Abstract
HIV-1, like all retroviruses, irreversibly inserts a viral DNA (vDNA) copy of its RNA genome into host target DNA (tDNA). The intasome, a higher-order nucleoprotein complex composed of viral integrase (IN) and the ends of linear vDNA, mediates integration. Productive integration into host chromatin results in the formation of the Strand Transfer Complex (STC) containing catalytically joined vDNA and tDNA. HIV-1 intasomes have been refractory to high-resolution structural studies. Here, using a soluble IN fusion protein, we present a high-resolution cryo-electron microscopy (cryoEM) structure of the core tetrameric HIV-1 STC and a higher-order form that adopts distinct domain rearrangements. The unique manner of STC assembly highlights how HIV-1 can use the common retroviral intasome core architecture to accommodate different IN domain modules for integration.
Catalytic integration of a vDNA copy of an RNA genome into host target tDNA represents the hallmark characteristic of all retroviruses, including HIV-1. Integration establishes a permanent infection in host cells and enables the newly inserted provirus to be replicated and transcribed in parallel with other genes of the host organism (1). This critical step in the HIV-1 replication cycle represents one of the underlying difficulties in combating the HIV/AIDS pandemic. Integration is catalyzed by the viral IN protein, which oligomerizes into a higher-order stable synaptic complex (SSC) containing the two vDNA ends. Following cleavage of the GT dinucleotide from both 3′ vDNA ends and nuclear entry, cleaved SSCs engage tDNA and catalyze irreversible DNA strand transfer into host chromatin to form the STC (2). Both retroviral SSCs and the post-catalytic STCs are collectively called intasomes. Clinically exploited HIV-1 IN strand transfer inhibitors (INSTIs) selectively bind cleaved SSCs and interfere with the formation of the STC. Therefore, high-resolution structures of key integration intermediate nucleoprotein complexes are required to further our understanding of the mechanisms of action of INSTIs and the evolution of drug resistant HIV-1 phenotypes (3). The mechanism of HIV-1 DNA integration has been extensively studied at the biochemical and cellular level, but progress with structural studies of nucleoprotein reaction intermediates has been slow; only structures of domains of HIV-1 IN are currently available (4–8), although intasome structures have been determined for related retroviruses (9–12) and predicted for HIV-1 through homology modeling (13, 14).
Structural studies of HIV-1 intasomes have been challenging due to the tendency of HIV-1 IN protein and assembled intasomes to aggregate. Fusion of the DNA binding protein Sso7d to the N-terminus of IN results in a protein that is hyperactive in vitro, has markedly improved solubility properties, and retains activity in vivo when incorporated into HIV-1 virions (15). We therefore used Sso7d-IN to assemble HIV-1 intasomes for structural studies. STC intasomes were assembled on branched DNA mimicking the product of DNA integration (Fig. S1A) using the strategy previously described for prototype foamy virus (PFV) (16) and Rous sarcoma virus (RSV) (12) intasomes. HIV-1 intasomes were first purified by Ni-affinity and anion exchange chromatography (Fig. S1B). Analytical ultracentrifugation following anion exchange chromatography indicated the presence of the tetrameric STC as well as larger discrete species (Fig. S2), in agreement with previous studies (17–19). An additional gel filtration step prior to cryoEM structural analysis yielded a preparation that was mainly tetrameric, but also included larger species, as evidenced by the broad and asymmetric peak shape (Fig. S1C).
Tetrameric HIV-1 STCs are relatively small by cryoEM standards (~200 kDa) and require the presence of high salt and glycerol to prevent aggregation, factors that negatively affect image contrast of individual particles. To overcome these problems, we employed a high-dose imaging strategy and an exposure filter that accounts for the effects of radiation damage while maximizing low-frequency contrast (20). Single-particle classification and refinement of exposure-filtered images produced a density map resolved to ~3.5–4.5 Å, with the highest resolution information characterizing the STC core, in and around the active site (Fig. S3–S4). This enabled derivation of a molecular model of an HIV-1 STC, which contained four IN protomers arranged with two-fold symmetry around the product of DNA strand transfer (Fig. 1 and Table S1).
Fig. 1. HIV-1 STC intasome structure.
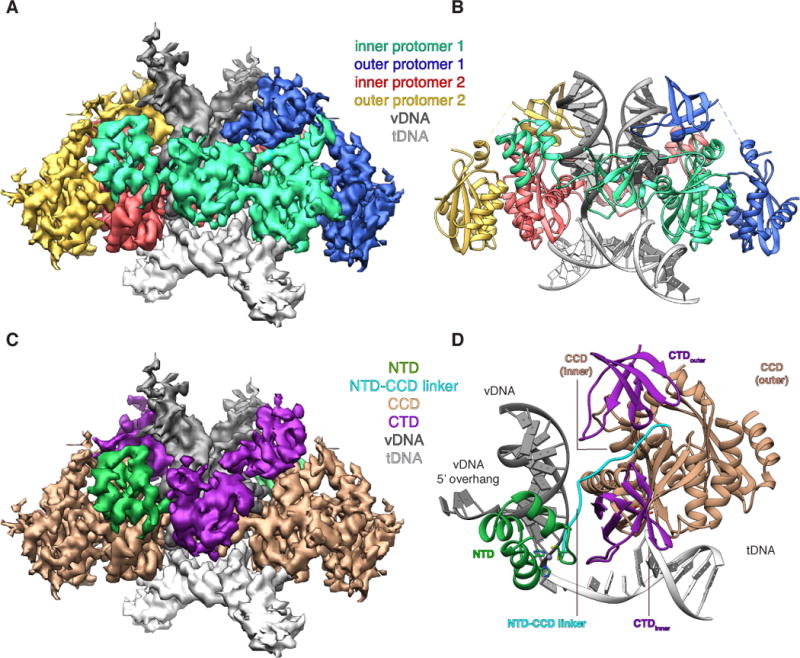
(A). CryoEM reconstruction of the STC, segmented by IN protomer (red, green, yellow, blue) and product DNA component (dark and light grey). (B) Atomic model derived from the cryoEM density, colored as in A. (C) segmented cryoEM density and (D) asymmetric subunit of the atomic model, colored by IN domain; NTD (green), CCD (beige), NTD-CCD linker (cyan), CTD (purple).
The tetrameric HIV-1 STC intasome is a dimer of dimers with a similar overall architecture to PFV intasomes (Fig. 1A–B). Each protomer contains an N-terminal (NTD), catalytic core (CCD), and C-terminal (CTD) domain. The inner protomers wrap their three functionally relevant domains around a pair of vDNA ends and dock onto tDNA, bringing two vDNA 3′-OH groups into proximity to catalyze concerted integration and form the STC. The inner protomers also make most of the contacts with vDNA and tDNA. The outer IN CTD (CTDouter) adopts a retracted configuration in the HIV-1 intasome, contributing partially to vDNA binding and positioning itself in proximity to the inner CTD (CTDinner) (Fig. 1C–D). The outer NTDs, as well as all Sso7d fusion domains, are disordered in the cryoEM density.
Retroviral intasomes recognize and cut target sites with a characteristic 4–6 bp spacing, generating equivalently sized target site duplications (TSDs) flanking either end of the integrated proviral DNA (2). To a large extent, the TSD sizes map onto the retroviral phylogenetic tree (21), although the precise TSD spacing can differ within an individual genus (22). In all the available STC structures, the target DNA is significantly distorted from B-form (Fig. S5), resulting in a 4 bp TSD for PFV which has the shortest distance between the active sites, and 5 bp and 6 bp for HIV-1 and RSV, respectively, which have a longer spacing.
The STC model substantiates and rationalizes much of the existing in vitro and in vivo data pertaining to HIV-1 IN residues involved in function, inhibitor binding, and mechanisms of drug resistance. In this regard, Figure 2 and Table S2 present a comprehensive analysis of predicted electrostatic protein-DNA and inter-domain interactions within these core components. Notably, the table includes all of the residues that were experimentally shown to affect IN function, but also provides additional details that were not captured by a homology model (13). Residues that interact with vDNA are distributed throughout all three IN domains and the NTD-CCD linker, whereas residues interacting with tDNA are largely clustered in the CCD (Fig. 2A). Multiple residues are involved in inter-domain interactions (Fig. 2B). Several residues deserve particular mention (displayed in Fig. 2C and Fig. S6). A cluster of basic residues is inserted into the vDNA minor groove next to the active site, including K46 that was not identified by a homology model, and K156, K159, and K160. The relevance of K46 is addressed below, while K156, K159, K160 play various roles in vDNA binding, sequence specificity, and catalysis (13, 23, 24). R231 – the only non-CCD residue that strongly interacts with tDNA – has previously been shown to affect nucleotide preferences within the target site, although the magnitude of this effect in HIV-1 IN mutants is significantly lower than analogous IN mutations in PFV (25). The weaker interaction between R231 of HIV-1 with target site nucleotides as compared to R329 of PFV, which contains a longer loop that can accommodate subtle structural changes, helps to explain the phenotype. Substitutions of HIV-1 IN S119 (25), similarly to analogous changes in RSV and PFV INs (10, 26), alter target site nucleotide specificity by perturbing interactions with tDNA. The model also provides important guidance for rationally improving clinically relevant inhibitors. Specifically, several residues around the vicinity of the active site, especially R231, are positioned differently in HIV-1 compared to PFV, which has been used as a model system to study mechanism of INSTI action (Fig. S7). Slight differences in the active site can be exploited to facilitate rational inhibitor design. Collectively, the current model provides a composite platform for both understanding IN function and elucidating modes of action of INSTIs.
Fig. 2. Network of interactions within the HIV-1 STC intasome.
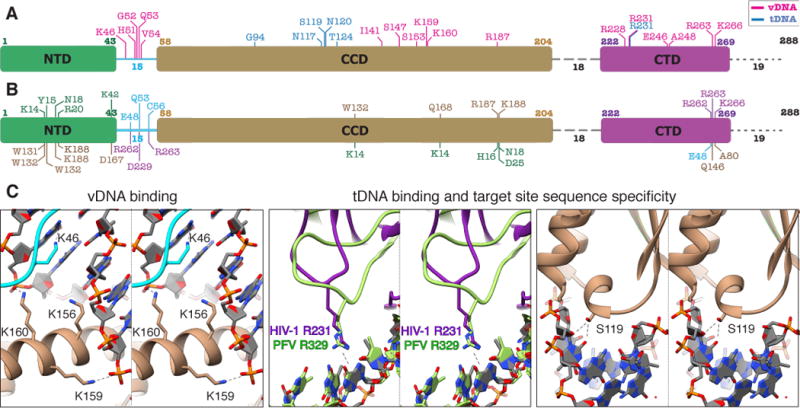
(A–C) Protein color scheme as in Fig. 1C–D. (A) Map of IN residues predicted to be involved in electrostatic protein-DNA interactions within the STC intasome structure. All three domains and the NTD-CCD linker participate in interactions with vDNA (pink), while tDNA interactions in blue are mostly restricted to the CCD. (B) Predicted inter-domain H-bond interactions within the STC. Residues designated below the domain schematic refer to the interacting domain and are colored accordingly. (C) Close-ups of selected regions involved in DNA interactions. For comparing HIV-1 R231 with PFV R329, the two structures were aligned to tDNA.
To gain a more thorough understanding of the heterogeneous STC data and improve regions of density outside of the core subunits, we employed a multi-step classification approach that revealed larger species containing flanking IN dimers (Fig. S4) positioned in trans, similarly to RSV and mouse mammary tumor virus (MMTV) (11, 12). We then included the IN-binding domain (IBD) of LEDGF/p75 in the STC preparation, based on the rationale that IBD preferentially binds and stabilizes multimeric IN (27, 28), and performed a cryoEM reconstruction of IBD-bound STCs (STCIBD). The resulting data contained a larger proportion of higher-order assemblies (Fig. S8), but was affected by substantial compositional heterogeneity; a cryoEM reconstruction of the largest and best resolved species clearly revealed 12 IN protomers within the map, with residual density contributed by a fraction of particles (Fig. 3A–B and Fig. S9–S11). IN can purify as tetramers from cells (29), and tetrameric INs constitute a portion of the higher-order assemblies (Fig. 3C, also below); it is therefore possible that the heterogeneous density corresponds to additional IN protomers that may collectively constitute a hexadecamer (or tetramer of tetramers). Possibly, the Sso7d fusion, which improves IN solubility (15), affects the assembly of fully formed higher-order species by mildly disrupting inter-protomer associations (Fig. S12). The higher-order assemblies utilize many of the principles underlying multimerization of IN protein in the absence of DNA (Fig. 3D–G and Fig. S13). For example, the isolated tetramer from each asymmetric unit contains positionally conserved CCDs and NTDs that were previously observed within a two-domain HIV-1 NTD-CCD (INNTD-CCD) structure (7) and Maedi-Visna virus INNTD-CCD bound to IBD (Fig. 3D) (28); individual dimers therein are also consistent with an HIV-2 INNTD-CCD structure (30) (Fig. 3E), while two of the CTDs interact in an identical manner to an NMR structure of a CTD dimer (Fig. 3F). Finally, the core CTDs adopt a configuration much like the two-domain INCCD-CTD (8) (Fig. 3G). The latter demonstrates an intriguing aspect of IN structure; whereas tetrameric HIV intasomes adopt a domain configuration much like PFV, higher-order intasomes reorganize their CTDs, utilizing them to form an inter-protomer CTD-CTD interface and to engage vDNA, but replacing their respective positions with additional CTDs donated by outer IN protomers (Fig. 3H). In sum, these alternative domain arrangements preserve the positional integrity of the catalytically competent intasome and demonstrate the structural plasticity of HIV-1 IN.
Fig. 3. HIV-1 STC intasomes form higher order oligomers through distinct mechanisms of assembly.
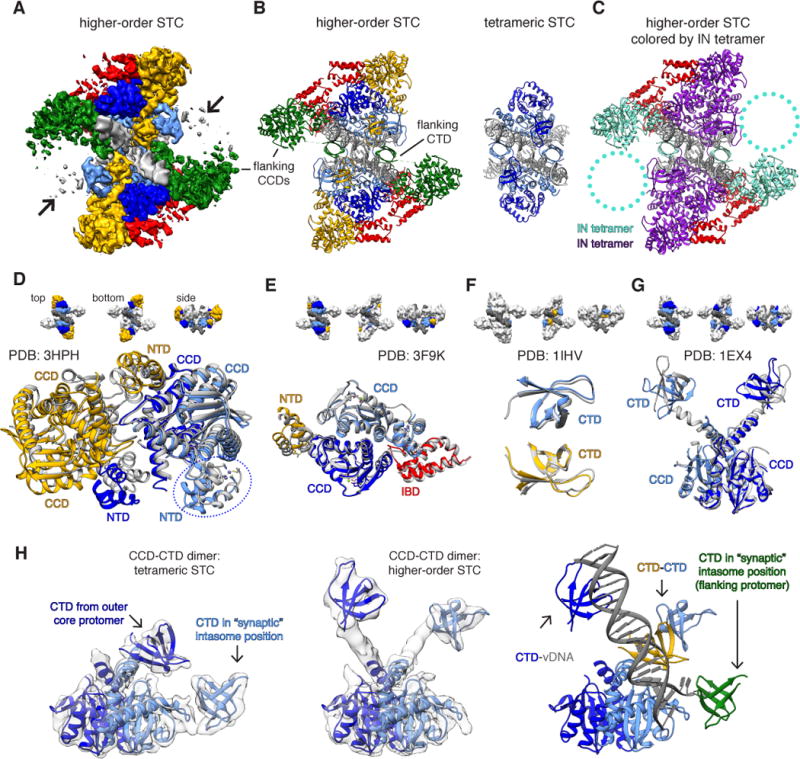
(A) CryoEM density map of IBD-bound STCs (STCIBD). Densities are segmented either by IN protomer (inner core: light blue, outer core: dark blue) or IN dimer (yellow and green), while IBD is in red. (B) Higher-order STC model assembled by rigid-body docking individual domain components, colored as in A. The higher-order STC (left) is shown side-by-side with the tetrameric STC from Fig. 1 (right). (C) Model as in B, colored by IN tetramer (28). Circled regions contain poorly resolved density that may harbor additional IN dimers. (D–G) Structural comparison of higher-order STC assembled through rigid-body docking of individual domains with prior multi-domain IN structures. The structural components of higher-order STCs are colored as in panels A–B, while the PDB structures used for comparison are gray. Comparisons include: (D) MVV INNTD-CCD tetramer (PDB: 3HPH, IBD has been omitted for clarity; the circled NTD arises from an IN protomer on the opposite side of vDNA), (E) HIV-2 INNTD-CCD dimer bound to IBD (PDB: 3F9K), (F) HIV-1 CTD dimer (PDB: 1IHV), and (G) HIV-1 INCCD-CTD dimer (PDB: 1EX4). (H) Conformational rearrangement within the core CCD-CTD dimer between (left) the tetrameric STC and (center) a higher-order STC, both overlaid on respective filtered experimental EM density. At right, the rearranged higher-order dimer is displayed in the context of additional CTDs and vDNA within the asymmetric unit. The “synaptic” position is required to form the conserved intasome core interface present in all retroviral intasomes.
HIV-1 intasomes assembled at lower protein and DNA concentrations than used in our cryo-EM study were reported to be tetrameric (17, 18). To test the relevance of the higher-order assemblies, we carefully selected IN residues that were predicted to disrupt formation of these species, but not the core tetramers. The most obvious candidates resided in the CTD-CTD interface – residues L242, I257, and V259 – which are solvent-exposed within tetrameric intasomes. Several other residues, including K14, E35, K240, K244, and R269 were predicted to be more relevant in the context of higher-order oligomers, although we cannot completely exclude their involvement in tetrameric intasomes (Fig. S14A–E). The selected residues were substituted in the context of both Sso7d and WT (NL4-3) INs, and the mutant proteins were assayed for concerted integration activity. Similar results were observed in the presence and absence of the Sso7d fusion protein, suggesting that Sso7d does not alter the nature of functional complexes, although it may influence their relative abundance. The selected mutants, especially those in the CTD-CTD interface, affected strand transfer activity to various extents (Fig. S14F–G). Furthermore, mildly disrupting or deleting many of the residues within the CCD-CTD linker region (aa ~206-220), which is disordered in tetrameric intasomes (and thus would not be expected to play a major role) but completely helical in higher-order assemblies, impaired catalytic activity (Fig. S14H). In addition, we tested the significance of select residues that have not been examined previously (31–33) for virus replication (Fig. S15). IN substitutions adversely affected virus replication, with >10-fold reductions observed for E212K, K240E, and I257D mutations, and relatively less detrimental effects (2-fold) seen for E35K. A mutation of K46, identified as a novel vDNA binding residue, was also included in functional assays, but was relevant for all oligomeric species. While the K46A substitution did not detectably affect viral growth (34), the K46E substitution reduced virus replication by ~5-fold and significantly reduced strand transfer activity in vitro. These data suggest that the higher-order HIV-1 intasomes are functionally relevant for efficient catalysis. However, since point mutations can affect protein structures and/or the intasome assembly pathway in unexpected ways, further systematic studies will be required to delineate the (likely pleiotropic) effects of single-site substitutions. The simplest explanation for the distinct structures is that tetrameric intasomes, containing intact core domains, illustrate the minimal form upon which higher-order complexes are built, although their exact relevance is not clear. They may represent minimally active species, or alternatively, may serve as structural scaffolds for higher-order assembly within the pre-integration complex (PIC) or during PIC nuclear import. Further work, especially in the context of IN dynamics, will be required to unravel the role(s) of the tetrameric and higher-order forms in vivo.
Retroviruses are closely related evolutionarily and would be expected to utilize similar nucleoprotein structures for DNA integration. In this regard, the structure of the PFV intasome (9, 10) presented a conundrum. The length of the linkers between the domains of PFV IN is longer than in most retroviral INs, and many retroviral INs have linkers that are too short to form a tetrameric intasome that is analogous to the PFV structure (11). HIV-1 IN has linker lengths that are intermediate between PFV and MMTV/RSV (Figure S16A). The recent structures of MMTV (11) and RSV (12) intasomes show that these viruses overcome this problem by assembling intasomes with the same set of “positionally conserved domains” in contact with DNA, but for MMTV and RSV, two of the CTDs are contributed by an additional pair of flanking dimers in an octameric arrangement. Whereas PFV intasomes assemble tetramers, MMTV/RSV intasomes assemble octamers, and HIV-1 intasomes can apparently form a range of oligomeric configurations (Figure S16B–C). The finding that HIV-1 IN can assemble intasomes in different ways while preserving the spatial arrangement of the key set of domains required for catalysis suggests that the evolutionary jump between retroviruses that assemble tetrameric and higher-order intasomes may not be as great as it appears at first sight.
Supplementary Material
One Sentence Summary.
We present structures of an HIV-1 strand transfer complex intasome, a critical structural intermediate produced from the integration of retroviral complementary DNA into the host genome.
Acknowledgments
D.L. acknowledges support from US National Institutes of Health (NIH) grant P50 GM103368 and the Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002. R.C. is supported by the Intramural Program of the National Institute of Diabetes and Digestive Diseases of the National Institutes of Health and by the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the NIH. These studies were also partly supported by NIH grant R01 AI062520 to M.K. Molecular graphics and analyses were performed with the USCF Chimera package (supported by NIH P41 GM103331). We thank B. Anderson and J-C. Ducom for help with EM data collection and network infrastructure, F. Dwyer for computational support, G. Lander and M. Herzik for help with ensemble refinements, and A. Engelman and M. Gellert for critical review of the manuscript. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. The EM maps of STC and STCIBD are deposited into the EMDB under accession codes EMD-8481 and EMD-8483, respectively. The STC model is deposited into the PDB under accession code 5U1C. The STC model ensemble or the composite model of the higher-order STCIBD oligomers are available upon request.
References and Notes
- 1.Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harb Perspect Med. 2012;2:a006890–a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lesbats P, Engelman AN, Cherepanov P. Retroviral DNA Integration. Chem Rev. 2016 doi: 10.1021/acs.chemrev.6b00125. acs.chemrev.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco JL, Whitlock G, Milinkovic A, Moyle G. HIV integrase inhibitors: a new era in the treatment of HIV. Expert Opinion on Pharmacotherapy. 2015;16:1313–1324. doi: 10.1517/14656566.2015.1044436. [DOI] [PubMed] [Google Scholar]
- 4.Dyda F, et al. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 5.Eijkelenboom AP, et al. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 6.Cai M, et al. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 7.Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JCH, et al. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: A model for viral DNA binding. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballandras-Colas A, et al. Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature. 2016;530:358–361. doi: 10.1038/nature16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Z, et al. Crystal structure of the Rous sarcoma virus intasome. Nature. 2016;530:362–366. doi: 10.1038/nature16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnan L, et al. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proceedings of the National Academy of Sciences. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson BC, Métifiot M, Ferris A, Pommier Y, Hughes SH. A homology model of HIV-1 integrase and analysis of mutations designed to test the model. J Mol Biol. 2013;425:2133–2146. doi: 10.1016/j.jmb.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Jurado KA, Lin S, Engelman A, Craigie R. Engineered hyperactive integrase for concerted HIV-1 DNA integration. PLoS ONE. 2014;9:e105078. doi: 10.1371/journal.pone.0105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Z, Lapkouski M, Yang W, Craigie R. Assembly of prototype foamy virus strand transfer complexes on product DNA bypassing catalysis of integration. Protein Sci. 2012;21:1849–1857. doi: 10.1002/pro.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Mizuuchi M, Burke TR, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotova S, Li M, Dimitriadis EK, Craigie R. Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy. J Mol Biol. 2010;399:491–500. doi: 10.1016/j.jmb.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bera S, Pandey KK, Vora AC, Grandgenett DP. Molecular interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J Mol Biol. 2009;389:183–198. doi: 10.1016/j.jmb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. elife. 2015;4 doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derse D, et al. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. Journal of Virology. 2007;81:6731–6741. doi: 10.1128/JVI.02752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballandras-Colas A, Naraharisetty H, Li X, Serrao E, Engelman A. Biochemical characterization of novel retroviral integrase proteins. PLoS ONE. 2013;8:e76638. doi: 10.1371/journal.pone.0076638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins TM, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen A, Weber IT, Harrison RW, Leis J. Identification of amino acids in HIV-1 and avian sarcoma virus integrase subsites required for specific recognition of the long terminal repeat Ends. Journal of Biological Chemistry. 2006;281:4173–4182. doi: 10.1074/jbc.M510628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serrao E, et al. Integrase residues that determine nucleotide preferences at sites of HIV-1 integration: implications for the mechanism of target DNA binding. Nucl Acids Res. 2014;42:5164–5176. doi: 10.1093/nar/gku136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harper AL, Sudol M, Katzman M. An amino acid in the central catalytic domain of three retroviral integrases that affects target site selection in nonviral DNA. Journal of Virology. 2003;77:3838–3845. doi: 10.1128/JVI.77.6.3838-3845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKee CJ, et al. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. 2008;283:31802–31812. doi: 10.1074/jbc.M805843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hare S, et al. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009;5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherepanov P. HIV-1 Integrase Forms Stable Tetramers and Associates with LEDGF/p75 Protein in Human Cells. Journal of Biological Chemistry. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 30.Hare S, et al. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu R, Ghory HZ, Engelman A. Genetic analyses of conserved residues in the carboxyl-terminal domain of human immunodeficiency virus type 1 integrase. Journal of Virology. 2005;79:10356–10368. doi: 10.1128/JVI.79.16.10356-10368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu R, Limón A, Ghory HZ, Engelman A. Genetic analyses of DNA-binding mutants in the catalytic core domain of human immunodeficiency virus type 1 integrase. Journal of Virology. 2005;79:2493–2505. doi: 10.1128/JVI.79.4.2493-2505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rihn SJ, Hughes J, Wilson SJ, Bieniasz PD. Uneven genetic robustness of HIV-1 integrase. Journal of Virology. 2015;89:552–567. doi: 10.1128/JVI.02451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu R, Vandegraaff N, Cherepanov P, Engelman A. Lys-34, dispensable for integrase catalysis, is required for preintegration complex function and human immunodeficiency virus type 1 replication. Journal of Virology. 2005;79:12584–12591. doi: 10.1128/JVI.79.19.12584-12591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorzano COS, et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol. 2010;171:197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyumkis D, Vinterbo S, Potter CS, Carragher B. Optimod – An automated approach for constructing and optimizing initial models for single-particle electron microscopy. J Struct Biol. 2013;184:417–426. doi: 10.1016/j.jsb.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheres SH. A Bayesian View on Cryo-EM Structure Determination. J Mol Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Ilca SL, et al. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat Commun. 2015;6:8843. doi: 10.1038/ncomms9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiMaio F, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. Journal of Virology. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrao E, Ballandras-Colas A, Cherepanov P, Maertens GN, Engelman AN. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology. 2015;12:39. doi: 10.1186/s12977-015-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quashie PK, et al. Characterization of the R263K Mutation in HIV-1 Integrase That Confers Low-Level Resistance to the Second-Generation Integrase Strand Transfer Inhibitor Dolutegravir. Journal of Virology. 2012;86:2696–2705. doi: 10.1128/JVI.06591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hare S, et al. Structural and Functional Analyses of the Second-Generation Integrase Strand Transfer Inhibitor Dolutegravir (S/GSK1349572) Mol Pharmacol. 2011;80:565–572. doi: 10.1124/mol.111.073189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hare S, et al. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci USA. 2010;107:20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lutzke RA, Plasterk RH. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. Journal of Virology. 1998;72:4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


