. fijiensis (75 Mb), was two and half times bigger than the smallest, M. populorum (30 Mb). This difference seems to be due to an acquisition of sequence in M. fijiensis rather than loss in M. populorum since these two species diverged from a common ancestor. Finally, extensive genome feature divergences were exhibited among them including genome size, genes number, gene density, gene structure and GC content. (Table 1, see next section for details). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
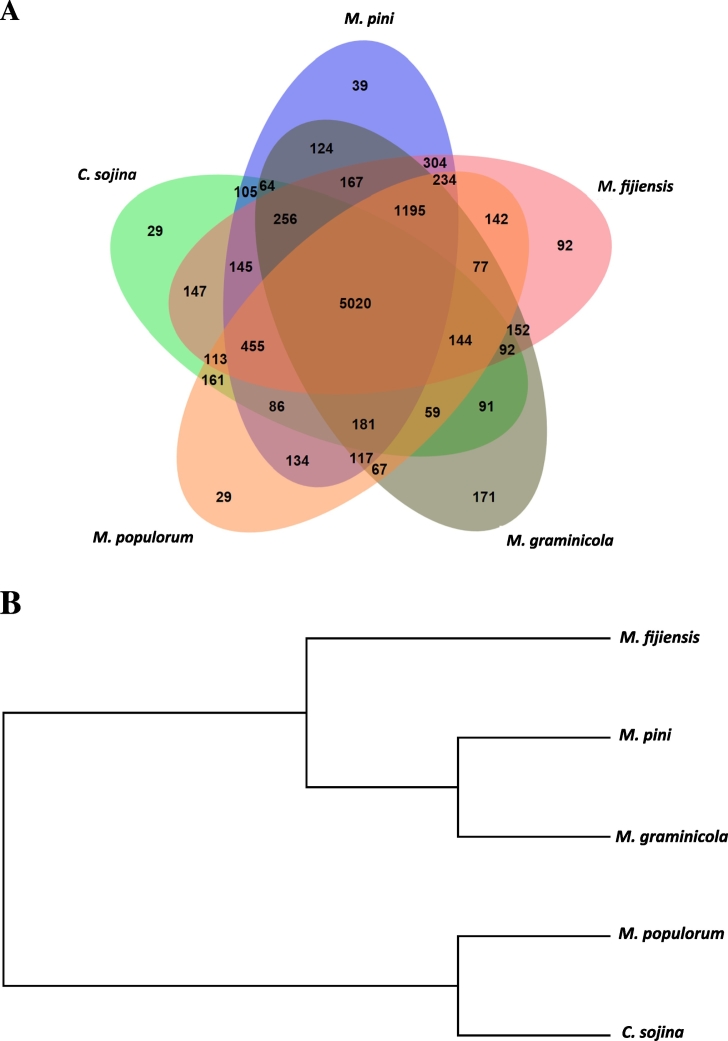
Whole-genome orthologous genes were comprehensively investigated in five close species. The overall comparison analysis result was displayed as Venn diagram in Fig. 2A. Total 5020 orthologous genes were shared among all five species, which highlights these five species are close relatives with considerable common coding gene. Whole genome phylogenetic analysis in our study reveals the evolutionary relationships of these five additional important close species which were not included in previous study of Goodwin SB, 2001 except specie of C. sojina [8]. Among the five relatives, M. populorum was found as the closest specie to C. sojina (Fig. 2B).
Fig. 2.
Whole-genome orthologous genes and evolutionary relationships among five close species.
(A) Venn-diagram of whole-genome orthologous genes in five close species. The numbers in the diagram indicate overlapped conserved genes or un-overlapped specific genes in those five species. (B) Basic cladogram for evolutionary relationships for five close species based on phylogenomic analysis.
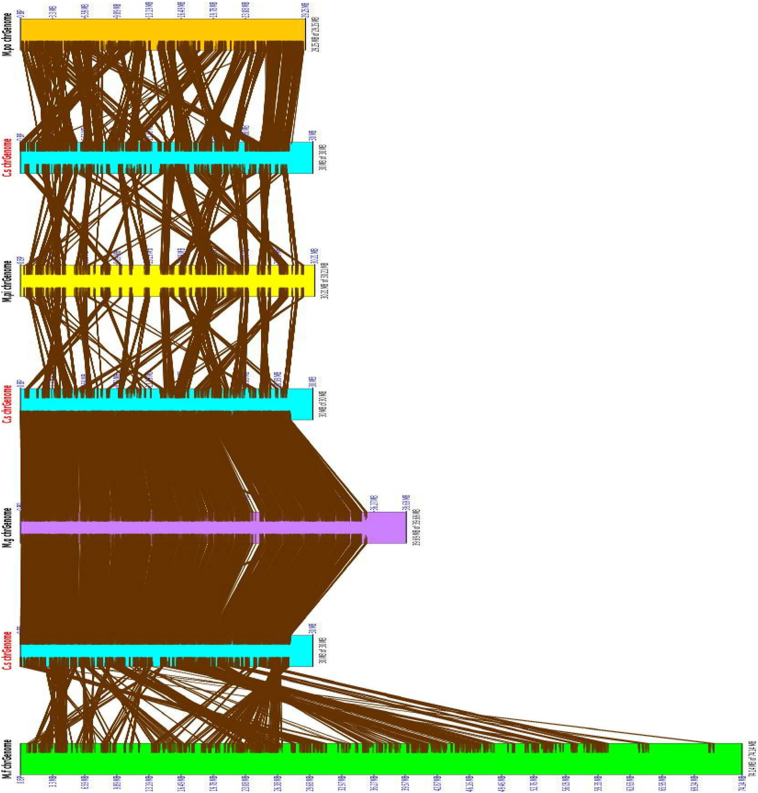
3.2. Genome synteny and genomic changes
The availability of these related fungal genome sequences has allowed comparisons of synteny among different species. These five fungi genomes provide an opportunity to study eukaryotic genome differences and similarities between the genomes. The genomes of C. sojina shared 92% overall synteny with that of Z. tritici (M. graminicola) (Additional file 1 Table S1). In contrast, only 46%, 63%, 66% of C. sojina genome assembly could be mapped to conserved syntenic blocks of the other three genomes of M. pini, M. fijiensis, and M. populorum, respectively (Additional file 1 Table S1). While the overall synteny between C. sojina and Z. tritici (M. graminicola) genomes was conserved, considerable rearrangements were detected among genomes of C. sojina and other three fungi (Fig. 3 and Additional file 2 Fig. S1). Previous studies suggested that synteny or collinearity were limited among genomes from different genera [18], [19], [20]. Our results mostly confirmed this notion with the exception of the C. sojina and Z. tritici (M. graminicola) genomes.
Fig. 3.
Syntenic relationship of C. sojina genome with four close pathogen relatives.
While the overall synteny between C. sojina and Z. tritici (M. graminicola) genomes is conserved, synteny or collinearity was limited between genomes of C. sojina and M. pini, M. populorum, M. fijiensis.
Consistent with the synteny result, genome-wide gene density and structure in C. sojina and Z. tritici (M. graminicola) shared higher average exon numbers per gene and gene density than those of the other three fungal genomes (Table 1). M. fijiensis, which is a pathogen of banana, displayed the unique genome feature with extremely high genome size, genes number but considerably low average exon number per gene and gene density, as well as low GC content (Table 1). For the pathogens of the two woody hosts of pine and poplar, M. pini and M. populorum also had the unique feature of the highest gene density and lowest average exon number per gene with smallest genome size among all pathogens in this study (Table 1).
Table 1.
General features of fungal genomes compared with that of C. sojina.
| Organism | Size | No. genes | Average exons No. per gene | Gene density (1 gene every n bp) | % GC |
|---|---|---|---|---|---|
| C. sojina | 31 Mb | 9099 | 3.2 | 3407 | 53.80 |
| Z. tritici | 40 Mb | 10933 | 2.6 | 3630 | 52.13 |
| M. pini | 31 Mb | 12580 | 2.1 | 2401 | 52.85 |
| M. populorum | 30 Mb | 10233 | 2.2 | 2859 | 50.40 |
| M.fijiensis | 75 Mb | 13107 | 1.8 | 5657 | 44.92 |
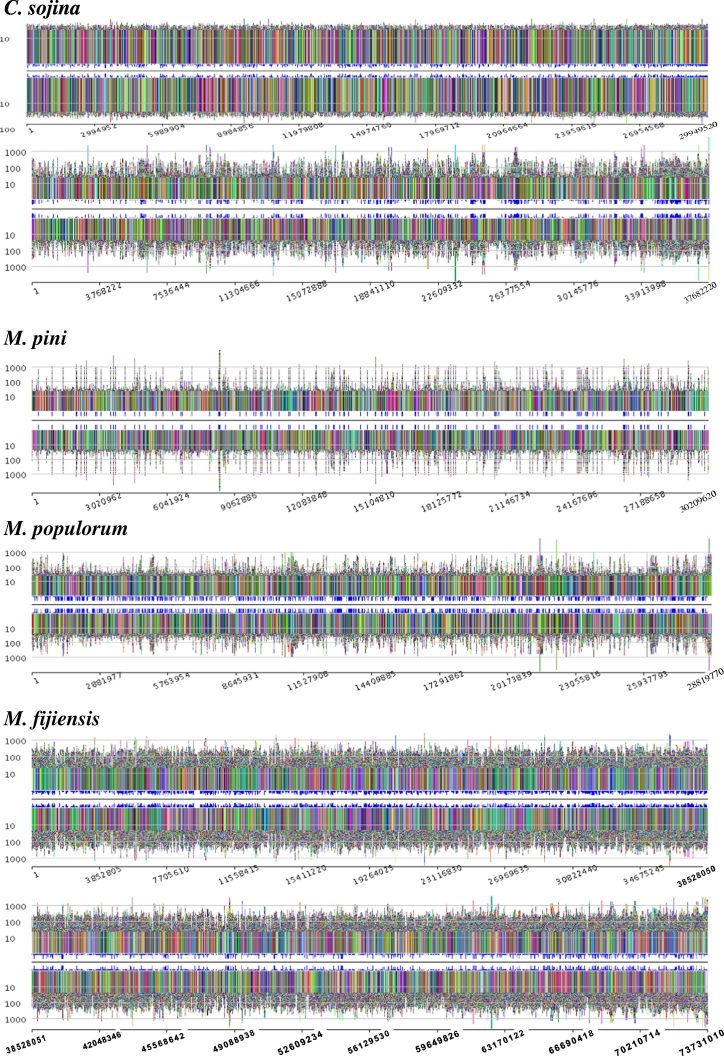
3.3. Genome repeat organization
Eukaryotic genomes contain many repetitive sequences. Many studies on genome sequences have revealed the major role played by repeated sequences in the structure, function, dynamics and evolution of genomes [21], [22], [23], [24], [25], [26]. Thus, understanding genome structure depends crucially on repeat organization and features.
Surprisingly, among the pathogen genomes analyzed in this study, the C. sojina genome displayed the most distinct repeat organization properties compared with other relatives, as shown in genome repeat profile map (Fig. 4). Uniform repeat size patterns present in C. sojina genome and most of the repeats were less than 100 bp on size. In contrast, significant variations of repeat size existed on all other fungi displayed as fluctuations in Fig. 4. Most important, considerable large repeats were contained in genomes of other species, some of which even reached around 10,000 bp.
Fig. 4.
Abstract visualization of genome-wide repeat organization pattern in C. sojina and four close pathogen relatives.
Furthermore, the pathogen of pine, M. pini represents a specific genome-wide repeat feature with very low repeat frequency detected through the whole genome as shown in Fig. 4. In contrast, the M. fijiensis genome had extremely high repeat density with a large size.
The organization of repeated structures in these five fungal genomes was de novo detected and generated by the Pygram pipeline. Extensive repeats present on the fungal genomes with distinct genome-wide repeat organization pattern. Repeats frequency are indicated by the small blue boxes proportional in size to the frequency located between the middle black line, and the plus(+) and minus (−)strand views at each occurrence of the repeat. The x-axis corresponds to the sequence strand coordinate position, the y-axis to repeat size and scale is logarithmic. Each repeat has its own specific color. All occurrences of the same repeat have the same color on both strands. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Comparison of genomic annotation and functional prediction of these close fungi species
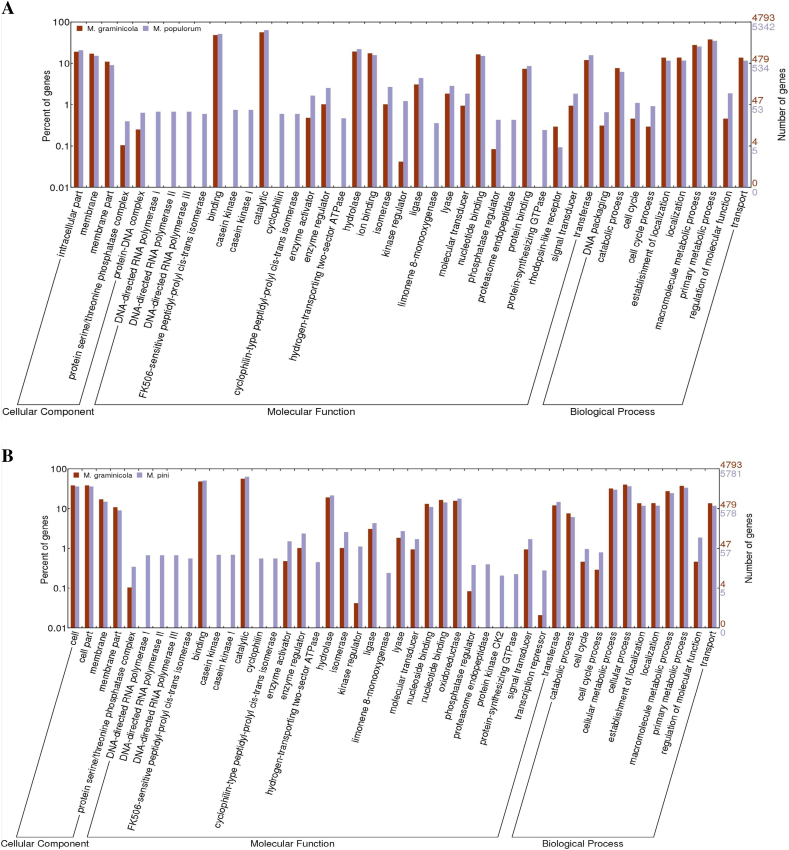
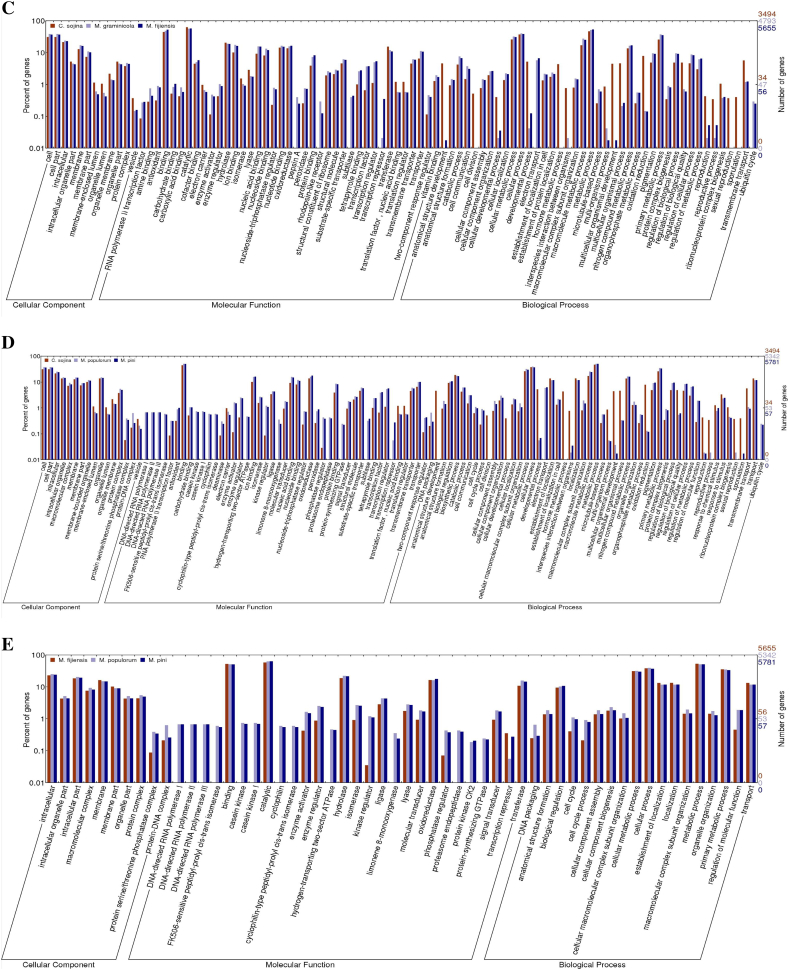
With systematic comparisons of functional prediction across all genomes in this study, we identified both specific and common genomic elements in these five fungi. A wide diversification of the biological association processes among these fungal genomes is illustrated in Fig. 5. Notably, highly abundant genes involved in cell division and oxidation reduction were identified in the C. sojina genome, compared with the other four genomes. At meantime, other fungi also have their preferential biological process versus C. sojina such as Cell Communication, Regulation of Cellular process/function, which reflect the genomic basis of pathogen specialization during evolution. For example, a higher number of two-component response regulators were found in the genome of Z. tritici (M. graminicola) and M. fijiensis, suggesting that these two fungi could act with more advanced social behavior than C. sojina.Genetic elements with function of transcription repressor and subtilase that act as “chemical weapons” were not found in C. sojina but in the other fungal genomes analyzed. And, pepsin A and Rhodopsin-like receptor only present in Z. tritici (M. graminicola) and M. fijiensis (Fig. 5A). In particular, as the pathogens of woody plant hosts, M. pini and M. populorum contain the factor functioned as casein kinase cyclophilin, limonene 8-monoxygenase and endopeptidase, not in the pathogens of herbaceous hosts in this study (Fig. 5B). Variation of secreted proteins and metabolites features displayed in genome of pathogens of woody versus herbaceous hosts suggests gene loss and acquisition across these fungi.
Fig. 5.
Genome-wide gene functional annotation and comparison with GO in five phytopathogen fungi.
Note: only the GO items with significant difference present here across all genomes compared in this study. While comprehensive comparison of all gene GO items are displayed as formats of outline and detail at different levels in Additional file 3 (Fig. S2) and Additional file 4 (Fig. S3) respectively. And y-axis in all the comparison histograms is displayed as log-scale.
Note: only the GO items with significant difference present here across all genomes compared in this study. While comprehensive comparison of all gene GO items are displayed as formats of outline and detail at different levels in Additional file 3 (Fig. S2) and Additional file 4 (Fig. S3) respectively. And y-axis in all the comparison histograms is displayed as log-scale.
Besides the wide divergence of pathogenic functional factors distribution among fungal genomes as mentioned above, genome comparison of phytopathogenic eukaryotes provides a powerful means of identifying conserved pathogenic processes from lineage-specific pathogenesis. Therefore, we also identified general functional elements and biological processes conserved among these species. Molecular Transducers are highly conserved among C. sojina, Z. tritici and M. fijiensis, and so do Molecular Catalytic events among C. sojina, M. pini and M. populorum. They are candidates as common targets for disease control and management. Moreover, Responses to Stimuli are widely conserved among all these five fungi. Similarly, Antioxidant, Transcription Regulator, Translation Regulator and Pigmentation Biology Process showed no significant differences among these species except C. sojina (Fig. 5 and Additional file 3 Fig. S2).
4. Discussion
4.1. Genome synteny comparison between C. sojina and four close phytopathogenic relatives
Comparative analysis with Mycosphaerella pathogens indicated considerable rapid rearrangements occur among these related fungal genomes, which form the genomic basis of the pathogen specialization. Previous studies suggested that synteny or collinearity were limited among genomes from different genera [18], [19], [20]. As indicated by genome comparison in our study, considerable conserved synteny between C. sojina and Z. tritici (M. graminicola), but less conserved synteny with the genomes of the other three fungal species in the genus Mycosphaerella, illuminating genomic basis of the localized polymorphism and pathogen specialization through the long term evolution during the interaction of fungal pathogens and their specific hosts. Moreover, C. sojina and Z. tritici (M. graminicola) shared higher average exon numbers per gene and gene density than those of the other three fungal genomes. These genome features form the molecular basis of the fact that the hosts of C. sojina and Z. tritici (M. graminicola), soybean and wheat, have similar characteristics of growing conditions and pathogen resistance, compared with perennial tree species pine, poplar, and banana as hosts of M. pini, M. populorum and M. fijiensis respectively. Our results mostly confirmed previous notion.
There are also large regions lacking synteny as reported in the fungal genus Aspergillus [27], in which it was thought to play a role in niche adaptation and virulence. Even for small-scale synteny between fungal species in some examples, there is no conservation of gene order or orientation [18], [19]. It can be hypothesized that rapid rearrangement of such regions lacking synteny or collinearity might facilitate species-specific evolution of pathogenicity determinant genes. These sequence divergence and genome rearrangements may result from selection due to interactions of fungi and their plant hosts. This study highlights the potential of comparative genomes of closely related species for identifying conserved pathogenic genomic events and differences involved in pathogenicity to different host plants [28]. It reveals the genomic basis of species-specific adaptation in Dothideomycete species.
4.2. Genome repeat organization and comparative analysis among related phytopathogenic fungi species
Very few large repeat sequences and uniform repeat size patterns observed in C. sojina genome represent its specific repeat properties through the growth and reproduction which limits the opportunity to acquire new repeats. It suggests the species-specific genome innovations during niche adaptation and pathogenicity evolution with the specific host of soybean which, as an herbaceous dicot, differs from other hosts. The lack of repetitive sequence has been previously thought due to operation of a genome-wide defense system known as the RIP (repeat-induced point mutation) like in some fungi such as F. graminearum [26], [29], [30], in which RIP identifies duplicated sequences [31] that are subject to extensive mutation. RIP could partially account for the reduced repeat content and apparent low number of paralogous genes [26], which may occur in C. sojina with fewer repeats and lower gene number. Moreover, three Aspergillus species' fungal genomes have a single predicted DNA methyltransferase gene that is essential for RIP in Neurospora. crassa [29]. Apart from it, no additional DNA methyltransferase genes were identified, which are required for methylation in these fungi [29], while a number of putative DNA methyltransferase genes were predicted in C. sojina genome. Although RIP has not been demonstrated in C. sojina, above features in the genome suggest that RIP as well as methylation might be active in C. sojina.
Furthermore, as the only gymnosperm among the hosts, pine differs from other hosts. Accordingly, the pathogen of pine, M. pini represents a specific genome-wide repeat feature with very low repeat frequency detected through the whole genome. In contrast, M. fijiensis genome presents extremely high repeat density with large size. It suggests the M. fijiensis specific genome innovation against its unique host banana which is a monocot tree in tropical area of the world. These findings further present the profound influences of repeats on genome innovation and pathogenic adaptation. Moreover, repeat density is correlated with genome AT richness in all these five species (Fig. 4 and Table 1). Extremely high repeat density in M. fijiensis go with remarkably low GC content. Consistently, lower abundance of repeat of in C. sojina and M. pini corresponding to higher GC content. The notable specific properties of genome-wide repeat profile in these related fungi pathogens represent the roles that repetitive elements have played, and are continuing to play, in the genome evolution and species-specific adaptation through the interactions of pathogens with their specific host.
Pepsin A and Rhodopsin-like receptor only present in Z. tritici (M. graminicola) and M. fijiensis, which implies that these secreted proteins and response factors to external cues are essential for the ability of these two close pathogens to decay the host material and adapt to fluctuating and competitive environments. Variety of secreted proteins and metabolites features displayed in genome of pathogens of woody versus herbaceous hosts suggests gene loss and acquisition across these fungi. These findings represent and extend the report that variety of fungi species-specific secreted proteins and metabolites, perhaps as effectors within plant cells, play roles in pathogenicity and determining the host range of the fungus [32]. Additionally, the relatively different number of transcription repressors between the wood pathogens M. pini and M. populorum as well as the herbaceous pathogens Z. tritici (M. graminicola) and M. fijiensis suggests the differential regulation action during species-specific pathogenesis (Fig. 5A, B). It implies that the various ecological niches and hosts occupied by different fungal species are reflected in both the gene family's class and abundance present in the genomes.
The identified specific and common genomic elements in C. sojina and other members of the genus Mycosphaerella represent a rich set of candidate targets for further investigation. Coupled with large-scale gene functional analysis studies, this will allow functional definition of these genetic elements that have the greatest potential in elucidating the phytopathogenesis.
5. Conclusions
Comparative genome analysis of C. sojina with M. pini, M. Populorum, Z. tritici (M. graminicola) and M. fijiensis on different plant hosts have shed new light on the genomic basis of pathogenicity diversity of these fungi likely to be common to all eukaryotic phytopathogens. The availability of genome data from C. sojina as well as the closely related species provides new insights into genomic basis of species-specific phytopathogenicity. From the analysis of comparative genomes, we identified the differences on genome features involved in pathogenicity to different plant hosts such as woody vs. herbaceous plants, small crop vs. large trees, tropical and temperate zones crops. These results represent the initial step in elucidating the evolution of pathogenicity. These efforts and ongoing sequencing projects for additional closely related particular phytopathogen isolates from the same species promise to reveal functional diversity of pathogenesis associated genetic elements, and will also facilitate to unveil the evolution of eukaryotic microbial pathogenicity. It will, ultimately, change our understanding of this important group of agronomically and scientifically relevant fungi.
The following are the supplementary data related to this article.
Summary of genome synteny analysis across C. sojina and series fungi.
Comparison of genome arrangements between C. sojina genome and four close pathogen relatives. (A) Genome synteny block detected between C. sojina and Z. tritici (M. graminicola), M. pini, M. populorum, M. fijiensis. (B) Cycle view of genome synteny relationship in C. sojina and Z. tritici (M. graminicola), M. pini, M. populorum, M. fijiensis.
Abstract function annotation comparison of phytopathogen fungal genomes at GO root level two.
Detailed function annotation comparison of phytopathogen fungal genomes at GO level three.
Transparency document
Transparency document.
Acknowledgments
We thank Jianfei Wu, Xinpeng Gao and Chaofan Wang for assistance on additional data analyses and MS revision. This project was supported by a grant from National Key R & D Program for Crop Breeding 2016YFD0100306 and the USDA-CSREES as part of the Soybean Disease Biotechnology Center at the University of Illinois. This work was also supported by NSFC 31401428, Fok Ying-Tong Foundation 151024 and Taishan Scholar Talent Project from China. The genome sequences used to compare with C. sojina were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community.
Footnotes
The Transparency document associated with this article can be found, in the online version.
Contributor Information
Fanchang Zeng, Email: fczeng@sdau.edu.cn.
Guirong Zhang, Email: grzhang@illinois.edu.
Carl A. Bradley, Email: carl.bradley@uky.edu.
Ray Ming, Email: rming@life.uiuc.edu, rming@life.illinois.edu.
References
- 1.Hane J.K., Williams A.H., Oliver R.P. Genomic and comparative analysis of the class Dothideomycetes. In: Pöggeler S., Wöstemeyer J., editors. TheMycota, XIV: Evolution of Fungi and Fungal-Like Organisms. Springer; 2011. pp. 205–229. [Google Scholar]
- 2.Stukenbrock E.H., Bataillon T., Dutheil J.Y., Hansen T.T., Li R., Zala M., McDonald B.A., Wang J., Schierup M.H. The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res. 2011;D21:2157–2166. doi: 10.1101/gr.118851.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver R. Genomic tillage and the harvest of fungal phytopathogens. New Phytol. 2012;196:1015–1023. doi: 10.1111/j.1469-8137.2012.04330.x. [DOI] [PubMed] [Google Scholar]
- 4.Ohm R.A., Feau N., Henrissat B., Schoch C.L., Horwitz B.A., Barry K.W., Condon B.J., Copeland A.C., Dhillon B., Glaser F. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grau C.R., Dorrance A.E., Bond J., Russin J.S. Fungal diseases. In: Boerma H.R., Specht J.E., editors. Soybeans: Improvement, Production and Uses. 3rd edition. Madison; ASA, CSSA, SSSA: 2004. pp. 679–763. [Google Scholar]
- 6.Palmer C.L., Skinner W. Mycosphaerella graminicola: latent infection, crop devestation and genomics. Mol. Plant Pathol. 2002;3:63–70. doi: 10.1046/j.1464-6722.2002.00100.x. [DOI] [PubMed] [Google Scholar]
- 7.Churchill A.C.L. Mycosphaerella fijiensis, the black streak pathogen of banana: progress towards understanding pathogen biology and detection, disease developmentand the challenges of control. Mol. Plant Pathol. 2011;12:307–328. doi: 10.1111/j.1364-3703.2010.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodwin S.B., Dunkle L.D., Zismann V.L. Phylogenetic analysis of Cercospora and Mycosphaerella based on the internal transcribed spacer region of ribosomal DNA. Phytopathology. 2001;91:648–658. doi: 10.1094/PHYTO.2001.91.7.648. [DOI] [PubMed] [Google Scholar]
- 9.Soderlund C., Nelson W., Shoemaker A., Paterson A. SyMAP: a system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006;16:1159–1168. doi: 10.1101/gr.5396706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soderlund C., Bomhoff M., Nelson W.M. SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Coleman-Derr D., Chen G., Gu Y.Q. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015;43:W78–W84. doi: 10.1093/nar/gkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2001;414:394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durand P., Mahé F., Valin A.S., Nicolas J. Browsing repeats in genomes: pygram and an application to non-coding region analysis. BMC Bioinf. 2006;7:477. doi: 10.1186/1471-2105-7-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Götz S., García-Gómez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., Robles M., Talón M., Dopazo J., Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 16.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean R.A., Talbot N.J., Ebbole D.J., Farman M.L., Mitchell T.K., Orbach M.J., Thon M., Kulkarni R., Xu J.R., Pan H. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 19.Hane J.K., Lowe R.G.T., Solomon P.S., Tan K.C., Schoch C.L., Spatafora J.W., Crous P.W., Kodira C., Birren B.W., Galagan J.E. Dothideomycete-plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell. 2007;19:3347–3368. doi: 10.1105/tpc.107.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soanes D.M., Richards T.A., Talbot N.J. Insights from sequencing fungal and oomycete genomes: what can we learn about plant disease and the evolution of pathogenicity? Plant Cell. 2007;19:3318–3326. doi: 10.1105/tpc.107.056663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X., Voytas D.F. A eukaryotic gene family related to retroelement integrases. Trends Genet. 2005;21:133–137. doi: 10.1016/j.tig.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Dooner H.K., Weil C.F. Give-and-take: interactions between DNA transposons and their host plant genomes. Curr. Opin. Genet. Dev. 2007;17:486–492. doi: 10.1016/j.gde.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Lai J., Li Y., Messing J., Dooner H.K. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9068–9073. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgante M., Brunner S., Pea G., Fengler K., Zuccolo A., Rafalski A. Gene duplication and exon shuffling by helitronlike transposons generate intraspecies diversity in maize. Nat. Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 25.Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 26.Cuomo C.A., Güldener U., Xu J.R., Trail F., Turgeon B.G., Di Pietro A., Walton J.D., Ma L.J., Baker S.E., Rep M. The Fusarium graminearum genome reveals a link betweenlocalized polymorphism and pathogen specialization. Science. 2007;317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- 27.Siriputthaiwan P., Jauneau A., Herbert C., Garcin D., Dumas B. Functional analysis of CLPT1, a Rab/GTPase required for protein secretion and pathogenesis in the plant fungal pathogen Colletotrichum lindemuthianum. J. Cell Sci. 2005;118:323–329. doi: 10.1242/jcs.01616. [DOI] [PubMed] [Google Scholar]
- 28.Sillo F., Garbelotto M., Friedman M., Gonthier P. Comparative genomics of sibling fungal pathogenic taxa identifies adaptive evolution without divergence in pathogenicity genes or genomic structure. Genome Biol. Evol. 2015;7:3190–3206. doi: 10.1093/gbe/evv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galagan J.E., Selker E.U. RIP: the evolutionary cost of genome defense. Trends Genet. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Selker E.U., Cambareri E.B., Jensen B.C., Haack K.R. Rearrangement of duplicated DNA in specialized cells of neurospora. Cell. 1987;51:741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- 31.Watters M.K., Randall T.A., Margolin B.S., Selker E.U., Stadler D.R. Action of repeat-induced point mutation on both strands of a duplex and on tandem duplications of various sizes in neurospora. Genetics. 1999;153:705–714. doi: 10.1093/genetics/153.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweeney M.J., Dobson A.D. Molecular biology of mycotoxin biosynthesis. FEMS Microbiol. Lett. 1999;175:149–163. doi: 10.1111/j.1574-6968.1999.tb13614.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of genome synteny analysis across C. sojina and series fungi.
Comparison of genome arrangements between C. sojina genome and four close pathogen relatives. (A) Genome synteny block detected between C. sojina and Z. tritici (M. graminicola), M. pini, M. populorum, M. fijiensis. (B) Cycle view of genome synteny relationship in C. sojina and Z. tritici (M. graminicola), M. pini, M. populorum, M. fijiensis.
Abstract function annotation comparison of phytopathogen fungal genomes at GO root level two.
Detailed function annotation comparison of phytopathogen fungal genomes at GO level three.
Transparency document.