Abstract
Filamins are essential, evolutionary conserved, modular, multi-domain, actin-binding proteins that organize the actin cytoskeleton and maintain extracellular matrix connections by anchoring actin filaments to transmembrane receptors. By crosslinking and anchoring actin filaments, filamins stabilize the plasma membrane, provide cellular cortical rigidity and contribute to the mechanical stability of the plasma membrane and the cell cortex. In addition to actin, filamins interact with over 90 other binding partners including intracellular signaling molecules, receptors, ion channels, transcriptions factors and cytoskeletal and adhesion proteins. Thus, filamins scaffold a wide range of signaling pathways and are implicated in the regulation of a diverse array of cellular functions including motility, maintenance of cell shape and differentiation. Here, we review emerging structural and functional evidence that filamins are mechanosensors and/or mechanotransducers, playing essential roles in helping cells to detect and respond to physical forces in their local environment.
Keywords: Actin-binding protein, mechanosensor, mechanotransduction, extracellular matrix, cytoskeleton
INTRODUCTION
Filamins are conserved, modular, multi-domain proteins that contribute to the link between the extracellular matrix (ECM) and the cytoskeleton and interact with a host of other structural and signaling proteins (86, 120, 143) . Thus, they are well positioned to serve as mechanosensors - proteins that receive mechanical cues and transform them into biochemical signals. Indeed, numerous studies point to roles for filamins in mechanical cell processes, including cell motility, membrane stability, mechanoprotection and ECM stiffness sensing (11-13, 34, 37, 55, 56, 113).
Filamins are potent F-actin cross-linking proteins (120), but in addition to binding actin, filamins act as scaffolds for a wide range of signaling pathways and interact with over 90 binding partners including intracellular signaling molecules, receptors, ion channels, transcription factors and cytoskeletal and adhesion proteins (86, 95, 119, 120, 144). Notably, in response to extrinsic (applied) or intrinsic (cell-generated) mechanical force, filamins undergo conformational changes and unfolding events that change their affinity for binding partners and may expose cryptic binding sites leading to recruitment of additional components (32, 37, 45, 52, 54, 64, 69, 92, 139). These changes provide a mechanism by which filamins may convert mechanical cues into biochemical signals. Here we review recent advances in understanding of filamin-ligand interactions and how applied forces may modulate these interactions. We also examine the evidence supporting in vivo roles for filamins as mechanosensors and consider the potential subcellular sites at which this mechanosensing may occur.
FILAMINS
In vertebrates, there are three filamin genes encoding filamin A (FLNa, ABP, ABP-280, FLN-1, α-FLN or non-muscle FLN), filamin B (FLNb, ABP-278, ABP-276, FH1, β-FLN or FLN-3) and filamin C (FLNc, ABP-L, FLN-2 or γ-FLN) (120, 127). Vertebrate filamins are large homodimers of 240-280kDa subunits that associate at their caboxy termini. Each filamin subunit consists of an amino-terminal actin-binding domain (ABD) followed by 24 β-pleated sheet immunoglobulin-like domains (IgFLN1-24) interrupted by two flexible hinge regions between IgFLN15-16 (H1) and IgFLN23-24 (H2) (39, 99, 120, 127). The dimerization is mediated by the 24th IgFLN domain (48, 99, 110) and majority of filamin interactions with receptors and signaling proteins are mediated through FLNaIg16-24 (119, 120, 143) (Figure 1).
Figure 1.
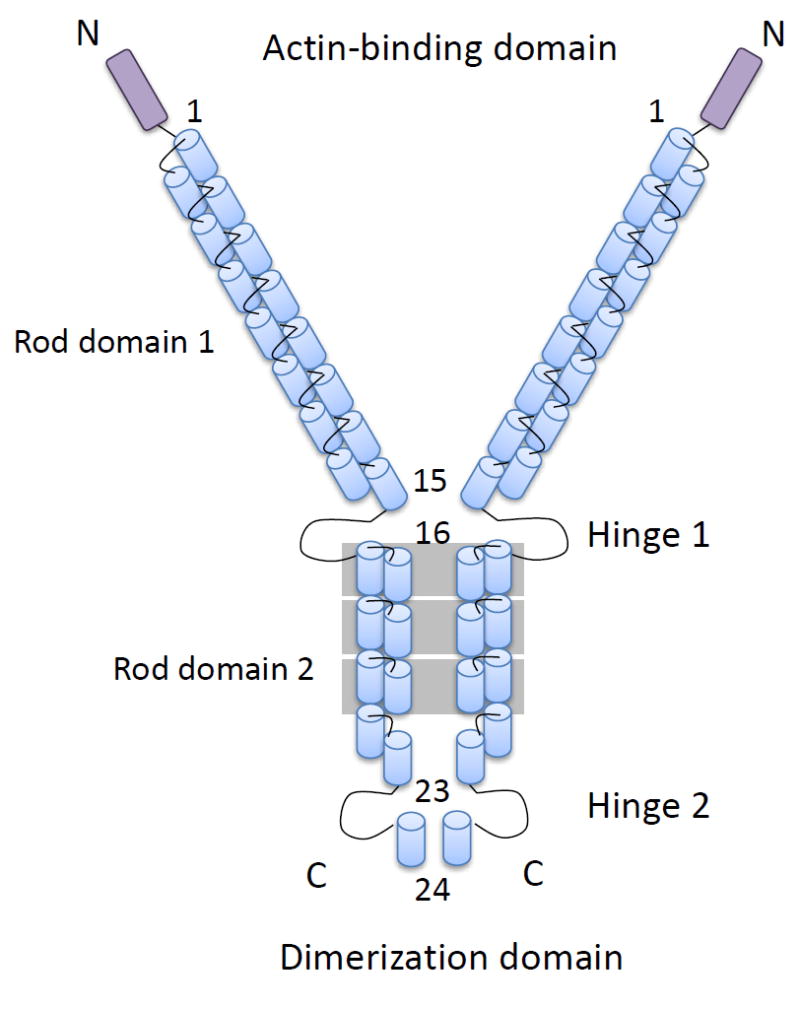
Schematic representation of human filamin. The N-terminal actin-binding domain containing two calponin-homology domains (CH1 and CH2) is followed by 24 Ig repeats of ~96 amino acids each. The Ig repeats are interrupted by two hinge regions (H1 and H2) and fold into anti-parallel β-sheets. The C-terminal 24th repeat is the dimerization domain. Repeats 1-15 make up rod domain 1, while repeats 16-24 make up rod domain 2. Domain pairs (16-17, 18-19 and 20-21) are boxed in gray.
The FLN ABD is composed of two calponin homology domains (CH1 and CH2) (9, 104, 109) and, based on similarity to other ABDs, is predicted to contain three actin-binding sites (ABS1, ABS2 and ABS3), two in CH1 and one in CH2 that together support binding to F-actin (36). However, while the ABD is necessary and sufficient for F-actin binding (100), FLNaIg9-15 contains an F-actin-binding domain that is necessary for high avidity F-actin binding (84). The H2 region, which precedes the dimerization domain, is present in all filamin isoforms, but H1 is absent in chicken filamin and in some splice variants of human FLNb and FLNc (2, 136, 138). With the exception of the H1 and H2 regions and an 82 amino acid insertion in IgFLNc20, the three filamin proteins share high sequence similarity and have both common and isoform-specific functions (57, 120, 127).
Filamins are essential to mammalian development and mutations in human filamins result in diverse congenital anomalies including defects in the brain, bone, cardiovascular system and many other organs (20, 28, 30, 62, 67, 102, 103, 106, 132, 143, 144). Consistent with these diverse phenotypes, FLNa and FLNb are widely expressed during development and show similar cellular and tissue expression patterns (127, 143). However, FLNc is largely restricted to cardiac and skeletal muscles (28, 38, 143). Nonetheless, some non-muscle cells express low but detectable levels of FLNc (1, 145) and knockdown of the other FLN isoforms leads to increased FLNc expression suggesting that a threshold level of filamins may be essential for cell viability (1).
Filamins are not restricted to vertebrates but are also found in a diverse array of organisms including amoeba, insects and nematodes, however the number of IgFLN domains varies across species. For example, Dictyostelium and Entamoeba histolytica filamins, are short, consisting of an ABD followed by only six and four Ig repeats respectively (31, 79, 87, 130). Differential splicing of the cheerio gene produces two Drosophila filamin variants, one composed of an ABD followed by 20 IgFLN domains with hinge regions between IgFLN11-12 and IgFLN19-20, and the other a truncated variant that lacks the ABD and contains domains IgFLN12-20 (42, 117, 118). The Caenorhabditis elegans filamin, FLN1, is also composed of an N-terminal ABD followed by 20 Ig domains, although two other filamin isoforms with C- and N-terminal truncations can also be formed (66). A second C. elegans filamin ortholog, FLN-2 (17), and the Drosophila filamin-related protein encoded by jitterbug (jbug) (118) have some distinct features not found in vertebrates including an ABD consisting of three CH-domains and several non-conserved stretches between IgFLNs (17, 65).
The distribution of filamins throughout the animal kingdom, their conservation across species, and the wide range of diseases and the complexity of phenotypes associated with filamin mutations highlight the importance of this class of actin-crosslinking and scaffolding proteins, and points to the involvement of filamins in a diverse array of interactions and processes.
A COMMON STRUCTURAL THEME FOR FILAMIN IG-LIKE DOMAIN INTERACTIONS
As noted above, filamins affect cellular activities through interactions with a range of binding partners. Consequently, there has been considerable interest in the structural basis of filamin interactions. Due to their large size and flexibility we still lack high-resolution structures of full-length filamin, but the full range of structural biology techniques, including X-ray crystallography, NMR spectroscopy, electron microscopy, X-ray scattering and molecular dynamics modeling, have been used to improve our understanding of individual filamin domains and short stretches of adjacent domains (8, 9, 45, 50, 51, 58, 69, 84, 91, 92, 104, 109).
For example, X-ray crystallography has confirmed that the FLNa and FLNb ABD is composed of two CH domains (9, 104, 109) and, while we still lack structures of the filamin ABD bound to F-actin, it appears to follow the general features of other double calponin homology actin-binding domains (9, 104, 109, 114). The bulk of filamin interactions are IgFLN domain-mediated and initial structures of complexes between IgFLN domains and peptides derived from the cytoplasmic tails of GPIbα and β7 integrins (58, 85) provided important information on factors affecting the filamin-ligand complex formation. Subsequent analysis of complexes with integrin β2, migfilin and cystic fibrosis transmembrane conductance regulator (CFTR) extended these findings (50, 51, 58, 68, 115, 121).
Strikingly, all available structures of IgFLN domains and their peptide ligands have very similar modes of interaction. The immunoglobulin-like β sandwich of IgFLN domains is composed of seven β-strands (A-G) that assemble into two β sheets (Figure 2). When bound, the ligand forms an additional anti-parallel β-strand next to β-strand C of the CFG sheet (Figure 2). Up to nine residues of the ligand contribute to the β-strand and form ten main chain hydrogen bonds. Although these main chain hydrogen bonds may be responsible for most of the interaction energy, specificity is determined by side chain interactions with the groove between IgFLN β-strands C and D (50, 58, 68, 85, 115, 121). Five out of the nine ligand residues interact with the groove via their side chains, forming in most cases a single hydrogen bond and multiple hydrophobic interactions (Figure 2 and Table 1). The first of these residues points towards the IgFLN CD-loop and is either aliphatic or aromatic. This is followed by a well-conserved serine residue that can form a hydrogen bond to a main chain carbonyl oxygen in the IgFLN D strand. Positions 3 and 4 are commonly occupied by either aliphatic or aromatic residues that are buried in the hydrophobic pocket in the groove. Sometimes threonine residues are found in these positions with terminal methyl groups facing the groove and the hydroxyl group pointing to the solvent. The last interacting amino acid points to the BC-loop of the filamin domain and is usually aliphatic (Figure 2 and Table 1).
Figure 2.
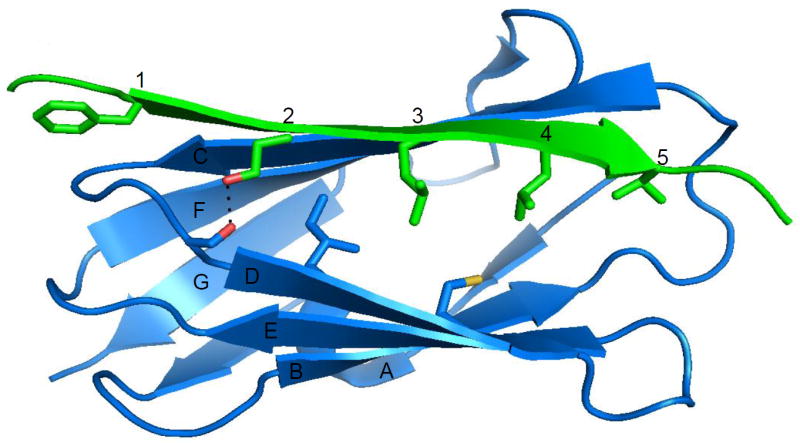
Details of the interaction between IgFLNa17 (blue) and the cytoplasmic peptide of platelet glycoprotein Ibα (Green) (85). Letters A-G indicate the β-stands of the filamin domain forming two sheets: GFC (above) and ABED (below). Note that the GPIbα forms an extra strand next to the strand C. The hydrophopic residues of the ligand extending towards the groove between stands C and D are shown as sticks and marked with numbers 1-5 (see Table 1).
Table 1.
Aligment of filamin Ig-domain binding sequences.
| Ligand | 1 | 2 | 3 | 4 | 5 | ||||
|---|---|---|---|---|---|---|---|---|---|
| GPIbα | F | R | S | S | L | F | L | W | V |
| β2-integrin | F | K | S | A | T | T | T | V | M |
| β7-integrin | Y | K | S | A | I | T | T | T | I |
| Migfilin | V | A | S | S | V | F | I | T | L |
| CFTR | V | V | S | K | L | F | F | S | W |
| FilGAP | F | S | T | F | G | E | L | T | V |
| pro-prion | L | I | S | F | L | I | F | L | I |
| ASB2α | Y | F | S | L | F | H | S | C | S |
| A-strand, domain 18 | R | M | S | H | L | K | V | G | S |
| A-strand, domain 20 | V | K | E | S | I | T | R | R | R |
1-5 Amino acids 1-5 shown in Figure 2.
1,3,4,5Shade in gray indicates the amino acid residues that face the binding groove in Ig-domain and are responsible for the specificity of the domain-ligand interaction.
2Shade in orange indicates the the most conserved amino acid residues that form hydrogen bond to the main chain carbonyl oxygen atom.
As the IgFLN domain-binding motif is a typical β-strand-forming sequence, with alternating hydrophopic residues pointing towards the interior of protein, it cannot be used to pick potential interaction sequences from databases. However, the motif has been successfully utilized to test for potential binding sequences within otherwise identified binding partners such as migfilin, FilGAP, CFTR, pro-prion and ASB2α proteins (Table 1). The interaction of migfilin and CFTR was subsequently verified by structural analysis and mutagenesis (50, 68, 93, 115). FilGAP binding was verified by mapping FilGAP induced NMR chemical shift changes to the CD-face of IgFLNa23 and by mutagenesis (83) while the pro-prion and ASB2α interaction mode was verified by mutagenesis (70, 72).
Available evidence suggests that the CD-face ligand-binding site provides a general mode of interaction for filamin ligands. However, complex structures have only been obtained with IgFLNa domains 17 and 21, while NMR and mutagenesis studies have confirmed similar interaction modes for IgFLNa domains 19 and 23 (68, 69, 83, 91). In addition to these domains, migfilin, GPIbα and integrin β7 peptides can also bind to FLNa domains 4, 9 and 12 (51) and the CFTR peptide binds to domains 9, 12, 17, 19 and 23 (94, 115). Thus, it appears that at least seven out of twenty four FLNa Ig-like domains use the CD face interaction mode to bind ligand. Nonetheless, many reported filamin-binding partners do not have sequences that obviously correspond to the β-strand forming binding motif and other interaction modes for IgFLN domains seem likely to exist. These alternative interactions await structural characterization.
MASKING OF FILAMIN INTERACTIONS VIA INTERDOMAIN CONTACTS AND UNMASKING BY MECHANICAL FORCE
Extrinsic or intrinsic forces impact ECM, adhesion and cytoskeletal proteins in a variety of ways. In most cases, force applied across bonds increases dissociation rates favoring disassembly of complexes (25), however in certain cases force-induced conformational changes can result in exposure of cryptic binding sites leading to recruitment of additional components and trigger a cascade of signal transduction events (21, 35, 49, 81). For instance, talin undergoes a force-induced conformational change leading to exposure of an otherwise cryptic vinculin binding site (15, 71) and force-induced unfolding of the focal adhesion protein p130CAS exposes phosphorylation sites that allows subsequent downstream signaling (108). Additionally, forced-induced unfolding of type III domains in the multi-domain ECM protein fibronectin permits extension of this molecule under stress and may play an important role in mechanotransduction (116, 131).
Similarly, IgFLN domains undergo conformational changes and unfolding events in response to mechanical force (32, 45, 52, 54, 64, 69, 92, 139). Using atomic force microscopy, Furuike and colleagues (32) showed that single molecules of FLNa unfold and stretch in response to external force and that refolding occurs upon removal of applied force. Thus, unfolding and stretching due to applied force allow filamin to extend its length and in turn protect the linkage between the membrane and the cytoskeleton (139). However, whether specific filamin-ligand or filamin-actin interactions are maintained under the forces required for domain unfolding is unclear, and structural studies on multi-domain filamin fragments suggest that filamins may participate in mechanotransduction at lower forces (and force loading rates) by altering domain-domain interactions that expose otherwise masked ligand binding sites.
It is now evident that IgFLNa16-17, 18-19 and 20-21 form three closely packed domain pairs. In these pairs the A strand of domains 18 binds to the CD face of domain 19 and the A strand of domain 20 to the CD face of domain 21. These interactions are almost identical to those of the ligand peptides (45, 69) and the amino acid compositions of the A strands of domains 18 and 20 resemble the other CD face binding ligands (Table 1). This interdomain interaction was first observed in the crystal structure of the three domain fragment IgFLNa19-21 and it was subsequently confirmed by NMR (69). The structure of the IgFLNa18-19 domain pair was solved by NMR (45). Biochemical experiments confirmed that the A-strand interactions inhibit ligand binding to domains 19 and 21 and that this inhibition can be released by mutations in the A strand (69). Thus, these interdomain interactions mask ligand binding to the CD face of IgFLNa19 and IgFLNa21 and provide a means to regulate filamin interactions. However, it seems that the masking interactions are not very tight, because excess ligand can effectively compete with the masking and cause conformation changes in filamin domain pairs (52, 91). The potential significance of the inhibitory interactions is highlighted by the observation that FLNa and FLNb splice variants (var-1), which lack a 41-amino acid sequence that includes the C-terminal part of IgFLN19 and N-terminal inhibitory strand from IgFLN20, exhibit enhanced binding to integrin β-tails (91, 126).
While alternative splicing provides one way of releasing the interdomain masking our structural studies suggest that external forces, transmitted along filamins, may displace the masking β-strand and so facilitate binding of other ligand proteins. As actin filament cross-linking proteins associated with a variety of intracellular actin structures, filamins are exposed to stresses and strains (22, 54, 80) and the mechanical displacement of the inhibitory strand provides a rapid and direct means for force to enhance filamin-ligand interactions. In steered molecular dynamic simulations, the unmasking for FLNa domain pairs 18-19 and 20-21 occurs much earlier, and at lower forces, than domain unfolding or even domain pair detachment (8, 92). The IgFLN domain pairs are organized such that their N- and C-termini locate to the same end of the domain pair, as a consequence, pulling forces transmitted through filamin will be locally targeted to the masking β-strand causing its opening in a zipper-like mechanism (92) (Figure 3). The situation is analogous to the titin kinase domain that is expected to be mechanically regulated (40, 98). Thus, mechanical disruption of domain-domain interactions provide a means to regulate the exposure of the ligand-binding CD faces on filamin domain 19 and 21 at lower, more physiologically relevant, forces than required for domain unfolding.
Figure 3.
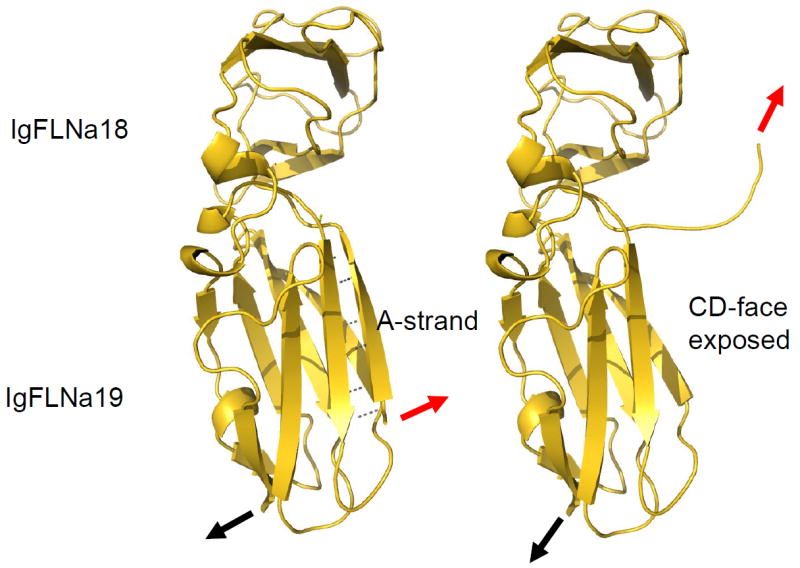
Mechanism of the ligand binding site opening in IgFLNa18-19 based on steered molecular dynamics simulations (92). The A-strand of IgFLNa18 unzips from the CD-face of IgFLNa19 under pulling forces that are much lower than those required for Ig-domain unfolding. The positions of some of the hydrogen bonds connecting the A-stand are indicated with dotted lines. Pulling-forces to the C-terminus and N-terminus are indicated by black and red arrows, respectively.
The two domain pairs (18-19 and 20-21) appear to be conserved in all vertebrate filamins (45), but sequence comparisons do not recognize similar domain pairs in other filamin regions, therefore it remains uncertain whether force can regulate interactions with other IgFLN domains. However, other types of domain-domain interactions that mask or sterically hinder protein binding to CD faces or to alternative binding sites in other IgFLN domains may occur. For example, it has been noticed that the N-terminal IgFLN domains show an alternating pattern of acidic and basic surface character (19) that may have consequences for domain packing. Thus, it seems clear that at least two IgFLN domains can be regulated by unmasking of ligand binding sites and additional mechanically regulated sites may be revealed as more multi-domain structures are solved.
EVIDENCE FOR MECHANOSENSING FUNCTIONS OF FILAMINS
The actin cytoskeleton is essential for maintenance of cell structure, resistance to mechanical stress and regulation of cellular processes including cell motility. Filamins are the most potent F-actin crosslinking proteins and provide mechanical stability to cells by promoting high-angle branching of actin filaments (84). Under force, FLNa and actin filaments accumulate at the cell cortex, reinforcing connections with extracellular adhesion sites and stabilizing the plasma membrane (37). Furthermore, force applied through clustered β1 integrins results in increased production of FLNa (12, 13) which promotes cell survival by stabilizing the cortical actin and preventing force-induced membrane depolarization (55). In response to fluid shear stress FLNa levels increase in osteoblasts and FLNa distributes more widely throughout the cell which may enhance the mechanical resistance of the cytoskeleton (53). Additionally, linkage between the platelet transmembrane receptor GPIbα and FLNa is essential for maintaining the mechanical stability of the plasma membrane. Disruption of the GPIbα-FLNa interaction, leads to development of unstable membrane tethers, defective platelet adhesion and loss of membrane integrity (10). Thus, as originally described by McCulloch’s group, filamins act as “mechanoprotective” elements under applied force and stabilize the plasma membrane and the cortex (13, 37, 55).
Filamins in cell migration
Cell migration is a complex mechanical process involving many force sensing and generating steps, including cell polarization, membrane protrusion, cytoskeletal re-arrangement, myosin-mediated contractility and cycles of adhesion and de-adhesion (101). There is an extensive literature on filamins in cell migration and as this has been reviewed in detail elsewhere (28, 86, 106, 120, 143) we will only consider it briefly here.
Filamins were first implicated in cell migration based on locomotion defects and plasma membrane blebbing in a FLNa-deficient human melanoma cell line (M2) (11). Re-expression of FLNa rescues the motility and plasma membrane blebbing defects, which are attributed cortical surface instability and weakening of the actin structure (11, 29). Support for a role of filamin in migration came with the finding that nonsense mutations in the X-linked FLNA gene are associated with the neuronal migration disorder, periventricular heterotopia (PVH) (30). Furthermore, filamin depleted cells exhibit impaired spreading (1, 61, 78), impaired initiation of migration (1), decreased migration capacity (16), decreased adhesion stability, decreased stress fibers and reduced transient traction forces (78).
Filamin overexpression also contributes to migration defects: overexpression of FLNa inhibits migration of M2 cells (11) and mouse cortical neurons (105), and increased filamin binding to β-integrin cytoplasmic domains inhibits cell migration (7). MEKK4-null mice also exhibit PVH associated with neurons that over-express FLNa (105). Finally, regulation of filamin levels through targeted degradation mediated by FILIP (Filamin-A Interacting Protein) or ASB2α (Ankyrin repeat containing protein with a suppressor of cytokine signaling box 2) modulates cell polarity, spreading and migration (1, 47, 82, 107).
Despite ample evidence pointing to roles for filamin in cell migration, FLNa-null mice do not exhibit evident migration defects or PVH (27, 44) and FLNa-null mouse embryonic fibroblasts (44), neural crest cells (27) and FLNa missense mutant human fibroblasts (9) display normal morphology and locomotion. Several studies suggest that the lack of overt migratory phenotypes in FLNa-deficient cells may be due to presence of other filamin isoforms that compensate for the loss of FLNa (1, 78, 112). Indeed, while in many cell types loss of FLNa or FLNb alone has no effect on migration, knockdown of both FLNa and FLNb, or proteasomal-induced degradation of all three filamin isoforms by ASB2α, results in impaired initiation of migration and spreading (1, 47).
The exact role that filamin plays during migration, and whether its mechanosensing activities are important, remains unclear. Our data show that cells deficient in filamins are impaired in initiation of migration, but are nonetheless capable of migrating at normal speeds (1). This suggests that filamins are not essential for core migration processes but rather that they contribute to its regulation, perhaps through formation of mechanosensitive links between integrins and the actin cytoskeleton.
Filamins in cell differentiation and morphogenesis
The importance of mechanical cues from the local environment during normal cell differentiation as well as tumor progression has been well established (5, 18, 49, 97, 134, 140). For example, the stiffness of the extracellular substrate in part determines lineage commitment of differentiating mesenchymal stem cells (MSCs) (23, 24) and regulates the formation and morphology of complex three-dimensional cell structures such as tubules and acini formed by breast epithelial cells (135, 137). Given the evidence that filamins are mechanosensitive cytoskeletal and signaling elements it is unsurprising that filamins have also been implicated in control of cell differentiation and morphogenesis.
In breast epithelial cells, FLNa is necessary to contract collagen gels and pull on collagen fibrils, leading to collagen remodeling and branching morphogenesis (34). Filamins are also implicated in substrate stiffness responses. FLNa-deficient M2 cells do not respond to substrate stiffness when linked to the substrate through collagen receptors (6) and exert much smaller contractile stresses on the substratum leading to decreased cell stiffness (56). In addition to responding to substrate stiffness, filamins interact with ion channel receptors such as the calcium-sensing receptor (CaR) and mediate transduction of extracellular signals into intracellular responses to regulate differentiation (123).
In mice, loss of FLNb causes impaired differentiation of chondrocyte precursors, severe skeletal malformation and ectopic bone formation in the cartilage (26, 75, 142, 145). FLNb is normally strongly expressed in chondrocytes and FLNb-deficient chondrocytes display diminished adhesion to the ECM suggesting that the link between the ECM and FLNb may contribute to differentiation (75). The skeletal defects and cartilage phenotype of FLNb-deficient mice closely resemble the human skeletal disorders associated with FLNb mutations (67) implicating FLNb as an important regulator of cartilage and bone development.
Filamins are also expressed in muscle cells and play a role in myogenic differentiation. In vitro, C2C12 myoblasts express all three filamin isoforms, however, during myogenic differentiation, expression of FLNb∆H1 and FLNc∆H1 splice variants lacking the H1 region is induced. Furthermore, ectopic expression of FLNbvar1∆H1, which lacks the H1 region and 41 amino acids between IgFLNb19 and IgFLNb20 (residues 2082-2122), accelerates C2C12 differentiation into myotubes (126). FLNc is largely restricted to the cardiac and skeletal muscles and is up-regulated during muscle differentiation (4, 38). In vitro knockdown of FLNc in C2C12 causes differentiation and fusion defects and FLNc-deficient mice exhibit severe skeletal muscle defects with less muscle mass and disorganized muscle structure (14). Consistent with these observations, FLNc mutations in human result in muscular dystrophies, myofibrillar myopathy and distal myopathy (20, 62, 111, 132).
Finally, recent data indicate that acute ASB2-mediated regulation of filamin levels by the ubiquitin-proteasomal machinery may be employed to control cell differentiation (4, 47). ASB2 is the specificity subunit of an E3 ubiquitin ligase complex (46, 63) and encodes two isoforms, ASB2α and ASB2β (4). ASB2α, the hematopoietic-specific isoform, is induced in response to retinoic acid (RA) treatment of myeloid leukemia cells and is involved in regulation of hematopoietic cell differentiation (41). ASB2α promotes proteasomal-mediated degradation of filamins (1, 47) and its up-regulation correlates with down-regulation of FLNa and FLNb in myeloid leukemia cells induced to differentiate (47). Furthermore, knockdown of ASB2α in leukemia cells delays differentiation and FLNa and FLNb degradation. ASB2α is proposed to regulate hematopoietic cell differentiation by modulating cell spreading, actin remodeling and cell adhesion through targeting of filamins for degradation (47, 70).
ASB2β, is expressed in muscle cells and is induced during myogenic differentiation (4). ASB2β selectively targets FLNb for degradation and ASB2β expression is transiently up-regulated during differentiation of C2C12 cells, concomitant with a loss of FLNb expression (4). Knockdown of ASB2β in C2C12 cells inhibits myoblast fusion and myotube formation and correlates with delayed degradation of FLNb (4). Notably, knockdown of FLNb in ASB2β knockdown cells restores myogenic differentiation. Thus, ASB2β regulates FLNb functions in myogenic differentiation by controlling its degradation (4).
Taken together, it is clear that filamins play important roles in cell differentiation and morphogenesis. However, this has only been linked to the mechanosensing activities of filamin in a few cases, most notably for breast epithelial cell tubulogenesis (34), where the levels of FLNa expression, and more specifically the extent of the integrin-filamin interaction, were shown to modulate the optimal matrix stiffness that supported tubulogenesis. In the future it will be important to assess how altering filamin levels impacts the effect of matrix stiffness on chondrocyte and muscle differentiation and ultimately to determine the role of specific force-sensitive filamin interactions in governing differentiation.
WHERE MIGHT FILAMINS MEDIATE MECHANOSENSING AND SIGNALING
In non-muscle cells, filamins co-localize with F-actin along stress fibers, at the cell cortex and along the leading edge (86, 120, 127). Filamins are also enriched at the end of actin stress fibers, the leading edge, and at trailing ends of mature focal adhesions (78) and concentrate in adhesion sites after application of force to cells (37). In migrating cells, filamins localize to membrane ruffles (125) and in spreading cells to filopodia, lamellipodia (61) and the endoplasm (78), the membranous organelle-rich region near the center of the cell. A small fraction of FLNa is also reported to be present in the nucleus where, through its interaction with BRCA2, it can participate in the DNA damage response (141). Filamin also binds androgen receptor and supports its nuclear targeting and transcriptional activity (74, 90). Given the diverse localization of filamins, it may mediate mechanosensing and signaling functions in a variety of regions including stress fibers, cell adhesion sites, the cell membrane, cell extensions and the nucleus.
Stress fibers and adhesions
While filamins are not evident in all adhesions they have been shown to co-localize with integrins in early adhesions (7) and force stimulates filamin recruitment to integrins (37). Indeed, force applied through β1 integrins results in increased production of FLNa (12, 13) which promotes cell survival and prevents force-induced membrane depolarization (55). In mouse embryonic fibroblasts (MEF) filamins localize to adhesions and stress fibers and play an important role in maintaining focal adhesions and stabilizing the actin cytoskeleton under force (78). MEF lacking FLNa and FLNb, have a disorganized actin cytoskeleton with decreased stress fibers, and smaller more dynamic focal adhesions that result in defects of force generation (78). Filamin-depleted cells also exhibit defective spreading (1, 47, 78).
Cell protrusions
FLNa concentrates in cell extensions, specifically filopodia and lamellipodia, of cells spreading on collagen (60, 61). The extension of filopodia and lamellipodia are early events in cell spreading (60) and knockdown of FLNa in human embryonic kidney (HEK-293) cells is sufficient to impair spreading, decrease the number of cell extensions and increase apoptosis (59-61). Furthermore, inhibition of β1 integrin recapitulates the spreading defects of FLNa-deficient cells and reduces localization of FLNa to cell extensions, suggesting that FLNa-β1 integrin interactions and recruitment of FLNa to cell extensions play important roles in cell spreading and survival (61). FLNa also interacts with the intermediate filament protein vimentin in cell extensions (60). Thus, through its interactions with integrin, F-actin and vimentin, FLNa bridges the ECM, the actin and intermediate filament cytoskeletons.
Cytoskeletal dynamics and cell protrusions are regulated by small GTPases of the Rho family (43, 101). FLNa associates with a wide range of signaling molecules including the small GTPases Rac, Rho, CdC42 and RalA and factors upstream and downstream of the GTPases (3, 83, 88, 89, 113, 124, 125). RalA induces filopodia formation downstream of CdC42 and recruits FLNa into the filopodial cytoskeleton (89). The interaction between FLNa and RalA is required for RalA-induced filopodia formation (89). The serine/threonine kinase p21-activated kinase 1 (Pak1), a downstream effector of CdC42 and Rac1, also colocalizes with FLNa in membrane ruffles and triggers FLNa phosphorylation, which is essential for Pak1-mediated cytoskeleton remodeling (125).
FLNa also binds ROCK, a downstream effector of the small GTPase Rho. In HeLa cells, ROCK and FLNa co-localize at protrusive cell membranes suggesting that ROCK-FLNa interactions may be important for cell migration (124). In addition to small GTPases and their effectors, filamins bind guanine nucleotide-exchange factors (GEFs) such as Trio (3), and GTPase-activating proteins (GAPs) such as FilGAP (88). Trio is concentrated in actin-rich ruffles where it co-localizes with filamins and activates the small GTPases RhoG and Rac1 (3).
FilGAP is recruited to sites of membrane protrusion (88) and sites of force transfer (113) by FLNa, where it antagonizes the small GTPase Rac. Redistribution of FilGAP to the cell periphery inhibits cell spreading and lamella formation by suppressing Rac activity (88, 113). In FLNa-null cells, mechanical forces applied through integrins result in Rac-mediated lamellae formation (113). Thus, FLNa suppresses force-induced lamellipodia formation and Rac activation by targeting FilGAP to sites of force transfer. Perturbations in FilGAP activity and disruption of FilGAP-FLNa interaction lead to increased force-induced apoptosis suggesting that force-induced recruitment of FLNa and FilGAP to the cell periphery plays an important role in mechanoprotection (113).
Nucleus and nuclear envelope
Mechanical forces are also transmitted into the nucleus and result in altered gene activity (133). While application of force through β1 integrins induces FLNa expression (12), it remains unknown whether filamins localizes to the nucleus in response to force to regulate transcriptional activity. Filamins are thought to regulate transcriptional activity through retention or accumulation of transcription factors in the cytoplasm and the nucleus (143). Furthermore, a small fraction of full-length FLNa (141) and a C-terminal fragment of FLNa are reported to reside in the nucleus (74, 90) suggesting that FLNa might regulate transcriptional activity in the nucleus.
Filamins also localize to the nuclear envelope and in complex with the F-actin bundling protein, Refilin (Regulator of FILamin ProteIN), promote formation of perinuclear parallel actin stress fibers (33). The interaction of refilin with FLNa is mediated through IgFLNa21 and refilin is proposed to act as a molecular switch that converts FLNa from an F-actin branching to an F-actin bundling protein (33). The Refilin-FLNa complex also stabilizes the perinuclear actin bundles anchored to the nuclear membrane, called transmembrae actin-associated nuclear (TAN) lines (33). The coupling of the perinculear actin network to the nuclear membrane is important for force transduction and nuclear positioning during cell migration and differentiation (76, 77) and it is plausible that filamin plays a role in this mechanical coupling.
Sarcomeres
Filamins have important roles in muscle differentiation and function and as highly mechanically active tissues, muscles are prime sites for filamin mechanotransduction. FLNc is the predominant isoform in muscle cells and it localizes to the sarcomeric Z-line complex and intercalated discs (96, 122, 128). While FLNc is 72% and 70% identical to FLNa and FLNb respectively, it has a unique 82 amino acid insertion in IgFLNc20 (136) which contains additional binding sites for other partners (129). The high expression of FLNc in muscles, its upregulation during myocyte differentiation and localization to Z-discs implicates an important role for FLNc during myofribrillogenesis (128).
In muscles, FLNc localizes at the periphery of the Z-disc and crosslinks thin filaments at their ends in the myofibrillar Z-dics. FLNc is also found at intercalated discs and is implicated in organization of thin filaments at sites that have to resist mechanical strain (128). Importantly, FLNc interacts with γ- and δ-sarcoglycans, members of the dystrophin-glycoprotein complex (122). The sarcoglycans are restricted to the sarcolemma, the cell membrane of muscle cells, and are responsible for connecting the actin cytoskeleton and the ECM (73, 122). In patients with FLNc mutations, several Z-disk-associated proteins and sarcolemmal proteins, including the sarcoglycans, are mis-localized and form intracellular aggregates (62, 73, 132). Perturbation in distribution of these proteins due to FLNc mutations may destabilize the muscle cell membrane and weaken the connection of myofibrillar cytoskeleton to the ECM leading to muscular defects.
Summary points.
By interacting with transmembrane receptors, filamins provide a mechanical link between the ECM, the plasma membrane and the actin cytoskeleton.
Filamins crosslink and anchor actin filaments, regulate actin cytoskeleton remodeling, stabilize the plasma membrane, and contribute to the mechanical stability of the plasma membrane and the cell cortex.
Filamins interact with over 90 binding partners and are implicated in the regulation of a diverse array of cellular functions including motility, maintenance of cell shape and differentiation.
At least seven out of twenty four filamin Ig-like domains interact so that the ligand forms an additional β strand next to strand C of the filmain domain.
Filamins are important for tuning cellular response to ECM stiffness and for responding to mechanical force.
Structural and functional studies suggest that filamins act as mechanosensors able to detect and transmit mechanical stimuli by undergoing conformational changes that expose otherwise masked ligand-binding sites.
The ability of cells to respond to physical cues and mechanical stresses in their environment is important for cellular processes such as cell migration, differentiation and development and mutations in filamins are associated with a wide range of developmental malformations and diseases.
Future Directions/Unresolved Issues.
Due to their large size and flexibility, there are still no high-resolution structures of full-length filamin. Structures of additional larger multi-domain fragments of filamin that reveal domain-domain interactions and dynamics remain an important future goal.
The domain pairs 18-19 and 20-21 appear to be conserved in all vertebrate filamins, but identification of other domain pairs or alternative domain-domain interactions will facilitate investigation of whether force can regulate interactions with other IgFLN domains.
Definitive evidence of altered filamin-ligand interaction in response to applied experimental forces is required to validate the proposed mechanisms of filamin mechanosensing.
The exact role that filamin plays during migration, and whether its mechanosensing activities are important, remains unclear. A detailed understanding of the role of filamins in mechanosensing will provide insight into how filamins function in cellular processes such as cell migration.
It will be important to assess how altering filamin levels impacts the effect of matrix stiffness on chondrocyte and muscle differentiation and ultimately to determine the role of specific force-sensitive filamin interactions in governing differentiation.
How acute ASB2-mediated proteosomal degradation of filamins contributes to leukocyte and myoblast differentiation remains an open question.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (RO1 GM-068600) to D.A.C., an award from the American Heart Association to ZR and by Academy of Finland grants 1147613, 135473, 138327 to J.Y. and T.M.
Definitions
- Mechanotransduction
The process through which cells sense the environment and respond to physical and mechanical signals and transmit these signals into intracellular signaling pathways.
- Mechanosensor
A molecule that is able to detect and respond to physical forces of the environment. Mechanosensors, undergo conformational changes in response to force. The conformational changes can result in exposure of cryptic binding sites leading to recruitment of additional components or regulate enzymatic or membrane channel activities and in turn trigger a cascade of signal transduction events.
Acronyms
- FLN
Filamin
- ECM
Extracellular matrix
- Ig
Immunoglobulin-like
- ABD
Actin-binding domain
- CH
Calponin homology domain
- PVH
Periventricular heterotopia
LITERATURE CITED
- 1.Baldassarre M, Razinia Z, Burande CF, Lamsoul I, Lutz PG, Calderwood DA. Filamins regulate cell spreading and initiation of cell migration. PLoS One. 2009;4:e7830. doi: 10.1371/journal.pone.0007830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry CP, Xie J, Lemmon V, Young AP. Molecular characterization of a multi-promoter gene encoding a chicken filamin protein. J Biol Chem. 1993;268:25577–86. [PubMed] [Google Scholar]
- 3.Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol. 2000;2:888–92. doi: 10.1038/35046533. [DOI] [PubMed] [Google Scholar]
- 4.Bello NF, Lamsoul I, Heuze ML, Metais A, Moreaux G, et al. The E3 ubiquitin ligase specificity subunit ASB2beta is a novel regulator of muscle differentiation that targets filamin B to proteasomal degradation. Cell Death Differ. 2009;16:921–32. doi: 10.1038/cdd.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9:108–22. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byfield FJ, Wen Q, Levental I, Nordstrom K, Arratia PE, et al. Absence of filamin A prevents cells from responding to stiffness gradients on gels coated with collagen but not fibronectin. Biophys J. 2009;96:5095–102. doi: 10.1016/j.bpj.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, et al. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3:1060–68. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- 8.Chen HS, Kolahi KS, Mofrad MR. Phosphorylation facilitates the integrin binding of filamin under force. Biophys J. 2009;97:3095–104. doi: 10.1016/j.bpj.2009.08.059. Describe steered molecular dynamic simulations that support force-induced exposure of cryptic ligand binding sites in FLNs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark AR, Sawyer GM, Robertson SP, Sutherland-Smith AJ. Skeletal dysplasias due to filamin A mutations result from a gain-of-function mechanism distinct from allelic neurological disorders. Hum Mol Genet. 2009;18:4791–800. doi: 10.1093/hmg/ddp442. [DOI] [PubMed] [Google Scholar]
- 10.Cranmer SL, Ashworth KJ, Yao Y, Berndt MC, Ruggeri ZM, et al. High shear-dependent loss of membrane integrity and defective platelet adhesion following disruption of the GPIbalpha-filamin interaction. Blood. 2011;117:2718–27. doi: 10.1182/blood-2010-07-296194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, et al. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–27. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 12.D’Addario M, Arora PD, Ellen RP, McCulloch CA. Interaction of p38 and Sp1 in a mechanical force-induced, beta 1 integrin-mediated transcriptional circuit that regulates the actin-binding protein filamin-A. J Biol Chem. 2002;277:47541–50. doi: 10.1074/jbc.M207681200. [DOI] [PubMed] [Google Scholar]
- 13.D’Addario M, Arora PD, Fan J, Ganss B, Ellen RP, McCulloch CA. Cytoprotection against mechanical forces delivered through beta 1 integrins requires induction of filamin A. J Biol Chem. 2001;276:31969–77. doi: 10.1074/jbc.M102715200. [DOI] [PubMed] [Google Scholar]
- 14.Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol. 2006;26:6522–34. doi: 10.1128/MCB.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–41. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Valle-Perez B, Martinez VG, Lacasa-Salavert C, Figueras A, Shapiro SS, et al. Filamin B plays a key role in vascular endothelial growth factor-induced endothelial cell motility through its interaction with Rac-1 and Vav-2. J Biol Chem. 2010;285:10748–60. doi: 10.1074/jbc.M109.062984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demaso CR, Kovacevic I, Uzun A, Cram EJ. Structural and Functional Evaluation of C. elegans Filamins FLN-1 and FLN-2. PLoS One. 2011;6:e22428. doi: 10.1371/journal.pone.0022428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 19.Djinovic-Carugo K, Carugo O. Structural portrait of filamin interaction mechanisms. Curr Protein Pept Sci. 2010;11:639–50. doi: 10.2174/138920310794109111. [DOI] [PubMed] [Google Scholar]
- 20.Duff RM, Tay V, Hackman P, Ravenscroft G, McLean C, et al. Mutations in the N-terminal Actin-Binding Domain of Filamin C Cause a Distal Myopathy. Am J Hum Genet. 2011;88:729–40. doi: 10.1016/j.ajhg.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol. 2011;12:308–19. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang HY, et al. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci. 2008;121:3794–802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 24.Engler AJ, Sweeney HL, Discher DE, Schwarzbauer JE. Extracellular matrix elasticity directs stem cell differentiation. J Musculoskelet Neuronal Interact. 2007;7:335. [PubMed] [Google Scholar]
- 25.Evans EA, Calderwood DA. Forces and bond dynamics in cell adhesion. Science. 2007;316:1148–53. doi: 10.1126/science.1137592. [DOI] [PubMed] [Google Scholar]
- 26.Farrington-Rock C, Kirilova V, Dillard-Telm L, Borowsky AD, Chalk S, et al. Disruption of the Flnb gene in mice phenocopies the human disease spondylocarpotarsal synostosis syndrome. Hum Mol Genet. 2008;17:631–41. doi: 10.1093/hmg/ddm188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, et al. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA. 2006;103:19836–41. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–38. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 29.Flanagan LA, Chou J, Falet H, Neujahr R, Hartwig JH, Stossel TP. Filamin A, the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J Cell Biol. 2001;155:511–17. doi: 10.1083/jcb.200105148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–25. doi: 10.1016/s0896-6273(00)80651-0. Links FLNa mutations to PVH providing the first in vivo evidence of a role for FLNs in migration or neuronal differentiation. [DOI] [PubMed] [Google Scholar]
- 31.Fucini P, Koppel B, Schleicher M, Lustig A, Holak TA, et al. Molecular architecture of the rod domain of the Dictyostelium gelation factor (ABP120) J Mol Biol. 1999;291:1017–23. doi: 10.1006/jmbi.1999.3046. [DOI] [PubMed] [Google Scholar]
- 32.Furuike S, Ito T, Yamazaki M. Mechanical unfolding of single filamin A ABP-280 molecules detected by atomic force microscopy. FEBS Lett. 2001;498:72–5. doi: 10.1016/s0014-5793(01)02497-8. [DOI] [PubMed] [Google Scholar]
- 33.Gay O, Gilquin B, Nakamura F, Jenkins ZA, McCartney R, et al. RefilinB (FAM101B) targets FilaminA to organize perinuclear actin networks and regulates nuclear shape. Proc Natl Acad Sci U S A. 2011;108:11464–9. doi: 10.1073/pnas.1104211108. FLNa is identified as a potential organizer of perinuclear actin network and regulator of nuclear shape. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gehler S, Baldassarre M, Lad Y, Leight JL, Wozniak MA, et al. Filamin A-beta1 integrin complex tunes epithelial cell response to matrix tension. Mol Biol Cell. 2009;20:3224–38. doi: 10.1091/mbc.E08-12-1186. Supports role for FLNa as a mechanosensitive complex capable of sensing and responding to the tension of the matrix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 36.Gimona M, Djinovic-Carugo K, Kranewitter WJ, Winder SJ. Functional plasticity of CH domains. FEBS Lett. 2002;513:98–106. doi: 10.1016/s0014-5793(01)03240-9. [DOI] [PubMed] [Google Scholar]
- 37.Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem. 1998;273:1689–98. doi: 10.1074/jbc.273.3.1689. Provides first evidence for role of FLNa in mechanoprotection against mechanical forces delivered through β1 integrins. [DOI] [PubMed] [Google Scholar]
- 38.Goetsch SC, Martin CM, Embree LJ, Garry DJ. Myogenic progenitor cells express filamin C in developing and regenerating skeletal muscle. Stem Cells Dev. 2005;14:181–7. doi: 10.1089/scd.2005.14.181. [DOI] [PubMed] [Google Scholar]
- 39.Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, et al. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990;111:1089–105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grater F, Shen J, Jiang H, Gautel M, Grubmuller H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys J. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guibal FC, Moog-Lutz C, Smolewski P, Di Gioia Y, Darzynkiewicz Z, et al. ASB-2 inhibits growth and promotes commitment in myeloid leukemia cells. J Biol Chem. 2002;277:218–24. doi: 10.1074/jbc.M108476200. [DOI] [PubMed] [Google Scholar]
- 42.Guo Y, Zhang SX, Sokol N, Cooley L, Boulianne GL. Physical and genetic interaction of filamin with presenilin in Drosophila. J Cell Sci. 2000;113(Pt 19):3499–508. doi: 10.1242/jcs.113.19.3499. [DOI] [PubMed] [Google Scholar]
- 43.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–14. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 44.Hart AW, Morgan JE, Schneider J, West K, McKie L, et al. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–67. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- 45.Heikkinen OK, Ruskamo S, Konarev PV, Svergun DI, Iivanainen T, et al. Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin. J Biol Chem. 2009;284:25450–8. doi: 10.1074/jbc.M109.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heuze ML, Guibal FC, Banks CA, Conaway JW, Conaway RC, et al. ASB2 is an Elongin BC-interacting protein that can assemble with Cullin 5 and Rbx1 to reconstitute an E3 ubiquitin ligase complex. J Biol Chem. 2005;280:5468–74. doi: 10.1074/jbc.M413040200. [DOI] [PubMed] [Google Scholar]
- 47.Heuze ML, Lamsoul I, Baldassarre M, Lad Y, Leveque S, et al. ASB2 targets filamins A and B to proteasomal degradation. Blood. 2008;112:5130–40. doi: 10.1182/blood-2007-12-128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Himmel M, Van Der Ven PF, Stocklein W, Furst DO. The limits of promiscuity: isoform-specific dimerization of filamins. Biochemistry. 2003;42:430–9. doi: 10.1021/bi026501+. [DOI] [PubMed] [Google Scholar]
- 49.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–23. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ithychanda SS, Das M, Ma YQ, Ding K, Wang X, et al. Migfilin, a molecular switch in regulation of integrin activation. J Biol Chem. 2009;284:4713–22. doi: 10.1074/jbc.M807719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ithychanda SS, Hsu D, Li H, Yan L, Liu DD, et al. Identification and characterization of multiple similar ligand-binding repeats in filamin: implication on filamin-mediated receptor clustering and cross-talk. J Biol Chem. 2009;284:35113–21. doi: 10.1074/jbc.M109.060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ithychanda SS, Qin J. Evidence for multisite ligand binding and stretching of filamin by integrin and migfilin. Biochemistry. 2011;50:4229–31. doi: 10.1021/bi2003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson WM, Jaasma MJ, Tang RY, Keaveny TM. Mechanical loading by fluid shear is sufficient to alter the cytoskeletal composition of osteoblastic cells. Am J Physiol Cell Physiol. 2008;295:C1007–15. doi: 10.1152/ajpcell.00509.2007. [DOI] [PubMed] [Google Scholar]
- 54.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–6. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kainulainen T, Pender A, D’Addario M, Feng Y, Lekic P, McCulloch CA. Cell death and mechanoprotection by filamin a in connective tissues after challenge by applied tensile forces. J Biol Chem. 2002;277:21998–2009. doi: 10.1074/jbc.M200715200. [DOI] [PubMed] [Google Scholar]
- 56.Kasza KE, Nakamura F, Hu S, Kollmannsberger P, Bonakdar N, et al. Filamin A is essential for active cell stiffening but not passive stiffening under external force. Biophys J. 2009;96:4326–35. doi: 10.1016/j.bpj.2009.02.035. Provides evidence for role of FLNa in altering cell stiffness and exerting internal contractile tension in response to mechanical stresses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kesner BA, Milgram SL, Temple BR, Dokholyan NV. Isoform divergence of the filamin family of proteins. Mol Biol Evol. 2009;27:283–95. doi: 10.1093/molbev/msp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, et al. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–47. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Kim H, Nakamura F, Lee W, Hong C, Perez-Sala D, McCulloch CA. Regulation of cell adhesion to collagen via beta1 integrins is dependent on interactions of filamin A with vimentin and protein kinase C epsilon. Exp Cell Res. 2010;316:1829–44. doi: 10.1016/j.yexcr.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 60.Kim H, Nakamura F, Lee W, Shifrin Y, Arora P, McCulloch CA. Filamin A is required for vimentin-mediated cell adhesion and spreading. Am J Physiol Cell Physiol. 2010;298:C221–36. doi: 10.1152/ajpcell.00323.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim H, Sengupta A, Glogauer M, McCulloch CA. Filamin A regulates cell spreading and survival via beta1 integrins. Exp Cell Res. 2008;314:834–46. doi: 10.1016/j.yexcr.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 62.Kley RA, Hellenbroich Y, van der Ven PF, Furst DO, Huebner A, et al. Clinical and morphological phenotype of the filamin myopathy: a study of 31 German patients. Brain. 2007;130:3250–64. doi: 10.1093/brain/awm271. [DOI] [PubMed] [Google Scholar]
- 63.Kohroki J, Nishiyama T, Nakamura T, Masuho Y. ASB proteins interact with Cullin5 and Rbx2 to form E3 ubiquitin ligase complexes. FEBS Lett. 2005;579:6796–802. doi: 10.1016/j.febslet.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 64.Kolahi KS, Mofrad MR. Molecular mechanics of filamin’s rod domain. Biophys J. 2008;94:1075–83. doi: 10.1529/biophysj.107.118802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korenbaum E, Rivero F. Calponin homology domains at a glance. J Cell Sci. 2002;115:3543–5. doi: 10.1242/jcs.00003. [DOI] [PubMed] [Google Scholar]
- 66.Kovacevic I, Cram EJ. FLN-1/filamin is required for maintenance of actin and exit of fertilized oocytes from the spermatheca in C. elegans. Dev Biol. 2010;347:247–57. doi: 10.1016/j.ydbio.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krakow D, Robertson SP, King LM, Morgan T, Sebald ET, et al. Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat Genet. 2004;36:405–10. doi: 10.1038/ng1319. Initial identification of mutations in the gene encoding FLNb associated with human skeletal disorders. [DOI] [PubMed] [Google Scholar]
- 68.Lad Y, Jiang P, Ruskamo S, Harburger DS, Ylanne J, et al. Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J Biol Chem. 2008;283:35154–63. doi: 10.1074/jbc.M802592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lad Y, Kiema T, Jiang P, Pentikainen OT, Coles CH, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 2007;26:3993–4004. doi: 10.1038/sj.emboj.7601827. Structural studies reveal packed domain arrangement of IgFLNa19-21 providing a potential mechanism for filamin mechanosensing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamsoul I, Burande CF, Razinia Z, Houles TC, Menoret D, et al. Functional and structural insights into ASB2{alpha}, a novel regulator of integrin-dependent adhesion of hematopoietic cells. J Biol Chem. 2011 doi: 10.1074/jbc.M111.220921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee SE, Kamm RD, Mofrad MR. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J Biomech. 2007;40:2096–106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 72.Li C, Yu S, Nakamura F, Pentikainen OT, Singh N, et al. Pro-prion binds filamin A, facilitating its interaction with integrin beta1, and contributes to melanomagenesis. J Biol Chem. 2010;285:30328–39. doi: 10.1074/jbc.M110.147413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lowe T, Kley RA, van der Ven PF, Himmel M, Huebner A, et al. The pathomechanism of filaminopathy: altered biochemical properties explain the cellular phenotype of a protein aggregation myopathy. Hum Mol Genet. 2007;16:1351–8. doi: 10.1093/hmg/ddm085. [DOI] [PubMed] [Google Scholar]
- 74.Loy CJ, Sim KS, Yong EL. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc Natl Acad Sci U S A. 2003;100:4562–7. doi: 10.1073/pnas.0736237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu J, Lian G, Lenkinski R, De Grand A, Vaid RR, et al. Filamin B mutations cause chondrocyte defects in skeletal development. Hum Mol Genet. 2007;16:1661–75. doi: 10.1093/hmg/ddm114. [DOI] [PubMed] [Google Scholar]
- 76.Luxton GG, Gomes ER, Folker ES, Worman HJ, Gundersen GG. TAN lines: A novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2011;2:173–81. doi: 10.4161/nucl.2.3.16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–9. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lynch CD, Gauthier NC, Biais N, Lazar AM, Roca-Cusachs P, et al. Filamin depletion blocks endoplasmic spreading and destabilizes force-bearing adhesions. Mol Biol Cell. 2011;22:1263–73. doi: 10.1091/mbc.E10-08-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCoy AJ, Fucini P, Noegel AA, Stewart M. Structural basis for dimerization of the Dictyostelium gelation factor (ABP120) rod. Nat Struct Biol. 1999;6:836–41. doi: 10.1038/12296. [DOI] [PubMed] [Google Scholar]
- 80.Meng F, Suchyna TM, Lazakovitch E, Gronostajski RM, Sachs F. Real Time FRET Based Detection of Mechanical Stress in Cytoskeletal and Extracellular Matrix Proteins. Cell Mol Bioeng. 2011;4:148–59. doi: 10.1007/s12195-010-0140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagano T, Morikubo S, Sato M. Filamin A and FILIP (Filamin A-Interacting Protein) regulate cell polarity and motility in neocortical subventricular and intermediate zones during radial migration. J Neurosci. 2004;24:9648–57. doi: 10.1523/JNEUROSCI.2363-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakamura F, Heikkinen O, Pentikainen OT, Osborn TM, Kasza KE, et al. Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS One. 2009;4:e4928. doi: 10.1371/journal.pone.0004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011–25. doi: 10.1083/jcb.200707073. Shows that IgFLNa9-15 contains a secondary F-actin-binding site establishing orientation of FLNa dimers in F-actin branching. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakamura F, Pudas R, Heikkinen O, Permi P, Kilpelainen I, et al. The structure of the GPIb-filamin A complex. Blood. 2006;107:1925–32. doi: 10.1182/blood-2005-10-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5:160–9. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Noegel AA, Rapp S, Lottspeich F, Schleicher M, Stewart M. The Dictyostelium gelation factor shares a putative actin binding site with alpha-actinins and dystrophin and also has a rod domain containing six 100-residue motifs that appear to have a cross-beta conformation. J Cell Biol. 1989;109:607–18. doi: 10.1083/jcb.109.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol. 2006;8:803–14. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 89.Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci USA. 1999;96:2122–28. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ozanne DM, Brady ME, Cook S, Gaughan L, Neal DE, Robson CN. Androgen receptor nuclear translocation is facilitated by the f-actin cross-linking protein filamin. Mol Endocrinol. 2000;14:1618–26. doi: 10.1210/mend.14.10.0541. [DOI] [PubMed] [Google Scholar]
- 91.Pentikainen U, Jiang P, Takala H, Ruskamo S, Campbell ID, Ylanne J. Assembly of a filamin four domain fragment and the influence of splicing variant-1 on the structure. J Biol Chem. 2011 doi: 10.1074/jbc.M110.195958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pentikainen U, Ylanne J. The regulation mechanism for the auto-inhibition of binding of human filamin A to integrin. J Mol Biol. 2009;393:644–57. doi: 10.1016/j.jmb.2009.08.035. Describe steered molecular dynamic simulations that support force-induced exposure of cryptic ligand binding sites in FLNs. [DOI] [PubMed] [Google Scholar]
- 93.Playford MP, Nurminen E, Pentikainen OT, Milgram SL, Hartwig JH, et al. Cystic fibrosis transmembrane conductance regulator interacts with multiple immunoglobulin domains of filamin A. J Biol Chem. 285:17156–65. doi: 10.1074/jbc.M109.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Playford MP, Nurminen E, Pentikainen OT, Milgram SL, Hartwig JH, et al. Cystic fibrosis transmembrane conductance regulator interacts with multiple immunoglobulin domains of filamin A. J Biol Chem. 2010;285:17156–65. doi: 10.1074/jbc.M109.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–19. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 96.Price MG, Caprette DR, Gomer RH. Different temporal patterns of expression result in the same type, amount, and distribution of filamin (ABP) in cardiac and skeletal myofibrils. Cell Motil Cytoskeleton. 1994;27:248–61. doi: 10.1002/cm.970270306. [DOI] [PubMed] [Google Scholar]
- 97.Provenzano PP, Keely PJ. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and Rho GTPase signaling. J Cell Sci. 2011;124:1195–205. doi: 10.1242/jcs.067009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schafer LV, et al. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci U S A. 2008;105:13385–90. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pudas R, Kiema TR, Butler PJ, Stewart M, Ylanne J. Structural basis for vertebrate filamin dimerization. Structure. 2005;13:111–19. doi: 10.1016/j.str.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 100.Razinia Z, Baldassarre M, Bouaouina M, Lamsoul I, Lutz PG, Calderwood DA. The E3 ubiquitin ligase specificity subunit ASB2{alpha} targets filamins for proteasomal degradation by interacting with the filamin actin-binding domain. J Cell Sci. 2011;124:2631–41. doi: 10.1242/jcs.084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–09. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 102.Robertson SP. Filamin A: phenotypic diversity. Curr Opin Genet Dev. 2005;15:301–07. doi: 10.1016/j.gde.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 103.Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, et al. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33:487–91. doi: 10.1038/ng1119. Initial identification of FLNa point mutations associated with human X-linked congenital malformations. [DOI] [PubMed] [Google Scholar]
- 104.Ruskamo S, Ylanne J. Structure of the human filamin A actin-binding domain. Acta Crystallogr D Biol Crystallogr. 2009;65:1217–21. doi: 10.1107/S0907444909037330. [DOI] [PubMed] [Google Scholar]
- 105.Sarkisian MR, Bartley CM, Chi H, Nakamura F, Hashimoto-Torii K, et al. MEKK4 signaling regulates filamin expression and neuronal migration. Neuron. 2006;52:789–801. doi: 10.1016/j.neuron.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sarkisian MR, Bartley CM, Rakic P. Trouble making the first move: interpreting arrested neuronal migration in the cerebral cortex. Trends Neurosci. 2008;31:54–61. doi: 10.1016/j.tins.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 107.Sato M, Nagano T. Involvement of filamin A and filamin A-interacting protein (FILIP) in controlling the start and cell shape of radially migrating cortical neurons. Anat Sci Int. 2005;80:19–29. doi: 10.1111/j.1447-073x.2005.00101.x. [DOI] [PubMed] [Google Scholar]
- 108.Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–26. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sawyer GM, Clark AR, Robertson SP, Sutherland-Smith AJ. Disease-associated substitutions in the filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance: Insights from the crystal structures of filamin B actin binding domains. J Mol Biol. 2009;390:1030–47. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 110.Seo MD, Seok SH, Im H, Kwon AR, Lee SJ, et al. Crystal structure of the dimerization domain of human filamin A. Proteins. 2009;75:258–63. doi: 10.1002/prot.22336. [DOI] [PubMed] [Google Scholar]
- 111.Shatunov A, Olive M, Odgerel Z, Stadelmann-Nessler C, Irlbacher K, et al. In-frame deletion in the seventh immunoglobulin-like repeat of filamin C in a family with myofibrillar myopathy. Eur J Hum Genet. 2009;17:656–63. doi: 10.1038/ejhg.2008.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sheen VL, Feng Y, Graham D, Takafuta T, Shapiro SS, Walsh CA. Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum Mol Genet. 2002;11:2845–54. doi: 10.1093/hmg/11.23.2845. [DOI] [PubMed] [Google Scholar]
- 113.Shifrin Y, Arora PD, Ohta Y, Calderwood DA, McCulloch CA. The role of FilGAP-filamin A interactions in mechanoprotection. Mol Biol Cell. 2009;20:1269–79. doi: 10.1091/mbc.E08-08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sjoblom B, Ylanne J, Djinovic-Carugo K. Novel structural insights into F-actin-binding and novel functions of calponin homology domains. Curr Opin Struct Biol. 2008;18:702–8. doi: 10.1016/j.sbi.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 115.Smith L, Page RC, Xu Z, Kohli E, Litman P, et al. Biochemical basis of the interaction between cystic fibrosis transmembrane conductance regulator and immunoglobulin-like repeats of filamin. J Biol Chem. 2010;285:17166–76. doi: 10.1074/jbc.M109.080911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, et al. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sokol NS, Cooley L. Drosophila filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr Biol. 1999;9:1221–30. doi: 10.1016/s0960-9822(99)80502-8. [DOI] [PubMed] [Google Scholar]
- 118.Sokol NS, Cooley L. Drosophila filamin is required for follicle cell motility during oogenesis. Dev Biol. 2003;260:260–72. doi: 10.1016/s0012-1606(03)00248-3. [DOI] [PubMed] [Google Scholar]
- 119.Stossel TP. Filamins and the potential of complexity. Cell Cycle. 2010;9:1456–65. [PubMed] [Google Scholar]
- 120.Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–45. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 121.Takala H, Nurminen E, Nurmi SM, Aatonen M, Strandin T, et al. Beta2 integrin phosphorylation on Thr758 acts as a molecular switch to regulate 14-3-3 and filamin binding. Blood. 2008;112:1853–62. doi: 10.1182/blood-2007-12-127795. [DOI] [PubMed] [Google Scholar]
- 122.Thompson TG, Chan YM, Hack AA, Brosius M, Rajala M, et al. Filamin 2 (FLN2): A muscle-specific sarcoglycan interacting protein. J Cell Biol. 2000;148:115–26. doi: 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tu CL, Chang W, Bikle DD. The calcium-sensing receptor-dependent regulation of cell-cell adhesion and keratinocyte differentiation requires Rho and filamin A. J Invest Dermatol. 2011;131:1119–28. doi: 10.1038/jid.2010.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ueda K, Ohta Y, Hosoya H. The carboxy-terminal pleckstrin homology domain of ROCK interacts with filamin-A. Biochem Biophys Res Commun. 2003;301:886–90. doi: 10.1016/s0006-291x(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 125.Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002;4:681–90. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- 126.van der Flier A, Kuikman I, Kramer D, Geerts D, Kreft M, et al. Different splice variants of filamin-B affect myogenesis, subcellular distribution, and determine binding to integrin [beta] subunits. J Cell Biol. 2002;156:361–76. doi: 10.1083/jcb.200103037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 128.van der Ven PF, Obermann WM, Lemke B, Gautel M, Weber K, Furst DO. Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil Cytoskeleton. 2000;45:149–62. doi: 10.1002/(SICI)1097-0169(200002)45:2<149::AID-CM6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 129.van der Ven PF, Wiesner S, Salmikangas P, Auerbach D, Himmel M, et al. Indications for a novel muscular dystrophy pathway. gamma-filamin, the muscle-specific filamin isoform, interacts with myotilin. J Cell Biol. 2000;151:235–48. doi: 10.1083/jcb.151.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vargas M, Sansonetti P, Guillen N. Identification and cellular localization of the actin-binding protein ABP-120 from Entamoeba histolytica. Mol Microbiol. 1996;22:849–57. doi: 10.1046/j.1365-2958.1996.01535.x. [DOI] [PubMed] [Google Scholar]
- 131.Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu Rev Biophys Biomol Struct. 2006;35:459–88. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 132.Vorgerd M, van der Ven PF, Bruchertseifer V, Lowe T, Kley RA, et al. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet. 2005;77:297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 134.Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583–95. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xie Z, Xu W, Davie EW, Chung DW. Molecular cloning of human ABPL, an actin-binding protein homologue. Biochem Biophys Res Commun. 1998;251:914–9. doi: 10.1006/bbrc.1998.9506. [DOI] [PubMed] [Google Scholar]
- 137.Xu R, Boudreau A, Bissell MJ. Tissue architecture and function: dynamic reciprocity via extra- and intra-cellular matrices. Cancer Metastasis Rev. 2009;28:167–76. doi: 10.1007/s10555-008-9178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xu W, Xie Z, Chung DW, Davie EW. A novel human actin-binding protein homologue that binds to platelet glycoprotein Ibalpha. Blood. 1998;92:1268–76. [PubMed] [Google Scholar]
- 139.Yamazaki M, Furuike S, Ito T. Mechanical response of single filamin A (ABP-280) molecules and its role in the actin cytoskeleton. J Muscle Res Cell Motil. 2002;23:525–34. doi: 10.1023/a:1023418725001. [DOI] [PubMed] [Google Scholar]
- 140.Yu H, Mouw JK, Weaver VM. Forcing form and function: biomechanical regulation of tumor evolution. Trends Cell Biol. 2011;21:47–56. doi: 10.1016/j.tcb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yuan Y, Shen Z. Interaction with BRCA2 suggests a role for filamin-1 (hsFLNa) in DNA damage response. J Biol Chem. 2001;276:48318–24. doi: 10.1074/jbc.M102557200. [DOI] [PubMed] [Google Scholar]
- 142.Zheng L, Baek HJ, Karsenty G, Justice MJ. Filamin B represses chondrocyte hypertrophy in a Runx2/Smad3-dependent manner. J Cell Biol. 2007;178:121–28. doi: 10.1083/jcb.200703113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhou AX, Hartwig JH, Akyurek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 2010;20:113–23. doi: 10.1016/j.tcb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 144.Zhou X, Boren J, Akyurek LM. Filamins in cardiovascular development. Trends Cardiovasc Med. 2007;17:222–9. doi: 10.1016/j.tcm.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 145.Zhou X, Tian F, Sandzen J, Cao R, Flaberg E, et al. Filamin B deficiency in mice results in skeletal malformations and impaired microvascular development. Proc Natl Acad Sci USA. 2007;104:3919–24. doi: 10.1073/pnas.0608360104. [DOI] [PMC free article] [PubMed] [Google Scholar]


