Abstract
Leukemias are highly immunogenic but have a low mutational load, providing few mutated peptide targets. Thus, the identification of alternative neoantigens is a pressing need. Here, we identify 36 MHC class I–associated peptide antigens with O-linked β-N-acetylglucosamine (O-GlcNAc) modifications as candidate neoantigens, using three experimental approaches. Thirteen of these peptides were also detected with disaccharide units on the same residues and two contain either mono- and/or di-methylated arginine residues. A subset were linked with key cancer pathways, and these peptides were shared across all of the leukemia patient samples tested (5/5). Seven of the O-GlcNAc peptides were synthesized and five (71%) were shown to be associated with multifunctional memory T-cell responses in healthy donors. An O-GlcNAc-specific T-cell line specifically killed autologous cells pulsed with the modified peptide, but not the equivalent unmodified peptide. Therefore, these post-translationally modified neoantigens provide logical targets for cancer immunotherapy.
Keywords: Leukemia, neoantigen discovery, cancer immunology, mass spectrometry, O-GlcNAc enrichment
Introduction
The role of antitumor immunity has been an intense focus of research for many decades (1–4). Although it is clear from strong correlative clinical data, combined with definitive experimental evidence from mouse cancer models, that T cells mediate this protection, the nature of the antigens targeted remains poorly characterized. Over the past decade the role of altered-self antigens, termed neoantigens, has become clear (5–9). Tumor-specific neoantigens act as targets for spontaneously arising adaptive immunity to cancer and thereby determine the ultimate fate of developing tumors (7). Nonsynonymous mutations in coding regions of expressed proteins are termed mutational neoantigens and, perhaps critically, are not subject to central tolerance. In patients with cancers that have high mutational loads, such as non-small cell lung cancer and melanoma, CD8+ T cells can be identified within the tumor that are specific for MHC class I–restricted neoantigens in response to immunotherapy (10, 11). However, tumor-resident immunity against mutational neoantigens occurs at very low frequencies and it would be surprising if this magnitude of immunity could be responsible for the dramatic reductions in tumor volume seen (7, 12). Additionally, some of the tumors with the best clinical responses to immunotherapy have some of the lowest mutational loads, for example, renal cell carcinomas and leukemias (12–14). Hematological malignancies, in particular, are known to be among the most immunogenic cancers (15). Therefore, it is likely that the antigens in these malignancies derive from other classes of antigens.
An alternative source of neoantigens may be the posttranslational modifications (PTMs) that occur in malignant and not healthy cells, particularly as dysregulated signaling is a hallmark of cancer (16). Indeed, a number of phosphorylated peptides have been identified as potent cancer antigens(17). Immunity to these antigens was seen in healthy donors, but lost in a subset of leukemia patients with poor clinical outcome and restored after stem cell transplant, suggesting a role for these antigens in the graft-versus-leukemia response. Dysregulation of cell signaling pathways in cancer is also caused by another PTM, β O-linked N-acetylglucosamine (O-GlcNAc), which is involved in cross-talk with phosphorylation (18–20). As such, aberrant O-GlcNAcylation can correlate with augmented cancer cell proliferation, survival, invasion, and metastasis (21). Synthetic O-GlcNAc–modified peptides can bind MHC class I complexes, and elicit glycopeptide-specific T-cell responses in mice, with X-ray structures confirming that the O-GlcNAc group was solvent exposed and accessible to the T-cell receptor (22–25). However, up until recently, limitations in proteomic technology made it impossible to characterize O-GlcNAcylated peptides from biological samples.
Here, we report three experimental approaches that allowed the detection and sequencing of O-GlcNAcylated peptides from a complex mixture of peptides presented by HLA-B*07:02 molecules on various primary leukemia samples. These methods allowed for the identification of 36 unique glycopeptides in several different states of glycosylation and, surprisingly, methylation. We go on to show that healthy donors have heterologous immunity to a number of these complex neoantigens and that T cells from these donors can specifically target and kill cells displaying only the modified peptide. Ultimately, we believe that these glycopeptide antigens will prove pivotal in the design of novel cancer immunotherapeutics.
Materials and Methods
Leukemia samples and cell lines
Leukemia samples were the same as those used previously (Supplementary Table S1)(17). All cell lines were grown at 37°C with 5% CO2 in medium consisting of RPMI 1640 supplemented with 10% fetal bovine serum (all from Sigma-Aldrich, St. Louis, MO). The JY cell line (ECACC – 94022533) was grown continually between 2006 and 2008 with its authenticity verified by HLA-typing and also in-house peptide profile.
Isolation of HLA-associated peptides
Class I MHC molecules were immunoaffinity-purified from cell lines or tumors and their associated peptides were extracted as described previously(17). Cells (108–109) were lysed in 10 mL of CHAPS buffer (Sigma-Aldrich, St. Louis, MO) and the lysate was centrifuged at 100,000 × g for 1 hour at 4°C. Supernatants were passed over protein A-sepharose preloaded with HLA-B7 specific antibody (ME1). Peptides were eluted from the purified MHC class I molecules with 10% acetic acid and separated by ultrafiltration (Ultrafree-MC, Millipore, Billerica, MA).
Enrichment of HLA-associated peptides
POROS20 AL beads (Applied Biosystems, Carlsbad, CA) were derivatized with amino-phenyl boronic acid (APBA; Thermo Fisher Scientific, Waltham, MA). Briefly, POROS20 beads (7 mg) were dispersed into 200 μL of PBS (pH 6–7) containing 40 μmol of APBA. Following the addition of NaCNBH3 (1.3 μmol in 1 μL of PBS), the reaction was allowed to proceed with agitation for 2 hr at RT and then quenched by washing the beads with water on a spin column (pore size < 20 μm). Water was removed under vacuum and the dried beads were stored at 4°C.
Class I MHC peptides from 2 ×108–5×108 cells in 0.1% acetic acid were desalted by loading the solution onto a fused-silica column (360 μm o.d. × 150 μm i.d.) packed in-house with 5 cm of irregular C18 (5–20 μm diameter) particles at a flow rate of 0.5 μl/min. After washing the column with 25 μL of 0.1% acetic acid, peptides were eluted into Eppendorf tubes with a 40 min gradient (0–80%) solvent B (A: 0.1M acetic acid, B: 70% acetonitrile, 0.1M acetic acid). Fractions were screened by MS and those that contained peptides, but not CHAPS detergent, were combined, taken to dryness and stored at −35°C.
APBA-beads were washed 3× with 100 μl of anhydrous DMF and then allowed to react with desalted peptides in 20 μl of anhydrous DMF for 1 hour with agitation. Solvent was removed by centrifugation and the beads were washed 2× with 100 μL of anhydrous acetonitrile. Bound peptides were released by agitating the beads in 20 μL of 0.1 M acetic acid for 30 min. Supernatant was collected, taken to dryness, and reconstituted in 10 μL of 0.1M acetic acid for loading onto an in-house packed C18 column for MS analysis.
RP-HPLC-MS
In-house, packed C18 columns were prepared as previously described(26). Peptides were eluted by a 2 hour 0–60% B gradient (A: 0.1M acetic acid, B: 70% ACN, 0.1M acetic acid). Without enrichment, samples were loaded directly onto the C18 column. The RP-HPLC elution was electrospray-ionized into an Orbitrap Velos, or Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, San Jose, CA), the former equipped with an in-house front-end ETD ion source. On the Orbitrap Velos, the instrument method was a top-10 CAD with ETD only when loss of dehydro-GlcNAc neutral loss of [203]+2 or [203]+3 was detected. On the Fusion, instrument method was a top speed HCD triggered ETD when three of six O-GlcNAc fingerprint ions (m/z 204, 186, 168, 144, 138, and 126) were detected at >5% relative abundance. Peptide sequences were determined by manual interpretation of HCD, CAD, and ETD mass spectra.
Synthetic Peptides
O-GlcNAc- and O-GalNAc-peptides were synthesized using Fmoc chemistry and purified by HPLC to >90% purity by Pierce Biotechnology, Rockford, Il. Sequences and purity for all synthetic peptides were confirmed by on-line HPLC MS/MS and manual interpretation of the resulting spectra. Immunodominant HLA-B*0702 restricted antigens from human cytomegalovirus (pp65417–426, TPRVTGGGAM); Epstein-Barr virus (EBNA-3A247–255, RPPIFIRRL) and Influenza A virus (PB1329–337, QPEWFRNVL) were synthesized at >90% purity by Genscript, Piscataway, NJ.
Selective transfer of N-azidoacetylgalactosamine (GalNAz) to O-GlcNAcylated peptides
A solution of the modified β1–4-galactosyltransferase, GalT1 (Invitrogen, Carlsbad, CA), was dried to 5 μL in a vacuum concentrator at 40°C. Excess synthetic glycopeptides or tumor peptides (3e8 cell equivalents) were taken to dryness, redissolved in a mixture containing 1 μL MnCl2, 5 μL uridinediphosphate N-azidoacetylgalactosamine, UDP-GalNAz (Invitrogen, Carlsbad, CA), and 5 μL enzyme and allowed to react for 5 hours at RT. After the reaction was quenched by addition of 0.2 μL glacial acetic acid, the solution volume was increased to 15 μL with 0.1% acetic acid, and then loaded directly onto an HPLC column for analysis by LC-MS/MS.
Intracellular cytokine staining
PBMCs were isolated from healthy donors and resuspended (106 cells/ml) in AIM-V medium (Invitrogen, Carlsbad, CA). Synthetic peptide antigens were added to the wells (10 μg/mL) and cells were expanded for 6 days. The positive control was stimulated with PHA (1 μg/ml). On day 6, cells were washed and re-stimulated with peptide antigen overnight or, for the positive control, with PMA/Ionomycin (4 ng/ml and 500 ng/ml respectively), in the presence of anti-CD107a–FITC. Cells were harvested, washed with PBS, and stained with fixable viability dye (APC-Cy7) and surface antibodies: anti-CD3and anti-CD8. Cells were fixed using 2% formaldehyde, permeablized using 0.5% saponin, and stained with anti-IFNγ–PE, anti-IL2–Pacific blue, and anti-TNFα–PE-Cy5.5 for 30 minutes at RT. Cells were washed, lightly fixed, and analyzed on the LSRFortessa ×20 flow cytometer (BD Bioscience, Oxford, UK). A list of antibodies used in the study is shown in Table S2
Establishment of a peptide-specific T-cell line
PBMCs (107) were stimulated with synthetic peptide and cultured for a week. They were subsequently restimulated overnight, in the presence of anti-CD107a–FITC and washed and labelled with anti-CD137–PE (Miltenyi Biotech, Bergisch Gladbach, Germany) and counterstained with anti-CD8–APC (Biolegend). Cells were sorted using a FACS Aria cell sorter (BD Bioscience), collected and expanded using the rapid expansion protocol previously described(27). The T-cell line was subsequently reassessed, using a similar protocol.
Europium Release Killing Assay
The Delfia EuTDA cytotoxicity assay (Perkin Elmer, Coventry, UK) was used according to the manufacturer’s instructions. Briefly, autologous transformed B-cell lines were used as target cells. These were washed and resuspended at 106 cells/ml in RPMI 10% FCS and, the relevant peptide antigen was added at 10 μg/ml and the mixture was incubated at 37°C, 5% CO2 in a humidified environment for 40 min. Subsequently, 2.5 μL/ml of the BATDA fluorescence enhancing ligand was added and the cells were incubated for a further 20 min. Cells were then washed 5× in excess medium. Target cells (104) were added to each well of a V-bottomed 96-well plate. T cells at varying effector to target (E:T) ratios were added to the test wells. All well volumes were made up to 200 μL. The plate was incubated for 2 hr at 37°C, 20 μl of each supernatant was transferred to a flat bottomed, white, 96 well plate and 200 μL of Europium solution was added, incubated for 15 min, with shaking, at room temperature. Fluorescence was measured with a time-resolved fluorometer (Tecan Infinite 200 PRO, Tecan, Switzerland).
Results
Experimental approaches that allow identification of O-GlcNAcylated peptide antigens
Three different experimental approaches were used to identify peptides with O-GlcNAc modifications from leukemia patient samples. The first approach used higher energy collision–induced dissociation (HCD) mass spectrometry (MS) to visualize the loss of a dehydro-N-acetyl-glucosamine moiety (203Th) from fragment ions. The HCD mass spectrum of the first O-GlcNAcylated class I MHC peptide, XPVsSHNSX (where X=I or L), detected during analysis of HLA B*07:02 peptides presented on ALL, is shown in Fig. 1A. The amino acid sequence, XPVsSHNSX, is uniquely present as IPVSSHNSL in a single human protein, myocyte-specific enhancer factor 2C. This approach is limited by the ability of nonglycosylated peptides to suppress electrospray ionization of co-eluting O-GlcNAcylated peptides(28).
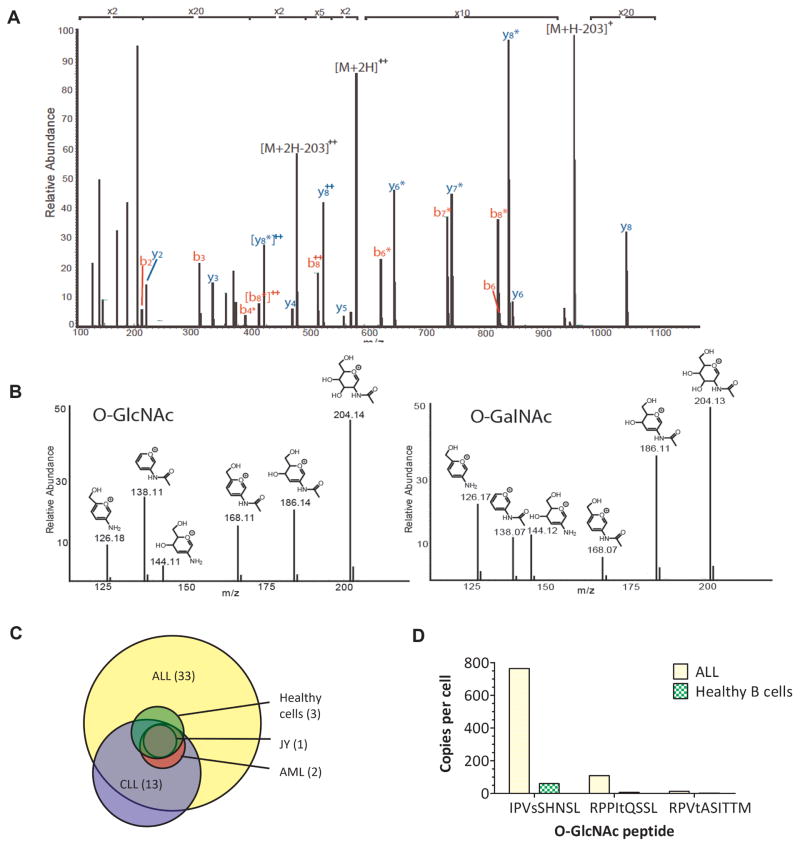
Figure 1. The discovery of MHC class I–associated glycopeptides on primary leukemia cells.
(A) HCD mass spectrum of the first O-GlcNAcylated peptide detected in ALL, IPVsSHNSL. Fragment ions that define the complete amino acid sequence are labelled as b and y. Those that have lost the O-GlcNAc moiety are labelled with an asterisk. (B) Fingerprint ions in the HCD spectra of O-GlcNAcylated and O-GalNAcylated peptides. Relative abundances of fragment ions derived from secondary fragmentation of the oxonium ion at m/z 204 are substantially different for O-GlcNAcylated and O-GalNAcylated peptides. (C) Distribution of 36 HLA-B*07:02–restricted glycopeptides among the different leukemia and healthy cells analysed. ALL = acute lymphoblastic leukemia. Healthy cells = healthy donor tonsil/spleen cells. LCL = lymphoblastoid cell line. AML = acute myeloid leukemia. CLL = chronic lymphocytic leukemia. (D) Number of copies per cell of the O-GlcNAcylated peptides identified on ALL versus healthy B cells (purified from a healthy spleen).
The second approach overcomes this limitation and allows detection and characterization of O-GlcNAcylated peptides from attomole quantities, using an enrichment procedure to selectively pull down the glycosylated peptides from the pool of unmodified peptides. The enrichment allowed selective esterification of glycosylated peptides, linking them to aminophenylboronic acid–derivatized POROS AL 20 beads. This O-GlcNAc enrichment procedure allowed us to achieve quantitative yields from femtomoles of starting material, and may be critical for efficient identification of these antigens from patient samples. This methodology was used in combination with an instrument method that selectively recorded electron transfer dissociation (ETD) when the collision-activated dissociation (CAD) spectrum contained a doubly charged ion corresponding to the loss of dehydro-N-acetyl-glucosamine (203Th)(29).
The third experimental approach extended the sensitivity of the method and allowed the user to obtain spectral information from extremely low-level peptide species. This was achieved using an instrument method that triggered an ETD spectrum whenever three of six O-GlcNAc fingerprint ions (m/z 204, 186, 168, 144, 138, and 126) were detected at >5% relative abundance in a particular HCD spectrum when the Orbitrap Fusion Tribrid was set to record HCD spectra at top speed. All of the fingerprint ions in Fig. 1B were the result of the O-GlcNAc oxonium ion at m/z 204 undergoing further fragmentation as a result of multiple collisions with the background nitrogen gas in the collision chamber. The use of multiple fingerprint fragments created a reliable trigger that minimized false positives.
36 O-GlcNAcylated peptides identified on HLA B*07:02 leukemia samples
In total, using the three experimental approaches outlined, 36 O-GlcNAcylated peptides were identified from leukemia samples (Table 1). 92% (33/36) of the O-GlcNAcs identified were only found on the leukemia samples tested and not the healthy tissue samples, making them potential leukemia neoantigens (Table 1, Fig. 1C, Supplementary Fig. S1). Just under a quarter (7/32) of the proteins that the O-GlcNAcylated peptides derived from were associated with key cancer pathways (as defined by the NCI pathway interaction database, which is now accessible via the NDEx database at http://www.ndexbio.org/#/). These pathways included many classical cancer signaling pathways, involving genes such as p38, p53, c-Myc, Notch, Wnt, Rb, ErbB1, and MAPK. Of note, IPVsSHNSL, which derives from Mef2c, a transcription factor implicated in leukemogenesis(30), was identified on nearly all of the leukemia samples tested (1/1 ALL, 3/3 CLL, 1/1 AML) and although it could be detected on healthy B cells, it was present in far lower amounts (Fig. 1D). An incidental, but significant, finding was that one of the O-GlcNAcylated peptides detected, RPPItQSSL, contained another PTM – a methylated Arg residue at P1 and was also found with an asymmetrically dimethylated Arg residue (Supplementary Fig. S2).
Table 1. O-GlcNAcylated peptides presented by HLA B*0702 Class I MHC molecules on leukemia.
Thirty-six peptides, often with multiple forms of glycosylation, were isolated from class I MHC molecules on several leukemias, cell lines, and healthy tissue. These sources are indicated as follows: CLL1,2; chronic lymphocytic leukemia samples 1 and 2, AML; acute myeloid leukemia, ALL; acute lymphoblastic leukemia, J; JY cell line, S; spleen and To; tonsil - see Supplementary Table S1. Small letters, s, t, and n specify Ser, Thr and Asn residues that are modified by O-GlcNAc unless otherwise indicated in a footnote. Parentheses enclose s and t residues that could be a site of GlcNAcylation. Samples were independently analyzed by MS at least 3 times.
| # | Sequence | Start - Stop | UniProt | Tumor | Source Protein |
|---|---|---|---|---|---|
| 1a | APP(sts)AAAL | 405–414 | Q86TM6 | ALL, CLL1 | E3 Ubiquitin-protein ligase synoviolin |
| 2b | APRGnVTSL | 60–68 | Q9NR96 | CLL1, CLL2 | Toll-like receptor 9 |
| 3 | APRtNGVAM | 187–195 | Q92567 | ALL, CLL1, CLL2 | Protein FAM168A |
| 4 | APTsAAAL | 1116–1123 | Q86Z02 | ALL | Homeodomain-interacting protein kinase 1 |
| 5 | APVsASASV | 1807–1815 | Q9Y520 | ALL | Protein PRRC2C |
| 6 | APVsSKSSL | 850–858 | Q86Z02 | ALL, CLL1, CLL2 | Homeodomain-interacting protein kinase 1 |
| 7 | EP(sst)VVSL | 1076–1085 | O75129 | ALL | Astrotactin-2 |
| 8 | HPMsTASQV | 345–353 | Q13492 | ALL | Clathrin assembly lymphoid myeloid leukemia |
| 9c | HP(sss)AAVL | 740–748 | Q86XN7 | ALL | Proline and serine-rich protein 1 |
| 10 | HP(sst)ASTAL | 3041–3050 | Q96T58 | ALL | Msx2-interacting protein |
| 11 | IPIsLHTSL | 1959–1967 | Q5JSZ5 | ALL | Protein PRRC2B |
| 12 | IPTsSVLSL | 710–718 | O15027 | ALL | Protein transport protein Sec16A |
| 13d | IPVsKPLSL | 104–112 | Q16621 | AML, ALL, CLL1 | Leucine zipper protein 1 |
| 14e | IPVsSHNSL | 147–155 | Q06413 | AML, ALL, CLL1, JY, S, To | Myocyte-specific enhancer factor 2C |
| 15f | KPP(ts)QSSVL | 411–420 | Q5T6F2 | ALL | Ubiquitin associated protein 2 |
| 16g | KPPVsFFSL | 95–103 | Q6PKC3 | ALL | Thioredoxin domain containing protein 11 |
| 17h | KPTLLYnVSL | 373–381 | P04220 | CLL1, CLL2 | Ig Mu heavy chain disease protein |
| 18 | LPRN(st)MM | 335–342 | Q9NPI6 | ALL | mRNA-decapping enzyme 1A |
| 19 | LPTsLPSSL | 2464–2472 | P46531 | ALL | Neurogenic locus notch homolog protein 1 |
| 20i | MPVRPTtNTF | 218–227 | Q7Z3K3 | ALL | pogo transposable element with ZNF domain |
| 21 | NPVsLPSL | 831–838 | Q6VMQ6 | ALL | Activating transcription factor 7-interacting protein |
| 22j | PPS(ts)AAAL | 405–414 | Q86TM6 | ALL | E3 Ubiquitin-protein ligase synoviolin |
| 23k | RPPItQSSL | 382–390 | Q9P2N5 | ALL, S | RNA binding protein 27 |
| 24l | RPPQsSSVSL | 937–946 | O15027 | ALL | Protein transport protein Sec16A |
| 25 | RPP(sss)QQL | 1758–1766 | Q8WYB5 | ALL | Histone acetyltransferase KAT6B |
| 26 | RPPVtKASSF | 341–350 | Q9Y2K5 | ALL, CLL1 | R3H domain containing protein 2 |
| 27 | RPVtASITTM | 927–936 | Q9ULH7 | ALL, CLL1, CLL2, S | MKL/myocardin-like protein 2 |
| 28 | TPASsRAQTL | 2320–2329 | Q01082 | CLL1 | Spectrin beta chain, non-erythrocytic 1 |
| 29 | TPAsSSSAL | 875–883 | Q9NPG3 | ALL, CLL1 | Ubinucleain 1 |
| 30 | TPIsQAQKL | 3024–3032 | Q96L91 | ALL | E1A-binding protein p400 |
| 31 | VPAsSTSTL | 576–584 | Q9NYV4 | ALL, CLL1 | Cyclin dependent kinase 12 |
| 32 | VPTtSSSL | 1284–1291 | Q14004 | ALL | Cyclin dependent kinase 13 |
| 33 | VPVsGTQGL | 93–101 | P23511 | ALL | Nuclear transcription factor Y subunit alpha |
| 34 | VPVsNQSSL | 146–154 | Q14814 | ALL | Myocyte-specific enhancer factor 2D |
| 35 | VPVsSASEL | 596–603 | Q7Z2W4 | ALL | Zinc finger CCCH-type, antiviral 1 |
| 36 | VPVsVGPSL | 1157–1164 | Q86Z02 | ALL | Homeodomain-interacting protein kinase 1 |
Footnotes are as follows:
Peptide was detected in a total of five forms: single GlcNac, double GlcNAc, single hexose-GlcNAc, single GlcNAc (S6) + hexose- GlcNac (T5), and double hexose-GlcNAc
N5 is modified by N-linked hexose-GlcNAc
Peptide was detected in two forms, GlcNAc on S4 and two GlcNAcs on S4 and S5.
Peptide was detected in two forms: GlcNAc (S4) and hexose-GlcNAc (S4)
Peptide was detected in four forms: GlcNAc (S4), double GlcNAc (S4, S5), single hexose-GlcNAc (S4), and an acetyl-GlcNAc (S4)
Peptide was detected in two forms: GlcNAc and hexose-GlcNAc (T4)
S5 is modified by O-linked hexose-GlcNAc
N7 is modified by N-linked hexose-GlcNAc
Peptide was detected in two forms: hexose-GlcNAc and asymmetric di-methyl (R4) + hexose-GlcNAc (T7)
T4 or S5 is modified by O-linked hexose-GlcNAc
Peptide was detected in four forms: GlcNAc (T5), mono-methyl (R1) + GlcNAc (T5), asymmetric di-methyl (R1) + GlcNAc (T5), and ), asymmetric di-methyl (R1) + acetyl-GlcNAc (T5)
S5 is modified by O-linked hexose-GlcNAc
Distinguishing O-GlcNAc from O-GalNAc
Because O-GlcNAc and O-GalNAc are isobaric, but have different biological properties, it was important to confirm that the peptides we identified indeed contained O-GlcNAc modifications, and not O-GalNAc. Furthermore, some MHC class I peptides we observed contained disaccharide units (see footnotes for Table 1), so they might have been derived from degradation of O- and N-linked glycans synthesized in the Golgi and ER, rather than true cancer neoantigens. To validate that the peptide antigens we had tested were O-GlcNAcylated, an in vitro enzyme reaction was used. β1–4-galactosyltransferase (GalT1) was shown to transfer N-azidoacetylgalactosamine (GalNAz) to four peptides (IPVsSHNSL and (me-)RPPItQSSL) in the ALL sample. Additionally, we found that synthetic O-GlcNAcylated vs. O-GalNAcylated peptides could be differentiated based on the relative ion abundances observed for fragments derived from the oxonium ion at m/z 204 in the corresponding fingerprint region of the HCD mass spectra (Fig. 1D)(29). All of the peptides observed (Table 1) produced HCD spectra with the necessary fingerprint region to confirm their identity as O-GlcNAc peptides.
Ten of the peptides detected were also found with disaccharide units attached to the same residues that were O-GlcNAcylated. It was determined that these correspond to a hexose bound to a HexNAc, because the oxonium ion observed for all of these peptides occurred at m/z 366 (204 +162). This was likely the result of the transfer of galactose to the O-GlcNAcylated peptide by a β-N-acetylglucosamine β1–4 galactosyltransferase; however, the remote possibility that this instead could involve the O-glycan synthetic pathway, in which the first residue to be added is a GalNAc and the second is either galactose or GlcNAc, needed to be excluded. Again, using synthetic peptides (IPVsSHNSL modified with Gal-GalNAc and Gal-GlcNAc) the fingerprint patterns for fragmentation of the oxonium ion at m/z 204 in HCD mass spectra could be distinguished, confirming that none of the disaccharide- modified peptides in Table 1 were derived from the O-glycan synthetic pathway.
Two of the glycosylated peptides in Table 1, APRGnVTSL and KPTLLYnVSL, have disaccharide units, Hexose-HexNAc, attached to Asn residues. Both peptides have consensus sequences, NX(S/T), for attachment of N-linked oligosaccharides. We conclude, therefore, that the observed Hexose-GlcNAc disaccharide units attached to Asn in these peptides probably result from degradation of the N-linked oligosaccharide structures to a single N-linked GlcNAc that then accepts a hexose such as galactose (from a β-N-acetylglucosamine β1–4 galactosyl- transferase). This is a novel finding, as the enzyme N-glycanase1 is responsible for removing all N-linked glycosylation prior to loading onto MHC class I molecules, potentially suggesting a new source of neoantigens in leukemia(31).
Leukemic glycopeptides elicited potent memory T-cell responses in healthy donors
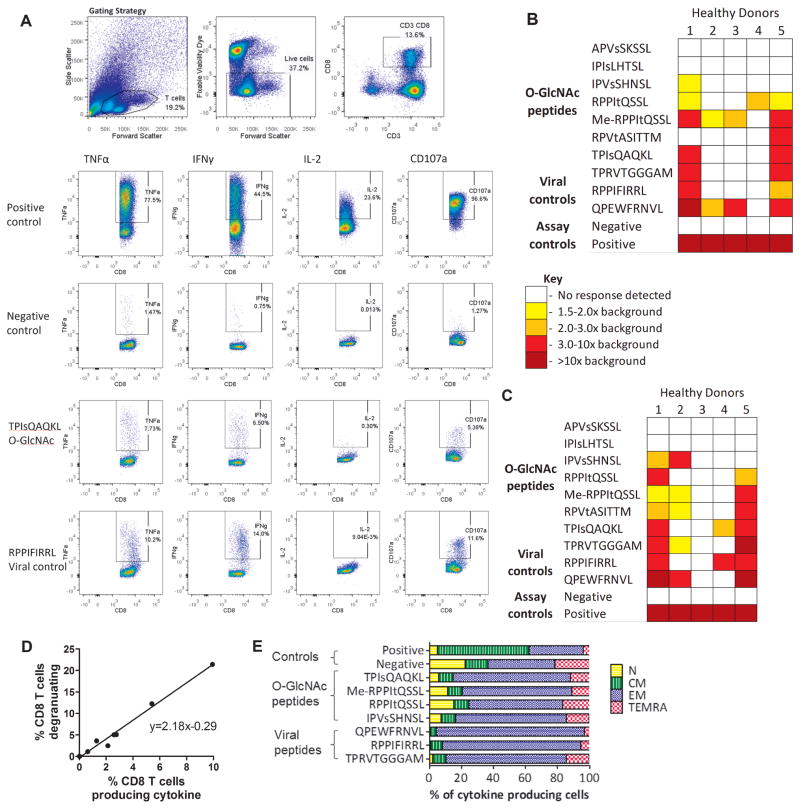
Previous studies have highlighted how post-translationally modified antigens can be immunogenic, with immunity against leukemia-associated MHC class I phosphopeptides having been shown to be present in healthy individuals. Immunity against naturally processed MHC class-I O-GlcNAc or methylated peptides has not been studied, but we hypothesized that it may exist in healthy individuals. Immunogenicity in healthy donors was assessed using seven of the O-GlcNAcylated peptides discovered on leukemic cells (Fig. 2A–E and Supplementary Figs. S2–S5). Five of the seven (71%) HLA-B*0702 glycopeptides were immunogenic—heterogeneous responses were seen, with both intra- and inter-donor variation (Fig. 2B and C). The responses were further validated using IFNγ ELISpot (Supplementary Fig. S5). All healthy donors had immunity to at least one of the glycopeptides and two had strong responses, similar to the magnitude of responses against chronic viral antigens. Degranulation was assessed as a proxy for killing (Fig. 2C) and despite some background staining, degranulation significantly correlated with multifunctional cytokine responses (Fig. 2D), suggesting that these T cells targeting O-GlcNAcylated peptide antigens have a cytotoxic phenotype. Furthermore, these T cells appeared to be largely the memory phenotypes (Fig. 2E).
Figure 2. Healthy donor immunity to leukemia-associated posttranslationally-modified neoantigens.
(A) Flow cytometry plots showing the gating strategy used in the ICS protocol to determine healthy donor immunity to the O-GlcNAcylated peptides (Fig. S4 contains additional plots). Immunity to viral antigens was used as an internal control, for comparison. Collated results of cytokine production (B) and degranulation (C) by healthy donor T cells in response to stimulation with posttranslationally-modified leukemia neoantigens. (D) The correlation between the percentage of cells producing cytokine and degranulating for HD1. (E) HD1 T cells that produced cytokine in response to stimulation with peptides were also stained with surface antibodies for phenotyping (CD27 and CD45RA; Supplementary Fig. S5). CM - central memory, N - naïve, EM - effector memory and TEMRA - terminal effector memory. Responses were independently verified at least twice.
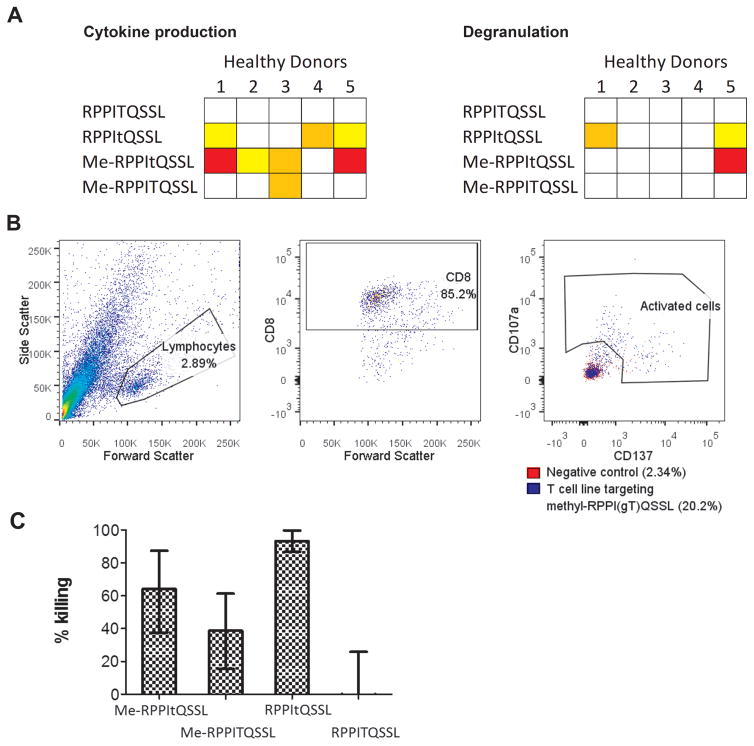
Modifications of a methylated glycopeptide specifically targeted by cytotoxic T cells
As responses were seen against the intriguing methylated glycopeptide ((me-R)PPI(GlcNAc-T)QSSL) in 4/5 (80%) of healthy donors tested, two being potent, these responses were further analyzed using peptides that were either methylated or glycosylated. Whereas no T-cell responses were seen against the unmodified peptide, responses in different individuals were seen targeting either the glycosylated or the methylated peptide (Fig. 3A and S6). In the two donors with potent responses to the methylated glycopeptides,\fewer T cells recognized the glycopeptide alone, suggesting that the methylation may somehow increase immunogenicity. To assess this further, a T-cell line was initiated using the methylated glycopeptide. After culture, around 18% of T cells were shown to be specific for the methylated glycopeptide (Fig. 3B and Supplementary Fig. S6). Autologous transformed B cells were pulsed with modified and unmodified peptides and killing by the T-cell line assessed. Specific killing was seen of the B cells pulsed with methylated, O-GlcNAcylated and doubly modified peptide, but not with the unmodified peptide (Fig. 3C). These results suggest that we may have identified modified peptides targeted by the endogenous anti-leukemia T-cell response, which may lead to fruitful targets for novel immunotherapeutics.
Figure 3. Investigating T-cell recognition of the methylated O-GlcNAc peptide.
(A) Healthy donor immunity to the unmodified, O-GlcNAcylated, methylated and both O-GlcNAcylated and methylated peptide, measured by cytokine production and degranulation. (B) A T-cell line was grown from HD5 against the methylated RPPItQSSL peptide. The percentage of cells recognizing the peptide were assessed by overnight stimulation with the peptide and detection of CD137 and CD107a surface markers. (C) This T-cell line was using a europium release killing assay to assess killing of autologous transformed B cells pulsed with different modifications of the peptide. Responses were independently verified at least twice.
Discussion
We outline here three methodologies for the identification of MHC class I peptides containing a little-known PTM, O-GlcNAc, a potential class of cancer neoantigens. Utilizing these methods, we identified 36 GlcNAcylated peptides from primary leukemia samples, and showed that a memory T-cell response against a subset of these antigens could be found in healthy donors. We also have identified peptides that contained other moieties—not previously seen on MHC class I peptides from cancer samples—namely methyl, disaccharide, and N-linked GlcNAc groups. Peptides containing these PTMs offer a hitherto untapped source of neoantigens in leukemia.
These neoantigens created by PTMs may be found on leukemic cells because of their aberrant cell signaling. This has been reported for phosphopeptide leukemia antigens and O-GlcNAcylation sites are usually identical, or in close proximity, to those that get phosphorylated(17, 20). Aberrant O-GlcNAcylation has been shown to correlate with augmented cancer cell proliferation, survival, invasion, and metastasis (21). The essential nature of these pathways to the leukemic cells suggests that these PTM neoantigens may not be patient-specific, as seen with the mutated neoantigens, but common across patients of the same HLA-type (7, 8). Indeed, we identified many of them on multiple samples from leukemia patients, even those with different clinical types. Around a quarter (7/32) of the proteins that the PTM peptides derived from are associated with key cancer pathways (as defined by the NCI pathway interaction database). Antigens from these key signaling pathways are ideal targets for immunotherapies since the leukemic cell is unlikely to be able to survive without these pathways, reducing the risk of immune escape. Although further work is required to ensure that these PTM peptides are truly cancer neoantigens and not found in healthy tissues, they may provide an attractive new avenue for immunotherapeutic targeting.
Not only are these neoantigens present on leukemia samples, but positional analysis indicates that the GlcNAc residues may be optimally positioned for T-cell recognition. The GlcNAc group is in the middle of the peptide (up to 34/36; 62% P4, 18% P5, 21% equivocal P4/P5; Supplementary Fig. S7), identical to the preferred position of phosphate groups in phosphopeptides, and where structural studies have revealed that the CDR3 regions of the TCR loops around the center of the peptide(32). Indeed, previous structural studies in mouse of TCR binding have demonstrated that GlcNAc modified antigens are recognized in this manner (22).
We saw potent multifunctional memory T-cell responses against these O-GlcNAcylated leukemia antigens in healthy donors, suggesting that these neoantigens may reflect an endogenous immunosurveillance system against leukemia). Not only did healthy donor T cells recognize the PTM neoantigen, but we also showed that they could specifically kill cells presenting modified peptides. Therefore, we would not expect targeting of these antigens to be compromised by self-tolerance, as may be seen with overexpressed antigens. What is more, if healthy donors have cytotoxic memory T cells targeting these PTM neoantigens without autoimmunity, targeted therapies against these neoantigens may have low toxicity. The most immunogenic peptide identified was me-RPPItQSSL, containing both a methylated arginine and O-GlcNAcylated serine. It is tempting to speculate that combined modifications lead to the most dramatic structural change and, therefore, peptides more antigenically distinct from self. We showed that T cells may recognize and kill cells presenting this peptide with either the methylation, or the O-GlcNAc modification, but not the unmodified peptide. This potent antigen, targetable by T cells from several healthy donors, is an attractive target for the development of immunotherapeutics. We are expanding this work to identify O-GlcNAcylated antigens from patients with other HLA-types and cancers and in the process of developing methods that allow for the identification of methylated peptide antigens from MHC class I.
Overall, this work identified both glycosylated and methylated residues as potent classes of tumor antigens, broadening the availability of immunotherapy targets, and potentially yielding safe and effective therapeutics for leukemia.
Supplementary Material
Acknowledgments
Financial support: This work was supported by National Institutes of Health Grants GM037537 and AI033993 (to D.F.H.). Kay Kendall Leukaemia Research grant KKL3227 and Bloodwise grant 08038 (to M.C.)
This work was supported by National Institutes of Health Grants GM037537 and AI033993 (to D.F.H.). Kay Kendall Leukaemia Research grant KKL3227 and Bloodwise grant 08038 (to M.C.)
Footnotes
Conflict of Interest: A patent application, entitled “Identification of Class I MHC Associated Glycopeptides as Targets for Cancer Immunotherapy,” has been filed U.S. Patent Application No. 62/202,359 which has been licensed to Agenus Inc.
D.F.H and M.C. receive research support and consultancy from Agenus Inc. The other authors declare no competing interests.
References
- 1.Gillis S, Smith KA. Long term culture of tumour-specific cytotoxic T cells. Nature. 1977;268(5616):154–156. doi: 10.1038/268154a0. [DOI] [PubMed] [Google Scholar]
- 2.Coulie PG, et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci U S A. 1995;92(17):7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox AL, et al. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264(5159):716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 4.Chen YT, et al. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci U S A. 1997;94(5):1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linnemann C, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015;21(1):81–85. doi: 10.1038/nm.3773. [DOI] [PubMed] [Google Scholar]
- 7.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segal NH, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68(3):889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 10.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ansell SM, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372(4):311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motzer RJ, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med. 2015;373(19):1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagorsen D, Scheibenbogen C, Marincola FM, Letsch A, Keilholz U. Natural T cell immunity against cancer. Clin Cancer Res. 2003;9(12):4296–4303. [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Cobbold M, et al. MHC class I-associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci Transl Med. 2013;5(203):203ra125. doi: 10.1126/scitranslmed.3006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart GW. Minireview series on the thirtieth anniversary of research on O-GlcNAcylation of nuclear and cytoplasmic proteins: Nutrient regulation of cellular metabolism and physiology by O-GlcNAcylation. The Journal of biological chemistry. 2014;289(50):34422–34423. doi: 10.1074/jbc.R114.609776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nature reviews Cancer. 2011;11(9):678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells L, Kreppel LK, Comer FI, Wadzinski BE, Hart GW. O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits. The Journal of biological chemistry. 2004;279(37):38466–38470. doi: 10.1074/jbc.M406481200. [DOI] [PubMed] [Google Scholar]
- 21.de Queiroz RM, Carvalho E, Dias WB. O-GlcNAcylation: The Sweet Side of the Cancer. Frontiers in oncology. 2014;4:132. doi: 10.3389/fonc.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glithero A, et al. Crystal structures of two H-2Db/glycopeptide complexes suggest a molecular basis for CTL cross-reactivity. Immunity. 1999;10(1):63–74. doi: 10.1016/s1074-7613(00)80007-2. [DOI] [PubMed] [Google Scholar]
- 23.Haurum JS, et al. Peptide anchor residue glycosylation: effect on class I major histocompatibility complex binding and cytotoxic T lymphocyte recognition. Eur J Immunol. 1995;25(12):3270–3276. doi: 10.1002/eji.1830251211. [DOI] [PubMed] [Google Scholar]
- 24.Haurum JS, et al. Recognition of carbohydrate by major histocompatibility complex class I-restricted, glycopeptide-specific cytotoxic T lymphocytes. J Exp Med. 1994;180(2):739–744. doi: 10.1084/jem.180.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kastrup IB, et al. Lectin purified human class I MHC-derived peptides: evidence for presentation of glycopeptides in vivo. Tissue Antigens. 2000;56(2):129–135. doi: 10.1034/j.1399-0039.2000.560203.x. [DOI] [PubMed] [Google Scholar]
- 26.Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat Protoc. 2008;3(11):1709–1717. doi: 10.1038/nprot.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother (1997) 2003;26(4):332–342. doi: 10.1097/00002371-200307000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, et al. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2010;9(1):153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao P, et al. Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J Proteome Res. 2011;10(9):4088–4104. doi: 10.1021/pr2002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cante-Barrett K, Pieters R, Meijerink JP. Myocyte enhancer factor 2C in hematopoiesis and leukemia. Oncogene. 2014;33(4):403–410. doi: 10.1038/onc.2013.56. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Huang C, Fujihira H. The cytoplasmic peptide:N-glycanase (NGLY1) - Structure, expression and cellular functions. Gene. 2016;577(1):1–7. doi: 10.1016/j.gene.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammed F, et al. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat Immunol. 2008;9(11):1236–1243. doi: 10.1038/ni.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.