Abstract
New antibiotics with novel mechanisms of action are urgently needed to overcome the growing bacterial resistance problem faced by clinicians today. PC190723 and related compounds represent a promising new class of antibacterial compounds that target the essential bacterial cell division protein FtsZ. While this family of compounds exhibit potent antistaphylococcal activity, they have poor activity against enterococci and streptococci. The studies described herein are aimed at investigating the molecular basis of the enterococcal and streptococcal resistance to this family of compounds. We show that the poor activity of the compounds against enterococci and streptococci correlates with a correspondingly weak impact of the compounds on the self-polymerization of the FtsZ proteins from those bacteria. In addition, computational and mutational studies identify two key FtsZ residues (E34 and R308) as being important determinants of enterococcal and streptococcal resistance to the PC190723-type class of compounds.
1. Introduction
Multidrug resistance has become increasingly prevalent among bacterial pathogens of acute clinical importance [1–4]. An ongoing global effort to develop new antibacterial agents has led to the identification of the bacterial protein FtsZ as an appealing new antibacterial target [5–29]. FtsZ is an essential cell division protein that self-assembles into polymers that form a cytokinetic ring (the Z-ring) at the site of division, while also serving to recruit other key protein constituents of the bacterial divisome [30–33]. Among the chemotypes that have been identified as promising new FtsZ-targeting agents is a family of heterocyclic compounds related to the benzamide PC190723 (PC) [10,14,19–20,34–35]. These compounds have been shown to act as inhibitors of bacterial cell division through enhancement of FtsZ self-polymerization and stabilization of FtsZ polymers [20,34]. They exhibit potent antistaphylococcal activity in vitro, including activity against multidrug-resistant (MDR) strains of methicillin-resistant Staphylococcus aureus (MRSA) [10,14,19].
While compounds related to PC have potent activity against certain Gram-positive bacteria like staphylococci and Bacillus subtilis, they are weakly active against others, such as enterococci and streptococci [36]. An important first step toward the design of next-generation compounds with broader spectra of activity that include enterococci and streptococci is to define the key molecular determinants for the resistance exhibited by these bacteria. Here we describe a series of studies aimed at addressing this goal, with PC and a related benzimide derivative (TXY536) being used as representative compounds (see structures in Figure S1). Our studies reveal that the poor activity of PC and TXY536 against enterococci and streptococci is correlated with a correspondingly weak impact on the polymerization of the FtsZ proteins from those bacteria. In addition, computational and mutational studies have enabled us to identify two key FtsZ residues (E34 and R308) as being important determinants of the enterococcal and streptococcal resistance phenotype.
2. Materials and Methods
2.1 Bacterial Strains
S. aureus 8325-4 was a gift from Dr. Glenn W. Kaatz (John D. Dingell VA Medical Center, Detroit, MI), and B. subtilis FG347 was a gift from Dr. Richard Losick (Harvard University, Boston, MA). B. subtilis 168 was obtained from the Bacillus Genetic Stock Center (Columbus, OH). All other bacterial strains were obtained from the American Type Culture Collection (ATCC).
2.2 PC, TXY536, and FtsZ proteins
PC was synthesized as previously described [36]. TXY536 was synthesized in the following two series of steps: (i) N-methylisonipecotic acid hydrochloride (0.5 g) was dissolved in dry SOCl2 (1.5 mL). The mixture was then heated at 80 °C for 2 hours under argon. Cooling and evaporation to dryness afforded the acyl chloride as a yellow solid, which was used without further purification. (ii) In a round bottom flask PC (25 mg, 0.07 mmol) was dissolved in 2.0 mL of dry THF, and the solution was cooled to 0 °C under nitrogen. This was followed by portion-wise addition of NaH (11 mg, 0.24 mmol, 60% dispersion in mineral oil). The mixture was stirred at 0 °C for 10 minutes and then at room temperature for 30 minutes. The mixture was then cooled to 0 °C, and a solution of the above acyl chloride (28 mg, 0.14 mmol) in 1 mL of THF was added drop-wise. The resulting reaction mixture was stirred at 0 °C for 10 minutes and then at room temperature overnight. After completion, the reaction was quenched by the addition of a few drops of 1N NaOH, and diluted with ethyl acetate. The organic phase was separated, washed successively with saturated NaHCO3 brine, and dried. The solvent was removed under reduced pressure, and the resulting residue was purified by ISCO using 10% MeOH in DCM + 1% NH4OH to afford a yellow solid (16 mg, 36% yield). m.p. 170–172 °C. 1H NMR (300 MHz, CDCl3) δ: 8.01 (d, J = 2.1 Hz, 1H), 7.82 (d, J = 8.7 Hz, 1H), 7.40 (dd, J = 6.6, 1.8 Hz, 1H), 7.25-7.14 (m, 1H), 6.93-6.87 (m, 1H), 5.51 (s, 2H), 2.94-2.85 (m, 3H), 2.28 (s, 3H), 2.09-1.69 (m, 6H). HRMS Calculated for C22H20ClF2N3O3S (M + H)+ 480.0955. Found 480.0875.
The FtsZ proteins from Enterococcus faecalis, B. subtilis, and Streptococcus pneumoniae were obtained from Cytoskeleton, Inc., and used without further purification. Wild-type and mutant FtsZ proteins from S. aureus were expressed in E. coli and purified as previously described [28].
2.3 Minimum Inhibitory Concentration (MIC) Assays
MIC assays were conducted in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines for broth microdilution [37]. Briefly, log-phase bacteria were added to 96-well microtiter plates (at 5 × 105 CFU/mL) containing two-fold serial dilutions of compound in broth at concentrations ranging from 64 to 0.125 µg/mL, with each concentration being present in duplicate. The final volume in each well was 0.1 mL, and the microtiter plates were incubated aerobically for 24 hours at 37 °C. Bacterial growth was then monitored by measuring OD600 using a VersaMax® plate reader (Molecular Devices, Inc.), with the MIC90 being defined as the lowest compound concentration at which growth was ≥90% inhibited. CAMH broth was used for the S. aureus, B. subtilis, Streptococcus pyogenes, Streptococcus agalactiae, and S. pneumoniae experiments, with brain heart infusion (BHI) broth being used for the E. faecalis and Enterococcus faecium experiments. CAMH broth was supplemented with 2% NaCl in the MRSA experiments, while being supplemented with 5% (v/v) defibrinated sheep’s blood (Becton Dickinson and Co.) in the S. pyogenes, S. agalactiae, and S. pneumoniae experiments.
2.4 FtsZ Polymerization Assays
FtsZ polymerization was monitored using a microtiter plate-based spectrophotometric assay in which polymerization is reflected by corresponding increase in absorbance at 340 nm (A340). PC and TXY536 (at concentrations ranging from 0 to 5 µg/mL) were combined with 5 µM FtsZ in 100 µL of reaction solution, which contained 50 mM Tris•HCl (pH 7.4), 50 mM KCl, and 10 mM magnesium acetate. Reactions were assembled in half-volume, flat-bottom, 96-well microtiter plates, and initiated by addition of 4 mM GTP. Polymerization was continuously monitored at 25 °C by measuring A340 in a VersaMax® plate reader over a time period of 60 minutes. In the stability studies of the SaFtsZ and BsFtsZ polymers formed in the presence of compound, acquisition was interrupted long enough for the addition of 40 mM GDP to the reaction mix and then resumed.
2.5 Phase Contrast Microscopy
Log-phase S. aureus 8325-4 cells were cultured in cation-adjusted Mueller-Hinton (CAMH) broth at 37 °C for 2–4 hours in the presence of vehicle (DMSO), 3 µg/mL TXY536, or 3 µg/mL PC. A 1 mL sample was withdrawn from each bacterial culture, and centrifuged at 16,000g for 3 minutes at room temperature. The supernatant was then removed and the bacterial pellet was washed with 1 mL of phosphate-buffered saline (PBS). The final bacterial pellet was then resuspended in 50 µL of PBS, with 5 µL of the resulting bacterial suspension being transferred onto a microscope slide together with 5 µL of 1% molten agarose (made in PBS). A cover slip was then applied and the slide was visualized with a Zeiss Axioplan 2 microscope equipped with a Plan-Apochromat 100× objective (NA = 1.40). Images were captured with a Zeiss Axiocam HR camera using the OpenLab software package.
2.6 Fluorescence Microscopy
Exponentially growing B. subtilis FG347 cells were cultured in CAMH broth for 1 hour at 37 °C with DMSO vehicle, 3 µg/mL TXY536, or 3 µg/mL PC. Expression of GFP-conjugated ZapA was then induced by addition of xylose to a final concentration of 0.25% (w/v), and the cells were incubated for 1 additional hour at 37 °C. The bacterial cultures were then treated and visualized as described above, with the additional incorporation of a standard GFP filter set.
2.7 Computational Studies
The structure of the S. aureus FtsZ used in the docking studies was derived from the crystal structure corresponding to PDB # 4DXD [38]. The structure of E. faecalis FtsZ used in the docking studies was a homology model built via the single template approach using the Modeller program (version 9.10) [39–40]. The model was built from the sequence of E. faecalis FtsZ [E. faecalis ARO1/DG] (NCBI Protein Accession # EEU87404.1). A BLAST search of non-redundant PDB sequences clustered at 95% identity gave the crystal structure of B. subtilis FtsZ (PDB # 2VXY) [10] as the optimum template, with a sequence identity of 81% and a resolution of 1.7 Å. The homology model with the lowest DOPE score was selected and the C-terminal residues from number 318 to the C-terminus were removed. Analysis of the model using PROCHECK revealed >95% of the residues to be in the most favored regions of the Ramachandran plot [41].
All docking of TXY536 and PC was performed using the Autodock Vina program (version 1.1.2) [42]. Missing sidechain atoms in the S. aureus FtsZ crystal structure were replaced using the Swiss PDB viewer [43]. In all dockings, the docking grid box size was kept at 24 Å × 18 Å × 18 Å, and was centered so as to include the two key residues flanking the binding pocket (residues 33 and 307 for S. aureus FtsZ and residues 34 and 308 for E. faecalis FtsZ). A Vina docking exhaustiveness of 12 was used. The small molecules were initially prepared and geometry optimized with the MMFF94 force field using Spartan 10 (Wavefunction, Inc.). All proteins and small molecules were prepared for Vina docking using AutoDock Tools (version 1.5.7rc1) [44].
3. Results and Discussion
3.1 MIC studies establish the resistance of enterococci and streptococci to TXY536 and PC
We evaluated the activities of TXY536 and PC against various Gram-positive bacteria, including methicillin-sensitive and methicillin-resistant S. aureus (MSSA and MRSA, respectively), B. subtilis, vancomycin-sensitive E. faecalis and E. faecium (VSE), vancomycin-resistant E. faecalis (VRE), S. pyogenes (Group A Streptococcus, GAS), S. agalactiae (Group B Streptococcus, GBS), and S. pneumoniae. While exhibiting potent activity against all the S. aureus (MIC = 0.25–1.0 µg/mL) and B. subtilis (MIC = 0.5–1.0 µg/mL) strains, neither compound exhibited significant activity (MIC = ≥32 µg/mL) against any of the Enterococcus or Streptococcus strains examined (Table 1).
Table 1.
MIC90 values of TXY536 and PC against various Gram-positive bacteriaa
| Organism | Strain | MIC90 (µg/mL) | |
|---|---|---|---|
|
| |||
| PC | TXY536 | ||
| S. aureus (MSSA) | 8325-4 | 0.25 | 0.5 |
| S. aureus (MSSA) | ATCC 19636 | 0.5 | 0.5 |
| S. aureus (MSSA) | ATCC 29213 | 1.0 | 1.0 |
| S. aureus (MSSA) | ATCC49951 | 0.5 | 1.0 |
| S. aureus (MRSA) | ATCC 33591 | 0.5 | 0.5 |
| S. aureus (MRSA) | ATCC 43300 | 0.5 | 1.0 |
| B. subtilis | 168 | 0.5 | 1.0 |
| E. faecalis (VSE) | ATCC 19433 | 32.0 | 32.0 |
| E. faecalis (VRE) | ATCC 51575 | 32.0 | 64.0 |
| E. faecium (VSE) | ATCC 19434 | 64.0 | >64.0 |
| S. pyogenes (GAS) | ATCC 19615 | >64.0 | >64.0 |
| S. agalactiae (GBS) | ATCC 12386 | >64.0 | >64.0 |
| S. pneumoniae | ATCC 49619 | >64.0 | 64.0 |
MIC90 values reflect the lowest compound concentrations at which growth was ≥90% inhibited.
3.2 TXY536 and PC enhance the polymerization of the FtsZ proteins from sensitive bacteria, while exerting little or no impact on the polymerization of the FtsZ proteins from resistant bacteria
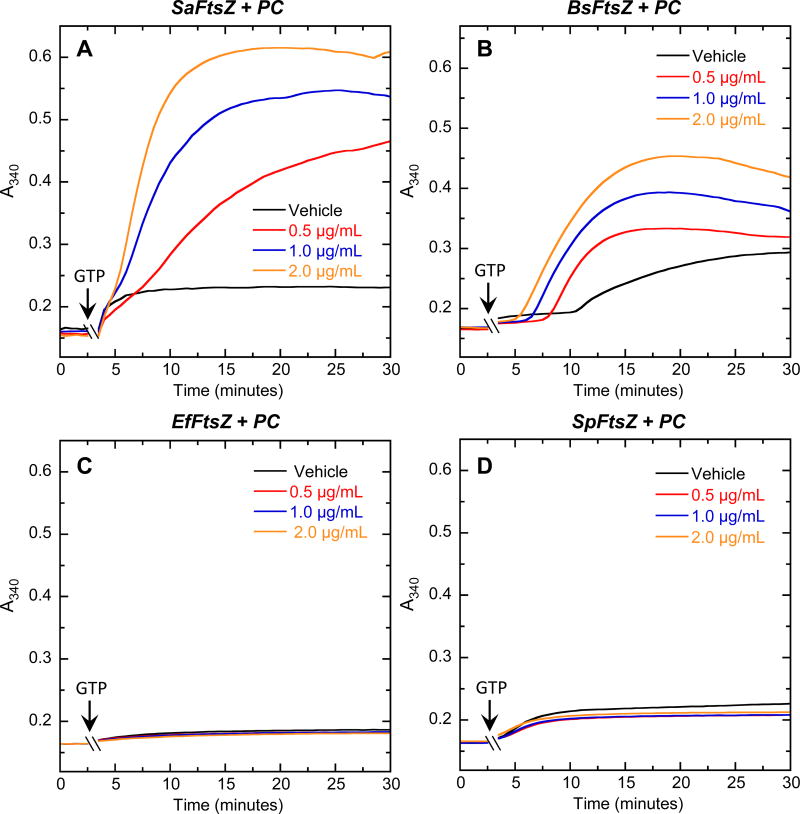
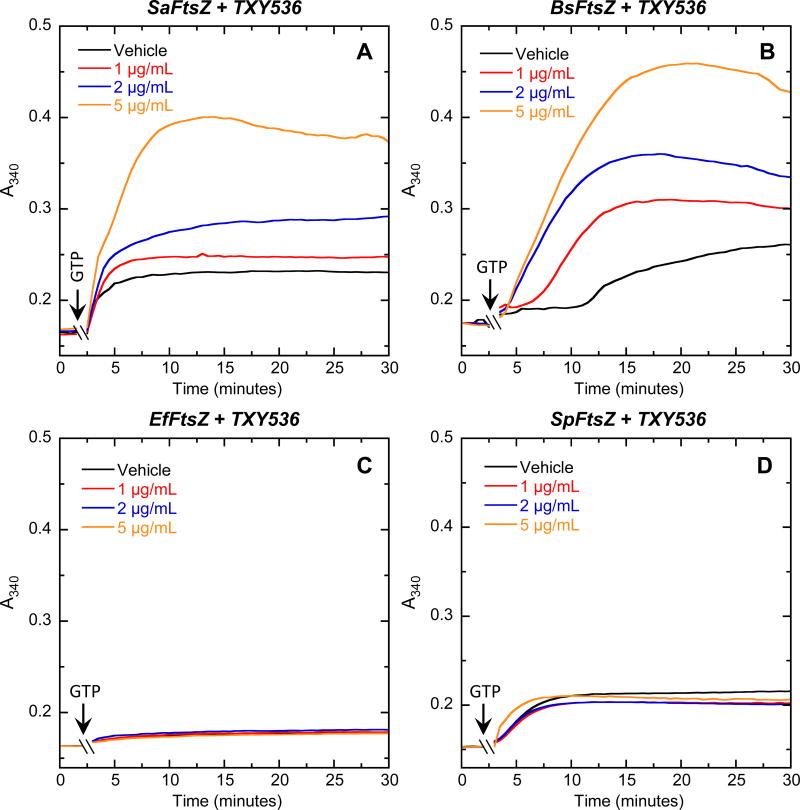
As an initial step toward exploring the molecular basis for the observed spectrum of activity of the TXY536 and PC, we compared the impacts of the compounds on the polymerization of purified S. aureus, B. subtilis, E. faecalis and S. pneumoniae FtsZ (henceforth denoted SaFtsZ, BsFtsZ, EfFtsZ, and SpFtsZ, respectively). To assay for the FtsZ self-polymerization function, we employed a microtiter plate-based spectrophotometric assay in which FtsZ polymerization is detected in solution by a time-dependent increase in solution absorbance at 340 nm (A340) after addition of GTP. Both compounds enhanced the extent of SaFtsZ (Figures 2A and 3A) and BsFtsZ (Figures 2B and 3B) polymerization, with the magnitude of these stimulatory effects increasing with increasing compound concentration. Note that the GTP-induced A340 profiles of SaFtsZ and BsFtsZ acquired in the presence of either compound were unaltered by subsequent addition of a 10-fold excess of GDP (see Figure S1 of the Supplementary Material for representative results with SaFtsZ). Thus, the SaFtsZ and BsFtsZ polymers induced by the presence of the compounds were stable to the depolymerizing effects of GDP. In striking contrast to the stimulatory impacts of PC and TXY536 on the polymerization of SaFtsZ and BsFtsZ, neither compound exerted a significant impact on the polymerization of EfFtsZ (Figures 2C and 3C) and SpFtsZ (Figures 2D and 3D). These differential FtsZ polymerization effects correlate well with the corresponding antibacterial activities of the compounds (Table 1). In short, the compounds enhance the polymerization of the FtsZ proteins from sensitive bacteria (S. aureus and B. subtilis), while exerting little or no impact on the polymerization of the FtsZ proteins from resistant bacteria (E. faecalis and S. pneumoniae).
Fig. 2.
Concentration dependence of the impact of PC on the polymerization of SaFtsZ (A), BsFtsZ (B), EfFtsZ (C), and SpFtsZ (D), as determined by monitoring time-dependent changes in absorbance at 340 nm (A340) at 25 °C. The time-dependent A340 polymerization profiles are shown for each target FtsZ in the presence of DMSO vehicle (black) or PC at a concentration of 0.5 (red), 1.0 (blue), or 2.0 (orange) µg/mL. Experimental conditions for all the FtsZ polymerization studies were 5 µM protein, 50 mM Tris•HCl (pH 7.4), 50 mM KCl, and 10 mM magnesium acetate. Polymerization reactions were initiated by addition of 4 mM GTP at the times indicated by the arrows.
Fig. 3.
Concentration dependence of the impact of TXY536 on the polymerization of SaFtsZ (A), BsFtsZ (B), EfFtsZ (C), and SpFtsZ (D). The time-dependent A340 polymerization profiles are shown for each target FtsZ in the presence of DMSO vehicle (black) or TXY536 at a concentration of 1.0 (red), 2.0 (blue), or 5.0 (orange) µg/mL. Experimental conditions were as described in the legend to Figure 2.
3.3 The stimulatory effect of TXY536 and PC on polymerization of the FtsZ proteins from sensitive bacteria correlates with inhibition of cell division and disruption of Z-ring formation in the target bacteria
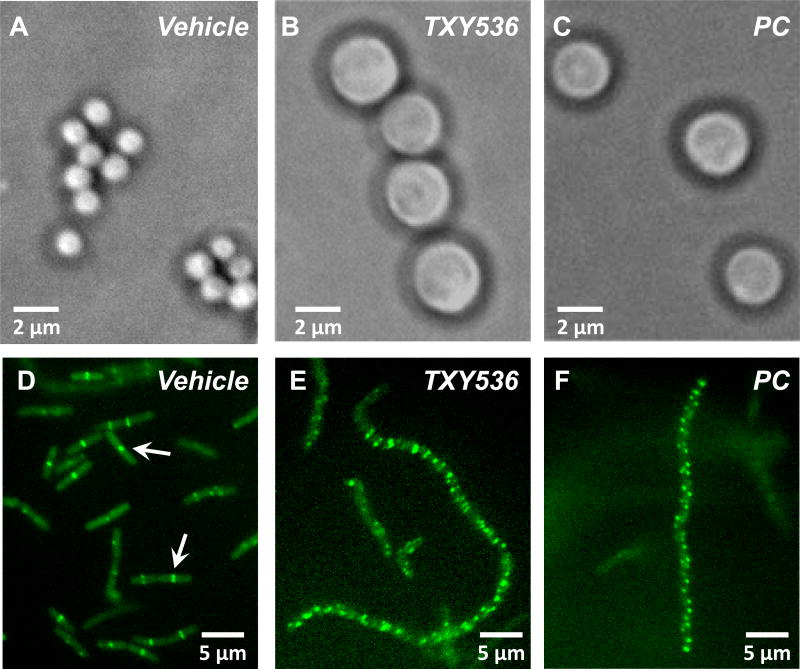
We next conducted a series of microscopy studies aimed at verifying that the compound-induced stimulatory impact on SaFtsZ and BsFtsZ polymerization observed in vitro correlated with disruption of FtsZ function in live bacteria. It has previously been shown that spherical bacteria (such as the S. aureus), which have impaired FtsZ function (either due to mutations or inhibitors like PC), exhibit an enlarged phenotype due to their inability to complete the cell division process [9–10,45]. In this connection, we examined the impact of TXY536 and PC treatment on the cellular morphology of S. aureus using phase contrast microscopy. Control S. aureus bacteria treated with vehicle (DMSO) alone are approximately 1 µm in diameter (Figure 4A), a value in agreement with previously reported diameters of typical S. aureus cocci [46]. In marked contrast, the diameter of the S. aureus bacteria treated with TXY536 or PC are 2.5 to 3.5-fold larger, ranging in diameter from 2.5 to 3.5 µm (Figures 4B and C). These observations are consistent with both compounds inhibiting S. aureus cell division through inhibition of FtsZ function in the bacteria.
Fig. 4.
(A–C) Phase-contrast micrographs of S. aureus 8325-4 bacteria treated with DMSO vehicle (A), 3 µg/mL TXY536 (B), or 3 µg/mL PC (C). (D–F) Fluorescence micrographs of B. subtilis FG347 bacteria that express a GFP-tagged, FtsZ Z-ring marker protein termed ZapA. The bacteria were treated with DMSO vehicle (D), 3 µg/mL TXY536 (E), or 3 µg/mL PC (F).
To verify that the compounds are indeed targeting FtsZ in bacteria, we used a strain of B. subtilis (FG347) that inducibly expresses GFP-tagged ZapA, a known protein marker for FtsZ Z-rings [47] FtsZ Z-rings bearing fluorescent marker proteins can be visualized in such strains using fluorescence microscopy [15,47–51]. We treated FG347 bacteria with TXY536 or PC and examined the impact of the compounds on FtsZ Z-ring formation. In the absence of compound, fluorescent foci corresponding to Z-rings are evident at midcell in the rod-shaped bacilli (highlighted by the arrows in Figure 4D). By contrast, PC- and TXY536-treated bacteria lack these midcell foci, instead exhibiting punctate fluorescent foci distributed throughout each cell (Figures 4E and F). It is likely that these punctate foci reflect non-functional FtsZ polymeric structures induced and stabilized by the compounds as a result of their stimulatory impacts on BsFtsZ polymerization (Figures 2B and 3B). Note that the compound-treated B. subtilis bacteria are also significantly more elongated (filamentous) than the vehicle-treated bacteria (compare Figures 4E and F with Figure 4D). This type of morphological change is characteristic of rod-shaped bacteria in which cell division has been disrupted through interference with FtsZ function by mutation or treatment with inhibitors, including PC [9–10,15,47–50]. Our collective microscopy results thus confirm that TXY536, like PC, acts as an inhibitor of cell division in sensitive bacteria through disruption of FtsZ Z-ring formation.
3.4 Computational studies highlight FtsZ residues E34 and R308 as potentially key contributors to the observed enterococcal and streptococcal resistance phenotype
We next sought to investigate the molecular basis for the differential impacts of TXY536 and PC on the FtsZ proteins from the sensitive versus the resistant bacteria. As a first step toward this end, we compared the amino acid sequences of the FtsZ proteins from the various bacteria listed in Table 1, with a particular emphasis on the residues in the vicinity of the FtsZ binding pocket that has been previously defined for PC [38,52] (see sequence alignment in Figure 5). Two key amino acid differences (highlighted in red) between the FtsZ proteins from the resistant versus the sensitive bacteria were noted among the residues bordering the compound binding site. Specifically, the FtsZ proteins from enterococci and streptococci, which are resistant to TXY536 and PC, possess a glutamic acid residue at position 34 and an arginine residue at position 308. By contrast, the identities of the residues at the corresponding positions in the FtsZ proteins from the sensitive bacteria are histidine (H33) and valine (V307) in the case of S. aureus and glutamine (N33) and valine (V307) in the case of B. subtilis.
Fig. 5.
Alignment of two regions of the amino acid sequences of the FtsZ proteins from S. aureus, B. subtilis, E. faecalis, E. faecium, S. pneumoniae, S. pyogenes, and S. agalactiae. Key amino acid positions in the area of the PC and TXY536 binding pocket of FtsZ are highlighted in red.
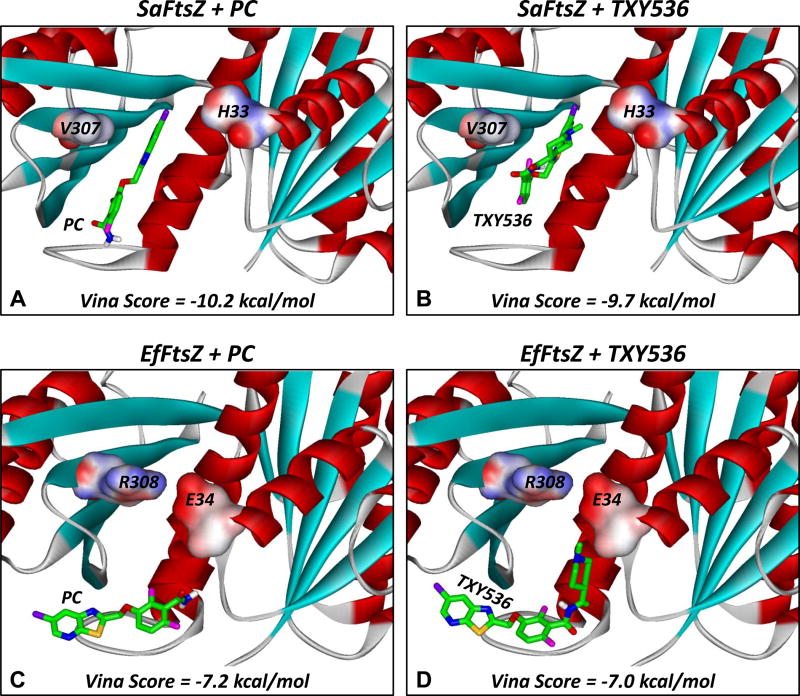
We reasoned that the E34 and R308 residues in the FtsZ proteins from the resistant bacteria may form a salt bridge that, in turn, could interfere with the binding of the compounds to the FtsZ target site. In this connection, we constructured a homolgy model of EfFtsZ for comparison with the recently reported crystal structure of SaFtsZ. We then docked TXY536 and PC to each of the target FtsZ structures using the AutoDock Vina docking algorithm, with the top scoring docking orientations resulting from these computational analyses being shown in Figure 6. PC docked into a narrow binding pocket on SaFtsZ, flanked by the H33 residue on one side and the V307 residue on the other (Figure 6A). Significantly, the orientation adopted by PC in the SaFtsZ binding pocket was similar to that observed in the recently reported crystal structure of the PC-SaFtsZ complex [38,52]. Despite its larger size (conferred by the methylpiperidine functionality), TXY536 was able to dock into the same SaFtsZ binding pocket as PC (Figure 6B), with the Vina docking scores of both compounds being similar (−10.2 kcal/mol for PC and −9.7 kcal/mol for TXY536).
Fig. 6.
Docked structures of PC (A,C) and TXY536 (B,D) in complex with SaFtsZ (A,B) and EfFtsZ (C,D). The compounds were docked using the AutoDock Vina algorithm, with the top scoring binding orientations being depicted and their associated scores indicated. The structure of SaFtsZ is derived from the reported crystal structure (PDB # 4DXD) [38], while the structure of EfFtsZ is a homology model built as described in the Materials and Methods. The FtsZ proteins are depicted according to their secondary structural elements (α-helices in red, β-strands in cyan, and loops in gray). The residues at positions 33/34 and 307/308 are portrayed in their solvent accessible surfaces and color-coded according to electrostatic potential (blue for positive, red for negative, and white for neutral). The compounds are shown in stick format and color-coded according to atom (carbon in green, nitrogen in blue, oxygen in red, sulfur in yellow, fluorine in magenta, and chlorine in violet).
The homolgy model of EfFtsZ did indeed reveal the presence of a salt bridge between residues E34 and R308 (Figures 6C and D), which, in turn, caused the compound binding site observed in SaFtsZ to be partially occluded in EfFtsZ. As a result, both PC (Figure 6C) and TXY536 (Figure 6D) were forced to bind outside of the compound target site in EfFtsZ, with the corresponding Vina docking scores (−7.2 kcal/mol for PC and −7.0 kcal/mol for TXY536) being significantly lower than those observed when SaFtsZ served as the target protein. In the aggregate, the computational docking results are consistent with FtsZ residues E34 and R308 contributing to enterococcal and streptococcal resistance to PC and TXY536 by antagonizing the abilities of the compounds to target the key FtsZ site.
3.5 Mutational studies confirm that FtsZ residues E34 and R308 are important determinants of enterococcal and streptococcal resistance
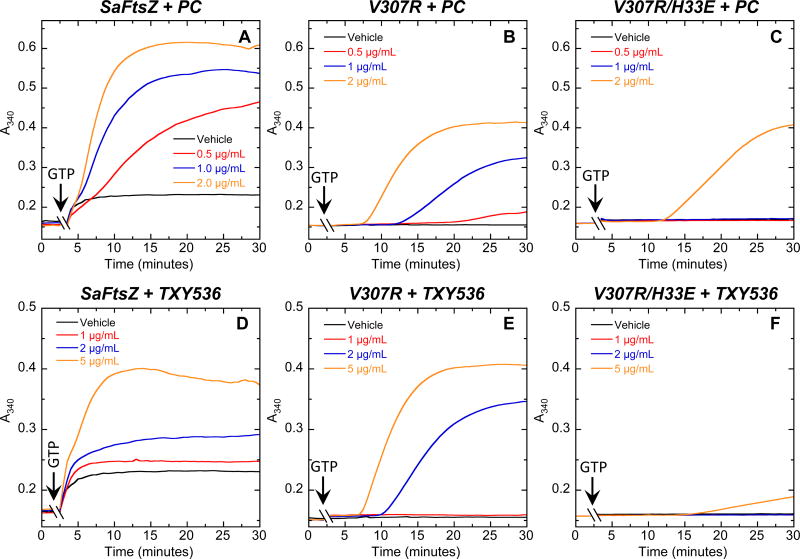
To experimentally test whether the E34 and R308 residues might indeed adversely affect the stimulatory impact of TXY536 and PC on FtsZ polymerization, we introduced the single V307R and double V307R/H33E mutations into SaFtsZ. We then examined how these mutations affect the abilities of increasing compound concentrations to enhance SaFtsZ polymerization. At the lower compound concentrations (0.5 – 1.0 µg/mL), the single V307R mutation introduces a modest reduction in the extent to which the compounds enhance SaFtsZ polymerization (compare the A340 profiles in Figures 7A and B as well as those in Figures 7D and E). However, this effect is not observed at the higher compound concentrations (2.0 – 5.0 µg/mL). It has been reported that a V307R mutation in BsFtsZ increases the MIC of PC against B. subtilis from 0.5 to 8.0 µg/mL [10]. Our mutational FtsZ polymerization results shown here provide a molecular explanation for this partial resistance behavior.
Fig. 7.
Concentration dependence of the impact of PC (A–C) and TXY536 (D–F) on the polymerization of wild-type (A,D), V307R single mutant (B,E), and V307R/H33E double mutant (C,F) SaFtsZ, as determined by monitoring time-dependent changes in absorbance at 340 nm (A340) at 25 °C. The time-dependent A340 polymerization profiles are shown for each target FtsZ in the presence of DMSO vehicle or the indicated concentrations of PC and TXY536. Experimental conditions were as described in the legend to Figure 2.
Unlike the single V307R mutation, the double V307R/H33E mutation introduces a pronounced reduction in the enhancement of SaFtsZ polymerization by the compounds at all the concentrations examined (compare Figures 7A and C as well as Figures 7D and F). This reduction in compound-induced enhancement of SaFtsZ polymerization afforded by the V307R/H33E double mutation is similar to the behavior we observed when comparing the impacts of the compounds on EfFtsZ and SpFtsZ polymerization to the corresponding impacts on SaFtsZ and BsFtsZ polymerization (Figures 2 and 3). In essence, incorporation of the V307R/H33E double mutation in SaFtsZ rendered the protein resistant to TXY536 and PC. Thus, confirming the hypothesis invoked by our computational studies, our mutational analysis identifies residues E34 and R308 as being important determinants of enterococcal and streptococcal resistance to the PC-type class of compounds.
In conclusion, our studies provide important insights into the molecular basis for the resistance of enterococci and streptococci to a promising new family of FtsZ-targeting antibacterial compounds. These findings should facilitate future design efforts geared toward development of next-generation broader spectrum compounds that can target not only staphylococcal, but also enterococcal and streptococcal FtsZ proteins.
Fig. 1.
Chemical structures TXY536 and PC190723 (PC).
Acknowledgments
This study was supported by research agreements between TAXIS Pharmaceuticals, Inc. and both the University of Medicine and Dentistry of New Jersey (D.S.P) and Rutgers - The State University of New Jersey (E.J.L.). We are indebted to Dr. Glenn W. Kaatz (John D. Dingell VA Medical Center, Detroit, MI) for providing us with the S. aureus 8325-4 strain and to Dr. Richard Losick (Harvard University, Boston, MA) for providing us with the B. subtilis FG347 strain.
References
- 1.Arias CA, Murray BE. Antibiotic-Resistant Bugs in the 21st Century--A Clinical Super-Challenge. New Engl. J. Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 2.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. Emergence of a New Antibiotic Resistance Mechanism in India, Pakistan, and the UK: a Molecular, Biological, and Epidemiological Study. Lancet Infect. Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, Lerner SA, Levy S, Lewis K, Lomovskaya O, Miller JH, Mobashery S, Piddock LJ, Projan S, Thomas CM, Tomasz A, Tulkens PM, Walsh TR, Watson JD, Witkowski J, Witte W, Wright G, Yeh P, Zgurskaya HI. Tackling Antibiotic Resistance. Nat. Rev. Microbiol. 2011;9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA. Tracking a Hospital Outbreak of Carbapenem-Resistant Klebsiella pneumoniae with Whole-Genome Sequencing. Sci. Transl. Med. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Läppchen T, Hartog AF, Pinas VA, Koomen GJ, den Blaauwen T. GTP Analogue Inhibits Polymerization and GTPase Activity of the Bacterial Protein FtsZ without Affecting its Eukaryotic Homologue Tubulin. Biochemistry. 2005;44:7879–7884. doi: 10.1021/bi047297o. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Galgoci A, Kodali S, Herath KB, Jayasuriya H, Dorso K, Vicente F, Gonzalez A, Cully D, Bramhill D, Singh S. Discovery of a Small Molecule that Inhibits Cell Division by Blocking FtsZ, a Novel Therapeutic Target of Antibiotics. J. Biol. Chem. 2003;278:44424–44428. doi: 10.1074/jbc.M307625200. [DOI] [PubMed] [Google Scholar]
- 7.Margalit DN, Romberg L, Mets RB, Hebert AM, Mitchison TJ, Kirschner MW, RayChaudhuri D. Targeting Cell Division: Small-Molecule Inhibitors of FtsZ GTPase Perturb Cytokinetic Ring Assembly and Induce Bacterial Lethality. Proc. Natl. Acad. Sci. USA. 2004;101:11821–11826. doi: 10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urgaonkar S, La Pierre HS, Meir I, Lund H, RayChaudhuri D, Shaw JT. Synthesis of Antimicrobial Natural Products Targeting FtsZ: (+/−)-Dichamanetin and (+/−)-2"'-hydroxy-5"-benzylisouvarinol-B. Org. Lett. 2005;7:5609–5612. doi: 10.1021/ol052269z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokes NR, Sievers J, Barker S, Bennett JM, Brown DR, Collins I, Errington VM, Foulger D, Hall M, Halsey R, Johnson H, Rose V, Thomaides HB, Haydon DJ, Czaplewski LG, Errington J. Novel Inhibitors of Bacterial Cytokinesis Identified by a Cell-Based Antibiotic Screening Assay. J. Biol. Chem. 2005;280:39709–39715. doi: 10.1074/jbc.M506741200. [DOI] [PubMed] [Google Scholar]
- 10.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, Baker PJ, Barynin VV, Rice DW, Sedelnikova SE, Heal JR, Sheridan JM, Aiwale ST, Chauhan PK, Srivastava A, Taneja A, Collins I, Errington J, Czaplewski LG. An Inhibitor of FtsZ with Potent and Selective Anti-Staphylococcal Activity. Science. 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 11.Reynolds RC, Srivastava S, Ross LJ, Suling WJ, White EL. A New 2-Carbamoyl Pteridine that Inhibits Mycobacterial FtsZ. Bioorg. Med. Chem. Lett. 2004;14:3161–3164. doi: 10.1016/j.bmcl.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Huang Q, Kirikae F, Kirikae T, Pepe A, Amin A, Respicio L, Slayden RA, Tonge PJ, Ojima I. Targeting FtsZ for Antituberculosis Drug Discovery: Noncytotoxic Taxanes as Novel Antituberculosis Agents. J. Med. Chem. 2006;49:463–466. doi: 10.1021/jm050920y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee S, Robinson CA, Howe AG, Mazor T, Wood PA, Urgaonkar S, Hebert AM, Raychaudhuri D, Shaw JT. N-Benzyl-3-sulfonamidopyrrolidines as novel inhibitors of cell division in E coli. Bioorg. Med. Chem. Lett. 2007;17:6651–6655. doi: 10.1016/j.bmcl.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czaplewski LG, Collins I, Boyd EA, Brown D, East SP, Gardiner M, Fletcher R, Haydon DJ, Henstock V, Ingram P, Jones C, Noula C, Kennison L, Rockley C, Rose V, Thomaides-Brears HB, Ure R, Whittaker M, Stokes NR. Antibacterial Alkoxybenzamide Inhibitors of the Essential Bacterial Cell Division Protein FtsZ. Bioorg. Med. Chem. Lett. 2009;19:524–527. doi: 10.1016/j.bmcl.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Beuria TK, Santra MK, Panda D. Sanguinarine Blocks Cytokinesis in Bacteria by Inhibiting FtsZ Assembly and Bundling. Biochemistry. 2005;44:16584–16593. doi: 10.1021/bi050767+. [DOI] [PubMed] [Google Scholar]
- 16.Domadia PN, Bhunia A, Sivaraman J, Swarup S, Dasgupta D. Berberine Targets Assembly of Escherichia coli Cell Division Protein FtsZ. Biochemistry. 2008;47:3225–3234. doi: 10.1021/bi7018546. [DOI] [PubMed] [Google Scholar]
- 17.Schaffner-Barbero C, Martin-Fontecha M, Chacón P, Andreu JM. Targeting the Assembly of Bacterial Cell Division Protein FtsZ with Small Molecules. ACS Chem. Biol. 2012;7:269–277. doi: 10.1021/cb2003626. [DOI] [PubMed] [Google Scholar]
- 18.Ma S. The Development of FtsZ Inhibitors As Potential Antibacterial Agents. ChemMedChem. 2012;7:1161–1172. doi: 10.1002/cmdc.201200156. [DOI] [PubMed] [Google Scholar]
- 19.Haydon DJ, Bennett JM, Brown D, Collins I, Galbraith G, Lancett P, Macdonald R, Stokes NR, Chauhan PK, Sutariya JK, Nayal N, Srivastava A, Beanland J, Hall R, Henstock V, Noula C, Rockley C, Czaplewski L. Creating an Antibacterial with in Vivo Efficacy: Synthesis and Characterization of Potent Inhibitors of the Bacterial Cell Division Protein FtsZ with Improved Pharmaceutical Properties. J. Med. Chem. 2010;53:3927–3936. doi: 10.1021/jm9016366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreu JM, Schaffner-Barbero C, Huecas S, Alonso D, Lopez-Rodriguez ML, Ruiz-Avila LB, Núñez-Ramírez R, Llorca O, Martín-Galiano AJ. The Antibacterial Cell Division Inhibitor PC190723 Is an FtsZ Polymer-Stabilizing Agent That Induces Filament Assembly and Condensation. J. Biol. Chem. 2010;285:14239–14246. doi: 10.1074/jbc.M109.094722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lock RL, Harry EJ. Cell-Division Inhibitors: New Insights for Future Antibiotics. Nat. Rev. Drug Discov. 2008;7:324–338. doi: 10.1038/nrd2510. [DOI] [PubMed] [Google Scholar]
- 22.Boberek JM, Stach J, Good L. Genetic Evidence for Inhibition of Bacterial Division Protein FtsZ by Berberine. PLoS One. 2010;5:e13745. doi: 10.1371/journal.pone.0013745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awasthi D, Kumar K, Ojima I. Therapeutic Potential of FtsZ Inhibition: A Patent Perspective. Expert Opin. Ther. Patents. 2011;21:657–679. doi: 10.1517/13543776.2011.568483. [DOI] [PubMed] [Google Scholar]
- 24.Huang Q, Tonge PJ, Slayden RA, Kirikae T, Ojima I. FtsZ: A Novel Target for Tuberculosis Drug Discovery. Curr. Top. Med. Chem. 2007;7:527–543. doi: 10.2174/156802607780059790. [DOI] [PubMed] [Google Scholar]
- 25.Kumar K, Awasthi D, Berger WT, Tonge PJ, Slayden RA, Ojima I. Discovery of Anti-TB Agents That Target the Cell-Division Protein FtsZ. Future Med. Chem. 2010;2:1305–1323. doi: 10.4155/fmc.10.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar K, Awasthi D, Lee SY, Zanardi I, Ruzsicska B, Knudson S, Tonge PJ, Slayden RA, Ojima I. Novel Trisubstituted Benzimidazoles, Targeting Mtb FtsZ, as a New Class of Antitubercular Agents. J. Med. Chem. 2011;54:374–381. doi: 10.1021/jm1012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor S, Panda D. Targeting FtsZ for Antibacterial Therapy: A Promising Avenue. Expert. Opin. Ther. Targets. 2009;13:1037–1051. doi: 10.1517/14728220903173257. [DOI] [PubMed] [Google Scholar]
- 28.Kaul M, Parhi AK, Zhang Y, LaVoie EJ, Tuske S, Arnold E, Kerrigan JE, Pilch DS. A Bactericidal Guanidinomethyl Biaryl That Alters the Dynamics of Bacterial FtsZ Polymerization. J. Med. Chem. 2012;55:10160–10176. doi: 10.1021/jm3012728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley C, Zhang Y, Parhi A, Kaul M, Pilch DS, LaVoie EJ. 3-Phenyl Substituted 6,7-Dimethoxyisoquinoline Derivatives as FtsZ-Targeting Antibacterial Agents. Bioorg. Med. Chem. 2012;20:7012–7029. doi: 10.1016/j.bmc.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bi EF, Lutkenhaus J. FtsZ Ring Structure Associated with Division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 31.Errington J, Daniel RA, Scheffers DJ. Cytokinesis in Bacteria. Microbiol. Mol. Biol. Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams DW, Errington J. Bacterial Cell Division: Assembly, Maintenance and Disassembly of the Z ring. Nat. Rev. Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 33.Kirkpatrick CL, Viollier PH. New(s) to the (Z-)ring. Current Opinions in Microbiology. 2011;14:691–697. doi: 10.1016/j.mib.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Adams DW, Wu LJ, Czaplewski LG, Errington J. Multiple Effects of Benzamide Antibiotics on FtsZ Function. Mol. Microbiol. 2011;80:68–84. doi: 10.1111/j.1365-2958.2011.07559.x. [DOI] [PubMed] [Google Scholar]
- 35.Stokes NR, Baker N, Bennett JM, Berry J, Collins I, Czaplewski LG, Logan A, Macdonald R, Macleod L, Peasley H, Mitchell JP, Nayal N, Yadav A, Srivastava A, Haydon DJ. An Improved Small-Molecule inhibitor of FtsZ with Superior in Vitro Potency, Drug-Like Properties, and in Vivo Efficacy. Antimicrob. Agents Chemother. 2013;57:317–325. doi: 10.1128/AAC.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorto NA, Olmstead MM, Shaw JT. Practical Synthesis of PC190723, an Inhibitor of the Bacterial Cell Division Protein FtsZ. J. Org. Chem. 2010;75:7946–7949. doi: 10.1021/jo101720y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.CLSI. Approved Standard-Eighth Edition. Clinical and Laboratory Standards Institute; Wayne, PA: 2009. Methods for Dilution Antimicrobial Sucseptibility Tests for Bacteria That Grow Aerobically. [Google Scholar]
- 38.Tan CM, Therien AG, Lu J, Lee SH, Caron A, Gill CJ, Lebeau-Jacob C, Benton-Perdomo L, Monteiro JM, Pereira PM, Elsen NL, Wu J, Deschamps K, Petcu M, Wong S, Daigneault E, Kramer S, Liang L, Maxwell E, Claveau D, Vaillancourt J, Skorey K, Tam J, Wang H, Meredith TC, Sillaots S, Wang-Jarantow L, Ramtohul Y, Langlois E, Landry F, Reid JC, Parthasarathy G, Sharma S, Baryshnikova A, Lumb KJ, Pinho MG, Soisson SM, Roemer T. Restoring Methicillin-Resistant Staphylococcus aureus Susceptibility to β-Lactam Antibiotics. Sci. Transl. Med. 2012;4:126ra135. doi: 10.1126/scitranslmed.3003592. [DOI] [PubMed] [Google Scholar]
- 39.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez R, Sali A. Comparative Protein Structure Modeling. Introduction and Practical Examples with Modeller. Methods Mol. Biol. 2000;143:97–129. doi: 10.1385/1-59259-368-2:97. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 42.Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An Environment for Comparative Protein Modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 44.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinho MG, Errington J. Dispersed Mode of Staphylococcus aureus Cell Wall Synthesis in the Absence of the Division Machinery. Mol. Microbiol. 2003;50:871–881. doi: 10.1046/j.1365-2958.2003.03719.x. [DOI] [PubMed] [Google Scholar]
- 46.Jorge AM, Hoiczyk E, Gomes JP, Pinho MG. EzrA Contributes to the Regulation of Cell Size in Staphylococcus aureus. PLoS One. 2011;6:e27542. doi: 10.1371/journal.pone.0027542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gueiros-Filho FJ, Losick R. A Widely Conserved Bacterial Cell Division Protein That Promotes Assembly of the Tubulin-Like Protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haeusser DP, Garza AC, Buscher AZ, Levin PA. The Division Inhibitor EzrA Contains a Seven-Residue Patch Required for Maintaining the Dynamic Nature of the Medial FtsZ Ring. J. Bacteriol. 2007;189:9001–9010. doi: 10.1128/JB.01172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA. EzrA Prevents Aberrant Cell Division by Modulating Assembly of the Cytoskeletal Protein FtsZ. Mol. Microbiol. 2004;52:801–814. doi: 10.1111/j.1365-2958.2004.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma X, Ehrhardt DW, Margolin W. Colocalization of Cell Division Proteins FtsZ and FtsA to Cytoskeletal Structures in Living Escherichia coli Cells by Using Green Fluorescent Protein. Proc. Natl. Acad. Sci. USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Handler AA, Lim JE, Losick R. Peptide Inhibitor of Cytokinesis During Sporulation in Bacillus subtilis. Mol. Microbiol. 2008;68:588–599. doi: 10.1111/j.1365-2958.2008.06173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsui T, Yamane J, Mogi N, Yamaguchi H, Takemoto H, Yao M, Tanaka I. Structural Reorganization of the Bacterial Cell-Division Protein FtsZ from Staphylococcus aureus. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2012;D68:1175–1188. doi: 10.1107/S0907444912022640. [DOI] [PubMed] [Google Scholar]