SUMMARY
As a central component in the maturation of Okazaki fragments, flap endonuclease 1 (FEN1) removes the 5′-flap and maintains genomic stability. Here, FEN1 was cloned as a suppressor of transcriptional gene silencing (TGS) from a forward genetic screen. FEN1 is abundant in the root and shoot apical meristems and FEN1-GFP shows a nucleolus-localized signal in tobacco cells. The Arabidopsis fen1-1 mutant is hypersensitive to methyl methanesulfonate and shows reduced telomere length. Interestingly, genome-wide chromatin immunoprecipitation and RNA sequencing results demonstrate that FEN1 mutation leads to a decrease in the level of H3K27me3 and an increase in the expression of a subset of genes marked with H3K27me3. Overall, these results uncover a role for FEN1 in mediating TGS as well as maintaining genome stability in Arabidopsis.
Keywords: FEN1, transcriptional gene silencing, H3K27me3, ChIP-seq, DNA damage repair, telomere, Arabidopsis thaliana
INTRODUCTION
Replication and repair of DNA, cell-cycle regulation and epigenetic systems ensure that genetic and epigenetic information is passed to the next generation accurately and genomic stability is maintained during cell proliferation (Alabert and Groth, 2012). Dysfunction of DNA replication or repair may result in replication stress and genome instability, which can cause cancer and cell death (Gaillard et al., 2015).
FEN1 is a structure-specific nuclease (Tsutakawa et al., 2011) that can remove 5′-flaps, the nucleic acid structures created by the primase and DNA polymerase during the process of Okazaki fragment (lagging strand) maturation (Balakrishnan and Bambara, 2013). FEN1 interacts with more than 30 proteins involved in different DNA metabolic pathways, including DNA replication, DNA repair, apoptotic DNA degradation and the maintenance of telomere stability (Zheng et al., 2011). FEN1 has single-stranded DNA endonuclease activity that is critical for resolving stalled replication forks (Zheng et al., 2005; Zhu et al., 2008). FEN1 also plays key roles in maintaining telomere homeostasis (Saharia et al., 2008). FEN1 has an important role as a tumor suppressor, and FEN1 mutations result in genomic instability and cancer (Henneke et al., 2003; Zheng et al., 2007).
FEN1 is highly conserved among species. Mice with a FEN1 deletion were embryonically lethal and hypersensitive to gamma radiation (Larsen et al., 2003). Conversely, homozygous fen1 null mutants in chicken DT40 cells were viable but sensitive to methyl methanesulfonate (MMS) and oxidative DNA-damaging agents (Matsuzaki et al., 2002). The yeast fen1 mutant was temperature-sensitive lethal and sensitive to MMS (Reagan et al., 1995). However, our understanding of FEN1 in the plant kingdom is limited. FEN-1a, but not the FEN-1b homologue from rice, could complement the Saccharomyces cerevisiae fen1 null mutant rad27 (Kimura et al., 2003). The Arabidopsis FEN1 mutant shade avoidance 6 (sav6) was hypersensitive to ultraviolet (UV)-C radiation and double-stranded DNA break-inducing agents (Zhang et al., 2016).
Epigenetic inheritance is important for development and in disease. We characterized several factors involved in mediating transcriptional gene silencing (TGS) using a specific transgenic system in Arabidopsis (the transgenic wild type in the C24 ecotype, referred to as TWT). This system contains 35S-NPTII (neomycin phosphotransferase II driven by the CaMV 35S promoter) and RD29A-LUC (a luciferase reporter driven by the stress-responsive RD29A promoter), which are two loci regulated by different mechanisms. Regulation of RD29A-LUC is primarily dependent on the small RNA-directed DNA methylation (RdDM) pathway, whereas 35S-NPTII is affected by DNA demethylation factors and DNA replication factors (Gong et al., 2002; Liu and Gong, 2011).
In this study, we identified Arabidopsis putative FEN1 as a suppressor of TGS in the dms3-4 mutant with silenced 35S-NPTII. We found that fen1-1 decreased the level of H3K27me3. The fen1-1 mutant was hypersensitive to MMS and exhibited shorter telomeres. Our results uncover a role for FEN1 in mediating TGS.
RESULTS
Map-based cloning of Arabidopsis FEN1
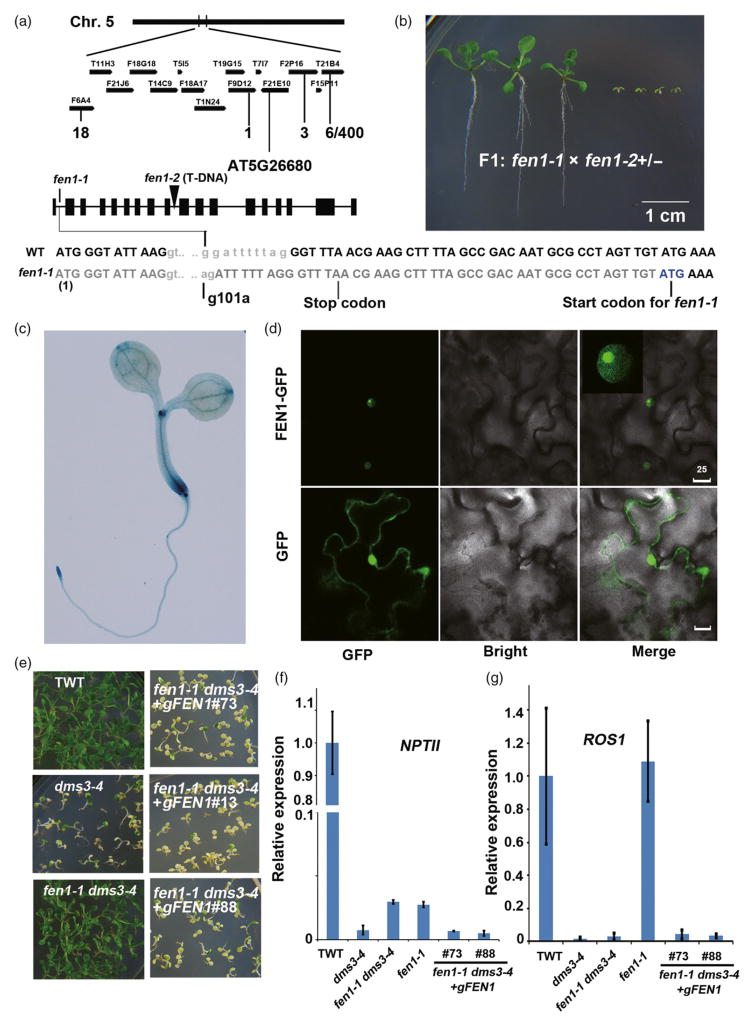
Defective in meristem silencing 3 (dms3-4) mutants are sensitive to kanamycin because this mutation reduces expression of ROS1 (Li et al., 2012). We isolated a kanamycin-resistant mutant (fen1-1) from an ethyl methanesulfonate (EMS)-mutagenized dms3-4 population containing approximately 12 000 M1 individuals. Genetic analysis indicated that fen1-1 mutation is recessive (Figure S1 in the Supporting Information). Using map-based cloning, the mutation was narrowed to the area between the F9D12 and F2P16 bacterial artificial chromosomes (BACs) on chromosome 5. We identified a G-to-A point mutation that led to abnormal splicing and the formation of a stop codon in the putative FEN1 (At5 g26680) (Figure 1a). An ATG downstream of the stop codon may serve as a potential start codon for the fen1-1 mutant. The mutation results in a 17-amino-acid deletion at the N-terminus of the FEN1 protein (Figure 1a). The putative Arabidopsis FEN1 is present in one copy (Shultz et al., 2007) and is essential because we could not obtain a homozygote from the progeny of a heterozygous fen1-2 T-DNA insertion mutant. When we crossed fen1-1 with the fen1-2+/− heterozygote, approximately half of the F1 progeny exhibited severe growth retardation (Figure 1b). These results demonstrate that fen1-1 and fen1-2 are allelic, that the severe growth phenotype of fen1-1 fen1-2 might be due to the lower level of FEN1 protein generated by a single copy of fen1-1 and that FEN1 is critical for plant development. β-Glucuronidase (GUS) staining in proFEN1::GUS transgenic lines indicated that FEN1 is ubiquitously expressed but more abundant in the root and shoot meristems where DNA replication is more active compared with the other tissues (Figure 1c). The transient expression of FEN1-GFP protein in tobacco cells indicated that FEN1-GFP is localized to the nucleus and enriched in the nucleoli (Figure 1d). The fen1-1 mutation partially restored NPTII expression and kanamycin resistance compared with TWT (Figure 1e,f). When we transferred wild-type genomic FEN1 DNA to the fen1-1 dms3-4 double mutant and randomly selected three independent transgene lines, these transgenic lines recovered the low expression level of NPTII and the kanamycin-sensitive phenotype that was present in dms3-4 (Figure 1e,f). These results confirm that the fen1-1 mutation causes the release of TGS in the dms3-4 mutant. The expression of ROS1 was greatly decreased by dms3-4 mutation, which is correlated with the reduced expression of NPTII (Figure 1f, g) (Li et al., 2012). The abundance of ROS1 in the fen1-1 single mutant was comparable to TWT, but the expression of NPTII was lower in the fen1-1 mutant than TWT (Figure 1f, g). fen1-1 also had no effect on the expression of ROS1 in the dms3-4 background (Figure 1g), suggesting that de-repression of NPTII caused by fen1-1 mutation in the dms3-4 background is not dependent on ROS1.
Figure 1.
Map-based cloning of FEN1 and complementation assay.(a) Map-based cloning of FEN1. The mutation was localized on chromosome 5 (Chr. 5) in the region between the F9D12 and F2P16 bacterial artificial chromosomes (BACs). The recombination rates were indicated under the matching BACs. A G to A mutation was identified 101 bp from ATG at gene At5 g26680. The transcript of the mis-splicing caused by the mutation led to the formation of a stop codon. An ATG downstream of the stop codon produced by the mutation might be used as a translation start site for the fen1-1 mutant. Therefore, fen1-1 produced a protein lacking 17 amino acids at the N-terminus. (b) Genetic analysis of the different fen1 alleles. Phenotype of the F1 progeny of fen1-1 crossed with fen1-2(+/−) heterozygotes. (c) Expression pattern of FEN1. GUS staining of the ProFEN1:GUS transgenic plant. (d) Subcellular localization of FEN1-GFP. FEN1 fused with GFP was transiently expressed in tobacco leaf cells. The 35S-GFP construct was used as a control. Bar = 25 μm. (e) fen1-1 complementation. Phenotype of the transgenic wild type (TWT), dms3-4, fen1-1 dms3-4 and three independent genomic DNA complementation lines (#76, #13 and #88) in the fen1-1 dms3-4 background on MS supplied with 50 mg L−1 kanamycin. (f) Relative expression of NPTII among TWT, dms3-4, fen1-1 dms3-4 and two independent genomic DNA complementation lines. (g) Relative expression of ROS1 among TWT, dms3-4, fen1-1 dms3-4 and two independent genomic DNA complementation lines.
fen1-1 suppresses 35S-NPTII silencing caused by the ros1 mutation
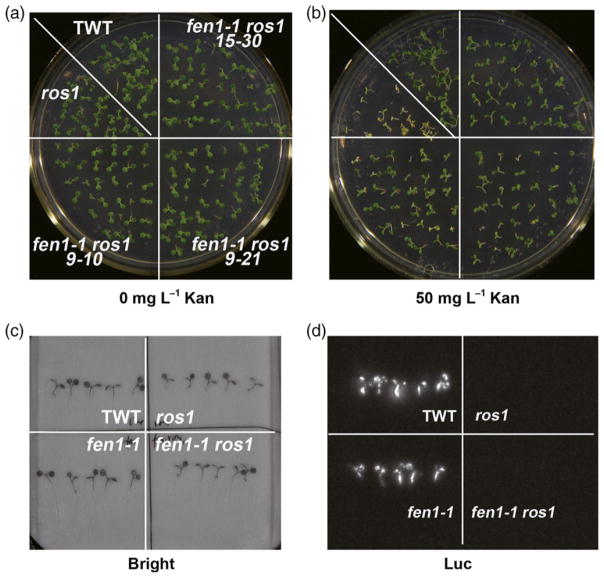
To confirm the involvement of FEN1 in the maintenance of TGS, we crossed fen1-1 with the ros1 mutant. ROS1 encodes a DNA demethylase, and its mutation causes silencing of 35S-NPTII (Gong et al., 2002; Zhu, 2009). We analyzed the phenotype of the fen1-1 ros1 double mutant. In contrast to the kanamycin-sensitive phenotype of the ros1 mutant, fen1-1 ros1 partially restored kanamycin resistance (Figure 2a,b). However, fen1-1 did not affect expression of RD29A::LUC in the ros1-1 mutant (Figure 2c, d). These results indicate that fen1-1 is a suppressor of ros1 on the 35S-NPTII locus similar to other DNA replication factors (Liu and Gong, 2011).
Figure 2.
fen1-1 is also a suppressor of ros1 at the 35S:NPTII locus but not the RD29A:LUC locus.
(a), (b) fen1-1 de-represses the silencing of 35S: NPTII in the ros1-1 mutant. Phenotype of the transgenic wild type (TWT), ros1-1, and three fen1-1 ros1-1 lines on a MS plate (a) or a MS plate supplied with 50 mg L−1 kanamycin (Kan) (b).
(c), (d) fen1-1 mutants could not release the silencing of RD29A:LUC caused by the ros1 mutant. Luminescence imaging of TWT, ros1-1, fen1-1, and fen1-1 ros1-1 samples. The bright-field image is shown in (c) and the LUC signal is presented in (d). The photos were taken after the treatment of the plants with 300 mM NaCl for 3 h.
The fen1-1 mutant is hypersensitive to MMS and exhibits shorter telomeres than TWT
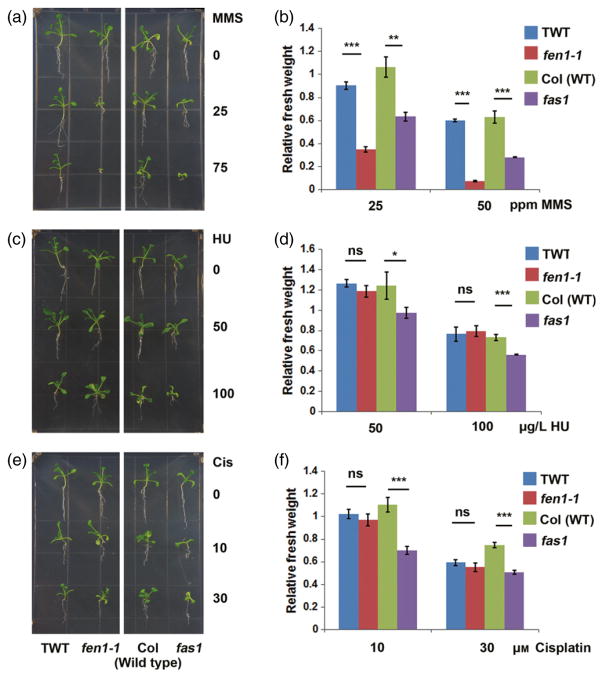
Previous results from yeast and animal cell lines suggested that FEN1 is an important factor in the DNA damage response. The fen1-1 mutant was hypersensitive to MMS (Figure 3a,b). The fen1-1 mutant is much smaller compared than TWT when exposed to 25 p.p.m. MMS. Furthermore, the fen1-1 mutant could not survive or develop in the presence of more than 75 p.p.m. MMS (Figure 3a,b). However, no significant growth differences were observed between the fen1-1 mutant and TWT exposed to hydroxyurea (HU, an inhibitor of DNA replication) (Figure 3c,d) or cisplatin (a DNA cross-linking agent) (Figure 3e,f). As a positive control, we demonstrated that the fasciata 1 (fas1, a chromatin assembly factor 1 in Arabidopsis) mutant was sensitive to MMS, cisplatin and HU (Figure 3). These results indicate that the fen1-1 mutant is specifically sensitive to MMS.
Figure 3.
fen1-1 is hypersensitive to methyl methanesulfonate (MMS).
Phenotype of 2-week-old seedlings on MS medium supplemented with different concentrations of DNA-damaging reagents: MMS (a); hydroxyurea (HU; c); and cis-diamineplatinum (II) dichloride (Cis; e). The fas1 mutant (Columbia-0 background) was used as a positive control. (b), (d), (f) Statistical analyses of the seedling fresh weight after treatment relative to no treatment as indicated in the left panel. Bars represent standard error from three biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001, ns, not statistically significant; paired t-test. TWT, transgenic wild type.
We performed RNA sequencing (RNA-seq) of TWT and the fen1-1 mutant with two biological replicates for each sample (Data S1). The expression levels of 18 genes involved in the DNA repair pathway were significantly upregulated in the fen1-1 mutant (Data S2), including RAS associated with diabetes protein 51 (RAD51), breast cancer susceptibility 1 (BRCA1) and poly (ADP-ribose) polymerase 2 (PARP2). The increased level of DNA damage-associated genes and the sensitivity to MMS indicated that the fen1-1 mutant suffered from a constitutive DNA stress response.
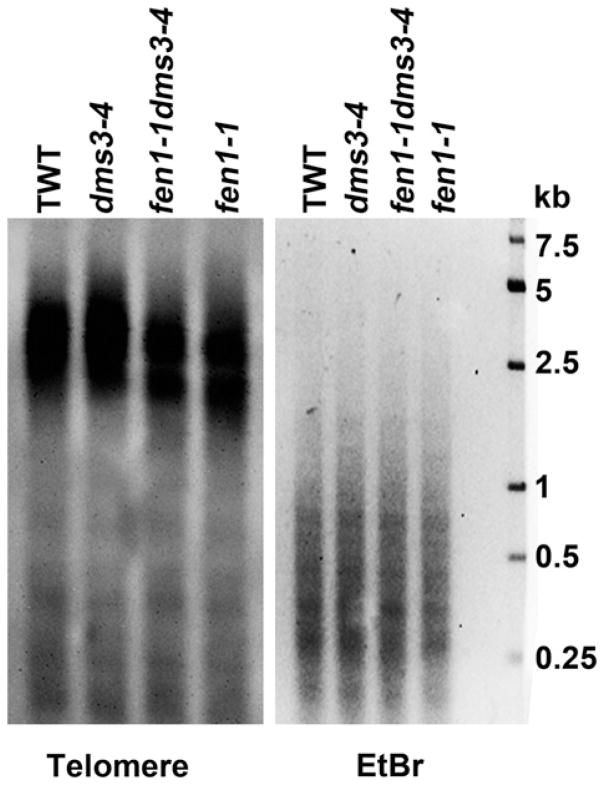
Telomere length disorder is another feature of genomic instability. Next, we investigated whether telomere length was altered due to the FEN1 mutation. Using a telomere-specific probe, we found that the telomere lengths of fen1-1 dms3-4 and fen1-1 were shorter than in TWT and the dms3-4 mutant (Figure 4). These results demonstrate that FEN1 maintains the telomere length in Arabidopsis and support a crucial role for FEN1 in genomic stability by maintaining telomere lengths and repairing DNA damage.
Figure 4.
FEN1 mutation results in shorter telomeres.
Southern blot detection of telomere length among the transgenic wild type (TWT) and the dms3-4, fen1-1 dms3-4 and fen1-1 mutants. Genomic DNA was isolated from 14-day-old seedlings and digested with BfuCI. A telomere-specific sequence was used as the probe (left panel). Digested DNA stained with ethidium bromide (EtBr) before transferring was used as the loading control (right panel).
fen1-1 releases NPTII silencing in dms3-4 by decreasing the level of H3K27me3
To elucidate the mechanism by which the fen1-1 mutants de-repressed the silencing locus, we performed chromatin immunoprecipitation sequencing (ChIP-seq) assays using H3K27m3, H3K4me3 and H3K9me2 antibodies to study changes in the epigenetic profile between TWT and fen1-1 mutant lines. The results obtained in this study correlated well with previous reports. We identified 7570 genes occupied by H3K27me3 (Data S3), which covered 76% (3881/5088) of the genes modified by H3K27me3 from a previous study (Lu et al., 2011) and 78% (3906/4980) of the genes reported from a second study (Zhang et al., 2007a) (Figure S2). We identified 17 548 genes that harbored H3K4me3 (Data S4), which represented a 97% (13788/14205) overlap with the genes obtained in a previous study (Luo et al., 2013) (Figure S2). We identified 3823 genes that contained H3K9me2 (Data S5), which covered 76% (872/1146) of the genes reported in Luo et al. (2013) (Figure S2). These results demonstrate that our ChIP-seq data are reliable.
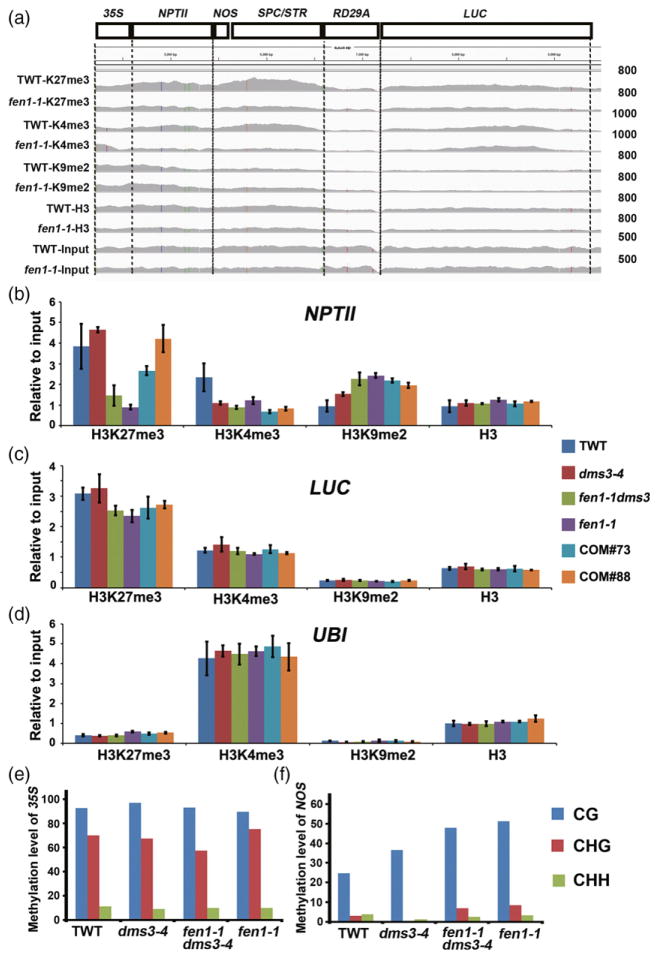
We analyzed histone modifications on the T-DNA locus between TWT and the fen1-1 mutant using the ChIP-seq data. We observed a decrease in H3K4me3 and H3K27me3, as well as an increase in H3K9me2 on the NPTII gene body in the fen1-1 mutant compared with TWT (Figure 5a). Then, we performed a ChIP-qPCR assay to confirm the changes in histone modification among TWT, dms3-4, fen1-1 and fen1-1 dms3-4 samples. Indeed, we identified a significant decrease in the H3K27me3 modification on the NPTII gene body in fen1-1 and fen1-1 dms3-4 compared with TWT and the dms3-4 mutant (Figure 5b), which was consistent with the ChIP-seq data. Two independent complementary lines showed a recovered H3K27me3 level similar to dms3-4 (Figure 5b), indicating that the decrease of H3K27me3 on the NPTII gene body is caused by the FEN1 mutation. Additionally, we identified an increase in H3K9me2 on the NPTII gene body in dms3-4 compared with TWT, which was consistent with the reduced expression of NPTII in the dms3-4 mutant (Figure 1f). However, the occupancy of H3K9me2 on NPTII in fen1-1 and fen1-1 dms3-4 was similar to the dms3-4 mutant, which exhibited an increased level relative to TWT (Figure 5b). We also observed a reduced level of H3K4me3 in the fen1-1, fen1-1 dms3-4 and dms3-4 mutants compared with TWT, which correlated with the reduced expression of NPTII in these samples compared with TWT (Figures 1f and 5b). The LUC region was also checked by ChIP-qPCR. The levels of H3K4me3, H3K9me2 and total H3 were comparable among these samples, while the H3K27me3 level was slightly decreased in fen1-1 dms3-4 and fen1-1 mutants relative to the other samples (Figure 5c). As a control, we demonstrated that the histone modification levels of the ubiquitin-conjugating enzyme 21 (UBI) gene were comparable among TWT, dms3-4, fen1-1 dms3-4 and fen1-1 samples (Figure 5d). We next examined DNA methylation level of some regions of the transgene. Consistent with previous results (Zhao et al., 2014), the 35S region harbored high levels of DNA methylation that were not changed by FEN1 mutation (Figure 5e). fen1-1 and the fen1-1 dms3-4 double mutant showed slightly increased DNA methylation in the NOS region compared with TWT and the dms3-4 mutant (Figure 5f). These results suggest that the release of NPTII silencing in dms3-4 by fen1-1 is not related to a change in DNA methylation. Together, these results suggest that derepression of NPTII in the dms3-4 background caused by the FEN1 mutation is most likely due to a decrease in H3K27me3 on the NPTII gene body.
Figure 5.
fen1-1 releases NPTII silencing in dms3-4 by decreasing the H3K27me3 level.
(a) Integrated Genome Viewer visualization of the epigenetic profile in H3K27me3 (K27me3), H3K4me3 (K4me3), and H3K9me2 (K9me2) at transgenic loci in the transgenic wild type (TWT) and fen1-1 mutant. (b) H3K27me3 levels at the NPTII gene body were reduced by the FEN1 mutation. Alterations in histone modification among TWT, dms3-4, fen1-1 dms3-4, fen1-1 and two complementary lines were verified by chromatin immunoprecipitation-qPCR. (c), (d) Histone modification status at the control region. UBI was used as a positive control for H3K4me3 and negative control for H3K27me3 and H3K9me2. (e), (f) fen1-1-released NPTII silencing in dms3-4 was not due to decreasing DNA methylation. DNA methylation levels of 35S (e) and NOS (f) were analyzed by bisulfite sequencing.
FEN1 modulates H3K27me3 and gene expression at the whole genome level
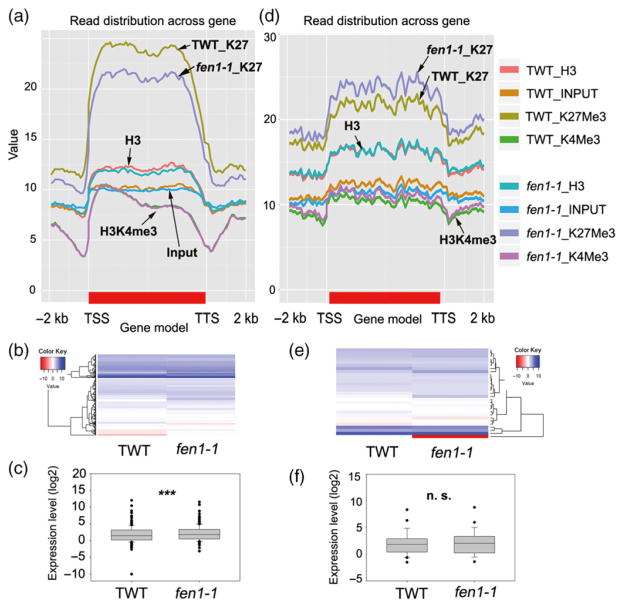
Using ChIP-seq data, we found 703 genes (Figure 6a, Data S6) with decreased and 381 genes (Figure 6d, Data S7) with increased H3K27me3 levels in the fen1-1 mutant compared with TWT [false discovery rate (FDR) <0.01, P < 0.01]. To determine whether the altered histone modifications correlated with gene expression, we performed RNA-seq analysis. The expression levels of the genes exhibiting reduced levels of H3K27me3 in the fen1-1 mutant were significantly increased compared with TWT (P < 0.0001, paired Student’s t-test; Figure 6b,c). In contrast, the expression levels of genes with increased H3K27me3 levels in the fen1-1 mutant were not significantly altered compared with TWT (P = 0.5742, paired Student’s t-test; Figure 6e,f). Although a good correlation was noted between high gene expression and high H3K4me3 histone modification or low gene expression and low H3K4me3 modification (Figure S3) in our RNA-seq and ChIP data, the fen1-1 mutant had a slight effect on H3K4me3. Only 26 (Figure S3a, Data S8) or 44 (Figure S3d, Data S9) genes harbored increased or reduced H3K4me3 levels, respectively, in the fen1-1 mutant relative to TWT (FDR < 0.01, P < 0.01). These results suggest that FEN1 primarily affects H3K27me3 and the expression of these H3K27me3-related genes.
Figure 6.
FEN1 mediates H3K27me3 modification and gene expression.
(a) Histone modification enrichment profiles for the transgenic wild type (TWT) and fen1-1 mutant among 703 genes with reduced H3K27me3 in the fen1-1 mutant. The genes were normalized to the same length [from transcription start site (TSS) to transcription termination site (TTS)] with a 2-kb extension upstream from the TSS or downstream from the TTS, respectively. (b), (c) Heat map and boxplot depicting the expression levels of the genes in (a) obtained from the RNA-seq data. The values are log2(FPKM)-transformed (FPKM, fragments per kilobase of transcript per million fragments mapped). ***P < 0.001, paired Student’s t-test. (d) Histone modification enrichment profiles for TWT and the fen1-1 mutant among 381 genes with an increased H3K27me3 level in the fen1-1 mutant. The genes were normalized to the same length (from TSS to TTS) with a 2-kb extension upstream from the TSS or downstream from the TTS, respectively. (e), (f) Heat map and boxplot depicting the expression of the genes in (d) obtained from the RNA-seq data. The values are log2(FPKM)-transformed. n.s., not statistically significant, paired Student’s t-test.
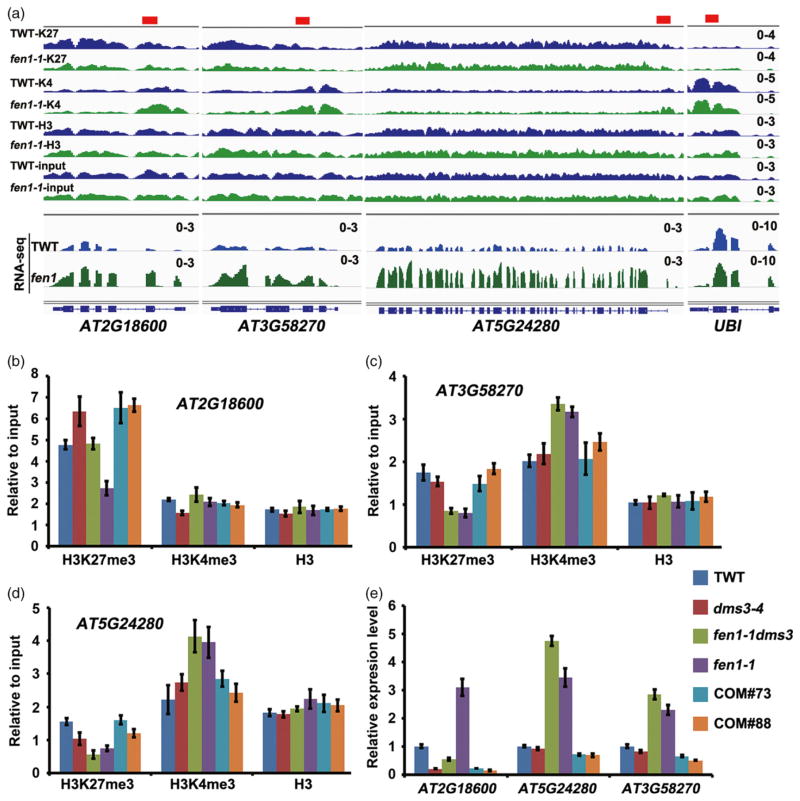
Next, we chose three H3K27me3-marked genes for validation by ChIP-qPCR (Figure 7a). For At2g18600, the H3K27me3 was increased and H3K4me3 was decreased in the dms3-4 mutant (Figure 7b) with a decreased gene expression level (Figure 7e), while in fen1-1 or fen1-1 dms3-4 mutation decreased H3K27me3 (Figure 7a,b) and increased gene expression (Figure 7e) compared with TWT or dms3-4, respectively. Two FEN1 complementary lines restored the levels of H3K27me3 and gene expression of fen1-1 dms3-4 to that of dms3-4 (Figure 7b,e). For At3g58270 and At5g24280, fen1-1 mutation resulted in lower H3K27me3 and higher H3K4me3 than in TWT or the dms3-4 mutant (Figure 7c,d). The expression levels of At3g58270 and At5g24280 were higher in fen1-1 and fen1-1 dms3-4 than TWT or the dms3-4 mutant (Figure 7e). Again, the two complementary lines restored the H3K27me3 level (Figure 7c,d) and repressed gene expression (Figure 7e). Collectively, these results point out the requirement for FEN1 in silencing H3K27me3-marked genes.
Figure 7.
FEN1 is required for silencing endogenous H3K27me3-marked genes.
(a) Integrated Genome Viewer visualization of the epigenetic profiles and mRNA levels in the affected genes in the fen1-1 mutant. The values were set to the same scale between the transgenic wild type (TWT) and the fen1-1 mutant.
(b)–(d) Histone modification changes of three endogenous genes examined by chromatin immunoprecipitation-qPCR. Values were related to input, bars are mean ± SE (n = 3).
(e) Relative gene expression level of indicated samples detected by qRT-PCR. Values were normalized to the UBI gene. Bars are mean ± SE (n = 3).
DISCUSSION
Roles of FEN1 in guarding genomic stability
FEN1 is essential in Arabidopsis. Indeed, we were not able to obtain a homozygote from the progeny of a heterozygous T-DNA insertion mutant, which is similar to the case reported from mice that are early embryonic lethal when FEN1 is completely deleted (Larsen et al., 2003). Arabidopsis FEN1 is most likely a DNA replication-coupled protein because it is highly expressed in the root and shoot meristems, consistent with the recent reports from the analysis of SAV6 (Arabidopsis FEN1) (Zhang et al., 2016). In rice, OsFEN1 was also reported to be abundant in the shoot apical meristem, young leaves and roots (Kimura et al., 2003). We found that FEN1-GFP is localized to the nucleus, which is consistent with the role of FEN1 in distinct DNA metabolic processes. Strikingly, FEN1 is enriched in the nucleolus in tobacco cells. This result is consistent with the results from HeLa cells, demonstrating that mammalian FEN1 is super-accumulated in the nucleolus (Guo et al., 2008). The nucleolus is the site of telomere clustering (Armstrong et al., 2001), and the fen1-1 mutation leads to telomere shortening. Previous work in mammalian cells demonstrated that FEN1 is localized to the telomere in a cell-cycle-dependent manner (Verdun and Karlseder, 2006) and that FEN1 depletion led to telomere shortening (Saharia et al., 2008; Sampathi et al., 2009). Our results suggest a critical role for FEN1 in dealing with telomere homeostasis in Arabidopsis.
The fen1-1 mutant in Arabidopsis is hypersensitive to MMS, which is consistent with the phenotype observed in chickens (Matsuzaki et al., 2002), mammalian cells (Shibata and Nakamura, 2002) and yeast (Reagan et al., 1995). In addition, it was reported recently that Arabidopsis FEN1 shows double-flap endonuclease activity and FEN1 mutation (sav6) results in sensitivity to UV-C and DNA double-strand break reagents, namely camptothecin (CPT) and Zeocin (Zhang et al., 2016). These results support the critical role of FEN1 in repairing DNA damage in Arabidopsis. Because FEN1 functions in the stability of triplex DNA (Liu et al., 2004), it is possible that Arabidopsis FEN1 contributes to the replication of structured DNA during the cell cycle. The shorter telomere length and the sensitivity to DNA-damaging reagents in the fen1 mutant appear to be a feature of the genomic instability caused by the FEN1 mutation. Moreover, several DNA replication factors such as RFC1, RPA2, Pol ε and Pol α have been characterized and participate in the maintenance of genomic stability (Liu and Gong, 2011). These results support a conserved role for FEN1 in DNA damage repair and maintenance of telomere length.
FEN1 in epigenetics
Although the enzyme activity, structure and biological function of FEN1 have been well established in other species, reports of a role for FEN1 in epigenetics are limited. In this study, FEN1 was isolated from a screen for TSG suppressors, and the combined ChIP-seq and RNA-seq data support a role for FEN1 in mediating H3K27me3 and gene silencing. We found that H3K27me3 levels are decreased in 703 genes but are increased in 381 genes in fen1-1 mutants, and that some of the genes with reduced H3K27me3 are indeed highly expressed. Several DNA replication and repair factors, including Pol α (Liu et al., 2010a; Hyun et al., 2013), Pol ε (Yin et al., 2009), RFC1 (Liu et al., 2010b), RPA2A (Elmayan et al., 2005; Kapoor et al., 2005; Xia et al., 2006), FAS1 (Elmayan et al., 2005) and BRU1 (Takeda et al., 2004), play roles in TGS. The FEN1 mutation resulted in a reduction in H3K27me3 on the NPTII gene body and an increase in NTPII expression, but had a minimal effect on the RD29A:LUC locus, whose expression was regulated by RdDM. Interestingly DNA damage generated from short telomeres in mice resulted in a reduction of H3K27me3 at the inactive X chromosome and de-repression of the X-chromosome-linked transgene, which may be silenced by H3K27me3 (Schoeftner et al., 2009). These results suggest that DNA replication and repair could affect H3K27me3 occupancy, thereby disturbing DNA replication and repair defects that would de-repress the silenced loci, especially those marked with H3K27me3. However, the mechanism by which DNA replication factors affect the level of H3K27me3 is unclear. Previous studies proposed that a mutation in the catalytic subunit of DNA polymerase α altered the binding of like heterochromatin protein 1 (LHP1) (Hyun et al., 2013), which was a crucial component of the polycomb repressive complex (PRC) for the maintenance of H3K27me3 in Arabidopsis (Zhang et al., 2007b; Shen et al., 2014). Recently, it was reported that DNA polymerase ε interacts with the components of PRC2, and dysfunction of DNA polymerase ε results in a decrease in the H3K27me3 level and increased expression of H3K27me3-marked genes (Del Olmo et al., 2016). It is likely that dysfunction of FEN1 might perturb the DNA replication machinery and thereby affect epigenetic hallmarks directly or indirectly. Further studies must be performed to uncover the mechanism underlying the maintenance of H3K27me3 and gene silencing by DNA repair factors.
EXPERIMENTAL PROCEDURES
Mutant materials and culture conditions
The transgenic wild-type line (TWT) used in this work was a stable transgenic plant containing the 35S:NPTII and RD29A:LUC loci in the C24 background (Li et al., 2012). dms3-1 was described in previous reports and kindly provided by Dr Matzke (Institute of Plant and Microbial Biology, Taipei, Taiwan) (Kanno et al., 2008). fen1-1 dms3-4 was isolated from EMS-mutagenized progeny of dms3-4 and backcrossed four times with dms3-4. The fen1-1 single mutant was obtained from an F2 population of fen1-1 dms3-4 crossed with TWT. fen1-2 (355B05) was a T-DNA insertion allele obtained from the stock center (Rosso et al., 2003). The fas1 mutant allele used in this study was isolated in our laboratory, and has a G to A substitution at 1989 bp from ATG. Seeds were sterilized and grown on MS plates containing 20 g L−1 sucrose and 8 g L−1 agar. The incubator was set to 21°C with an illumination period of 23 h light and 1 h dark.
Map-based cloning of FEN1 and complementary assay
fen1-1 dms3-4 was crossed with dms3-1 (Col-0), and 200 DNA samples from kanamycin-resistant plants selected from the F2 progeny were analyzed with simple sequence length polymorphism markers (Data S10). The mutation was narrowed to an area between BAC clones F9D12 and F2P16. The putative FEN1 gene was selected and sequenced, and a point mutation was characterized. For complementation, an approximately 7-kb FEN1 genomic DNA fragment containing 2832 bp upstream of the putative ATG and 468 bp downstream of the TAA was amplified using primers (FEN1genomic-Sal1F and FEN1genomic-Spe1R; Data S10) and cloned into pCAMBIA1391. The plasmid was introduced into the fen1-1 dms3-4 mutants by Agrobacterium tumefaciens strain GV3101. Three independent transgenic lines were selected for further analysis.
Expression pattern and subcellular localization of FEN1
A 4390-bp fragment containing 2832 bp upstream of the putative ATG and 1558 bp downstream of the ATG was amplified from genomic DNA using the proFEN1-Sal1F and proFEN1-BamHIR primers (Data S10). The fragment was cloned into pCAMBIA1391 containing a GUS reporter gene. The plasmid was introduced into TWT by A. tumefaciens strain GV3101. At least eight independent transgenic lines were selected for GUS staining. The chosen line was one of the eight transgenic lines that exhibited similar GUS staining patterns.
For subcellular location detection, the FEN1 coding sequence was obtained from cDNA using paired primers (FEN1GFP-Xba1F and FEN1GFP-Sal1R; Data S10). Then, the product was cloned into the 35S-GFP vector derived from pCAMBIA1300 to generate a C-terminal fused GFP (35S:FEN1-GFP) vector. The vector was introduced into tobacco leaves using A. tumefaciens strain GV3101. After 3 days, the GFP signal was collected using a confocal laser scanning microscope (Leica SP5). The construct with 35S-GFP was used as the control.
DNA damage assay
Seeds were sterilized and grown on MS medium or MS medium supplemented with different concentrations of DNA-damaging reagents as indicated: 10 or 30 μM CIS (cis-diamineplatinum (II) dichloride, Sigma no. 479306; http://www.sigmaaldrich.com/); 25 or 75 p.p.m. (0.0075%) MMS (Sigma no. 129925); or 100 or 200 mg L−1 HU (Sigma no. H8627). After growth for 2 weeks, the seedlings were imaged and weighed. The DNA damage was calculated using the fresh weight of seedlings treated with DNA-damaging reagents relative to the untreated controls. Three biological replicates were performed.
Telomere length detection
Genomic DNA was extracted from 14-day-old seedlings using a DNeasy Plant Mini Kit (Qiagen, http://www.qiagen.com/) according to the manufacturer’s instructions. One microgram of DNA was digested with BfuCI in a 50-μl system at 37°C. Southern blotting was performed with the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche, http://www.roche.com/). The synthesized telomere repeat (TTTAGGG)7 labeled with digoxigenin was used as the probe.
DNA methylation analysis
Genomic DNA was extracted from 10-day-old seedlings using a DNeasy Plant Mini Kit (Qiagen). Five hundred nanograms of DNA was treated using an EZ DNA Methylation-Gold Kit (Zymo Research, http://www.zymoresearch.com/). The DNA fragments were eluted with 10 μl of elution buffer, and 2 μl was used for amplification by PCR. The product of PCR was ligated into pMD18-T vector (Takara, http://www.takara-bio.com/). At least 10–15 clones were sequenced for analysis. Primers used were the same as previously reported (Zhao et al., 2014).
RNA-seq data analysis
Total RNA was extracted from 10-day-old seedlings using an RNeasy Plant Mini Kit (Qiagen). Libraries were generated from 3 μg of total RNA using NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, https://www.neb.com/). Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation and first-strand and second-strand cDNA synthesis, the products were end repaired, dA-tailed and NEBNext adaptors ligated. cDNA fragments of preferentially 150–200 bp in length were selected using an AMPure XP system (Beckman Coulter, https://www.beckmancoulter.com/). The library was amplified using Phusion High-Fidelity DNA polymerase (New England Biolabs). After cluster generation using a TruSeq PE Cluster Kit v3-cBot-HS (Illumina, http://www.illumina.com/), the library was sequenced on an Illumina Hiseq platform. Two biological replicates were performed for each fen1-1 and TWT. Paired-end reads were aligned to the Arabidopsis reference genome (TAIR10) using TopHat (version 2.1.0) with read-mismatches 2 and bowtie1. Cufflinks (version 2.2.1) was used to calculate the FPKM (fragments per kilobase of transcript per million fragments mapped) value as an expression value for each gene (Trapnell et al., 2012).
Chromatin immunoprecipitation assay
A 2-g sample of the 10-day-old seedlings was crosslinked with 1% formaldehyde. Chromatin was isolated using nuclei extraction buffer [1 M hexylene glycol, 20 mM 2-amino-2-(hydroxymethyl)-1,3-propanediol (TRIS)-HCl, pH 8.0, 0.15 mM spermine, 5 mM 2-mercaptoethanol, 1% Triton X-100, 0.1 mM PMSF and protease inhibitor cocktail] and precipitated by centrifugation at 2000 g for 10 min at 4°C. After resuspension in 300 μl of nuclei lysis buffer (50 mM TRIS-HCl, pH 8.0, 10 mM EDTA, 1% SDS and protease inhibitor cocktail) at 4°C, the chromatin was sonicated into fragments of approximately 300 bp using a Bioruptor (15 cycles of 30 sec on and 30 sec off at high intensity). The following antibodies were used: anti-H3K27me3 (ab6002; Abcam, http://www.abcam.com/), anti-H3 (ab1791, Abcam), anti-H3K4me3 (ab8580, Abcam) and anti-H3K9me2 (ab1220, Abcam). Dynabeads (10001D, Thermo Fisher Scientific, https://www.thermofisher.com) were used for immunoprecipitation. The DNA was treated with RNase A and Proteinase K and then recovered using a QIAquick PCR purification kit (Qiagen). The DNA concentration was quantified using the Qubit 3.0 system (Life Technologies).
High-throughput sequencing libraries were generated using the NEBNext® ChIP-Seq Library Prep Master Mix Set for Illumina (E6240, New England Biolabs) from a total amount of 10 ng DNA per sample. Briefly, the DNA was end repaired, dA-tailed and adaptor ligated. The products were amplified using NEBNext Q5 Hot Start HiFi PCR Master Mix (New England Biolabs). The index primers used for each sample were as follows: AGTCAA for TWT-H3K27me3; GTCCGC for TWT-H3K4me3; AGTTCC for TWT-H3K9me2; ATGTCA for TWT-H3; CCGTCC for TWT-Input; GTGGCC for fen-H3K27me3; GTGAAA for fen-H3K4me3; GTTTCG for fen-H3K9me2; CGTACG for fen-H3; and GAGTGG for fen-Input. The libraries were sequenced on the Illumina NextSeq 500 System with single-end 75-bp reads.
For ChIP-qPCR, the ChIPed or input samples were adjusted with distilled H2O to a concentration of 50 pg μl−1. Then, 1 μl of immunoprecipitated or input DNA was used for amplification in a 20-μl reaction using the SYBR Green Master Mix (Takara). The real-time PCR reaction was performed as follows: 95°C for 5 min and 40 cycles of 95°C for 15 sec and 60°C for 1 min. The values were calculated using 2(Ct input − Ct chiped).
ChIP-seq analysis
The reads were mapped to the Arabidopsis genome (TAIR10) using Bowtie1 (version 0.12.9) (Langmead et al., 2009) allowing two nucleotide mismatches with the following parameters: bowtie -q -k 1 -n 2 -l 36 –best -S -p 2. The mapped reads were de-duplicated and sorted by samtools (version 0.1.19-44428 cd) (Li et al., 2009). The igvtools count was used to generate files for visualization with the Integrative Genomics Viewer (Robinson et al., 2011). Histone modification peaks were identified using macs software (version 1.4.2) (Zhang et al., 2008) with H3 as the background and 1 × 10−5 as the P-value threshold. Genes were annotated by BEDTools (Quinlan and Hall, 2010) according to TAIR10. Changes in histone occupancy were calculated using sicer software (version 1.1) (Zang et al., 2009). The window size and gap size were set at 200 bp according to the guidelines. The results were filtered by P < 0.01 and FDR < 0.01. For H3K4me3, the fold change was set to greater than 1.5.
Supplementary Material
Figure S1. fen1-1 mutation is recessive.
Figure S2. Validation of the chromatin immunoprecipitation sequencing results.
Figure S3. The FEN1 mutation affects some H3K4me3-marked genes.
Data S1. RNA sequencing data.
Data S2. DNA repair-associated genes upregulated in the fen1-1 mutant.
Data S3. H3K27me3 peaks identified in this study.
Data S4. H3K4me3 peaks identified in this study.
Data S5. H3K9me2 peaks identified in this study.
Data S6. Genes with reduced H3K27me3 levels in the fen1-1 mutant.
Data S7. Genes with increased H3K27me3 levels in the fen1-1 mutant.
Data S8. Genes with increased H3K4me3 levels in the fen1-1 mutant.
Data S9. Genes with reduced H3K4me3 levels in the fen1-1 mutant.
Data S10. Primers used in this study.
Acknowledgments
This research is supported by National Science Foundation of China (project no. 31330041). We thank Dr Marjori Matzke (Institute of Plant and Microbial Biology, Taipei, Taiwan) for providing dms3-1 seeds.
Footnotes
Accession Numbers: FEN1 (At5 g26680), DMS3 (At3 g49250), ROS1 (At2 g36490), FAS1 (At1 g65470), UBI (At5 g25760), At2 g18600, At5 g24280 and At3 g58270. The ChIP-seq and RNA-seq data have been submitted to Gene Expression Omnibus (GEO) with an accession number: GSE79738.
AUTHOR CONTRIBUTIONS
ZG conceived the original research plans. JZ performed most of the experiments. SX provided the bioinformatics analysis. J-KZ gave comments on writing and experiments. JZ and ZG wrote the article with contributions from all authors.
Additional Supporting Information may be found in the online version of this article.
References
- Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Franklin FC, Jones GH. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci. 2001;114:4207–4217. doi: 10.1242/jcs.114.23.4207. [DOI] [PubMed] [Google Scholar]
- Balakrishnan L, Bambara RA. Flap endonuclease 1. Annu Rev Biochem. 2013;82:119–138. doi: 10.1146/annurev-biochem-072511-122603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Olmo I, Lopez JA, Vazquez J, Raynaud C, Pineiro M, Jarillo JA. Arabidopsis DNA polymerase recruits components of Polycomb repressor complex to mediate epigenetic gene silencing. Nucleic Acids Res. 2016;44:5597–5614. doi: 10.1093/nar/gkw156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T, Proux F, Vaucheret H. Arabidopsis RPA2: a genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr Biol. 2005;15:1919–1925. doi: 10.1016/j.cub.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Gaillard H, Garcia-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Guo Z, Qian L, Liu R, Dai H, Zhou M, Zheng L, Shen B. Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol Cell Biol. 2008;28:4310–4319. doi: 10.1128/MCB.00200-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke G, Friedrich-Heineken E, Hubscher U. Flap endonuclease 1: a novel tumour suppresser protein. Trends Biochem Sci. 2003;28:384–390. doi: 10.1016/S0968-0004(03)00138-5. [DOI] [PubMed] [Google Scholar]
- Hyun Y, Yun H, Park K, Ohr H, Lee O, Kim DH, Sung S, Choi Y. The catalytic subunit of Arabidopsis DNA polymerase alpha ensures stable maintenance of histone modification. Development. 2013;140:156–166. doi: 10.1242/dev.084624. [DOI] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, Huettel B, Bohmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJM. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Agarwal M, Andreucci A, Zheng X, Gong Z, Hasegawa PM, Bressan RA, Zhu JK. Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr Biol. 2005;15:1912–1918. doi: 10.1016/j.cub.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Kimura S, Furukawa T, Kasai N, Mori Y, Kitamoto HK, Sugawara F, Hashimoto J, Sakaguchi K. Functional characterization of two flap endonuclease-1 homologues in rice. Gene. 2003;314:63–71. doi: 10.1016/s0378-1119(03)00694-2. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Proc GPD. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Qian WQ, Zhao YS, Wang CL, Shen J, Zhu JK, Gong ZZ. Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:11425–11430. doi: 10.1073/pnas.1208557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Gong Z. The coupling of epigenome replication with DNA replication. Curr Opin Plant Biol. 2011;14:187–194. doi: 10.1016/j.pbi.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang H, Veeraraghavan J, Bambara RA, Freudenreich CH. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol Cell Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ren X, Yin H, Wang Y, Xia R, Wang Y, Gong Z. Mutation in the catalytic subunit of DNA polymerase alpha influences transcriptional gene silencing and homologous recombination in Arabidopsis. Plant J. 2010a;61:36–45. doi: 10.1111/j.1365-313X.2009.04026.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang J, Miki D, Xia R, Yu W, He J, Zheng Z, Zhu JK, Gong Z. DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell. 2010b;22:2336–2352. doi: 10.1105/tpc.110.076349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu FL, Cui X, Zhang SB, Jenuwein T, Cao XF. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet. 2011;43:715–U144. doi: 10.1038/ng.854. [DOI] [PubMed] [Google Scholar]
- Luo C, Sidote DJ, Zhang Y, Kerstetter RA, Michael TP, Lam E. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 2013;73:77–90. doi: 10.1111/tpj.12017. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Adachi N, Koyama H. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 2002;30:3273–3277. doi: 10.1093/nar/gkf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, Stewart SA. Flap endonuclease 1 contributes to telomere stability. Curr Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathi S, Bhusari A, Shen BH, Chai WH. Human flap endonuclease I Is in complex with telomerase and is required for telomerase-mediated telomere maintenance. J Biol Chem. 2009;284:3682–3690. doi: 10.1074/jbc.M805362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blanco R, Lopez de Silanes I, Munoz P, Gomez-Lopez G, Flores JM, Blasco MA. Telomere shortening relaxes X chromosome inactivation and forces global transcriptome alterations. Proc Natl Acad Sci USA. 2009;106:19393–19398. doi: 10.1073/pnas.0909265106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Thong Z, Gong X, Shen Q, Gan Y, Yu H. The putative PRC1 RING-finger protein AtRING1A regulates flowering through repressing MADS AFFECTING FLOWERING genes in Arabidopsis. Development. 2014;141:1303–1312. doi: 10.1242/dev.104513. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Nakamura T. Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J Biol Chem. 2002;277:746–754. doi: 10.1074/jbc.M109461200. [DOI] [PubMed] [Google Scholar]
- Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WF. Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 2007;144:1697–1714. doi: 10.1104/pp.107.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Tadele Z, Hofmann I, et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004;18:782–793. doi: 10.1101/gad.295404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Xia R, Wang J, Liu C, et al. ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell. 2006;18:85–103. doi: 10.1105/tpc.105.037507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Zhang X, Liu J, Wang Y, He J, Yang T, Hong X, Yang Q, Gong Z. Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell. 2009;21:386–402. doi: 10.1105/tpc.108.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang CZ, Schones DE, Zeng C, Cui KR, Zhao KJ, Peng WQ. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007a;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007b;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wen C, Liu S, Zheng L, Shen B, Tao Y. Shade avoidance 6 encodes an Arabidopsis flap endonuclease required for maintenance of genome integrity and development. Nucleic Acids Res. 2016;44:1271–1284. doi: 10.1093/nar/gkv1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xie S, Li X, Wang C, Chen Z, Lai J, Gong Z. REPRESSOR OF SILENCING5 encodes a member of the small heat shock protein family and is required for DNA demethylation in Arabidopsis. Plant Cell. 2014;26:2660–2675. doi: 10.1105/tpc.114.126730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Chai Q, Parrish J, Xue D, Patrick SM, Turchi JJ, Yannone SM, Chen D, Shen B. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 2005;6:83–89. doi: 10.1038/sj.embor.7400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- Zheng L, Jia J, Finger LD, Guo Z, Zer C, Shen B. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 2011;39:781–794. doi: 10.1093/nar/gkq884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. fen1-1 mutation is recessive.
Figure S2. Validation of the chromatin immunoprecipitation sequencing results.
Figure S3. The FEN1 mutation affects some H3K4me3-marked genes.
Data S1. RNA sequencing data.
Data S2. DNA repair-associated genes upregulated in the fen1-1 mutant.
Data S3. H3K27me3 peaks identified in this study.
Data S4. H3K4me3 peaks identified in this study.
Data S5. H3K9me2 peaks identified in this study.
Data S6. Genes with reduced H3K27me3 levels in the fen1-1 mutant.
Data S7. Genes with increased H3K27me3 levels in the fen1-1 mutant.
Data S8. Genes with increased H3K4me3 levels in the fen1-1 mutant.
Data S9. Genes with reduced H3K4me3 levels in the fen1-1 mutant.
Data S10. Primers used in this study.