Abstract
Adult stem cell proliferation rates are precisely regulated to maintain long-term tissue homeostasis. Defects in the mechanisms controlling stem cell proliferation result in impaired regeneration and hyperproliferative diseases. Many stem cell populations increase proliferation in response to tissue damage and reacquire basal proliferation rates after tissue repair is completed. Although proliferative signals have been extensively studied, much less is known about the molecular mechanisms that restore stem cell quiescence. Here we show that Tis11, an Adenine-uridine Rich Element (ARE) binding protein that promotes mRNA degradation, is required to re-establish basal proliferation rates of adult Drosophila intestinal stem cells (ISC) after a regenerative episode. We find that Tis11 limits ISC proliferation specifically after proliferation has been stimulated in response to heat stress or infection, and show that Tis11 expression and activity are increased in ISCs during tissue repair. Based on stem cell transcriptome analysis and RNA immunoprecipitation, we propose that Tis11 activation represents an integral part of a negative feedback mechanism that limits the expression of key components of several signaling pathways that control ISC function and proliferation. Our results identify Tis11 mediated mRNA decay as an evolutionarily conserved mechanism of re-establishing basal proliferation rates of stem cells in regenerating tissues.
Introduction
To maintain tissue homeostasis, stem and progenitor cell proliferation has to be tightly controlled. In response to tissue damage or during tissue growth, stem cell (SC) proliferation has to increase, while basal proliferation rates have to be re-established when growth or regeneration ceases. Defects in this regulation are likely to cause degenerative or hyper-proliferative syndromes, including cancer (Liu and Rando, 2011).
A comprehensive model for the dynamic control of SC proliferation is starting to emerge in the Drosophila intestine, where tissue damage by infection or stress is accompanied by a transient regenerative response fueled by Intestinal Stem Cells (ISCs). In response to infection by the mild enteropathogen Erwinia carotovora carotovora (Ecc15), for example, ISC proliferation rates rapidly increase, regenerating the epithelium, before returning to basal levels within 24 hours (Ayyaz et al., 2015; Buchon et al., 2010). The induction of ISC proliferation is governed by complex interactions between damaged enterocytes (ECs), the visceral muscle, hemocytes, and ISCs, and involves stress, growth factor and cytokine signaling, including the JNK, EGFR, Jak/Stat and Insulin signaling pathways (Amcheslavsky et al., 2009; Ayyaz and Jasper, 2013; Biteau et al., 2008, 2011; Biteau and Jasper, 2011; Buchon et al., 2013; Jiang et al., 2009). Return to ISC quiescence, in turn, is promoted by the BMP-like decapentaplegic (Dpp), which is induced in the visceral muscle after regeneration has concluded (Ayyaz et al., 2015; Guo et al., 2013). In addition, Nrf2/CncC signaling inhibits ISC proliferation (Hochmuth et al., 2011). However, the molecular mechanism(s) that negatively regulate proliferation in ISCs remain largely unexplored.
Activation of ISC proliferation is associated with and requires a broad transcriptional program that includes induction of cell cycle and replication genes (Dutta et al., 2015; Zeng et al., 2015). Engagement of post-transcriptional regulatory mechanisms is likely to e required for efficient control of this transcriptional response when ISCs return to quiescence. Post-transcriptional regulation has recently emerged as critical for the control of SC function and maintenance in a range of organisms. In mice, for example, microRNAs, 3′ untranslated region (3′UTR) switching and regulation of mRNA translation have been shown to regulate pluripotent and tissue stem cell function by affecting the biology of mRNAs encoding key self-renewal factors (Cheung and Rando, 2013; Ji et al., 2009; Ji and Tian, 2009; Zismanov et al., 2016).
Here we show that Tis11, an Adenine-uridine Rich Element (ARE) binding protein that promotes mRNA degradation, is required to re-establish basal proliferation rates in the intestinal epithelium after regenerative episodes. The Tis11 protein family binds AREs present in the 3′UTR of many mRNAs (Baou et al., 2009). It is composed of four members in mammals, ZFP36L1/Tis11b, ZFP36L2/Tis11d, ZFP36L3 and Tis11/TTP, and one member in Drosophila, Tis11 (Baou et al., 2009). Tis11 binding is a critical step for the recruitment of the mRNA degradation machinery, leading to what is known as ARE-mediated decay (AMD). Through AMD, Tis11-related proteins negatively regulate the expression of a large number (up to 8–16% of all mRNAs) of genes in mammals and flies (Baou et al., 2009; Spasic et al., 2012).
Dynamic control of gene expression by AMD has been shown during inflammatory responses in mammalian macrophages and Drosophila cultured cells. In macrophages, activation of TNF-α signaling induces Tis11 expression, which in turn binds to the ARE contained in the 3′UTR of the TNF-α mRNA, establishing a negative feedback mechanism to resolve the inflammatory response (Carballo et al., 1998). In Drosophila S2 cells, the innate immune response promotes the expression of Tis11, which targets select antimicrobial peptide (AMP) mRNAs for AMD, thereby resolving the innate immune response (Lauwers et al., 2009).
We find that Tis11 expression and activity are increased in ISCs during tissue repair, negatively regulating the proliferative activity of ISCs. Based on transcriptome and RNA binding data, we propose that Tis11 activation represents a negative feedback mechanism that limits the expression of several components of critical signaling pathways controlling ISC function. Our results identify AMD as an evolutionarily conserved mechanism of SC regulation in regenerating tissues.
Results
Tis11 is expressed in all intestinal progenitor cells
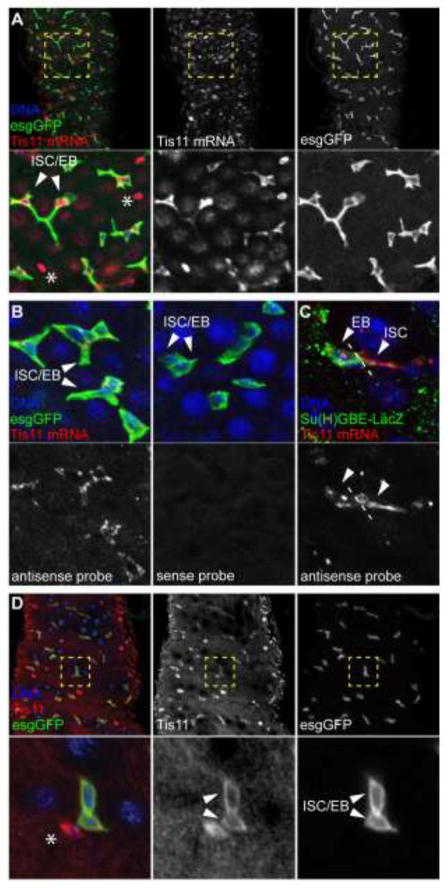
Recent expression profiling experiments from sorted ISCs, performed by us and others, have identified Tis11 expression in ISCs (Dutta et al., 2015; Wang et al., 2015). This observation, and the fact that Tis11 is a critical regulator of AMD, prompted us to investigate the function of Tis11 in the ISC lineage. To confirm the expression of Tis11 in the intestinal epithelium, we performed RNA in situ hybridization of dissected guts in which progenitor cells were labeled using GFP expressed under the control of the ISC/EB driver esgGal4. Tis11 mRNA was specifically detected in all GFP-positive cells, as well as in basally located esg-negative cells that are likely to be enteroendocrine cells (Figure 1A, B). We confirmed expression in EBs using the EB-specific marker Su(H)GBE-LacZ (Figure 1C). We further confirmed the Tis11 expression pattern using a recently described anti-Tis11 antibody (Twyffels et al., 2013), and found Tis11 protein in all esg-positive cells (Figure 1D).
Figure 1. Tis11 is expressed in stem cells and progenitors in the adult Drosophila intestine.
(A–B) Confocal images of the posterior midgut of esgGal4>UAS-GFP (A, B) or Su(H)GBE-LacZ (C) flies show that in situ hybridization (red) specifically detects Tis11 mRNA expression in all ISCs and EBs. Note that basally located esg-negative diploid cells also show high Tis11 expression (asterisks in panel A). (D) Tis11 protein is also detected in esg-positive cells by immunohistochemistry using an anti-Tis11 antibody (red). In all panels, DNA is stained with Hoestch (blue).
Tis11 functions to limit ISC proliferation
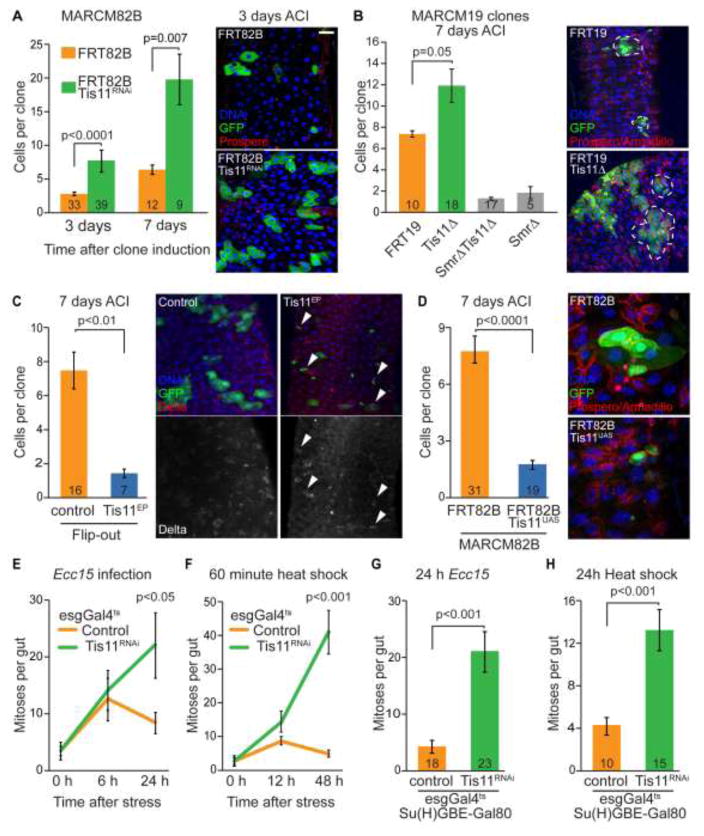
To characterize Tis11 function in the ISC lineage, we generated GFP-marked stem cell clones expressing a dsRNA directed against Tis11 (Tis11RNAi) using MARCM (Mosaic Analysis with a Repressible Cell Marker Clones; (Lee and Luo, 1999)). We confirmed that the dsRNA used is sufficient to knock-down Tis11 expression (Figure S1A). As early as 3 days after clone induction, Tis11RNAi expressing clones are significantly larger than control clones, indicating that Tis11 is required to limit ISC proliferation (Figure 2A). This notion was confirmed in ISC lineages homozygous for a Tis11 null allele: we generated MARCM clones using a deletion (Df(1)G0124; Tis11G0124,SmrG0124 (Figure S1B) that covers both Tis11 and the neighboring gene Smrter (Smr), and rescued the Smr mutation with the duplication Dp(1;3)DC258. These Tis11 mutant ISC clones are approximately two-fold larger than controls (Figure 2B). Based on this analysis, Tis11 does not appear to govern ISC differentiation: the ratio of enteroendocrine (EE) to enterocytes (EC) in control and Tis11 deficient clones is not significantly different (Figure S1C). We asked how increased Tis11 expression would affect ISC function, and generated Flip-out clones driving P{XP}Tis11d00685, a UAS-containing allele of Tis11 allowing the over-expression from the endogenous Tis11 locus (Figure S1D). Seven days after clone induction, Tis11d00685 expressing clones are significantly smaller than controls (1–2 cells per clone as compared to an average of seven cells) (Figure 2C). Importantly, these small clones express the ISC-specific marker Delta (Figure 2C), demonstrating that Tis11 over-expression impairs the proliferative capacity of ISCs without affecting their identity or survival. We confirmed Tis11-mediated inhibition of ISC proliferation in MARCM clones over-expressing Tis11 from a newly constructed Tis11 transgene (UAS-Tis11) (Figure 2D).
Figure 2. Tis11 is required in ISCs to promote the re-entry into quiescence after stimulation.
(A) MARCM clones expressing a dsRNA construct directed against Tis11 grow larger than controls. Enteroendocrine (EEs) are identified by prospero staining (nuclear, red). (B) MARCM clones homozygous Tis11 mutant are larger than wild-type clones. Detailed genotypes used are Control: FRT19, Tis11Δ: FRT19 Df(1)G0124;DC257; Tis11ΔSmrΔ: FRT19 Df(1)G0124; SmrΔ: FRT19 Df(1)G0124;DC258. Df(1)G0124 is a deletion covering both Smr and Tis11, DC257 and DC258 duplications respectively rescue the Smr and Tis11 loci. GFP expression labels clones (green), enteroendocrine (EEs) are identified by prospero staining (nuclear, red), cell boundaries are indicated by armadillo staining (membrane, red) and DNA is stained with Hoestch (blue). (C and D) Growth of Flip-out and MARCM clones over-expressing Tis11 is significantly impaired. An EP line, P{XP}Tis11d00685, was used to over-express the endogenous Tis11 gene in C and a UAS-Tis11 transgene was used in D. Delta staining is used to identify ISCs (arrowheads, red, C) and prospero/armadillo staining labels EEs and cell boundaries (red, D). (E and F) Time course analysis of intestinal proliferation in response to Ecc15 infection (E) and heat shock (F). While the number of mitoses per gut transiently increases in control flies, high proliferation rates remain at later time points when Tis11 is knocked-down in esg-positive ISCs and EBs. (G and H) ISC-specific knock-down of Tis11, using the esgGal4ts; Su(H)GBE-Gal80 driver, is sufficient to cause ISC hyper-proliferation 24 hours after Ecc15 infection (G) or heat shock (H). In A–D, GFP expression marks clones (green) and DNA is stained with Hoestch (blue). Number of clones (A–D) or guts (G, H) are indicated. In all panels, values are presented as average +/− s.e.m. All p-values are calculated using unpaired two- tailed Students t-test. See also Figure S1 and S2 for additional experiments.
Tis11 over-expression has previously been shown to induce defects in the developing eye (Yeh et al., 2012). To test whether Tis11 universally inhibits proliferation, we generated Flip-out clones expressing P{XP}Tis11d00685 and Tis11RNAi in the larval brain and wing imaginal discs. Cell proliferation was not significantly affected in these clones (Figure S1E), demonstrating that Tis11 does not function as a general regulator of cell proliferation and suggesting that this function may be restricted to ISCs.
Tis11 functions to promote the re-entry into quiescence
While clonal analysis allows detailed characterization of mutant phenotypes in marked ISC lineages, the sustained genetic perturbation in cells of the clone, as well as complex non-autonomous interactions between these cells preclude a distinction between selective effects of Tis11 in specific cell types of the ISC lineage. To gain further insight into the exact role of Tis11 and into its dynamic control of ISC proliferation during regeneration, we perturbed Tis11 selectively in ISC and/or EBs in an inducible manner (Figure 2).
Knocking down Tis11 in ISCs and EBs using esgGal4 (in combination with Gal80ts) did not impact the number of mitotic ISCs in the entire midgut (quantified using phospho-HistoneH3 (pH3) as mitotic marker) in the absence of a proliferative stimulus (Figure 2E, 2F, S2A). To further test this notion, we expressed Tis11RNAi in ISCs and EBs for 13 days and found no significant difference in the number of pH3 positive cells between esg>Tis11RNAi and control intestines (Figure S2B). When ISC proliferation was induced by infection with Ecc15, however, Tis11RNAi expression in ISCs and EBs perturbed the return to quiescence (Figure 2E, S2A): While Tis11 deficient progenitors respond to Ecc15 infection at the same rate as wild-type ISCs, they fail to re-establish basal proliferation rates at 24 hours after infection. Similar results were observed in Df(1)G0124/+ heterozygotes (Figure S2C) and after heat-shock, which also induces a temporary increase in ISC proliferation (Figure 2F, S2D). Our results suggest that Tis11 functions to restore low ISC proliferation rates after diverse stimuli.
To demonstrate that Tis11 functions cell-autonomously in ISCs to elicit these phenotypes, we combined esgGal4,tub-Gal80ts with Su(H)GBE-Gal80, which expresses the Gal4 inhibitor Gal80 in EBs, and thus allows ISC-specific induction of UAS-linked transgenes (Wang et al., 2015). Knockdown of Tis11 with this driver combination phenocopied the effects of knockdown with esgGal4, tub-Gal80ts, confirming that Tis11 activity in ISCs alone is required to re-establish their quiescence (Figure 2G, 2H).
Tis11 activity is regulated during the regenerative response
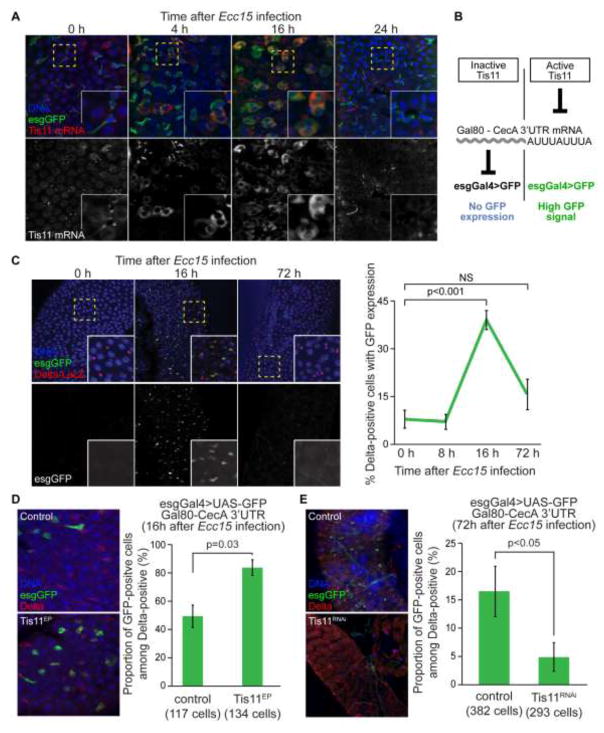
Across species, Tis11 proteins have been shown to be regulated in response to mitogenic stimuli and during the innate immune response (Baou et al., 2009; Twyffels et al., 2013). We tested whether such regulation would also be observed in the fly intestinal epithelium. Using in situ hybridization, we found elevated levels of tis11 expression 4 and 16 hours after Ecc15 infection (in esg-positive progenitors as well as in enterocytes) and a return to basal levels at 24 hours (Figure 3A). To confirm that this dynamic regulation of Tis11 expression leads to changes in Tis11-mediated mRNA degradation activity, we developed a GFP-based transgenic reporter to directly measure Tis11 activity in ISCs during the regenerative response. Building on a previously described luciferase-based Tis11 activity reporter used in cultured cells (Cairrao et al., 2009), we fused the Cecropin A 3′UTR (ARE rich) sequence to GFP or Gal80 coding sequences. We confirmed that the CecA-3′UTR is sensitive to Tis11 levels, as Drosophila S2 cells co-expressing Tis11 and GFP-CecA-3′UTR show reduced GFP expression compared to controls (Figure S3). We generated transgenic flies carrying a hsGal80-CecA-3′UTR construct, in which the Tis11-sensitive Gal80 cassette is expressed using a minimal Hsp70 promoter. When combined with esgGal4, UAS-GFP, this reporter identifies ISCs and EBs with high Tis11 activity by their GFP expression (Figure 3B). To validate this system, we introduced it into wild-type, P{XP}Tis11d00685 and Tis11RNAi genetic backgrounds and infected these flies in order to induce expression of the endogenous Tis11 gene and initiate the expression of our reporter system. In these conditions, around half of all ISCs express GFP in control animals, while the vast majority of ISCs are GFP-positive when Tis11 is over-expressed (Figure 3D). Conversely, when Tis11 is knocked-down, the number of ISCs with active Tis11 was significantly decreased (Figure 3E).
Figure 3. Tis11 expression and activity are regulated during the intestinal regenerative response.
(A) Time course analysis of Tis11 expression in the posterior midgut by in situ hybridization. Tis11 mRNA levels are increased in esg-positive cells as well as large polypoid ECs 4 and 16h after infection, compared to unstressed condition. Tis11 messenger returns to basal levels or lower 24h after Ecc15 feeding. (B) Graphic representation of the reporter system used to detect Tis11 activity in esg-positive cells. Low Tis11 activity represses GFP expression, while high Tis11 activity results in high GFP signal in ISCs and EBs. C) Time course analysis of Tis11 activity in Delta-LacZ-positive cells in the posterior midgut epithelium, using the esgGal4>GFP;Gal80-ARE reporter. The proportion of ISCs expressing GFP significantly increases 16 hours after Ecc15 feeding, and returns to low basal level 3 days after infection. (D, E) The esgGal4>GFP;Gal80-ARE reporter is sensitive to Tis11. GFP expression in Delta-positive ISCs was scored after imaging of the posterior midgut epithelium using confocal microscopy. The proportion of ISCs expressing GFP is significantly higher in flies over-expressing Tis11 (esgGal4>Tis11EP) compared to control animals (D). Conversely, the fraction of GFP-positive ISCs is significantly lower in flies expressing Tis11RNAi (esgGal4>Tis11RNAi). In C, D and E values are presented as average +/− s.e.m, p-values are calculated using unpaired two-tailed Students t-test. NS, not significant.
We then used this reporter to analyze Tis11 activity during the intestinal response to Ecc15 (Figure 3C). Before infection, 7.5% of ISCs (identified by Delta-lacZ expression) express GFP, suggesting that Tis11 is largely inactive under basal conditions. This proportion strongly increases 16 hours after the infection (GFP is expressed in 40% of Delta+ cells) and decreases 72 hours after infection (15% of Delta+ cells express GFP).
Collectively, these experiments demonstrate that Tis11 expression and activity are transiently induced in ISCs and EBs during the regenerative response of the intestine.
Tis11 targets mRNA for degradation in ISCs
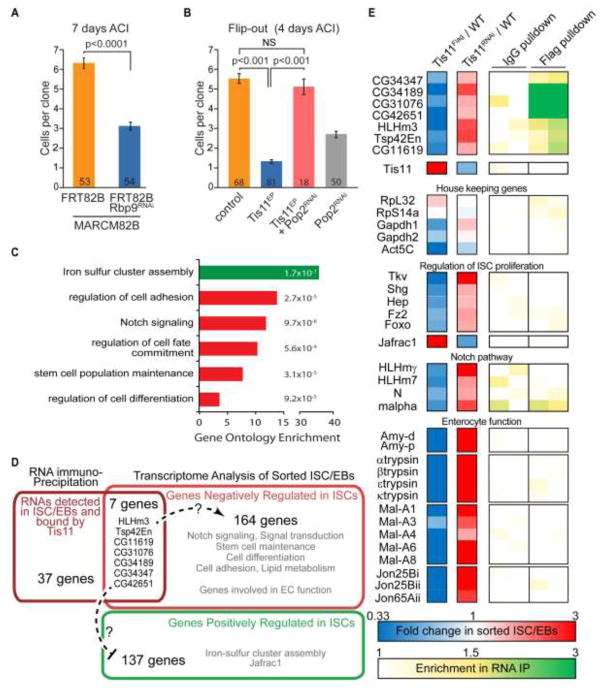
Our reporter analysis strongly suggests that Tis11 functions in ISC in its canonical fashion, namely to control the stability of AU-rich mRNAs. To test this notion further, we assessed how ISC function is affected when ARE-containing mRNAs are regulated in a Tis11-independent fashion. The protein HuR also binds AREs located in the 3′UTR of mRNA to regulate stability (Hinman and Lou, 2008). However, unlike Tis11, HuR mediates the stabilization and translation of its targets. The Drosophila homolog of HuR is the RNA binding protein 9 (Rbp9) (Kim-Ha et al., 1999). We generated ISC clones expressing Rbp9RNAi to test whether failure to stabilize mRNA was sufficient to limit ISC proliferation (Figure 4A). We found that MARCM clones expressing Rbp9RNAi are 50% smaller than control clones, consistent with a role of AMD in promoting ISC quiescence.
Figure 4. Tis11 controls the expression of genes involved in many aspects of ISC function and regulation.
(A) MARCM clones expressing a dsRNA construct directed against Rbp9 are smaller than controls. (B) Knocking-down Pop2 is sufficient to rescue the impaired clone growth associated with Tis11 over-expression. (C) Selected Gene Ontology categories enriched among genes positively (green) and negatively (red) regulated by Tis11 in esg-positive cells. Enrichment score and p values calculated using GOrilla are indicated. (D) Diagram representing the numbers of genes regulated by Tis11 and mRNAs expressed in ISC/EBs bound by Tis11, as well as selected Gene Ontology categories enriched in each gene group. (E) Representation of changes in gene expression in ISC/EBs and enrichment in Tis11Flag RIP-seq for selected genes. In A and B, values are presented as average +/− s.e.m, p-values are calculated using unpaired two-tailed Students t-test. NS, not significant.
We further asked whether preventing mRNA decay downstream of Tis11 would rescue the inhibition of ISC proliferation caused by increased Tis11 expression. The protein Pop2 (CAF1) is a deadenylase required for poly(A) shortening, the first step in mRNA decay (Temme et al., 2010) and is required for AMD in Drosophila (Lauwers et al., 2009). We generated Flip-out clones expressing both P{XP}Tis11d00685 and Pop2RNAi, and found that, although knocking down Pop2 alone has a small anti-proliferative effect, Pop2RNAi rescues clone growth defects from Tis11 over-expressing ISCs to a size not significantly different from wild-type clones (Figure 4B). Combined, these experiments support the notion that Tis11 functions in ISCs to negatively regulate cell proliferation by promoting AMD.
Tis11 controls the expression of a core set of mRNAs in ISCs
Our reporter analysis and genetic interaction with the AMD pathways strongly suggests that Tis11 regulates the stability of target mRNAs to control ISC proliferation. In order to identify these messages, we first determined ISC/EB-specific transcriptional changes induced by Tis11 manipulation. We used a previously established cell sorting protocol (Wang et al., 2015) to isolate wild-type, Tis11 over-expressing (expressing a functional Flag-tagged Tis11, Figure S4A) and Tis11 deficient (expressing Tis11RNAi) ISCs and EBs at 24 hours after Ecc15 infection. RNA-sequencing (RNA-seq) revealed 171 genes that are down-regulated in response to Tis11 activity (1.5 fold induction in Tis11RNAi expressing ISC/EBs compared to controls and 2/3 fold depletion in UAS-Tis11Flag compared to controls). These genes include several components of key signaling pathways that control ISC proliferation, such as Hep/JNKK, Tkv (Ayyaz et al., 2015; Biteau et al., 2008; Guo et al., 2013), as well as genes involved in stem cell differentiation and enterocyte function (e.g. Notch, HLHm3, digestive enzymes; (Dutta et al., 2015; Ohlstein and Spradling, 2007)) (Figure 4C–E, Table S1). Conversely, 137 genes are positively regulated in response to Tis11 activity (1.5 fold induction in UAS-Tis11Flag expressing ISC/EBs verses controls and 2/3 fold depletion in Tis11RNAi compared to controls), including genes involved in iron-sulfur cluster assembly and the peroxidase Jafrac1, which we have previously shown to negatively affect ISC proliferation (Hochmuth et al., 2011) (Figure 4C–E, Table S2). We confirmed by qPCR in sorted ISCs that Tis11 gene dose strongly influences the mRNA levels of Jafrac1 and HLHm3 (Figure S4E), two genes that we have shown control ISC proliferation (see below for HLHm3).
To determine which of these mRNAs are directly bound by Tis11 in vivo, we performed RNA-immunoprecipitation (RIP) followed by RNA-seq. We expressed UAS-Tis11Flag combined with Pop2RNAi, using esgGal4ts, in order to stabilize Tis11-bound messengers in ISC/EBs (Figure S4A–C). In this RIP-seq experiment, we found 320 RNAs reproducibly enriched in anti-Flag pull-downs compared to controls (Table S3). Among these transcripts, 44 were detected in our ISC/EBs RNA-seq experiment (Figure S4D). Specifically, 7 genes are down regulated by active Tis11 in ISC/EBs, and none are among the genes we described as positively regulated by Tis11, suggesting that these seven genes (HLHm3, Tsp42En, CG34189, CG31076, CG42651, CG11619 and CG34347) might be directly destabilized by Tis11 in esg-positive stem and progenitor cells.
We next asked whether any of the genes identified by our genome-wide analysis influence ISC proliferation. We generated Flip-out clones expressing HLHm3RNAi and found that this was sufficient to block proliferation (Figure S4F), suggesting that HLHm3 is one of the transcripts destabilized by Tis11 to control ISC quiescence re-entry.
Discussion
Our study establishes that Tis11-mediated RNA decay is essential to halt the proliferative response in the Drosophila intestinal epithelium after periods of tissue damage. Although much is known about the signaling pathways that contribute to the increased ISC proliferation in response to a variety of stimuli, only Tkv and Nrf2/CncC signaling have been found to prevent intestinal proliferation (Ayyaz et al., 2015; Hochmuth et al., 2011). It is interesting to note that most proliferative signaling pathways (e.g. JNK, JAK/Stat, Hpo/Yki) are self-limiting, their activation inducing the expression of pathway specific negative regulators. Our data suggest that, unlike these built-in negative feedback loops, Tis11 acts as a critical regulator promoting the restoration of basal proliferation rates in response to multiple stimuli by simultaneously inhibiting key components of several signaling pathways. Supporting this model, disrupting Tis11 activity does not appear to affect proliferation of cells in developing tissues, indicating that it does not target the general cell cycle machinery. Rather, our transcriptional analysis suggests that Tis11 impacts the ISC-specific ability to respond to stress and maintain an appropriate balance between self-renewal and differentiation. Indeed, among the transcriptional changes seen in progenitor cells in response to Tis11 activity, several genes identified have been previously shown to control ISC proliferation and differentiation in the adult intestinal lineage. For example, we found that the peroxiredoxin Jacfrac1 is upregulated in response to Tis11, an induction that is sufficient to inhibit ISC proliferation (Hochmuth et al., 2011). We also found that the expression of numerous components of the Notch signaling pathway, an essential regulator of the self-renewal/differentiation balance (Ohlstein and Spradling, 2007), to be affected by Tis11 activity. One of these Tis11 targets, HLHm3 is required in ISCs to instigate the regenerative response. These changes indicate that the dynamics of Tis11 activity modulate Notch signaling to promote stem cell identity as part of the reestablishment of basal tissue turn-over conditions after repair. Likewise, Tis11 family proteins have been shown to regulate the self-renewal and differentiation of erythroid progenitor cells by modulating the Notch signaling pathway (Vignudelli et al., 2010; Zhang et al., 2013). Finally, the expression level of several other signaling molecules, such as the transcriptional repressor Aop or the FGF receptor Btl, is also influenced by Tis11. Further studies will be necessary to test their potential role in the control of ISC function.
We find that Tis11 expression and activity are transiently induced in ISCs in response to Ecc15 infection. This is reminiscent of Tis11 regulation in mammalian macrophages, where its expression is induced by TNF signaling, which in turn is negatively regulated by Tis11 (Carballo et al., 1998). The vertebrate Tis11 transcript contains an ARE in its 3′UTR and has been proposed to be a target of AMD (Brooks et al., 2004; Tchen et al., 2004). A similar regulation may account for the temporal control of Drosophila Tis11 expression in ISCs during intestinal regeneration. However, additional experiments will be required to test whether Tis11 is a transcriptional target of one or multiple proliferative signaling pathways (e.g. JNK, ERK, Dpp, JAK/Stat or Hpo/Yki signaling). In addition to a direct regulation by stress signaling pathways, it is possible that Tis11 activity is regulated by the ISC cell cycle. This hypothesis remains to be tested. Interestingly, the ARE-containing 3′UTR is a conserved feature also found in Drosophila Tis11 transcripts and could explain that Tis11 transcript is not detectable any more at 24h after infection. This feedback regulation of Tis11 mRNA stability may ensure that ISCs are capable of responding to successive periods of tissue damage.
Consistent with our findings that Tis11 limits ISC proliferation in Drosophila, TTP has recently been shown to regulate murine satellite cell quiescence (Hausburg et al., 2015). Tis11 proteins have also been implicated in the development of multiple cancers and shown to regulate progenitors in vertebrates. Loss of TTP activity or expression is associated with hyper-proliferation and metastasis in the brain, cervix, breast, colon and liver (Brennan et al., 2009; Sohn et al., 2010; Suswam et al., 2008; Young and Dixon, 2010). Similarly, deletion of ZFP36L1 and ZFP36L2 in mice results in over-proliferation and the development of T-lymphoblastic leukemia, caused by the failure to post-transcriptionally regulate the expression of the Notch receptor (Hodson et al., 2010). These similarities between the function and regulation of Tis11-like proteins in fly and mammalian stem cells highlight the critical, evolutionarily conserved role for regulated mRNA decay in the control of stem cell function and activity.
Materials and Methods
Drosophila stocks and culture
The following strains were obtained from the Bloomington Drosophila Stock Center: w1118, Df(1)G0124/FM7c (#11915), Dp(1;Y)BSC257 (#33019), Dp(1;Y)BSC258 (#33020), UAS-Rbp9RNAi (#28669), UAS-Pop2RNAi (#30492), UAS-HLHm3RNAi (#25977 and 55302) and Dl05151/TM3 (#11651).
P{XP}Tis11[d00685] was obtained from Exelixis collection at Harvard Medical School.
The following fly lines were used: MARCM82B from B. Ohlstein (Columbia University, USA), esgGal4NP5130 from S. Hayashi (RIKEN, Japan), esgGal4,Su(H)GBEGal80,UAS-2XEYFP,tubGal80ts from S. Hou (NIH, USA), Su(H)GBE-LacZ from S. Bray (Cambridge, UK), MARCM19A from M. Uhlirova (University of Cologne, Germany).
All flies were raised on standard yeast and molasses-based food, at 25°C and 65% humidity, on a 12-hour light/dark cycle, unless otherwise indicated.
Tis11 knock-down and expression constructs
To construct the UAS-Tis11RNAi, a 396bp fragment of Tis11 coding region was amplified from genomic DNA of w1118 flies using the primers Forward:5′-CACGATGCCCTGCAGCATGG-3′ and Reverse: 5′-TTAGAGTCCCAAATTGGACTG-3′ and cloned twice tail-to-tail in the pWIZ vector. For UAS-Tis11, most of the Tis11 cDNA was amplified using the primers Forward: 5′-TGCCGGGCAACCGAATCGAAA-3′ and Reverse: 5′-CCGTAGTAGTCCGTAATTCGGTCCTT-3′ and cloned in the pUAST vector using EcoRI and XbaI sites. For UAS-Tis11Flag, the following reverse primer 5′-GAGTCCCAAATTGGACTGC-3′ was used to remove the stop codon and clone the Tis11 coding sequence in frame with a C-terminal 3xFlag tag in a modified pUAST.
Tis11 activity reporters construction
The coding sequences of GFP and Gal80 proteins, as well as the Cecropin A 3′UTR were amplified from genomic DNA of MARCM82B flies using the following primers: GFP Forward 5′-AAAAGCGGCCGCATGGTGAGCAAGGGCGAGGAGCTG-3′, GFP Reverse 5′-TAAATAATTATAAATAATCATCGTGGTTACTTGTACAGCTCGTCCATGCCG-3′, Gal80 Forward 5′-AAAAGCGGCCGCATGGACTACAACAAGAGATCTTCGG-3′ and Gal80 Reverse 5′-TAAATAATTATAAATAATCATCGTGGTTATAAACTATAATGCGAGATATTG-3′,CecA3′UTR Forward 5′-CCACGATGATTATTTATAATTATTTATTTAAAGATCTATTTATTCTGTTGCTCCC-3′, CecA3′UTR Reverse 5′-AAAGGATCCTTCTTTAAATTTTTAAAATTGTTTTATTTACAGGGAGCAACAGAATAAA TA-3′. GFP- CecA3′UTR and Gal80-CecA3′UTR fusions were generated by PCR sewing and cloned into the pAc5.1 vector, using NotI and BamHI sites, for expression
To create reporter expressing transgenic flies, pAct5C-GFP/Gal80-CecA-3′UTR fusions were amplified with the following primers: Forward 5′-AAAATCTAGATCTAAAAACACAAATGATACTTCT-3′ and Reverse 5′-AAAATCT AGAGTTGAAGGCTCTCAAGGGCATCG-3′ and cloned in pCasper3-hs (pJZ3) using XbaI sites. Transgenic animals were made by standard microinjection of pCasper3-hs into the Drosophila germline (Genetic Services).
Conditional expression of UAS-linked transgenes
The TARGET system was used to conditionally express UAS-linked transgenes in ISCs and EBs (McGuire et al., 2003). The esgGal4 was combined with UAS-GFP and a ubiquitously expressed temperature-sensitive Gal80 inhibitor (tubGal80ts). This conditional driver is termed esgGFPts. Crosses and flies were kept at 18–20°C (permissive temperature), then shifted to 29°C to allow expression of the transgenes.
Erwinia carotovora carotovora infection and heat shock treatments
Previously described procedures were followed for oral bacterial challenge (Buchon et al., 2009; Buchon et al., 2010; Jiang et al., 2009). Briefly, Ecc15 was cultured in LB medium overnight at 30°C. Flies were fed on 500 μl of concentrated bacteria (OD100) in 5% sucrose for the time indicated.
For heat shock treatment, flies were heat shocked at 37°C for one hour and transferred back at 25°C until dissection.
Mosaic analysis with a repressible cell marker clones and Flip-out lineage tracing
Positively marked clones were generated using the following fly stocks: hsFLP,tubGal80,FRT19A;T80Gal4,UAS-GFP (MARCM19); hsFlp,UAS-GFP;;tubGal4,FRT82B,tubGal80 (MARCM82B) and hsFlp;act5c>FRT>Stop>FRT>Gal4,UAS-GFP (Flip-out). 3–5 day old mated female flies were heat-shocked for 45 minutes at 37°C to induce somatic recombination. Flies were returned back to 25°C until dissection.
Fluorescent RNA in situ hybridization and immunochemistry in Drosophila guts
Primers used to amplified genomic DNA to make Tis11 specific probe: Forward 5′-CTCACACGAACCATTTCACAGCC-3′ and Reverse 5′-TGCGGCAGTATTCCGTCTTGTACT-3′. The 335 base pair PCR product was cloned into a TOPO-TA vector and directionality of insert confirmed by PCR. Sp6 or T7 polymerases were used to synthesize sense and antisense probes.
In situ hybridization was performed using a protocol adapted from Lecuyer et al. (Lecuyer et al., 2008).
For immuno-staining, fly intestines were dissected in PBS and fixed at room temperature for 45 minutes in 100 mM glutamic acid, 25 mM KCl, 20 mM MgSO4, 4 mM Sodium Phosphate, 1 mM MgCl2, 4% formaldehyde. All subsequent incubations were done in PBS, 0.5% BSA, 0.1% TritonX-100 at 4°C. For Delta staining, guts were fixed by the methanol-heptane fixation method as previously described (Lin et al., 2008).
The following primary antibodies were obtained from the Developmental Studies Hybridoma Bank: anti-Prospero, anti-Armadillo and anti-β-galactosidase and used 1:250, 1:100 and 1:500 respectively. Rabbit anti-pH3 from Millipore, 1:1000. The rabbit anti-Tis11 was obtained from C. Gueydan and used 1:500. The rat anti-Delta antibody was obtained from M. Rand and used 1:200. The Fluorescent secondary antibodies were obtained from Jackson Immunoresearch. Hoechst was used to stain DNA.
Confocal images were collected using a Leica SP5 confocal system or a Zeiss 510 confocal system and processed using the Leica software or Zeiss software and Adobe Photoshop CS5.
Reporter analysis in cultured cells
S2 cells were seeded at a density of 1 × 106 cells/ml in a 6 well plate. The next day transfections were performed with 500ng of total DNA in cells using the calcium phosphate method. When the pMT copper-inducible expression system (Invitrogen) was used to express Tis11, medium was supplemented with +/− 500μM CuSO4. Cells were harvested 48hrs after transfection, lysed and proteins were analyzed via Western Blot.
FACS sorting and RNAseq
The protocol for FACS sorting and transcriptional profiling of Drosophila intestinal progenitor cells has been previously described (Wang et al., 2015). Briefly, Wild-type Flies (esgGal4ts,UAS-GFP > +), flies with progenitor cells expressing dsRNA against Tis11 (esgGal4ts,UAS-GFP > UAS-Tis11RNAi) and flies with progenitor cells over-expressing Tis11 (esgGal4ts,UAS-GFP > UAS-Tis11FLAG) were exposed to Ecc15 for 24 hours (5% sucrose solution), followed by GFP+ labeled ISCs/EBs FACS sorting. Total RNA was then extracted using Trizol (Invitrogen) and used as template to generate RNA-seq libraries for Illumina sequencing. Expression was normalized as reads per kbp per million reads (RPKPM) using the Illumina software. Only genes reliably detected in sorted cells (RPKPM ≥ 5 in at least one library) were analyzed further. Genes were defined as negatively regulated by Tis11 when Tis11Flag/Control ≤ 2/3 and Tis11RNAi/Control ≥ 1.5. Conversely, genes were considered positively regulated by Tis11 when Tis11Flag/Control 1.5 ≥ and Tis11RNAi/Control ≤ 2/3. Gen Ontology enrichment analysis was performed using the GOrilla online software. For Tis11, relative expression was calculated using read counts for exon, which is not included in the hairpin construct used for RNAi.
RIP-Seq
RIP-Seq was performed by the previously described RNA-immunoprecipitation protocol (Keene et al., 2006) with the following modifications: esgGAL4ts,UAS-GFP > UAS-Tis11FLAG, UAS-Pop2RNAi were infected with Ecc15 after 3 day at 29°C. 24 hours after infection, flies were snap frozen in liquid nitrogen and crushed with a tissue homogenizer. A volume equal to the volume of pulverized fly tissue of polysome lysis buffer was added to the tissue homogenate and lysed as directed by protocol. Protein G Dynabeads (Invitrogen) were coated with either 2.5 μg Mouse anti-FLAG antibodies (Sigma-Aldrich) or 2.5 μg Mouse IgG2A proteins (Sigma-Aldrich). Fly lysates were added to prepared beads, in duplicate (FLAG1, FLAG2, IgG1, IgG2), and incubated as directed. Following the IP, beads were washed and a fraction was set aside to analyze for the enrichment of Tis11 protein. The remainder was treated with Trizol (Invitrogen) to extract the Tis11-bound mRNAs and generate RIP-Seq libraries for Illumina sequencing. Expression was normalized as reads per kbp per million reads (RPKPM) using the Illumina software. RNAs were defined as reproducibly enriched in Flag pull-down when at least 3 out of the 4 comparisons (FLAG1 vs IgG1, FLAG1 vs IgG2, FLAG2 vs IgG1, FLAG2 vs IgG2) ≥ 1.5. Only mRNAs detected in sorted cells (RPKPM ≥ 5 in at least one RNAseq library) were considered as potential Tis11 targets in ISC/EBs. For enrichment calculation, RPKPM values were divided by the average RPKPM value of the 2 IgG beads samples. All sequencing data have been deposited to the Gene Expression Omnibus (GSE97216).
Supplementary Material
Highlights.
Tis11 RNA binding protein is expressed in Drosophila adult intestinal progenitors
Tis11-mediated mRNA decay is essential for the re-entry of ISC into quiescence
Tis11 expression and activity are dynamically regulated during tissue repair
Tis11 influences the expression of several essential proliferative pathways
Acknowledgments
We are grateful to Yanyan Qi for technical assistance. This work was funded by the National Institute of General Medical Sciences (5R01GM108712 to B.B) and National Institute on Aging (5F31AG044097 to L.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell stem cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyaz A, Jasper H. Intestinal inflammation and stem cell homeostasis in aging Drosophila melanogaster. Front Cell Infect Microbiol. 2013;3:98. doi: 10.3389/fcimb.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyaz A, Li H, Jasper H. Haemocytes control stem cell activity in the Drosophila intestine. Nat Cell Biol. 2015;17:736–748. doi: 10.1038/ncb3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baou M, Jewell A, Murphy JJ. TIS11 family proteins and their roles in posttranscriptional gene regulation. J Biomed Biotechnol. 2009 doi: 10.1155/2009/634520. 634520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell stem cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell stem cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Jasper H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development (Cambridge, England) 2011;138:1045–1055. doi: 10.1242/dev.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan SE, Kuwano Y, Alkharouf N, Blackshear PJ, Gorospe M, Wilson GM. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA, Connolly JE, Rigby WF. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J Immunol. 2004;172:7263–7271. doi: 10.4049/jimmunol.172.12.7263. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes & development. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Kuraishi T, Lemaitre B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 2010;8:152. doi: 10.1186/1741-7007-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Lemaitre B. Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nature reviews Microbiology. 2013;11:615–626. doi: 10.1038/nrmicro3074. [DOI] [PubMed] [Google Scholar]
- Cairrao F, Halees AS, Khabar KS, Morello D, Vanzo N. AU-rich elements regulate Drosophila gene expression. Molecular and cellular biology. 2009;29:2636–2643. doi: 10.1128/MCB.01506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science (New York, NY. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nature reviews. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Dobson AJ, Houtz PL, Glasser C, Revah J, Korzelius J, Patel PH, Edgar BA, Buchon N. Regional Cell-Specific Transcriptome Mapping Reveals Regulatory Complexity in the Adult Drosophila Midgut. Cell reports. 2015;12:346–358. doi: 10.1016/j.celrep.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Guo Z, Driver I, Ohlstein B. Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J Cell Biol. 2013;201:945–961. doi: 10.1083/jcb.201302049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausburg MA, Doles JD, Clement SL, Cadwallader AB, Hall MN, Blackshear PJ, Lykke-Andersen J, Olwin BB. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. eLife. 2015;4:e03390. doi: 10.7554/eLife.03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochmuth CE, Biteau B, Bohmann D, Jasper H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell stem cell. 2011;8:188–199. doi: 10.1016/j.stem.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K, et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol. 2010;11:717–724. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kim J, Kim YJ. Requirement of RBP9, a Drosophila Hu homolog, for regulation of cystocyte differentiation and oocyte determination during oogenesis. Molecular and cellular biology. 1999;19:2505–2514. doi: 10.1128/mcb.19.4.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers A, Twyffels L, Soin R, Wauquier C, Kruys V, Gueydan C. Post-transcriptional Regulation of Genes Encoding Anti-microbial Peptides in Drosophila. J Biol Chem. 2009;284:8973–8983. doi: 10.1074/jbc.M806778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Parthasarathy N, Krause HM. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol Biol. 2008;420:289–302. doi: 10.1007/978-1-59745-583-1_18. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;193:257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science (New York, NY. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science (New York, NY. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Sohn BH, Park IY, Lee JJ, Yang SJ, Jang YJ, Park KC, Kim DJ, Lee DC, Sohn HA, Kim TW, et al. Functional switching of TGF-beta1 signaling in liver cancer via epigenetic modulation of a single CpG site in TTP promoter. Gastroenterology. 2010;138:1898–1908. doi: 10.1053/j.gastro.2009.12.044. [DOI] [PubMed] [Google Scholar]
- Spasic M, Friedel CC, Schott J, Kreth J, Leppek K, Hofmann S, Ozgur S, Stoecklin G. Genome-wide assessment of AU-rich elements by the AREScore algorithm. PLoS genetics. 2012;8:e1002433. doi: 10.1371/journal.pgen.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suswam E, Li Y, Zhang X, Gillespie GY, Li X, Shacka JJ, Lu L, Zheng L, King PH. Tristetraprolin down-regulates interleukin-8 and vascular endothelial growth factor in malignant glioma cells. Cancer Res. 2008;68:674–682. doi: 10.1158/0008-5472.CAN-07-2751. [DOI] [PubMed] [Google Scholar]
- Tchen CR, Brook M, Saklatvala J, Clark AR. The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. J Biol Chem. 2004;279:32393–32400. doi: 10.1074/jbc.M402059200. [DOI] [PubMed] [Google Scholar]
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyffels L, Wauquier C, Soin R, Decaestecker C, Gueydan C, Kruys V. A masked PY-NLS in Drosophila TIS11 and its mammalian homolog tristetraprolin. PLoS One. 2013;8:e71686. doi: 10.1371/journal.pone.0071686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignudelli T, Selmi T, Martello A, Parenti S, Grande A, Gemelli C, Zanocco-Marani T, Ferrari S. ZFP36L1 negatively regulates erythroid differentiation of CD34+ hematopoietic stem cells by interfering with the Stat5b pathway. Mol Biol Cell. 2010;21:3340–3351. doi: 10.1091/mbc.E10-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ryoo HD, Qi Y, Jasper H. PERK Limits Drosophila Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress. PLoS genetics. 2015;11:e1005220. doi: 10.1371/journal.pgen.1005220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh PA, Yang WH, Chiang PY, Wang SC, Chang MS, Chang CJ. Drosophila eyes absent is a novel mRNA target of the tristetraprolin (TTP) protein DTIS11. Int J Biol Sci. 2012;8:606–619. doi: 10.7150/ijbs.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LE, Dixon DA. Posttranscriptional Regulation of Cyclooxygenase 2 Expression in Colorectal Cancer. Curr Colorectal Cancer Rep. 2010;6:60–67. doi: 10.1007/s11888-010-0044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Han L, Singh SR, Liu H, Neumuller RA, Yan D, Hu Y, Liu Y, Liu W, Lin X, et al. Genome-wide RNAi Screen Identifies Networks Involved in Intestinal Stem Cell Regulation in Drosophila. Cell reports. 2015;10:1226–1238. doi: 10.1016/j.celrep.2015.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Prak L, Rayon-Estrada V, Thiru P, Flygare J, Lim B, Lodish HF. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature. 2013;499:92–96. doi: 10.1038/nature12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, Koromilas AE, Crist C. Phosphorylation of eIF2alpha Is a Translational Control Mechanism Regulating Muscle Stem Cell Quiescence and Self-Renewal. Cell stem cell. 2016;18:79–90. doi: 10.1016/j.stem.2015.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.