Abstract
Genome-wide association studies have identified numerous genetic variants conferring autoimmune disease risk. Most of these genetic variants lie outside protein-coding genes hampering mechanistic explorations. Numerous mRNAs are also differentially expressed in autoimmune disease but their regulation is also unclear. The majority of the human genome is transcribed yet its biologic significance is incompletely understood. We performed whole genome RNA-sequencing [RNA-seq] to categorize expression of mRNAs, known and novel long non-coding RNAs [lncRNAs] in leukocytes from subjects with autoimmune disease and identified annotated and novel lncRNAs differentially expressed across multiple disorders. We found that loci transcribing novel lncRNAs were not randomly distributed across the genome but co-localized with leukocyte transcriptional enhancers, especially super-enhancers, and near genetic variants associated with autoimmune disease risk. We propose that alterations in enhancer function, including lncRNA expression, produced by genetics and environment, change cellular phenotypes contributing to disease risk and pathogenesis and represent attractive therapeutic targets.
1. Introduction
We now appreciate that the majority of the human genome is transcribed and many new RNA classes, such as long non-coding RNAs (lncRNAs) [1], enhancer-associated RNAs, micro-RNAs and piwi-interacting RNAs have been identified [2–4]. Whole genome profiling of mRNAs from a common tissue source has been employed to analyze an array of diseases, including autoimmune diseases, and shows potential for medical application including diagnosis, assessment of disease activity and determining or predicting response to therapy [5]. Functions of these new RNA classes are incompletely understood and it is even argued that pervasive transcription may represent biologic ‘noise’ [6–8]. Arguments against the ‘noise’ hypothesis may be to ask if these RNA classes are induced or repressed by presence of idiopathic disease, the extent to which they are regulated similarly in related idiopathic diseases, such as autoimmune diseases [9, 10], how expression changes during disease progression and initiation of therapies, their distributions across the genome, and their proximity to and regulation by genetic variants associated with these diseases.
Genome-wide association studies (GWAS) have also identified numerous genetic variants (single nucleotide polymorphisms, SNPs) that confer autoimmune disease risk. The vast majority of these genetic polymorphisms lie outside protein-coding genes, which hampers our understanding of how they may contribute to disease risk. An alternate possibility is that these genetic variants may also regulate expression of these newly discovered classes of RNAs that, in turn, may regulate expression of protein-coding genes, either in cis or trans, and alter cellular phenotypes and disease risk [9, 11–16].
We identified annotated and novel lncRNA, as well as mRNAs, differentially expressed in whole blood obtained from subjects with various autoimmune diseases by whole genome RNA-sequencing (RNA-seq). We found that loci transcribing novel lncRNAs were not randomly distributed across the genome but were localized near leukocyte transcriptional enhancers, especially super-enhancers (SEs) compared to typical enhancers [16, 17]. Further, genomic positions of both annotated and novel lncRNA loci were near GWAS-identified SNPs that confer risk for developing autoimmune disease. We propose that both genetic and environmental effects may alter lncRNA expression profiles and contribute to onset and pathogenesis of autoimmune disease and may represent attractive therapeutic targets.
2. Materials and Methods
2.1 Subject populations
We obtained blood samples in PAXGENE tubes from age and gender matched healthy controls (CTRL, N=8), subjects with ulcerative colitis (UC, N=6), Crohn’s disease (Cr, N=6), irritable bowel syndrome (IBS, N=6), Celiac disease (Ce, N=6), fibromyalgia (FMS, N=6), rheumatoid arthritis (N=6), systemic lupus erythematosus (SLE, N=3), psoriasis (Ps, N=3), psoriatic arthritis (PsA, N=3), and Sjogren’s (Sj, N=3). Subjects with relapsing remitting multiple sclerosis (MS, N=18) were divided into 1) subjects after a clinical event suggestive of demyelination (clinically isolated syndrome, CIS) at the time of their blood draw but before diagnosis of MS (MS-C, N=6), subjects at the time of diagnosis of MS but prior to onset of therapies (MS-N, N-6) and subjects with established MS of 1–3 years’ duration (MS-E, N=6). MS-E subjects were not on disease-modifying therapies such as interferon-beta or Tysabri. Diagnoses were made by specialists in the field using established criteria. Subjects with RA were divided into those on current methotrexate therapy (RA+MTX) and those not on current methotrexate therapy (RA-MTX). All samples were obtained with informed consent after Vanderbilt institutional review board approval.
2.2 RNA-seq Sample Preparation and Data Analysis
RNA was isolated from PAXGENE tubes using standard protocols, including DNase digestion. Using this procedure there is <1% DNA contamination in these RNA samples. Poly(A)+ RNAs were isolated by mixing RNA samples with poly(T) oligomers covalently attached to magnetic beads using standard procedures. Library preparation was performed using the Illumina Tru-Seq Stranded RNA kit. RNA sequencing was performed by the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core facilities. An Illumina HiSeq2500 instrument was used to generate 100bp paired-end reads. Average sequencing depth of all samples was 35 million mapped reads +/− 9 million (S.D.). RNA-seq was performed consecutively on all samples. Quality control steps were performed at all stages of sequencing analysis including raw data, alignment and expression quantification [18–20]. The RNA data were aligned with TopHat 2 and gene expression levels were quantified using Cufflinks [21, 22]. Differentially expressed genes measured using Cufflinks/Cuffdiff were expressed as FPKM (fragments per kilobase per million reads) and a cutoff of 0.5 FPKM was employed for all RNA measurements. False discovery rate (FDR < 0.025) was used to correct for multiple testing. De novo transcriptome assembly was performed on whole genome RNA-sequencing data from Illumina Tru-Seq Stranded Total RNA libraries using TopHat 2 and Cufflinks using upper quartile normalization (-N) and fragment bias correction (-b). As a quality control step, we also compared values obtained from the upper quadrant methods used for normalization here and values obtained from DESeq procedures [23] across all RNA-seq samples and found that the correlation between the two methods was > 0.85. All normalization was performed the same way across the entire analysis of all RNA-seq samples. Novel transcripts were assembled from reads prealigned to human genome 19 (GRCh37/hg19) using TopHat. Identification of novel lincRNA transcripts was accomplished using established methodologies including getorf to analyze open reading frames as described [6]. PhyloCSF, Coding Potential Calculator (CPC), and Coding-Potential Assessment Tool (CPAT) were employed to remove RNAs with protein-coding potential. The summarized pipeline for discovery of novel lincRNAs and prediction of transcripts with protein-coding potential has been previously reported [24]. The overall normalization and filtering processes were performed identically for all samples at the same time.
2.3 Bin definition
Genomic locations of loci producing novel lncRNAs (> 0.5 FPKM in ≥ 2/3 of samples from at least one cohort) were sorted based upon chromosomal location from the p-terminus to the q-terminus, e.g. chromosome 1, basepair 1 to chromosome 1, basepair 249,000,000, etc. Genomic distances between each novel lncRNA producing loci were determined. Bins were delimited if there was > 20 kb space between loci encoding a novel lncRNA.
2.4 Statistical Analysis
1) Comparison of bin and enhancers locations
Genomic locations of super enhancers and typical enhancers found in leukocytes subsets were obtained from published data [17]. We wrote a program in ‘R’ to determine overlaps of these enhancers with novel lncRNA genomic bins identified here. The starting point for determining correlations between the lncRNA bins, enhancers for the varieties of CD cells, SNPs associated specific diseases, and expressions levels in the lncRNA bins for a spectrum of auto-immune diseases are four databases: (i) a list of bins that contain the lncRNA bin identifiers and their positions on 24 chromosomes; a list of enhancers and super-enhancers for 16 CD cell types (CD3 TE, CD3 SE, CD4 memory TE, CD4 memory SE, CD4 Naïve TE, CD4 Naïve SE, CD8 memory TE, CD8 memory SE, CD14 TE, CD14 SE, CD19 TE, CD19 SE, CD20 TE, CD20 SE, CD56 TE, CD56 SE); (iii) a GWAS catalog (hg19 version) for 25,742 SNPs associated with 1,457 disease classes such as Cr, MS, and SLE; and a list of FPKM expression levels in the lncRNA bins for 72 individuals having one of the auto-immune disease considered in paper (Cr, MS, RA, Ps, SLE, UC, etc.). The first task was to establish the correlations between the 6,431 lncRNA bins and the 107,492 enhancers for the 16 CD cell types. This calculation was done in various ways. For a given lncRNA bin (location: chromosome, start, stop), the bin was extended on each end by x bp, x = 0, 10K, 20K, …, 50K, and intersections (as location intervals) of the extended lncRNA bins with the enhancer locations (chromosome, start, stop) were determined. When a non-empty intersection was found, the lncRNA bin (and its extension) and the enhancer type (e.g., CD4 memory TE) were recorded. With an extension of 20K bp, there were 5,378 lncRNA bins that overlapped the enhancer locations. For each extended lncRNA bin, a count of the number of enhancer types was tabulated. Generally speaking, for a particular lncRNA bin extension, the number of enhancer types was 0, 1, or 2. Next, we established correlations between the lncRNA bins and disease SNPs. The frequency of SNPs associated with a particular disease class varies. For example, there are 165 SNPs for UC, 239 for Cr, 215 SNPs for MS, 316 for RA, 160 for SLE, and 136 for PS + PsA. Using the same strategy as bin-enhancer correlations, lncRNA bins were extended and SNPs were found that lay within their extensions. Then disease specific correlations between bins and SNPs were found. The number of SNPs and their identities (rs-identifier, hg 19 position, mapped genes) for each lncRNA bin extension was recorded. The final correlation brings together the lncRNA bins, the enhancer locations, and the expression data for 72 individuals each with a specific disease. Hence, for each lncRNA bin, we have the enhancers and SNPS near the bin, and the FPKM expressions levels for each bin.
2) Comparison of bin and GWAS SNP locations
Genomic locations of GWAS SNPs were obtained from the GWAS catalog [https://www.ebi.ac.uk/gwas]. We wrote a program in ‘R’ to determine overlaps of these SNPs with novel lncRNA genomic bins identified here. Methods for statistical calculations have been described. Next, we established correlations between the lincRNA bins and disease SNPs. The frequency of SNPs by disease varies. For example, there are 220 SNPs for Crohn’s Disease, 177 SNPs for Multiple Sclerosis, 117 SNPs for Inflammatory Bowel Disease, and 163 SNPs for Ulcerative Colitis. Using the same strategy as bin-enhancer correlations, lincRNA bins were extended and SNPs were found that lay within their extensions. Then disease specific correlations between bins and SNPs were found. The number of SNPs and their identities (rs-identifier, hg 19 position, mapped genes) for each lincRNA bin extension was recorded. The final correlation brings together the lincRNA bins, the enhancer locations, and the expression data for 72 individuals each with a specific disease.
3) GREAT analysis
GO enrichment for novel lncRNA bin genomic coordinates was determined using GREAT with default settings [25]. Binomial FDR Q values are reported in Supplementary Table 1.
4) SNP χ2 analysis
First, we compared total bp present in all ‘bins’ or ‘bins + extensions’ to the number of bp in the genome not contained in ‘bins’. Second, we compared total number of GWAS identified SNPs for a given disease present in bins or bins + extensions to the number of GWAS identified SNPs for a given disease in the genome not contained in bins and performed χ2 analysis to determine if differences were or were not random.
| Disease | # GWAS SNPs |
|---|---|
| IBD | 121 |
| Cr | 139 |
| UC | 165 |
| Ce | 88 |
| MS | 215 |
| RA | 315 |
| SLE | 160 |
| PD | 139 |
3. Results
3.1 Expression profiles of mRNAs, annotated lncRNAs and novel lncRNAs in idiopathic disease
We obtained blood samples from healthy control [HC] subjects, subjects with the following autoimmune diseases: ulcerative colitis [UC], Crohn’s [Cr], relapsing remitting multiple sclerosis [MS], rheumatoid arthritis [RA], systemic lupus erythematosus [SLE], psoriasis [Ps], psoriatic arthritis [PsA], and Sjogren’s Syndrome [Sj], and subjects with the following syndromes, irritable bowel syndrome [IBS] and fibromyalgia [FMS]. The MS cohorts were sub-divided into subjects with early disease [MS-C, after a clinical event suggestive of MS but prior to diagnosis], at the time of diagnosis but before onset of therapy or treatment naive [MS-N], and with established disease of 1–3-years duration and on therapies [MS-E] [26, 27]. Subject demographics are shown in Table 1. We performed whole-genome RNA-sequencing (RNA-seq) to identify differentially expressed mRNAs, known or annotated lncRNAs and novel lncRNAs [18–20, 22, 24]. Known mRNAs and lncRNAs were assessed using Gencode.v17 annotation lists. Novel lncRNAs were determined via a de novo search of RNA-seq data. We defined expressed RNAs as having RNA levels ≥ 0.5 fragments per kilobase per million reads [FPKM] in >67% of samples in at least one subject cohort. Using these criteria, we identified 12,852 expressed mRNAs (total = 20345), 2,338 expressed annotated lncRNAs (total = 13870), and 41,087 expressed novel lncRNAs [Fig. 1A]. Average FPKM of these mRNAs, annotated lncRNAs, and novel lncRNAs were 33+/−81, 7+/−8, and 25+/−2237, respectively. Thus, average expression levels of all novel lncRNAs were between that of the mRNAs and annotated lncRNAs but expression levels of the novel lncRNAs exhibited much greater variability across subject cohorts than did other RNAs (see also note S1). Sum of FPKM expression of all novel lncRNAs, known lncRNAs, and mRNAs expressed in these samples was 1,042,398, 15,315, and 421,004, respectively. Thus, most transcripts detected in leukocytes represented the novel lncRNA class.
Table 1.
Subject Demographics
| AGE±S.D.a | % F b | Therapy c | CD14 d | CD4 | CD8A | CD19 | CD56 | FCGR3A | |
|---|---|---|---|---|---|---|---|---|---|
| UC | 30±4 | 67 | + | 0.95 | 0.83 | 0.85 | 0.86 | 0.88 | 0.98 |
| Cr | 31±4 | 67 | + | 0.86 | 0.81 | 0.83 | 0.76 | 1.06 | 0.86 |
| Ce | 38±5 | 67 | + | 1.03 | 0.96 | 1.06 | 1.04 | 0.94 | 1.09 |
| IBS | 37±4 | 83 | − | 1.05 | 0.88 | 0.81 | 0.85 | 0.98 | 1.17 |
| FMS | 43±4 | 83 | − | 1.22 | 1.24 | 1.21 | 1.21 | 1.07 | 0.89 |
| MS-C | 32±3 | 67 | − | 1.29 | 1.03 | 0.81 | 1.14 | 1.09 | 1.02 |
| MS-N | 34±3 | 83 | − | 1.26 | 1.04 | 0.88 | 1.16 | 0.89 | 1.13 |
| MS-E | 39±3 | 67 | + | 1.08 | 0.84 | 0.86 | 0.80 | 1.09 | 1.05 |
| HC | 38±11 | 75 | − | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| RA-MTX | 47±4 | 67 | + | 0.96 | 1.06 | 0.93 | 1.27 | 1.11 | 1.14 |
| RA+MTX | 48±5 | 83 | + | 0.93 | 0.89 | 0.83 | 0.80 | 0.97 | 1.11 |
| SLE | 36±5 | 83 | + | 1.15 | 1.05 | 1.23 | 1.11 | 0.86 | 1.09 |
| Ps | 38±4 | 67 | + | 0.91 | 0.87 | 1.21 | 1.30 | 1.15 | 0.82 |
| PsA | 43±5 | 67 | + | 0.90 | 1.17 | 0.94 | 1.25 | 0.86 | 1.06 |
| Sj | 48±5 | 67 | + | 1.26 | 0.90 | 0.90 | 0.89 | 1.06 | 1.17 |
average age ± standard deviation, P>0.05 compared to HC
P>0.05 compared to HC, all subjects were Caucasian
Except for the RA±MTX group, subjects within each disease group were not on the same treatment therapies in an attempt to reduce effects treatments may have on RNA profiles.
from RNA-seq data, ratios of expression of the different cell-type specific cell surface markers in each disease cohort compared to HC, P>0.05 compared to HC; CD14, monocytes; CD4, helper T cells; CD8A, cytotoxic T cells; CD19, B cells; CD56, NK cells; FCGR3A, CD16, neutrophils.
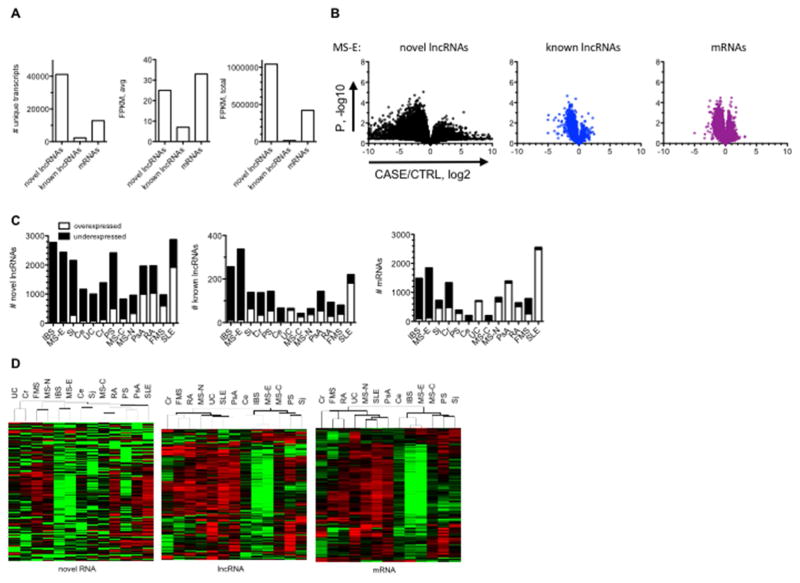
Fig. 1.
Differential expression of RNA classes in idiopathic disease. A, Distribution of RNA classes, novel IncRNAs, known IncRNAs, and mRNAs, determined by RNA-seq. B, ‘Volcano plots’ showing differential expression of different RNA classes in MS-E compared to CTRL. C, Numbers of over- and under-expressed novel IncRNAs, known IncRNAs. and mRNAs in different idiopathic disease cohorts after FDR correction. D, Hierarchical clustering of RNA classes across idiopathic disease cohorts after FDR correction.
Certain autoimmune diseases are associated with mild to severe lymphopenia and we considered that this property could influence the analysis if lymphopenia altered frequency of certain lymphoid or myeloid populations in whole blood [28, 29]. To test this, we calculated expression levels of genes encoding standard markers of monocytes, CD14, T cells, CD4 and CD8A, B cells, CD19, and neutrophils, CD16 or FCGR3A in the case cohorts relative to the HC cohort. We found that expression levels of these cell surface proteins in the case cohorts were not statistically different from the HC cohort (Table 1).
To determine differential expression of these RNA classes, we determined CASE/CTRL average ratios, log2, X-axis and P, −log10, student’s T-test, of the difference, Y-axis [Fig. 1B, MS-E and Supplementary Fig. 1, all cohorts]. The range of differential expression of novel lncRNAs between CASE/CTRL cohorts was 22–210 or 4–1000-fold. In contrast, the range of differential expression of annotated lncRNAs and mRNAs typically did not exceed 22–23. After correcting for false discovery rates [FDR] [30], we enumerated numbers of novel and known lncRNAs and mRNAs. IBS, as well as several autoimmune diseases were characterized by preferential loss of expression of novel and known lncRNAs as well as mRNAs while SLE was characterized by preferential gain of expression of these RNA classes [Fig. 1C and Supplementary Fig. 2]. Hierarchical clustering demonstrated a high degree of similarity between IBS and MS-E as well as between SLE and PsA and these patterns were conserved across RNA classes [Fig. 1D].
3.2 Genomic loci transcribing novel lncRNAs overlap with leukocyte transcriptional enhancers
We found that genomic loci transcribing these novel lncRNAs were not randomly distributed across the genome but were localized in small genomic regions we refer to as ‘bins’ [Fig. 2A]. Certain ‘bins’ transcribed a high density of lncRNAs differentially expressed in multiple disease cohorts while other ‘bins’ transcribed a high density of lncRNAs that were not differentially expressed across multiple disease cohorts. The majority of novel lncRNAs we detected were transcribed from these ‘bins’ and distances between ‘bins’ were large compared to ‘bin’ size. When examined as bin loci rather than novel lncRNA loci, 85% of bins were differentially expressed in at least one disease cohort compared to the HC cohort after FDR correction (Supplementary Data File 1). These genomic ‘bins’ overlapped with genomic leukocyte transcriptional enhancers defined by H3K27Ac marks, both typical- and super-enhancers [17] [Fig. 2B]. Taken together, these results are consistent with the notion that novel lncRNAs identified are enhancer-associated lncRNAs. Enhancer RNAs [eRNAs] represent an additional class of long non-coding RNAs sub-divided into 1-directional [1D-eRNA] or 2-directional [2D-eRNA] RNAs according to whether or not they are transcribed from one DNA strand or in both sense and antisense directions, respectively [31]. The 1d-eRNAs are not easily distinguished from lncRNAs but are transcribed from transcriptional enhancers that may be defined by epigenetic markings. Thus, novel lncRNAs described here bear certain similarities to 1d-eRNAs
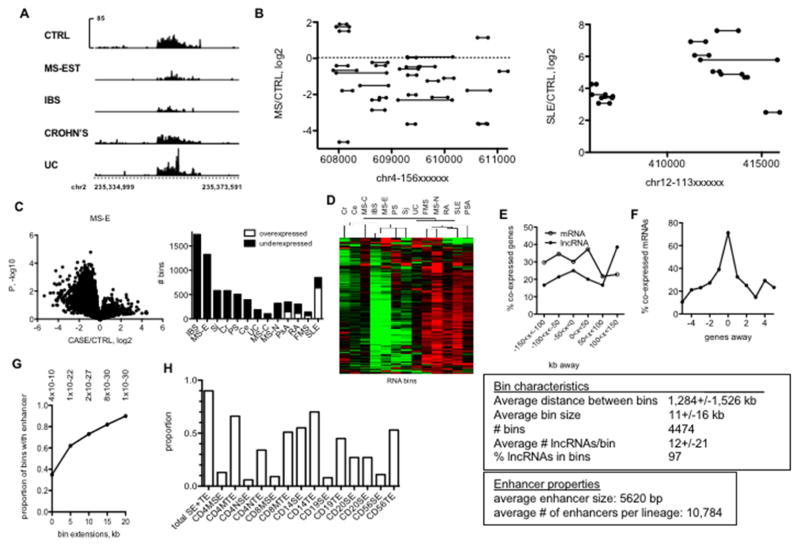
Fig. 2.
Novel IncRNAs are transcribed from discrete locations in the genome (‘bins’) overlapping with known typical and super-enhancers. A, Examples of two genomic regions transcribing a high density of novel IncRNAs (bins). Spikes indicate individual IncRNAs. Y-axes aremean expression of IncRNAs and X-axes are genomic positions on chr1. B, Example of IncRNA bin differentially expressed in the indicated disease cohorts. Y axes are RPKM and X axis is genomic position on chr2. C, Left panel: ‘Volcano’ plots illustrating differential bin activity in MS-E versus CTRL. Right panel: Total numbers of under- and over-expressed ‘bins’ in different idiopathic disease cohorts relative to CTRL after FDR correction. D, Hierarchical clustering of RNA bins across idiopathic disease cohorts after FDR correction. E, Co-expression of bin activity and expression of neighboring IncRNA and mRNA genes. Y-axis is the % of co-expressed genes relative to all genes within the indicated genomic distances of a ‘bin’, X axis is genomic distances from bins. F, Correlation of co-expression of protein-coding genes as a function of distance from a known IncRNA gene. The text box describes overall properties of bins in the genome. G, Relationship between genomic positions of bins and myeloid and lymphoid enhancers. Y-axis is the proportion of bins that contain a myeloid or lymphoid enhancer defined by H3K27Ac levels. X-axis: effect of increasing bin size by the indicated kb in both 5′ and 3′ directions.* = P-values determined by χ2 analysis. H, Proportion of bins that contain enhancers in the indicated lymphoid and myeloid lineages using the bin + 20 kb extension.
We re-analyzed RNA-seq data using bin genomic loci as the definition list to determine total FPKM expressed by bin genomic loci and whether these bin loci also exhibited markedly different activities in the disease cohorts compared to control cohorts. We found that many disease cohorts were characterized by marked loss of bin activity while SLE singularly displayed overall gain of activity [Fig. 2C]. Disease-specific differential bin activity was observed in bins with both high and low overall FPKM [Supplementary Fig. 3]. Expression patterns of bins were similar in IBS and MS-E as seen with the other RNA classes [Fig. 2D]. Bin activity also correlated with expression of neighboring protein-coding and lncRNA genes spanning about 300 kb across all disease and CTRL cohorts suggesting bin activity may determine expression of these neighboring genes [Fig 2E]. In contrast, co-expression of protein-coding genes and lncRNA genes were most enriched when the two genes were overlapping in the genome [Fig. 2F]. GWAS-associated single nucleotide polymorphisms [SNPs] are enriched at enhancer loci defined by either DNase hypersensitivity [DNase HS] or H3K27-acetylation [16, 32, 33]. Therefore, we asked if bin genomic loci also overlapped with transcriptional enhancer loci found in hematopoietic cells defined by H3K27-acetylation marks (19). We found that genomic bin loci as defined here were also enriched at transcriptional enhancer loci [Fig. 2G]. Presence of these enhancers was not restricted to one hematopoietic lineage but dispersed among several distinct lineages (Fig. 2H).
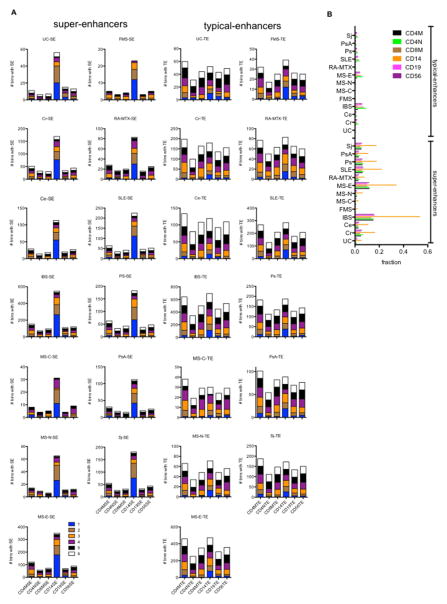
To explore the relationship between ‘bin’ activity and enhancers in further detail, we identified ‘bins’ differentially expressed in the different disease cohorts after FDR correction [Fig 3]. We segregated enhancers into super-enhancers and typical-enhancers by cell type. Differentially expressed “bins” were further sub-divided according to whether they contained enhancers present in 1–6 cell types, e.g. SE or TE in memory CD4 T+ cells (CD4MSE, CD4MTE), SE or TE in naïve CD4 T+ cells (CD4NSE, CD4NTE), SE or TE in memory CD8 T+ cells (CD8MSE, CD8MTE), SE or TE in CD14+ cells (CD14SE, CD14TE), SE or TE in CD19+ cells (CD19SE, CD19TE), or SE or TE in CD56+ cells (CD56SE, CD56TE). We also found that certain SE and TE were present in only one cell type (blue in the stack plots) while others were present in multiple cell types (see color-coding) and some were present in all cell types (white in the stack plots). A disproportionate number of CD14SEs were present in differentially expressed ‘bins’ for each disease type relative to SEs in the other cell types [Fig. 3A]. This was not the case for the TEs. TEs within disease-regulated ‘bins’ were present in the different cell types in similar proportions. We also compared the total number of SEs or TEs present in the individual cell types to the fraction of SEs or TEs found in disease-regulated ‘bins’. For example, > 50% of all CD14SEs were present in IBS-regulated ‘bins’ and > 30% of all CD14SEs were present in MS-E-regulated ‘bins’ [Fig. 3B]. Lower proportions were observed in the other diseases but overall the proportion of total SEs in disease-regulated ‘bins’ was much greater than the proportion of total TEs in disease-regulated ‘bins’. Taken together, these results argue that a much higher proportion of SEs than TEs are present in disease-regulated ‘bins’ and CD14+ cells are enriched with altered disease-regulated bins and SEs compared to other hematopoietic cell types.
Fig. 3.
Genomic ‘bins’ are enriched with CD14 super-enhancers. (A) We determined the number of ‘bins’ differentially expressed in the indicated idiopathic diseases that contained an SE or TE present in the indicated cell types. Numbers from 1–6 indicate if ‘bins’ contain SEs or TEs present in one or more cell types (from ref. 23). Left column: stack plots showing number of bins with an SE in the indicated cell types, X-axis; right column, stack plots showing number of bins with an TE in the indicated cell types, X-axis. (B) Fraction of total TEs or SEs present in the indicated cell types contained within a differentially expressed ‘bin’ in the indicated idiopathic diseases.
3.3 Enhancer-associated lncRNA activity and idiopathic disease
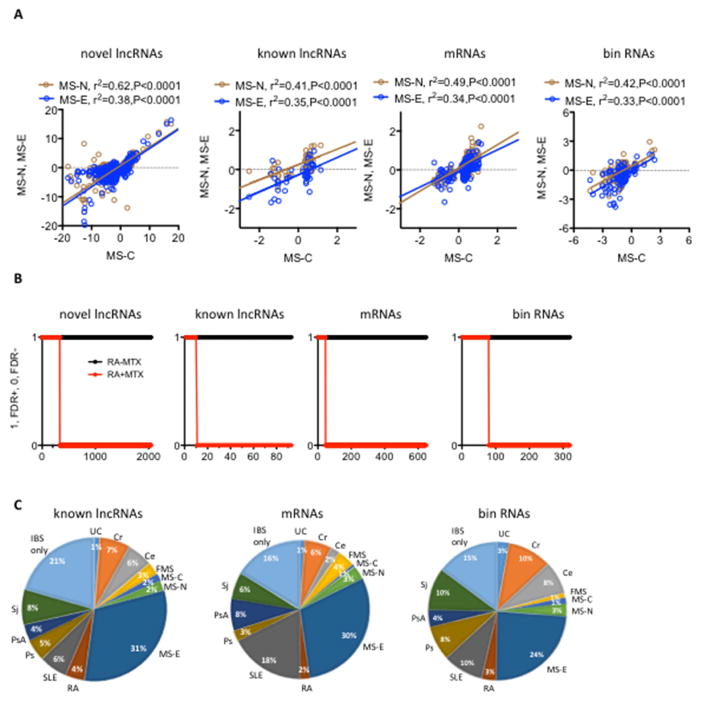
We employed linear regression analysis to determine if differences in expression of RNA classes observed early in disease [MS-C] were sustained later in disease [MS-N and MS-E]. Differential expression of all RNAs in MS-C was largely sustained in both MS-N and MS-E cohorts in this cross-sectional analysis [Fig. 4A]. Results were replicated in a larger cohort using RT-PCR [Supplementary Table 1]. Low-dose methotrexate [MTX] is an effective therapy for RA and via multiple pathways is known to regulate expression of certain mRNAs and lincRNA-p21, a known lncRNA [1, 34–36]. We compared expression of novel lncRNAs, known lncRNAs, mRNAs and bins in subjects with RA who were [RA-MTX] or were not [RA+MTX] on current MTX therapy in a cross-sectional analysis. We found that the majority of these RNA classes differentially expressed in the RA-MTX cohort were not different differentially expressed in the RA+MTX cohort [Fig. 4B]. Thus, expression levels of all classes of RNAs regulated by presence of RA were impacted or ‘corrected’ by MTX therapy.
Fig. 4.
Regulation of ‘bin’ activity. A, Differentially expressed MS-C RNA classes were determined after FDR adjustment. Expression levels of these MS-C RNAs were compared to their expression in MS-N and MS-E cohorts by linear regression. P values are the probability that the regression line is non-zero. B, RNAs differentially expressed in subjects with RA who were not on methotrexate therapy were identified by adjusting for CASE/CTRL FDR. Expression levels of these RNAs were determined ins subjects on current methotrexate therapy (RA+MTX) by determining RA-MTX/CTRL FDR+. Y-axis, FDR+=1, FDR−=0, X-axis: number of discrete transcripts in each of the RNA classes C, Proportion of differentially expressed RNAs unique to a single cohort or shared among different cohorts. Results for IBS are shown, see also Supplements Table 2.
We determined the extent to which differential expression of known lncRNAs, mRNAs and bin RNAs were unique to an individual disease or shared among multiple diseases. IBS serves as an example [Fig. 4C]. The majority of RNAs differentially expressed in IBS were also differentially expressed in other diseases and the most striking overlap was seen in MS-E. Patterns of sharing of these different RNA classes between IBS and the different diseases were relatively similar [also see Supplementary Table 2 for a complete comparison of all disease cohorts and RNA classes]. ‘GREAT’ [genomic regions enrichment of annotations tool] is a software tool that attempts to assign biological meaning to non-coding elements in the genome by analyzing annotations of neighboring protein coding genes and we employed this tool to explore potential functions of genomic loci that encode bin RNAs [25]. Overwhelmingly, most predominant pathways identified were those impacting functions of innate and adaptive immunity [Supplementary Table 3]. Similar pathways were identified in the different autoimmune diseases as well as in IBS, but not FMS.
3.4 Genomic positions of enhancer-associated RNAs and disease-specific genetic polymorphisms
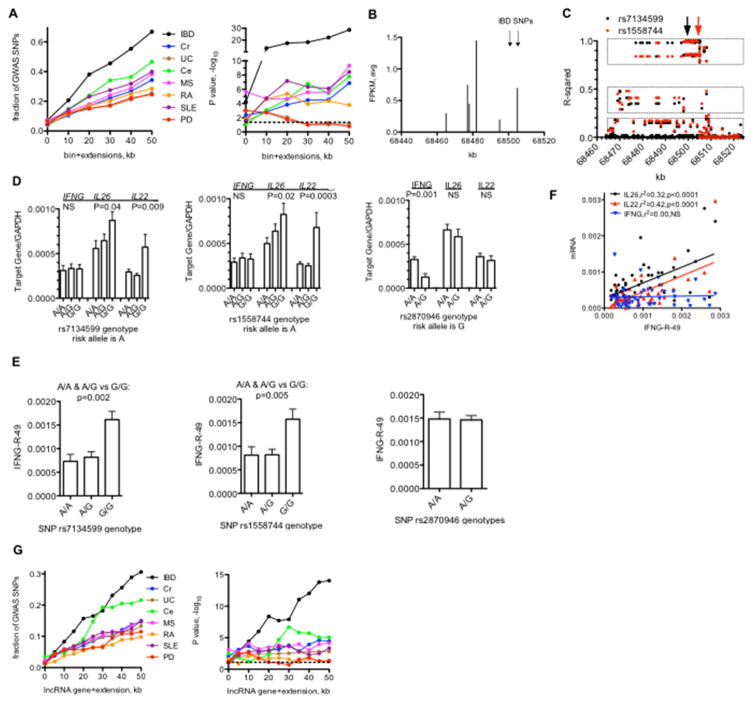
Most genetic variants (single nucleotide polymorphisms, SNPs) associated with complex diseases identified by genome-wide association studies (GWAS) lie outside protein-coding genes. We asked if they may be enriched in bin genomic regions identified here. To do so, we identified the fraction of disease specific GWAS-identified SNPs near a genome bin and asked if these SNPs were nearer a genomic bin than expected by chance. Depending upon disease, 20–60% of disease-specific GWAS-identified SNPs were near a genomic bin (Fig. 5A). We identified one bin on chr12 with six associated lncRNAs and two SNPs associated with risk for IBD upstream of IFNG (Fig. 5B). We next constructed a more extended haplotype map using data from the 1000 genomes project. We identified SNPs in high linkage disequilibrium with rs7134599 or rs1558744 and found that SNPs in high linkage disequilibrium with rs7134599 and rs1558744 spanned a region of 30–40 kb (Fig. 5C) [37]; average size of a haplotype block in the human genome is 30–70 kb. These results argue that rs7134599 or rs1558744 with other SNPs within this region define a haplotype block with at least three haplotypes in high, moderate, or low linkage equilibrium with rs7134599 and rs1558744. Genotypes of rs7134599 and rs1558744 were associated with levels of IL26 and IL22 in human peripheral leukocytes but not IFNG while the genotype of an IBD associated SNP in the IL26 intron, rs2870946, was associated with levels of IFNG, but not IL26 and IL22 (Fig. 5D). We also measured levels of one of the bin RNAs we named IFNG-R-49 (R for RNA, -49 as it is 49 kb upstream of IFNG) in human peripheral leukocytes and found that IFNG-R-49 levels were also associated with rs7134599 and rs1558744 genotypes but not rs2870946 genotype (Fig 5E). We performed linear regression analysis and found that levels of IFNG-R-49 correlated with levels of IL26 and IL22, but not IFNG (Fig. 5F). We interpret these results to suggest that rs7134599 and rs1558744 haplotypes are associated with IFNG-R-49 levels and IFNG-R-49 levels contribute to control of IL26 and IL22 expression levels.
Fig. 5.
LncRNA bins and disease-specific genetic polymorphisms. A, Left panel: Fraction of disease-associated SNPs identified by GWAS studies near novel IncRNA ‘bins’ per disease. Y-axis is the fraction of total SNPs per indicated disease near a IncRNA bin and the X-axis is the bin (0) + indicated extensions in kb from 5′ and 3′ ends. Right panel: P values. −log10 determined by C2 analysis. B, Genomic positions on chr12 transcribing unique IncRNAs (X-axis) relative to transcript level (FPKM avg. of each IncRNA in total sample set). Positions of SNPs associated with IBD are indicated by the arrows. C Linkage disequilibrium across the same genomic region on chr12. Y-axis: Linear regression relative to the indicated IBD-associated SNPs. filled black circles=linear regression compared to rs7134599, filled red circles=linear regression compared to rs1558744. Boxes indicate haplotypes in high (upper), moderate (middle), and low (bottom) linkage disequilibrium with the indicated SNPs. D, Association of IFNG. IL26, and IL22 transcript levels with indicated genotypes. Y-axes are transcript levels in whole blood relative to GAPDH for the given genotypes, rs 7134599 or rs1558744: A/A, N=40, A/C, N=64, G/G, N=32: rs2870946: A/A, N=120, A/G, N=22. E, Association of IFNG-R-49 transcript levels with indicated genotypes, Y-axes are transcript levels in whole blood relative to GAPDH for the given genotypes, subject #s and genotypes as in (d). F, Association between IFNG-R-49 and IFNG, IL26 and IL22 transcript levels in whole blood determined by linear regression analysis. Indicated transcripts normalized to GAPDH, p values are probability that the regression line is non-zero. G, Left panel: Fraction of disease-associated SNPs identified by GWAS studies near annotated IncRNA genes per disease. Y-axis is the fraction of total SNPs per indicated disease near a IncRNA bin and the X-axis is the bin (0) + indicated extensions in kb from 5′ and 3′ ends. Right panel: P values, −log10 determined by C2- analysis. Dashed lines indicate P<0.05.
Using a similar strategy as described above (Fig. 5A), we found that annotated lncRNA genes were nearer disease-specific GWAS-identified SNPs than expected by chance (Fig. 5G). Overall, the fraction of disease-specific GWAS SNPs near bins or enhancers was somewhat greater than the fraction of SNPs near annotated lncRNA genes. By comparison, ~3% of GWAS-identified SNPs have been mapped to protein-coding sequences [15].
4. Discussion
We performed whole genome RNA-seq using whole blood samples to identify mRNAs as well as annotated and novel lncRNAs differentially expressed in autoimmunity. The vast majority of novel lncRNAs we identified co-localize with leukocyte transcriptional enhancers suggesting that these RNAs can be broadly classified as enhancer RNAs (eRNAs). Differential expression of these RNA classes is sustained during disease progression in MS and responds to MTX therapy in RA indicating they are biologically regulated and may be therapeutic targets. Regulation of many eRNA clusters, as well as lncRNAs and mRNAs is shared among multiple autoimmune diseases. Most notably, MS-E and IBS, which is usually not considered an autoimmune disease, exhibited the highest degree of sharing. eRNA clusters and annotated lncRNA genomic loci co-localize or are near SNPs that confer risk for developing these diseases and we show that SNP genotypes associate with eRNA expression. We interpret these results to indicate that genetic regulation eRNA clusters and lncRNAs may confer disease risk. In summary, our results are consistent with a model where differential expression of bin lncRNAs is pervasive in autoimmune diseases, as well as IBS, and this may reflect altered enhancer function.
How altered enhancer-associated lncRNA expression arises is not clear but it should produce altered cellular phenotypes reflected by both basal and inducible expression of mRNAs that generate changes in immunologic function observed in autoimmune disease. Alterations in the epigenetic machinery at the level of transcription factor binding, corresponding epigenetic modifications, recruitment of additional regulatory proteins and/or recruitment of RNA polymerase II to enhancers are some possibilities as have been described in certain cancers [38]. In many diseases, most notably MS-E and IBS, presence of disease is associated with loss of expression of eRNA clusters, which does suggest certain approaches to future investigations to explore underlying mechanisms. These patterns of shared and unique lncRNA expression in different idiopathic diseases/syndromes may also provide an alternative way of subgrouping diseases according to alterations in enhancer-associated lncRNA expression that are independent of affected target tissues. Bin RNAs/enhancers that are unique to individual diseases or shared among multiple diseases may also represent unique targets for therapeutic intervention.
SEs (or stretch-enhancers) [10, 12] are distinguished from TEs by their breadth of epigenetic modifications such as H3K27-Ac modifications or recruitment of histone acetyltransferases (HATs) and a higher level of modification. Proponents have also argued that SEs more than TEs play important roles in determining cell-specific identity and may contribute to onset of disease. Our results generally support this notion. Disease-specific differentially regulated ‘bins’ are enriched with SEs compared to TEs. These SEs are also enriched in CD14+ cells compared to other hematopoietic cells.
There are also strong associations between eRNA cluster genomic loci, novel lncRNA genomic loci and genetic variants that confer disease risk and in one locus we studied in detail, SNP genotypes correlate with expression of eRNAs expressed within the haplotype block as well as expression of nearby protein coding genes, IL26 and IL22. It should be possible to determine if other genetic variants that confer disease risk also regulate activity of eRNA clusters in a similar fashion to determine if this may be a general property of these genetic variants and identify both associated eRNA clusters and associated variations in expression of protein-coding genes, which may lead to a broader understanding of the genetics of complex diseases.
A limitation of our studies is that we do not really address if the eRNAs clusters produced by leukocyte enhancers have biologic function or if simply the act of transcription alters the epigenetic machinery to affect expression of protein-coding genes and this is a subject that is generally debated and the two possibilities are not mutually exclusive [8]. However, the locus transcribing the eRNA, IFNG-R-49, is associated with expression of both IL26 and IL22 at a distance of over 100 kb, which may be more consistent with the eRNA exhibiting biologic function rather than the act of transcription though additional studies will be required to actually determine which eRNA clusters have functions as RNA molecules and which do not.
A general view is that expression of annotated lncRNAs and enhancer-associated lncRNAs may show greater cell-type specificity than expression of mRNAs and as such may also show greater disease specificity. Thus, analysis of these RNA classes in larger populations may help identify biomarkers to aid in diagnosis and management of subjects with autoimmune diseases as well as syndromes such as IBS and FMS. Further, targeting the epigenetic machinery has begun to emerge as an attractive therapeutic strategy and if underlying mechanisms that give rise to altered expression of annotated lncRNA and enhancer associated lncRNAs in autoimmune disease can be identified, it should be possible to develop targeted therapies that correct these defects and these may produce beneficial outcomes. This is notable when it is considered that 3–5% of the population has an autoimmune disease and perhaps >10% has IBS or FMS [39–41]. Finally, our results support the notion that many genetic variants that confer disease risk may alter expression of enhancer-associated lncRNAs, which, in turn, affects expression of target protein coding genes leading to altered cellular phenotypes.
Supplementary Material
Supplementary Table 1. PCR validation of lncRNA expression differences between MS and HC cohorts
Supplementary Table 2. Extent that differential expression of RNA classes is shared among different subject cohorts compared to CTRL.
Supplementary Fig. 1. Differential expression of RNA classes in different disease/syndrome cohorts.
Supplementary Table 3. GREAT pathways analysis of bin genomic locations.
Supplementary Data File 1. Expression profiles of RNAs in disease cohorts
Supplementary Fig. 2. Differential expression of ‘bins’ in disease/syndrome cohorts compared to CTRL.
Supplementary Fig. 3. Disease-specific differential bin activity (FDR+) relative to total bin FPKM.
Highlights.
Changes in expression of super-enhancer-associated long noncoding RNAs are pervasive in human autoimmune disease.
These genomic loci are in linkage with genetic variants that confer autoimmune disease risk.
Acknowledgments
We wish to thank the individuals who provided blood samples. Funding: This work was supported by grants from the National Institutes of Health (NIAID: R01AI44924, NIAMS: R21AR068247, NIGMS T32 GM007569) Funding sources had no role in study design. Vanderbilt’s VANTAGE core facility was supported in part by grants from the National Institutes of Health (P30 CA68485, P30 EY08126 and G20 RR030956).
Footnotes
Competing interests: TMA and CFS are co-founders of IQuity Labs. JTT has a financial interest in IQuity Labs.
Data and materials availability: Deposition of RNA-seq data into GEO is complete. Accession number is 92472.
Author contributions: TMA, PSC AEP, and CFS analyzed data, NJO provided clinical samples and data, JTT performed PCR and SNP analysis, TMA wrote the paper with critical support and insights from the other authors, and all authors edited and approved the final version of the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014;30:439–52. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu GQ, Mattick JS, Taft RJ. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle. 2013;12:2061–72. doi: 10.4161/cc.25134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–37. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pascual V, Chaussabel D, Banchereau J. A Genomic Approach to Human Autoimmune Diseases. Annu Rev Immunol. 2010;28:535–71. doi: 10.1146/annurev-immunol-030409-101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hangauer MJ, Vaughn IW, McManus MT. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. Plos Genet. 2013;9 doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen TH, Jacquier A, Libri D. Dealing with Pervasive Transcription. Mol Cell. 2013;52:473–84. doi: 10.1016/j.molcel.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Li WB, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–23. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 9.Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, et al. Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression. Plos Genet. 2013;9 doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SCJ, Erdos MR, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corradin O, Cohen AJ, Luppino JM, Bayles IM, Schumacher FR, Scacheri PC. Modeling disease risk through analysis of physical interactions between genetic variants within chromatin regulatory circuitry. Nat Genet. 2016;48:1313–20. doi: 10.1038/ng.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quang DX, Erdos MR, Parker SCJ, Collins FS. Motif signatures in stretch enhancers are enriched for disease-associated genetic variants. Epigenet Chromatin. 2015;8 doi: 10.1186/s13072-015-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mells GF, Hirschfield GM. Making the most of new genetic risk factors - genetic and epigenetic fine mapping of causal autoimmune disease variants. Clin Res Hepatol Gas. 2015;39:408–11. doi: 10.1016/j.clinre.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Farh KKH, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ricano-Ponce I, Wijmenga C. Mapping of Immune-Mediated Disease Genes. Annu Rev Genom Hum G. 2013;14:325–53. doi: 10.1146/annurev-genom-091212-153450. [DOI] [PubMed] [Google Scholar]
- 16.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Ye F, Sheng QH, Clark T, Samuels DC. Three-stage quality control strategies for DNA re-sequencing data. Brief Bioinform. 2014;15:879–89. doi: 10.1093/bib/bbt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Zhao SL, Sheng QH, Ye F, Li J, Lehmann B, et al. Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics. 2014;103:323–8. doi: 10.1016/j.ygeno.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, Zhao SL, Ye F, Sheng QH, Shyr Y. MultiRankSeq: Multiperspective Approach for RNAseq Differential Expression Analysis and Quality Control. Biomed Res Int. 2014 doi: 10.1155/2014/248090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spurlock CF, Tossberg JT, Guo Y, Collier SP, Crooke PS, Aune TM. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun. 2015;6 doi: 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–U155. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, et al. Recommended diagnostic criteria for multiple sclerosis: Guidelines from the International Panel on the Diagnosis of Multiple Sclerosis. Ann Neurol. 2001;50:121–7. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 27.Lennardjones JE. Classification of Inflammatory Bowel-Disease. Scand J Gastroentero. 1989;24:2–6. doi: 10.3109/00365528909091339. [DOI] [PubMed] [Google Scholar]
- 28.Storek J, Zhao Z, Lin E, Berger T, McSweeney PA, Nash RA, et al. Recovery from and consequences of severe iatrogenic lymphopenia (induced to treat autoimmune diseases) Clin Immunol. 2004;113:285–98. doi: 10.1016/j.clim.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulze-Koops H. Lymphopenia and autoimmune diseases. Arthritis Res Ther. 2004;6:178–80. doi: 10.1186/ar1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y. Discovering the false discovery rate. J R Stat Soc B. 2010;72:405–16. [Google Scholar]
- 31.Natoli G, Andrau JC. Noncoding Transcription at Enhancers: General Principles and Functional Models. Annu Rev Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 32.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. P Natl Acad Sci USA. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peeters JGC, Vervoort SJ, Tan SC, Mijnheer G, de Roock S, Vastert SJ, et al. Inhibition of Super-Enhancer Activity in Autoinflammatory Site-Derived T Cells Reduces Disease-Associated Gene Expression. Cell Rep. 2015;12:1986–96. doi: 10.1016/j.celrep.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 34.Cronstein BN. Low-dose methotrexate: A mainstay in the treatment of rheumatoid arthritis. Pharmacol Rev. 2005;57:163–72. doi: 10.1124/pr.57.2.3. [DOI] [PubMed] [Google Scholar]
- 35.Spurlock CF, Tossberg JT, Fuchs HA, Olsen NJ, Aune TM. Methotrexate increases expression of cell cycle checkpoint genes via JNK activation. Arthritis Rheum-Us. 2012;64:1780–9. doi: 10.1002/art.34342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spurlock CF, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate Inhibits NF-kappa B Activity Via Long Intergenic (Noncoding) RNA-p21 Induction. Arthritis Rheumatol. 2014;66:2947–57. doi: 10.1002/art.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P, et al. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayliss J, Mukherjee P, Lu C, Jain SU, Martinez D, Margol AS, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. J Neuropath Exp Neur. 2016;75:592. doi: 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooper GS, Bynum MLK, Somers EC. Recent insights in the epidemiology of autoimmune diseases: Improved prevalence estimates and understanding of clustering of diseases. J Autoimmun. 2009;33:197–207. doi: 10.1016/j.jaut.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clauw DJ. Fibromyalgia A Clinical Review. Jama-J Am Med Assoc. 2014;311:1547–55. doi: 10.1001/jama.2014.3266. [DOI] [PubMed] [Google Scholar]
- 41.Makker J, Chilimuri S, Bella JN. Genetic epidemiology of irritable bowel syndrome. World J Gastroentero. 2015;21:11353–61. doi: 10.3748/wjg.v21.i40.11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. PCR validation of lncRNA expression differences between MS and HC cohorts
Supplementary Table 2. Extent that differential expression of RNA classes is shared among different subject cohorts compared to CTRL.
Supplementary Fig. 1. Differential expression of RNA classes in different disease/syndrome cohorts.
Supplementary Table 3. GREAT pathways analysis of bin genomic locations.
Supplementary Data File 1. Expression profiles of RNAs in disease cohorts
Supplementary Fig. 2. Differential expression of ‘bins’ in disease/syndrome cohorts compared to CTRL.
Supplementary Fig. 3. Disease-specific differential bin activity (FDR+) relative to total bin FPKM.