Abstract
The spread of mcr-1 in the United States remains poorly defined. mcr-1-producing Escherichia coli that also carried blaSHV-12 was detected in a hospitalized patient. No additional cases were identified during screening of 801 Gram-negative isolates. Genomic sequencing identified an IncX4 mcr-1- harboring plasmid and ST117 clonal background associated with avian pathogenic E coli.
Keywords: avian pathogenic E coli, colistin, IncX4 plasmid, mcr-1, polymyxin B
Polymyxins (including polymyxin B and colistin) are considered antibiotics of “last resort” for treating infections with multidrug-resistant Gram-negative bacteria (MDR-GNB). mcr-1, a plasmid-associated gene for colistin resistance, was described in China in 2015 [1]. The possibility of horizontal transfer of colistin resistance has raised significant concern regarding spread of mcr-1. After initially being described in isolates from food animals, human carriage of mcr-1 was noted in Escherichia coli and Klebsiella pneumoniae in urine and blood samples from hospitalized patients. By January 2016, cases had been reported from 19 countries worldwide [2]. In the United States, 12 human mcr-1 cases have been reported in 9 states to date [3–7]. Most mcr-1 infections have occurred in patients that had not received polymyxins, leading to speculation that colonization with mcr-1 may have occurred after ingestion of contaminated meat products [2]. Underdetection of colistin resistance is possible because many microbiology laboratories do not perform routine colistin susceptibility testing on carbapenem-susceptible isolates [8]. An mcr-1-producing E coli isolate was detected during our center’s participation in the SENTRY Antimicrobial Surveillance Program [3]. In this study, we describe the clinical characteristics, molecular survey, and bacterial genomics of this mcr-1 infection from May 2015.
METHODS
After detection of the mcr-1-positive isolate, we conducted a molecular survey for mcr-1-positive isolates with retrospective and prospective components. Our institution is a 2200-bed urban campus in Northern Manhattan. The microbiology laboratory processes samples from both inpatients and outpatients from our institution and a smaller affiliated community hospital also situated in Northern Manhattan. For the retrospective survey, we screened routinely collected MDR-GNB clinical isolates from 2011 to 2016 for the presence of mcr-1 by polymerase chain reaction (PCR) [1]. These included (1) all carbapenem-resistant isolates that had undergone polymyxin B susceptibility testing and were found to be resistant to polymyxin B (polymyxin B mean inhibitory concentration [MIC], >2 mcg/mL [9]) as well as (2) all E coli isolates resistant to third-generation cephalosporins (whether polymyxin B susceptibility was performed or not). For the prospective survey, we conducted polymyxin B susceptibility testing on all Gram-negative isolates resistant to third-generation cephalosporins for 1 month. Gram-negative bacteria known to be intrinsically polymyxin resistant were not tested. Polymyxin B susceptibility testing was performed with E-test (bioMerieux, Durham, NC). Approval was granted by the Columbia University Institutional Review Board (IRB-AAAQ9741).
Whole-genome sequencing (WGS) of the mcr-1-positive isolate was performed using the HiSeq 2500 sequencer (Illumina, San Diego, CA). Sequences obtained were de novo assembled in contigs using Velvet after optimization of settings with VelvetOptimiser [10, 11]. Automatic annotation of the genome was performed using RAST [12]. The multilocus sequence type (MLST), resistome, and plasmid type were characterized using ResFinder 2.1, PlasmidFinder 1.3, and MLST 1.8, respectively [13–15]. Additional characterization of chromosomal resistance determinants was performed using CARD Resistance Gene Identifier [16]. Findings were confirmed by performing pairwise alignment of the relevant genes to wild-type E coli K-12 MG1655 (GenBank accession no. U00096) in Geneious 9.0.2 [17]. The GenBank Genome database was searched for complete E coli genomes, and these were characterized using MLST 1.8 [15]. Multiple sequence alignment of the study isolate and reference E coli ST117 genomes was performed using Mauve in the progressiveMauve setting [18, 19]. Genomes were visualized with BRIG [20]. Plasmid sequences were aligned using Geneious 9.0.2 and visualized using EasyFig [17, 21].
The complete nucleotide sequences of plasmid pKP1749_MCR1 has been deposited as GenBank accession no. CP0000000. The draft genome of E coli strain KP1749 was deposited as GenBank accession no. MAJK00000000.
RESULTS
The patient was a man in his 50s with a past history of sickle cell anemia, recurrent cholangitis, and cholecystectomy who was admitted for elective biliary surgery in May 2015. The patient had no prior documented history of receiving polymyxins but had received a course of ciprofloxacin for cholangitis during an admission in another institution 4 months prior. He had a history of travel to the Dominican Republic in 2013. The patient was noted to be febrile at 38.2°C on postoperative day 1. Vancomycin 1 gram q12h and meropenem 500 mg q8h were commenced empirically according to the patient’s renal function. The patient’s modified Pitt bacteremia score was 3 [22].
Blood cultures taken during the febrile episode and intraoperative biliary fluid cultures were positive for E coli with the following MICs on the Vitek-2 instrument (bioMerieux): ceftriaxone 2 mcg/mL, ceftazidime 16 mcg/mL, piperacillin-tazobactam <4 mcg/mL, cefepime <1 mcg/mL, meropenem <0.25 mcg/mL, amikacin <2 mcg/mL, tobramycin <1 mcg/mL, gentamicin <1 mcg/mL, levofloxacin >8 mcg/mL, trimethoprim/sulfamethoxazole >320 mcg/mL, and minocycline 4 mcg/mL. The isolate was resistant to fosfomycin on disk diffusion susceptibility testing with a zone of 18 mm noted [23]. Ertapenem E-test indicated an MIC of 0.006 mcg/mL (bioMerieux). Polymyxin B susceptibility testing was not initially performed due to susceptibility of the isolate to many β-lactam antibiotics. Polymyxin B susceptibility testing and mcr-1 PCR were performed retrospectively after detection of mcr-1 in the SENTRY program. Both isolates had a polymyxin B MIC of 4 mcg/mL and were mcr-1 positive by PCR [24]. On postoperative day 5, the patient’s antibiotic regimen was switched to cefepime 2 grams q8h, metronidazole 500 mg q8h, and fluconazole 400 mg q24h, and he completed 2 weeks of therapy with that regimen. The patient was discharged home on postoperative day 14 having had no further fever, abdominal symptoms, or cultures positive for E coli.
For the retrospective survey, mcr-1 PCR was performed on 633 E coli isolates (selected as described above) and 171 polymyxin B-resistant isolates (163 K pneumoniae, 1 E coli, and 7 Acinetobacter baumanii isolates, respectively). Median polymyxin B MIC for polymyxin B-resistant isolates was 16 mcg/mL (interquartile range, 12–32 mcg/mL). All isolates tested were negative for mcr-1. For the prospective survey, 94 Gram-negative isolates were identified over the 1-month period, and all were tested for polymyxin B resistance according to criteria described above. No isolate had a polymyxin MIC >2 mcg/mL, hence none underwent further mcr-1 PCR.
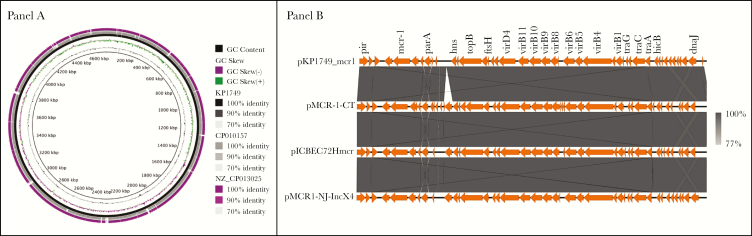
We then carried out WGS on the blood culture isolate (named KP1749). In addition to mcr-1, the isolate harbored resistance genes for β-lactams (blaSHV-12, blaTEM-1B), aminoglycosides [aph(3’)-IIa, aph(6)-Ic, aadA2, aph(3’)-Ia, strA, strB], sulphonamides (sul1, sul2), trimethoprim (dfrA12), chloramphenicol (catA1), macrolides (mphA), fosfomycin (fosA), and tetracycline [tet(A)]. Of note, blaSHV-12 codes for an extended-spectrum β-lactamase [25]. Chromosomal fluoroquinolone resistance mutations were detected in gyrA (S83L and D87N) and parC (S80I) [26]. KP1749 was compared with the 2 E coli ST117 isolates with complete genomes available on GenBank (GenBank accession nos. CP010157.1 from China and NZ_CP013025.1 from New York) (see Figure 1, Panel A). KP1749 shared 99.7% and 99.65% nucleotide identity, respectively, with the isolates across the core genome. Multilocus sequence type analysis indicated that the isolate was ST117, an important avian pathogenic E coli lineage also noted to cause disease in humans [27].
Figure 1.
(Panel A) Comparative genomic analysis of KP1749 compared with 2 reference Escherichia coli ST117 genomes identified in GenBank. (Panel B) Backbone of pKP1749_MCR1 plasmid (GenBank accession no. CP0000000) of size 33.3 kb carrying mcr-1 from a human E coli strain isolated in New York. pKP1749_MCR1 was compared with plasmid pMCR-1-CT (GenBank accession no. CP018773) from an E coli strain isolated in Connecticut, pICBEC72Hmcr IncX4 plasmid (GenBank accession no. CP015977) carrying the mcr-1 gene in a human E coli strain isolated in Brazil, and plasmid pMCR1-NJ-IncX4 (GenBank accession no. KX447768) from a human E coli strain isolated in New Jersey.
The mcr-1 gene was carried on an IncX4 plasmid (named pKP1749_MCR1) 33.3 kbp in length. There was a high level of architectural conservation with mcr-1-carrying IncX4 plasmids from Brazil, Connecticut, and New Jersey (GenBank accession nos. CP015977, CP018773, and KX447768, respectively) (Figure 1, Panel B) [5, 28]. pKP1749_MCR1 lacked an IS26 insertion sequence present in the other plasmids but otherwise differed 8 single-nucleotide polymorphisms (SNPs) to pMCR-1-CT (Connecticut), 10 SNPs to pICBEC72Hmcr (Brazil), and 95 SNPs to pMCR1-NJ-IncX4 (New Jersey). An IncI1 plasmid ~113.2 kbp in length was found to harbor blaSHV-12 and resistance genes for multiple other antibiotic classes: aminoglycosides, aph(3’)-IIa, aph(6)-Ic, aadA2, aph(3’)-Ia, strA, and strB; trimethoprim, dfrA12; sulphonamides, sul1; macrolides, mphA.
DISCUSSION
We describe a human infection with an mcr-1-producing E coli belonging to the ST117 lineage. This is one of the first cases of mcr-1 noted in a US hospital setting and preceded the initial description of mcr-1 in 2015 [1]. We were unable to identify further cases during a molecular survey for mcr-1, suggesting lack of widespread local circulation of the plasmid. In addition, other mcr-1 isolates from the United States are of different clonal background (ST405 and ST457) [4, 5].
The ST117 clonal background of this isolate raises the possibility that it was acquired in the community from an avian source. Escherichia coli ST117 is a major pathogen in the poultry industry and may form a reservoir of human extraintestinal pathogenic E coli and antimicrobial resistance [27]. mcr-1 and multiple mcr-1-carrying IncX4 plasmids have also been identified in poultry isolates from Asia, Europe, Latin America, and Africa [28, 29]. mcr-1 was detected in 12 of 980 isolates in a collection of avian pathogenic E coli [30]. There are now several reports that suggest a possible association between E coli ST117 and mcr-1 carriage. Initially, 2 of 3 isolates carrying mcr-1 during a 2016 survey in the Netherlands were E coli ST117 from retail chicken meat [31]. Two recent Chinese studies reported 3 and 2 mcr-1 carrying E coli ST117 in human samples, respectively [32, 33].
Therefore, poultry may be a reservoir of E coli carrying mcr-1, particularly in regions where colistin is routinely used in animal husbandry such as China and Latin America [34]. This patient had traveled to the Dominican Republic, and much of our center’s patient population is of Dominican background, suggesting a possible epidemiological link [35]; however, no cases of mcr-1 have yet been reported from that country. A human mcr-1 case from Connecticut reported recent travel to the Caribbean and consumption of chicken and goat meat purchased from a live animal market [6]. The high degree of similarity between pKP1749_MCR1 and IncX4 mcr-1 carrying plasmids from Connecticut, New Jersey, and Brazil suggests the contribution of this plasmid in disseminating mcr-1 through the Americas [28].
CONCLUSIONS
The spread of mcr-1 to carbapenem-resistant Enterobacteriaceae-endemic regions such as the north-eastern United States is an active concern. An E coli isolate harboring both mcr-1 and blaNDM-5 has already been noted in New Jersey [5]. Fortunately, isolate KP1749 retained susceptibility to multiple β-lactam agents, thus allowing adequate treatment and a satisfactory outcome. Since the description of mcr-1, several other transferable mechanisms of colistin resistance have been identified [2]. Therefore, we need to develop more active surveillance approaches for detecting resistance to colistin. Although phenotypic susceptibility testing forms the cornerstone, there are limitations in accuracy depending on the method used [8]. This problem is further compounded by the finding that most mcr-1-producing isolates have lower colistin MICs in the range of 2–4 mcg/mL, and some isolates that are phenotypically susceptible may produce mcr-1 [2]. Therefore, our study may have underdetected mcr-1, both due to issues with polymyxin susceptibility testing and because our screening strategy required isolates to initially have resistance to third-generation cephalosporins. Integrating molecular techniques such as WGS may have an increasingly important role in stopping the spread of transferable colistin resistance by providing valuable epidemiological data and allowing detection of multiple resistance mechanisms.
Acknowledgments
Disclaimer. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Financial support. This work was funded by grants from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01 AI116939; to A-C. U.). Merck provided funding for research.
Potential conflicts of interest. All authors: No reported conflicts of interest.All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed
References
- 1. Liu YY, Wang Y, Walsh TR et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16:161–8. [DOI] [PubMed] [Google Scholar]
- 2. Caniaux I, van Belkum A, Zambardi G et al. MCR: modern colistin resistance. Eur J Clin Microbiol Infect Dis 2017; 36:415–20. [DOI] [PubMed] [Google Scholar]
- 3. Castanheira M, Griffin MA, Deshpande LM et al. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob Agents Chemother 2016; 60:5623–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McGann P, Snesrud E, Maybank R et al. Escherichia coli harboring mcr-1 and blaCTX-M on a Novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 2016; 60:4420–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mediavilla JR, Patrawalla A, Chen L et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio 2016; 7:pii: e01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vasquez AM, Montero N, Laughlin M et al. Investigation of Escherichia coli harboring the mcr-1 resistance gene—Connecticut, 2016. MMWR Morb Mortal Wkly Rep 2016; 65:979–80. [DOI] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention. Tracking mcr-1 2017 Available at: https://www.cdc.gov/drugresistance/tracking-mcr1.html. Accessed 22 April 2017.
- 8. Humphries RM. Susceptibility testing of the polymyxins: where are we now? Pharmacotherapy 2015; 35:22–7. [DOI] [PubMed] [Google Scholar]
- 9. European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters 2013 Available at: http://www.eucast.org. Accessed 14 December 2016.
- 10. VelvetOptimiser 2012 Available at: http://www.vicbioinformatics.com/software.velvetoptimiser.shtml. Accessed 21 December 2016.
- 11. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aziz RK, Bartels D, Best AA et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carattoli A, Zankari E, García-Fernández A et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58:3895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zankari E, Hasman H, Cosentino S et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67:2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larsen MV, Cosentino S, Rasmussen S et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 2012; 50:1355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia B, Raphenya AR, Alcock B et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017; 45:D566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kearse M, Moir R, Wilson A et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 2004; 14:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 2010; 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011; 12:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics 2011; 27:1009–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paterson DL, Ko WC, Von Gottberg A et al. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum beta-lactamases. Clin Infect Dis 2004; 39:31–7. [DOI] [PubMed] [Google Scholar]
- 23. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard M2-A10. 10th ed Wayne, PA: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 24. National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Testing. NCCLS Approved Standard M100-S9. Wayne, PA: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 25. Nüesch-Inderbinen MT, Kayser FH, Hächler H. Survey and molecular genetics of SHV beta-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob Agents Chemother 1997; 41:943–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bagel S, Hüllen V, Wiedemann B, Heisig P. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob Agents Chemother 1999; 43:868–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manges AR, Johnson JR. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin Infect Dis 2012; 55:712–9. [DOI] [PubMed] [Google Scholar]
- 28. Fernandes MR, McCulloch JA, Vianello MA et al. First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli sequence type 101 isolate from a human infection in Brazil. Antimicrob Agents Chemother 2016; 60:6415–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill 2016; 21:30155. [DOI] [PubMed] [Google Scholar]
- 30. Lima Barbieri N, Nielsen DW, Wannemuehler Y et al. mcr-1 identified in avian pathogenic Escherichia coli (APEC). PLoS One 2017; 12:e0172997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kluytmans-van den Bergh MF, Huizinga P, Bonten MJ et al. Presence of mcr-1-positive Enterobacteriaceae in retail chicken meat but not in humans in the Netherlands since 2009. Euro Surveill 2016; 21:30149. [DOI] [PubMed] [Google Scholar]
- 32. Wang Y, Tian GB, Zhang R et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis 2017; 17:390–9. [DOI] [PubMed] [Google Scholar]
- 33. Quan J, Li X, Chen Y et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis 2017; 17:400–10. [DOI] [PubMed] [Google Scholar]
- 34. Fernandes MR, Moura Q, Sartori L et al. Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro Surveill 2016; 21: doi: 10.2807/1560-7917.ES.2016.21.17.30214. [DOI] [PubMed] [Google Scholar]
- 35. Uhlemann AC, McAdam PR, Sullivan SB et al. Evolutionary dynamics of pandemic methicillin-sensitive Staphylococcus aureus ST398 and its international spread via routes of human migration. MBio 2017; 8:pii: e01375-16. [DOI] [PMC free article] [PubMed] [Google Scholar]