Abstract
Insulin-like peptides (ILPs) play important roles in growth and metabolic homeostasis, but have also emerged as key regulators of stress responses and immunity in a variety of vertebrates and invertebrates. Furthermore, a growing literature suggests that insulin signaling-dependent metabolic provisioning can influence host responses to infection and affect infection outcomes. In line with these studies, we previously showed that knockdown of either of two closely related, infection-induced ILPs, ILP3 and ILP4, in the mosquito Anopheles stephensi decreased infection with the human malaria parasite Plasmodium falciparum through kinetically distinct effects on parasite death. However, the precise mechanisms by which ILP3 and ILP4 control the response to infection remained unknown. To address this knowledge gap, we used a complementary approach of direct ILP supplementation into the blood meal to further define ILP-specific effects on mosquito biology and parasite infection. Notably, we observed that feeding resulted in differential effects of ILP3 and ILP4 on blood-feeding behavior and P. falciparum development. These effects depended on ILP-specific regulation of intermediary metabolism in the mosquito midgut, suggesting a major contribution of ILP-dependent metabolic shifts to the regulation of infection resistance and parasite transmission. Accordingly, our data implicate endogenous ILP signaling in balancing intermediary metabolism for the host response to infection, affirming this emerging tenet in host–pathogen interactions with novel insights from a system of significant public health importance.
Introduction
Insulin/insulin-like growth factor signaling (IIS) is highly conserved from nematodes to mammals and insulin-like peptides (ILPs) regulate a wide array of physiological processes [1–3]. Our previous work demonstrated that IIS modulates diverse facets of mosquito biology, including lifespan, response to oxidative stress, autophagy, midgut stem cell activity, host-seeking behavior, and immunity [4–15]. Other groups have examined the role of IIS in controlling mosquito reproduction, blood meal digestion, and metabolism [16–20]. Despite this understanding, little is understood about the roles of endogenous mosquito ILPs in resistance to infection.
In the midgut of Anopheles stephensi, expression of ILP genes is induced in response to human insulin and infection with the human malaria parasite Plasmodium falciparum [21]. We previously showed that knockdown of either of two infection-induced ILPs, ILP3 or ILP4, in the midgut decreased P. falciparum infectivity through kinetically distinct effects on A. stephensi innate immune defenses [13]. Specifically, knockdown of ILP4 increased early expression of antiparasite genes (1–6 h post-infection) and increased killing of ookinetes prior to invasion, whereas knockdown of ILP3 increased anti-parasite gene expression at a later time (24 h post-infection), boosting killing of parasites during and after invasion [13]. While we predicted that decreased P. falciparum infectivity following ILP knockdown was due, at least in part, to increased expression of antiparasite effector genes in the midgut, the specific mechanisms by which ILPs regulate mosquito resistance to infection remained unconfirmed.
In Drosophila melanogaster, antimicrobial responses are largely controlled by NF-κB-dependent transcription downstream of the Toll and immune deficiency pathways, as well as JAK/STAT signaling [22,23]. These pathways are networked with IIS-associated phosphatidylinositol 3-kinase/Akt signaling and the downstream transcription factor FOXO as well as c-Jun N-terminal kinase ( JNK) and extracellular signal-regulated kinase (ERK) [12,24–29]. In addition to directly regulating antiparasite responses, IIS controls a variety of cellular processes that are intrinsically linked to infection resistance and that could contribute to the effects of ILPs on P. falciparum infection. For example, IIS is involved in the regulation of midgut epithelial barrier homeostasis through control of autophagy and cell renewal [20,30,31], two processes implicated in pathogen resistance in both mammals and invertebrates [8,32–37]. Furthermore, IIS is widely known to control carbohydrate and lipid metabolism across the same range of organisms [16–17,38], and a growing body of literature suggests that alterations in central metabolism are driving forces in the control of inflammatory responses to infection [39–45]. In particular, metabolic shifts may occur to maximally allocate available resources to immunity [39], but may also be due to pathogenic processes stemming from pathogen colonization [40–42]. Accordingly, changes in mosquito metabolism by ILPs produced in the midgut during P. falciparum infection could contribute to their effects on parasite resistance and transmission.
Given the aforementioned possibilities, we sought to confirm our understanding of ILP regulation of P. falciparum infectivity and to identify the effects of ILP3 and ILP4 on the broader host response to infection. To this end, we examined various outputs of immunity, cell signaling, and intermediary metabolism in the midgut of mosquitoes provisioned with ILPs. Our results suggest that ILP3 and ILP4 differentially regulate P. falciparum development in the mosquito through diverse effects on midgut physiology, with metabolic shifts acting as key drivers of infection resistance.
Results
ILP3 and ILP4 differentially affect P. falciparum infectivity in A. stephensi
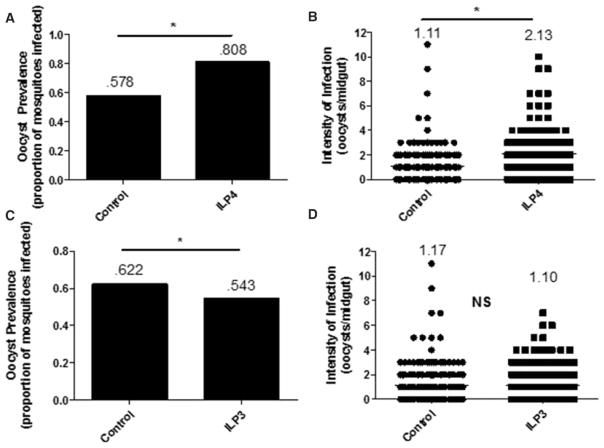
We have previously shown that knockdown of either ILP3 or ILP4 can significantly reduce the prevalence and intensity of P. falciparum infection in A. stephensi, suggesting that mosquito ILPs are immunosuppressive [13]. While the primary aim of the present study was to determine the mechanisms by which these ILPs modulate resistance to infection, we first sought to extend previous findings by characterizing the phenotypic effects of these peptides using ILP provisioning as a complement to our knockdown studies. To this end, we quantified P. falciparum oocysts in mosquitoes fed ILP3 or ILP4 in an infected blood meal using a design based on our previous studies with human insulin. Specifically, we showed that provision of 170 pM human insulin can activate IIS in the mosquito midgut to facilitate parasite development [12]. Accordingly, we used this concentration of ILPs in our feeding assays. Provision of 170 pM ILP4 significantly increased the prevalence (proportion of mosquitoes infected) of infection (Figure 1A) from 57.8 to 80.8% and the intensity (oocysts/midgut) of infection (Figure 1B) from 1.11 to 2.13 oocysts/midgut when compared with controls, as predicted by our recent work [13]. Surprisingly, feeding of 170 pM ILP3 significantly decreased the prevalence of infection from 62.2 to 54.3% (Figure 1C), but had no effect on the intensity of infection relative to controls (Figure 1D). Therefore, whereas both ILP3 and ILP4 are necessary for optimal parasite development [13], only ILP4 is sufficient to increase parasite growth. Together, these data suggested that ILP3 and ILP4 differentially regulate mosquito physiology, possibly through the activation of distinct cell signaling pathways to control resistance to P. falciparum infection.
Figure 1. ILP3 and ILP4 differentially affect P. falciparum infectivity in A. stephensi.
Cohorts of A. stephensi were fed (A and B) 170 pM ILP4 or (C and D) 170 pM ILP3 in a P. falciparum-infected blood meal. Ten days later, midguts were dissected and stained for visualization of oocysts to determine the prevalence (percentage of mosquitoes infected) and intensity of infection (number of oocysts/midgut). All experiments were independently replicated with 3–7 cohorts of 60–120 mosquitoes each. Infection prevalence and intensity data were analyzed by χ2 goodness-of-fit test and nonparametric Mann–Whitney test, respectively, to determine differences between groups. Numbers above the graphs indicate the mean prevalence or intensity. *P-values were deemed significant when P ≤ 0.05.
ILP3 and ILP4 reduce NF-κB-dependent immune gene expression in response to P. falciparum
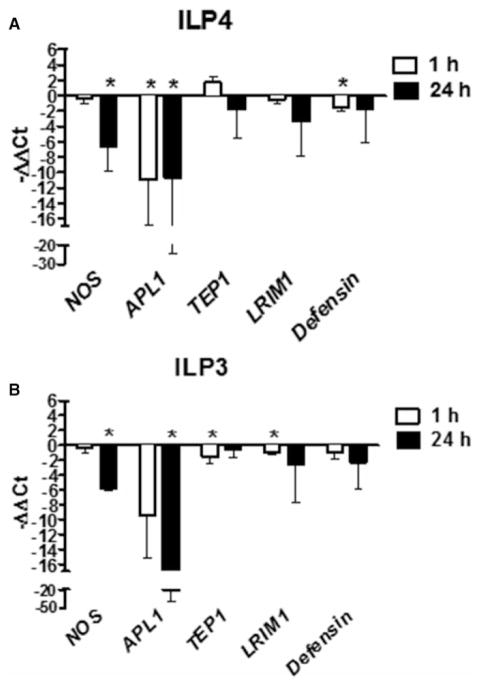
Our infection data did not preclude the regulation of immunity and resistance to infection by ILP3, but rather suggested a difference in the mechanism of action between ILP3 and ILP4. To address this, we analyzed the expression of known antiparasite effector genes [46] in the midgut of ILP-fed mosquitoes at 1 and 24 h post-feeding of a blood meal containing soluble P. falciparum products (PfPs) [12,13] and supplemented with ILP3 or ILP4 (Figure 2). We found that both ILP3 and ILP4 repressed transcript levels of NF-κB-dependent immune genes to varying degrees. Specifically, expression of nitric oxide synthase (NOS) was significantly reduced at 24 h after feeding of ILP3 or ILP4. Expression of APL1 was also significantly decreased at 1 and 24 h post-ILP4 feeding and at 1 h post-ILP3 feeding. Although expression levels of TEP1 and LRIM1 at 1 h postfeeding were unaffected by ILP4, and both were significantly reduced by ILP3 at this time point. In addition, expression of defensin was significantly reduced by ILP4 feeding at 1 h postfeeding. While all other changes were not significant, all trended toward reduced expression with the exception of a small increase in TEP1 expression levels at 1 h post-ILP4 feeding. These results demonstrate that while ILP3 and ILP4 have distinct effects on parasite infection, both peptides suppressed transcript levels of NF-κB-dependent immune genes to a similar degree. These data suggested that the differential effects of ILP3 and ILP4 on parasite infection are not due solely to effects on host immunity.
Figure 2. ILP3 and ILP4 reduce NF-κB-mediated immune gene expression in response to P. falciparum.
Cohorts of A. stephensi were fed either (A) 170 pM ILP4 or (B) 170 pM ILP3 in a meal of human RBCs supplemented with soluble PfPs. Midguts were dissected at 1 and 24 h postfeeding for RNA extraction and qRT-PCR analysis of NOS, Defensin, LRIM1, APL1, and TEP1 gene expression. Data from 3 to 7 independent cohorts of 45–60 mosquitoes each are represented as mean −ΔΔCt values ± SEM on a Log2 scale (1 Ct value = 2-fold change, PfPs-treated controls set at 0). Data were analyzed by t-test to determine differences between ILP-treated and control groups (PfPs only; set at 0). *P-values were deemed significant when P ≤ 0.05.
ILP3 and ILP4 differentially induce MAPK phosphorylation in the midgut
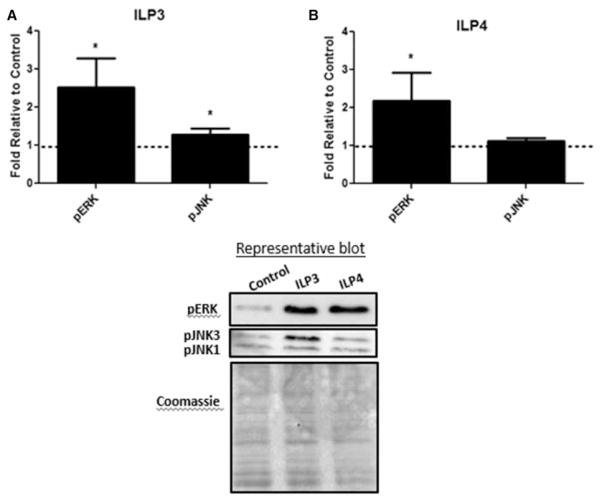
To understand the physiological mechanisms by which ILPs might regulate host resistance to P. falciparum, we provided ILP3 or ILP4 (170 pM) in a meal of human red blood cells (RBCs) to A. stephensi and examined signaling protein activation in the midgut at 15 min after feeding. Intriguingly, neither ILP3 nor ILP4 activated IIS-associated Akt or FOXO in vivo (data not shown). This result was in contrast with stimulation with human insulin which induced Akt and FOXO phosphorylation within this timeframe in parallel experiments and in previous work [12]. However, both ILPs induced ERK phosphorylation relative to control, while ILP3 induced moderate, but significant levels of JNK1/3 phosphorylation relative to control (Figure 3A,B). In D. melanogaster, homeostasis of the midgut epithelium is controlled by ILPs and the integration of IIS with several signaling pathways, including Ras-ERK and JNK [47], suggesting that the observed signaling differences could contribute to differences in ILP effects on infection.
Figure 3. ILP3 and ILP4 signaling in the A. stephensi midgut.
Cohorts of A. stephensi were fed (A) 170 pM ILP3 or (B) 170 pM ILP4 in a meal of uninfected human RBCs. Midguts were collected 15 min after feeding and protein extracts were used for Western blotting to detect protein phosphorylation. Values from ILP-treated samples were normalized first to Coomassie brilliant blue stain for total protein and then to untreated controls (unsupplemented human RBCs; set at 1) for calculation of fold change. Experiments were independently replicated with 3–5 cohorts of 60–90 mosquitoes each and data were analyzed by t-test to determine differences between ILP-treated and control groups. *P-values were deemed significant when P ≤ 0.05.
ILP3 and ILP4 alter nutrient intake and intermediary metabolism in A. stephensi
In D. melanogaster, moderate activation of JNK in midgut stem cells of young flies is associated with expression of cytoprotective genes and tissue regeneration [48], physiology that we have observed recently [7], and that could explain ILP3-associated resistance to infection. In particular, provision of physiologically low concentrations of human IGF1 in blood to A. stephensi induced moderate activation of JNK in the midgut that was associated with resistance to P. falciparum infection in the absence of NF-κB-mediated immunity [7]. Although these data were consistent with JNK signaling regulation of cellular functions ranging from apoptosis to cyto-protection and homeostasis in the fruit fly gut, biochemical data from both the fruit fly and the mosquito to confirm these assertions are limited [7,48,49].
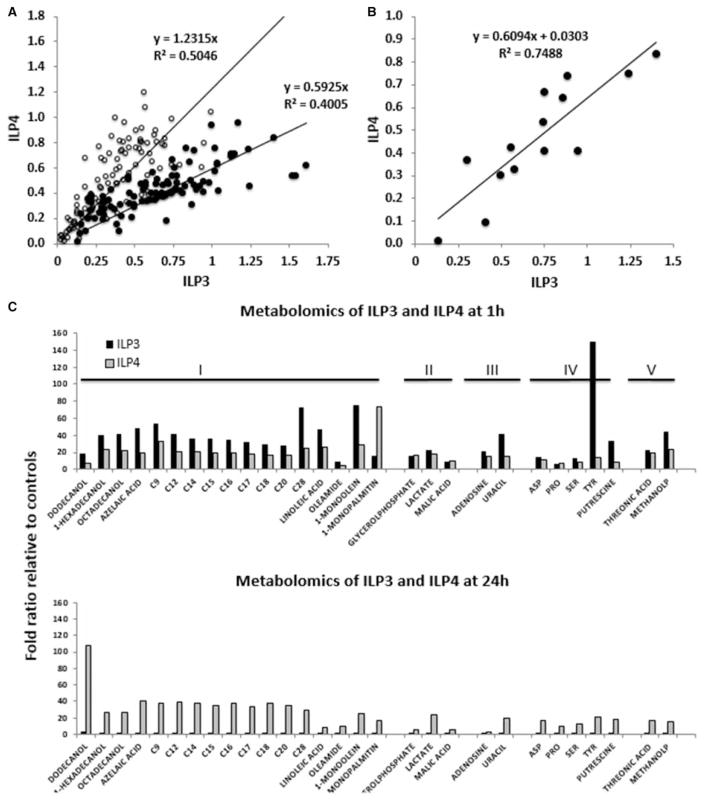
Accordingly, to address whether differential regulation of intermediary metabolism was associated with ILP-specific patterns of resistance, we performed untargeted metabolomics analyses with LC–MS/MS of intra-cellular metabolites present in the midgut at 1 and 24 h after provision of a blood meal containing 170 pM ILP3 or ILP4. Through this approach, we identified 106 metabolites in both treatments that showed different abundance in the midgut relative to blood-fed controls not provisioned with ILP3 or ILP4. The majority of these metabolites (92 of 106) were determined to be of mosquito origin based on exclusion of 14 exogenous metabolites of human origin (e.g. cholesterol), plant origin (e.g. β-sitosterol, 3-cholestanol, and stigmasterol), and present in food or blood additives or of iatrogenic origin (e.g. acetophenone, salicylaldehyde, methyltetra-hydrophenanthrenone, lactose, lactulose, maltotriose, mannitol, galactose and benzoic acid). The relative concentrations of these 92 metabolites with ILP3 and ILP4 at each time point showed a strong linear and positive correlation (Pearson’s P < 0.01; Figure 4A). Eight metabolites identified at 1 h were excluded from this analysis because they were outside the 95% CI calculated for all metabolites. The slope of the line obtained at 1 h indicated that those metabolites associated with ILP3 were 1.69-fold of those with ILP4, whereas at 24 h postmeal, the slope and, hence, fold change were 1.23 (black circles; Figure 4A). This indicated that the majority of meta-bolites in ILP3-fed mosquitoes had higher concentrations than that in ILP4-fed mosquitoes at 1 h. This effect was reversed at 24 h (white circles; Figure 4A). A separate correlation was performed with the 14 metabolites at 1 h considered to be exogenous to A. stephensi (Figure 4B). Metabolites present in the sugar chase after the blood meal and not yet digested were also included (e.g. sucrose) among these metabolites. This correlation was also significant (Pearson’s P < 0.01) with the metabolites in ILP3-fed mosquitoes present at higher levels than those in ILP4-fed mosquitoes (1.64-fold of ILP4). Consistent with the above results, a higher content of hemoglobin was obtained in midguts of ILP3-fed relative to ILP4-fed mosquitoes (Drabkin’s assay; 1.25 relative to control for ILP3 vs. 0.71 relative to control for ILP4). Thus, endogenous metabolites that are produced and/or absorbed in the midgut as well as exogenous metabolites in the blood meal tracked with higher hemoglobin content. We also evaluated the ratio of total amino acids over that of sucrose in the midguts of ILP3-fed relative to ILP4-fed mosquitoes at 1 h postmeal. The ratios were 20.3 and 8.5 for ILP3 and ILP4, respectively, suggesting a preference for a protein-rich blood meal over a carbohydrate-rich sugar meal (provided ad libitum in cotton pads after the blood meal) in ILP3-fed relative to ILP4-fed mosquitoes at 1 h. These observations suggested that ILP3 induces an orexigenic response to blood ingestion relative to ILP4.
Figure 4. ILPs differentially have an impact on intermediary metabolism in A. stephensi.
Mosquitoes were fed either 170 pM ILP3 or ILP4 in a meal of uninfected human RBCs. (A) Identified metabolites were normalized to control levels at each of the two time points (1 h shown as black circles; 24 h shown as white circles). (B) Correlation of metabolites (n = 14) identified as exogenous to the mosquitoes (present in the blood meal or sugar chase) at 1 h with each of the ILP treatments. Metabolites included here are of plant origin (β-sitosterol, 3-cholestanol, and stigmasterol), are iatrogenic or food additives (acetophenone, benzoic acid, salicylaldehyde, methyltetrahydrophenanthrenone, lactose, lactulose, maltotriose, mannitol, and galactose), present in sugar chase (sucrose), or derived from RBCs (cholesterol). (C) Compounds were analyzed using the BRITE hierarchy algorithm within the KEGG database to categorize them according to their biological roles: I, lipids, fatty acids and derivatives; II, carbohydrates and derivatives; III, nucleic acids and derivatives; IV, amino acids and biogenic amines; V, others.
To further understand ILP-mediated metabolic shifts, we focused on endogenous metabolites that were significantly different from controls with either treatment (with P < 0.05; Figure 4C). These metabolites were analyzed by using Brite from the KEGG database to classify them into their biological roles. Several important features were noted in the metabolic profile and are described below. For both ILPs, most metabolites (17 of 29) belonged to the ‘lipids’ category (including fatty acids, fatty alcohols, dicarboxylic acids, fatty amides, and monoacylglycerols), followed by the amino acid and biogenic amine category (5 of 29), carbohydrates and derivatives (3 of 29), and metabolites of nucleic acids and derivatives and ‘others’ category (2 of 29 each; Figure 4C). ILP3 treatment resulted in higher levels of metabolites than observed for ILP4 treatment at 1 h (25 of 29 metabolites; Figure 5C), again reflecting the orexigenic response of ILP3-fed mosquitoes with only a single metabolite (monopalmitin) higher than that of ILP4-fed mosquitoes. However, we cannot exclude the effect of amino acid ingestion, which could induce the synthesis of trypsin coincidentally with the activation of the TOR pathway [50]. At 24 h, most metabolites in ILP3-fed mosquitoes returned to control levels, whereas for ILP4-fed mosquitoes, 20 metabolites were higher than controls but, more importantly, had higher levels than those at 1 h (6 were lower and the remainder were comparable to control levels; Figure 4C).
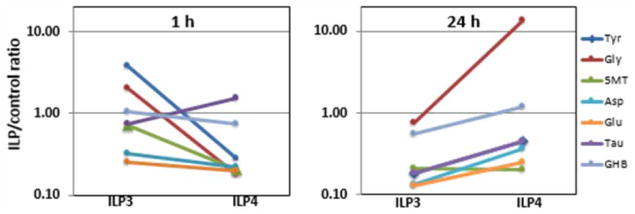
Figure 5. ILP3 and ILP4 alter neurotransmitter levels in A. stephensi.
Neurotransmitters and derivatives identified by metabolomics for each treatment and at each time point were normalized to control values (i.e. mosquitoes fed blood only). To fit the ratios for all compounds and time points, the ratios were plotted in a log scale. Tyr, tyrosine; Gly, glycine; 5MT, 5-methoxytryptamine (as a proxy for serotonin); Asp, aspartate; Glu, glutamate; Tau, taurine; GHB, γ-hydroxybutyrate (as a proxy for GABA).
In ILP3-fed mosquitoes, the increase in midgut metabolites at 1 h followed by their decrease at 24 h appears to reflect increased biosynthetic activities (esterification of fatty acids as triacylglycerides and synthesis of nucleic acids and proteins). Conversely, the persistent and, in some cases, increased levels of metabolites at 24 h in ILP4-fed mosquitoes, especially those represented by the lipid category, along with increases in lactate, are consistent with an increased use of carbohydrates (glycolysis) with a significant decrease in fatty acid oxidation. In an attempt to compensate for this energy imbalance, increased lipolysis and glycolysis (increased lactate over glucose) are triggered. Further experimental evidence for this scenario in ILP4-fed mosquitoes is increased glycolysis not accompanied by oxidative phosphorylation. This is supported by the higher levels of both Ala (the transamination product of pyruvate) and lactate (reduced product of pyruvate via lactate dehydrogenase) normalized to that of glucose (to account for the difference in blood meal size), which were 2.6 and 5.8 at 1 and 24 h for ILP4-fed mosquitoes, respectively, compared with 1.9 and 4.0 for ILP3-fed mosquitoes. This finding is also supported by the higher sucrose-to-protein ratio ingested by ILP4-fed mosquitoes compared with ILP3 as stated above. Energy deficits in ILP4 were supported by the increased levels of AMP normalized to glucose at both 1 h (1.3 in ILP4 relative to 0.5 in ILP3) and 24 h (2.9 in ILP4 relative to 1.5 in ILP3) and the higher steady state of free fatty acids (FFAs), which act as uncouplers of mitochondrial electron transport and ATP synthesis [51–53]. Decreased mitochondrial fatty acid β-oxidation in ILP4-fed mosquitoes was supported by (i) the sustained levels of pelargonic acid (C9) at both 1 and 24 h, which suggests decreased mitochondrial utilization of fatty acids, even those that are carnitine-independent; (ii) increased dicarboxylic acid (azelaic acid) between 1 and 24 h suggesting an increase in extra-mitochondrial ω-oxidation of accumulated (not processed by mitochondria) fatty acids [54]; and (iii) increased levels of 3-hydroxyvalerate [55] which result from deficits in the mitochondrial pathway from odd-chain fatty acids and some amino acids such as Ile, Val, and Met to form succinyl CoA for its oxidation in the TCA cycle. Increased lipolysis is reflected by the lower fat content of ILP4-fed mosquitoes (30% of controls) relative to ILP3-fed mosquitoes (80% of controls) and the increased levels of monoacylglycerols (monostearin and monopalmitin) in ILP4-fed mosquitoes relative to ILP3-fed mosquitoes at 24 h. Decreased synthesis of critical biomolecules (such as proteins and nucleic acids) is supported by the accumulation of adenosine, uracil, and amino acids as well as fatty alcohols (dodecanol, hexadecanol, and octadecanol) that are needed for the synthesis of cuticular wax. Taken together, these results indicated that ILP3 induces a metabolic state in the mosquito midgut with increased capability to synthesize biomolecules and, more importantly, to sustain this demand with increased energy capability from mitochondria (lower lactate). In contrast, ILP4 induces an ‘energy-deficient state’, which cannot match the energy demand.
ILP3 and ILP4 alter neurotransmitter levels in A. stephensi
In addition to the above metabolites, we identified amino acids and derivatives that have been classified either as neurotransmitters (NTs) or modulators of insect NTs for which receptors have been reported in mosquitoes [56]. Among these were Tyr (precursor for the NT dopamine and the biogenic amine tyramine), Gly, 5-methoxytryptamine (5MT; a derivative closely associated with the NTs serotonin and melatonin, which in vertebrates is synthesized by the deacetylation of melatonin in the pineal gland), Asp, Glu, γ-hydroxybutyrate (GHB, a derivative of GABA), and taurine. At 1 h, higher levels of Tyr (13-fold), Gly (11.2-fold), and 5MT (3.5-fold) were observed in ILP3-fed mosquitoes relative to ILP4-fed mosquitoes (Figure 5). These amino acid values could be interpreted as reflecting directly the proteolysis of human hemoglobin (the most abundant protein in the blood meal), but the most abundant amino acids of the α- and β-chains of the tetramer (Ala, Leu, and Val at >10% calculated from primary sequences) were not proportionally represented.
The only NT or neuromodulator higher in ILP4-fed relative to ILP3-fed mosquitoes at 1 h was taurine. Taurine may act as a neuromodulator and cell survival factor in Drosophila in clearing excess Asp within the synaptic cleft to ensure excitatory amino acid concentrations below excitotoxicity levels [57], consistent with the various roles of taurine in developmental plasticity and the long-term potentiation of synaptic transmission in vertebrates [58–60]. At 24 h, most of these NTs/neuromodulators were higher in ILP4-fed relative to ILP3-fed mosquitoes (except 5MT with equal levels for both ILPs) and higher in ILP3-fed relative to ILP4-fed mosquitoes at 1 h; the most notable changes included Gly (18-fold) followed by Tyr, Asp, and GHB (2-fold for each). While the first three are considered excitatory NTs, GABA (or GHB) is considered the predominant inhibitory neurotransmitter in mammalian and insect brains. The GABA-B family receptor GPRGBB1 [61] has been found associated with the Serratia marcescens infection phenotype in Anopheles gambiae and the D. melanogaster ortholog of GPRGBB1 has been implicated in behavioral responses to alcohol sensitivity [62–64].
Taken together, these results suggested that in ILP3-fed relative to ILP4-fed mosquitoes, there is an early and higher concentration of excitatory NTs (Tyr, Gly, and 5MT) relative to taurine and GHB. At 24 h, most of these NTs decrease in ILP3-fed mosquitoes (at about half of the ratio observed at 1 h), while in ILP4-fed mosquitoes a significant increase in excitatory NTs (Gly, Tyr, and Asp) is observed along with that of a potent inhibitory NT (GHB). Accordingly, the orexigenic response along with the preference for a protein-rich diet triggered by ILP3 seems to follow an increase in excitatory NTs.
ILP-dependent shifts in A. stephensi metabolism regulate P. falciparum infectivity
To determine the potential role of ILP-mediated metabolic alterations (Figure 4) in regulating resistance to P. falciparum, we utilized small-molecule inhibitors to reverse the metabolic effects of ILP3 and ILP4 (Supplementary Figures S1–S3). Specifically, we used heptelidic acid (HA), an inhibitor of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), to inhibit glycolysis in ILP3-fed mosquitoes [65], and 4-hydroxy-L-phenylglycine (4H-PG), an inhibitor of carnitine palmitolytransferase-1 (CPT1) to inhibit β-oxidation of fatty acids [66]. Our rationale was that by inhibiting glycolysis with HA in ILP3-fed mosquitoes, we would induce a shift in metabolism that would result in more amino acids and fatty acids being utilized for energy, while the carbon skeletons of amino acids would be used as anaplerotic substrates for the TCA. While this switch is more energy efficient, mobilization of fat and protein for energy use is significantly slower than for carbohydrates and requires an adaptation period. Thus, this treatment would mimic a ‘low-energy state’. The midgut metabolome in this case would show a decrease in FFA (increased fatty acid oxidation or esterification to be shipped elsewhere for use) with increased free amino acids derived from proteolysis, and increased AMP and decreased lactate due to increased pyruvate utilization by mitochondria. On the other hand, the use of 4H-PG in ILP4-fed mosquitoes would reduce the content of FFA from intake by forcing their esterification and storage as triacylglycerol (and decreasing their uncoupling effect on mitochondria) while deriving energy mainly from carbohydrates followed by amino acids Thus, this treatment would mimic a ‘high-energy state’. The metabolome in this case would show decreased FFA with increased free amino acids from proteolysis, increased AMP, and decreased lactate due to increased pyruvate utilization by mitochondria. The confirmation that each of the inhibitors was acting at the desired step was obtained from the metabolomes of midguts from mosquitoes fed either inhibitor at 24 h (Supplementary Figures S2 and S3).
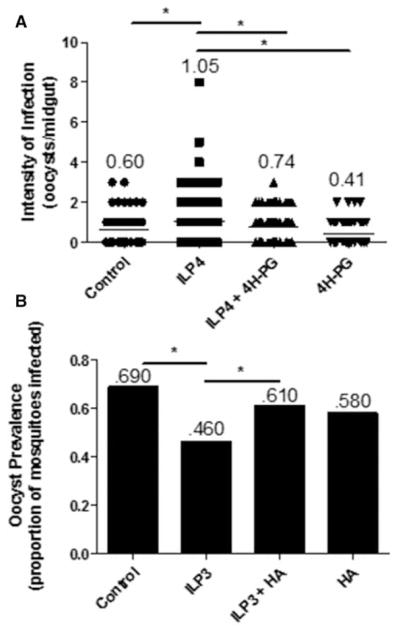
Inhibition of CPT1 with 4H-PG prior to feeding of a parasite-infected blood meal reversed the effects of ILP4 on infection (Figure 6A). That is, ILP4 alone significantly increased infection intensity (oocysts/midgut) relative to diluent-fed controls, but pretreatment with 4H-PG prior to feeding of ILP4-supplemented infected blood significantly decreased infection intensity relative to an identical meal without prior inhibition of CPT1 to a level that was indistinguishable from that of controls. Similarly, inhibition of GAPDH prior to feeding of an ILP3-supplemented infected blood meal reversed the effects of ILP3 on infection prevalence (Figure 6B). That is, feeding of ILP3 alone significantly decreased the prevalence of infection relative to diluent-fed controls, while pretreatment with HA prior to feeding of an ILP3-supplemented infected blood meal resulted in infection prevalence that was not significantly different from controls and that was increased relative to an ILP3-supplemented meal without prior inhibition of GAPDH. Pretreatment of A. stephensi with 4H-PG had no effect on NF-κB-dependent transcript levels in the midgut (Supplementary Figure S4A). Pretreatment with HA had no significant effect on transcript levels of NF-κB-dependent immune genes (Supplementary Figure S4B). Accordingly, our results suggest that ILP modulation of metabolism, in the absence of prototypical NF-κB-dependent antiparasite responses, appears to be key in regulating parasite infection.
Figure 6. ILP-mediated metabolic shifts control P. falciparum infectivity in A. stephensi.
Cohorts of A. stephensi were pretreated with water alone or water supplemented with inhibitors of metabolism for 24 h and then fed a P. falciparum-infected blood meal supplemented with 170 pM ILP3 or ILP4 or supplemented with an equivalent volume of ILP diluent. Ten days later, midguts were dissected and stained for visualization of oocysts to determine the prevalence of infection (percentage of mosquitoes infected) or intensity of infection (number of oocysts/midgut). (A) Mosquitoes were pretreated with 1 mM 4H-PG to inhibit β-oxidation or (B) 10 μM HA to inhibit GAPDH. Experiments were independently replicated with 3–4 cohorts of 60–90 mosquitoes each. Infection prevalence and intensity data were analyzed by χ2 goodness-of-fit test and nonparametric ANOVA (Kruskal–Wallis) followed by Dunn’s multiple comparisons test, respectively, to determine differences between groups. *P-values were deemed significant when P ≤ 0.05.
ILP3 and ILP4 differentially regulate A. stephensi feeding and flight behavior
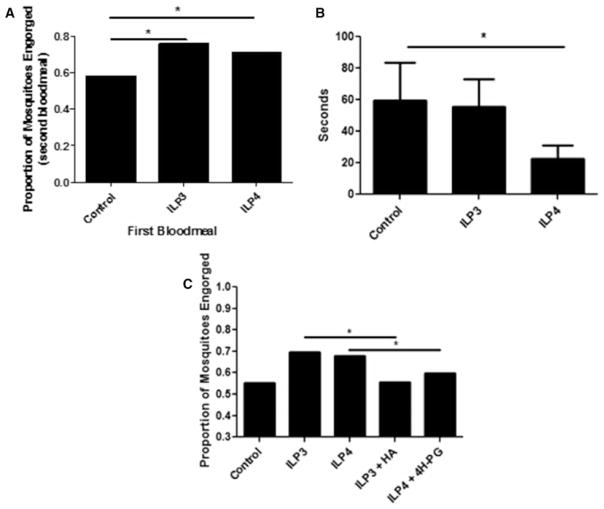
Changes in intermediary metabolism are not only strongly linked to changes in the host response to infection [43–45] (Figure 6), but are also a driving force behind nutrient sensing [67–69] and the balance between energy supply (food intake and mobilization of energy storage) and energy output (activity). Our metabolic analyses indicated that ILP3 induces a high-energy state, whereas ILP4 induces energy deficiency (Figure 4). Furthermore, both ILP3 and ILP4 can regulate NT activity (Figure 5), whereas ILP3 appears to induce an orexigenic response. As such, we sought to directly examine mosquito blood-feeding behavior and flight activity after provision of ILP3 and ILP4. We found that feeding of either ILP in a blood meal significantly increased the proportion of mosquitoes relative to controls that engorged when offered a second blood meal 3 days later (Figure 7A). Furthermore, ILP4, but not ILP3, in the blood meal significantly decreased the amount of time spent flying relative to controls during a 10-min period 24 h after feeding (Figure 7B). Interestingly, the effects of ILP3 and ILP4 on blood feeding appear to be due to their effects on metabolism, as inhibiting GAPDH in the context of ILP3 feeding and inhibiting CPT1 in the context of ILP4 feeding both significantly decreased blood-feeding propensity of mosquitoes offered a second blood meal (Figure 7C); the inhibitors alone had no effect relative to control on the proportions of mosquitoes that engorged (Supplementary Figure S5). Notably, these findings were consistent with increased host-seeking activity associated with increased ILP3 and ILP4 expression in the midgut during P. falciparum infection in A. stephensi [4].
Figure 7. ILP3 and ILP4 regulate A. stephensi feeding and flight behavior.
Mosquitoes were fed either 170 pM ILP3 or ILP4 in a meal of uninfected human RBCs and the effects of this meal on (A) the propensity of mosquitoes to take a second blood meal 3 days later and (B) flight activity (average number of seconds spent flying in 10 min) at 24 h after feeding were measured. (C) Mosquitoes-fed ILPs in blood were treated with small-molecule inhibitors of GAPDH (HA) or CPT1 (4H-PG) in water for 3 days then assayed for the propensity to take a second blood meal. Blood-feeding experiments were (A) replicated three times with cohorts of 84–126 mosquitoes each or (C) replicated three times with cohorts of 356–428 mosquitoes each and data were analyzed by χ2 test to determine significant differences between groups. Flight activity experiments (B) were replicated with A. stephensi females from six cohorts (n = 5 mosquitoes per replicate, 30 mosquitoes total) and data were analyzed using a zero-inflated negative binomial regression model to determine significant differences between groups. *P-values were deemed significant when P ≤ 0.05.
Discussion
In the previous work, we showed that knockdown of either ILP3 or ILP4 in A. stephensi reduced the prevalence and intensity of P. falciparum infection while also increasing the expression of antiparasite genes [13]. As a follow-up to these studies, here we show that feeding of ILP4 in a blood meal increased infection prevalence and intensity. Unexpectedly, feeding of ILP3 decreased the prevalence of parasite infection, despite intake of more blood, with no effect on the intensity of infection. Collectively, these data indicate that ILP4 is necessary for optimal parasite development and sufficient for increased development, whereas ILP3 is necessary for optimal parasite development, but not sufficient to increase development. The insufficiency of ILP3 may be due to the requirement of additional signaling components to achieve its effects. For example, ILP3 may act in concert with other ILPs during a natural infection. An alternative explanation is that the receptor for ILP3 may be expressed at a low level and therefore signal saturation occurs rapidly, somewhat mitigating the effect of ILP3 added to the blood meal. Both ILP4 and ILP3 decreased transcript levels of NF-κB-dependent immune genes, suggesting that ILP effects on infection are not exclusively dependent on prototypical antiparasite responses. Accordingly, we investigated the roles of ILP3 and ILP4 in cell signaling and intermediary metabolism in A. stephensi and observed differences in the regulation of these processes by ILP3 and ILP4.
From our studies, intermediary metabolism was significantly modulated by ILP3 and ILP4. Importantly, ILP3 induced moderate induction of JNK and a metabolic shift that could enhance tissue homeostasis [7,48] and provide metabolic fuel [43,44] for successful resistance to P. falciparum infection. In contrast with ILP3, provision of ILP4 appeared to induce an ‘energy-deficient’ state, which could alter the balance of biochemical intermediates for early host defense responses [43,44]. Indeed, the effects of inhibitor pretreatment provide complementary arguments that ILP-dependent changes in intermediary metabolism and likely energy balance regulate parasite development in the mosquito host.
The processes of seeking a blood meal and ingestion of a second blood meal are critical features that determine the likelihood of successful parasite transmission. Accordingly, provision of ILP3 and ILP4 both increased the propensity to take a second blood meal, but probably for very different reasons. ILP3 appears to be orexigenic with early activation (1 h) of excitatory NTs, physiology that may be sustained through to a second blood meal, while the low-energy state of ILP4-fed mosquitoes limited their flight activity, but increased NT activity and refeeding because of the need for replenishment of energy stores.
While ILP3 is orexigenic compared with ILP4, the differential effects of the two peptides may also derive from differences in receptor-binding affinity. In the mosquito Aedes aegypti, ILP3 and ILP4 do not compete for a receptor, but instead exert differential effects on hormone synthesis and metabolism by binding to the single insulin receptor and to an uncharacterized 44 kDa membrane protein, respectively [70]. Thus, as in A. aegypti, A. stephensi ILP3 and ILP4 may activate cell signaling from multiple receptors, thereby leading to the differential effects on intermediary metabolism and behavior that we have observed.
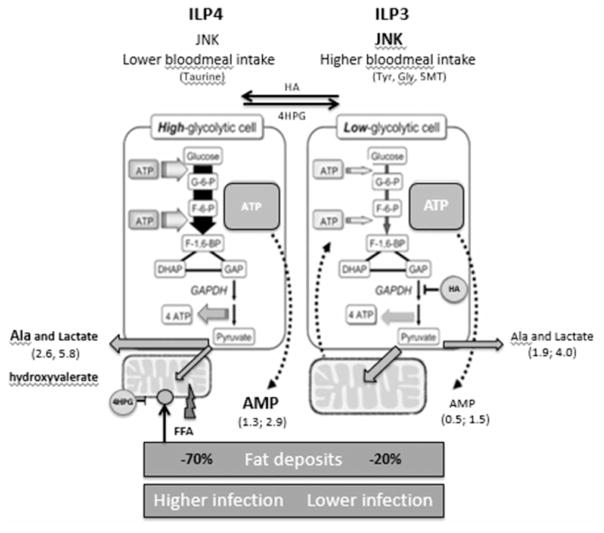
Ultimately, our results demonstrate a major contribution of ILP-mediated metabolic changes to the regulation of P. falciparum development in the mosquito host (Figure 8). Our work also extends the understanding of fundamental connections among intermediary metabolism, behavior, and infection, while providing novel mechanistic insights from this medically important species with unique mitochondrial physiology [71]. We not only show that two ILPs differentially regulate intermediary metabolism, likely altering homeostasis in the midgut, but we also provide evidence that these metabolic changes have an impact on P. falciparum infection independently of canonical NF-κB-mediated immunity. Additionally, we suggest that ILP regulation of metabolism is a driving force behind behavioral alterations as has been observed in organisms from nematodes to mammals [66,68,69,72,73] and that these changes are particularly important for successful parasite transmission. Taken together, our data implicate ILP3 and ILP4 as key mediators of an array of physiological processes in the mosquito that influence its capacity to act as an efficient vector arthropod.
Figure 8. Proposed model: ILP3 and ILP4 differentially regulate behavior and P. falciparum infection in A. stephensi through effects on intermediary metabolism.
ILP3 induces moderate JNK activation and up-regulates excitatory NTs, resulting in increased nutrient uptake and a high-energy state that can reduce parasite infection. In contrast, ILP4 does not activate JNK signaling and results in an energy-deficient state that increases parasite infection and leads to increased feeding behavior to replenish nutrient stores.
Materials and methods
P. falciparum culture and mosquito infections
The NF54 strain of P. falciparum was initiated at 1% parasitemia in 10% heat-inactivated human serum and 6% washed RBCs in RPMI 1640 with HEPES (Gibco, Manassas, VA) and hypoxanthine. At days 15–17, stage V gametocytes were evident and exflagellation was evaluated the day before and day of feeding by observation of blood smears before the addition of fresh media at 200× magnification with phase-contrast or modified brightfield microscopy. Female A. stephensi were provided water only (no sucrose) for 24 h and then fed a meal of either unsupplemented, P. falciparum-infected RBCs, or P. falciparum-infected RBCs supplemented with 170 pM ILP3 or ILP4 (CPC Scientific, Sunnyvale, CA), using a Hemotek artificial circulation system (Discovery Workshops, Accrington, UK). To inhibit GAPDH, mosquitoes were supplemented with 10 μM HA (Biovisions, Milpitas, CA) in water-soaked cotton pads for 24 h before blood feeding. To inhibit CPT1, mosquitoes were supplemented with 1 mM 4H-PG (Sigma-Aldrich, St Louis, MO) in water-soaked cotton pads for 24 h before blood feeding. Treatments were performed in this manner to minimize direct effects on parasites in the blood meal. Mosquitoes were allowed to feed on infected RBCs for 30 min and were then maintained on cotton pads soaked in 10% sucrose until day 10 postinfection. To quantify infection levels, midguts were dissected on day 10 postinfection and stained with 0.5% mercurochrome for visualization of P. falciparum oocysts by microscopy. Experiments were independently replicated with 3–7 cohorts of 60–120 mosquitoes. Data were pooled for analysis as one-way ANOVA determined that infections in control groups were not significantly different among replicates. Infection prevalence and intensity data were analyzed by χ2 test, Mann–Whitney test, or nonparametric ANOVA (Kruskal–Wallis) followed by Dunn’s multiple comparisons test to determine differences between groups. P-values were deemed significant when P ≤ 0.05.
Mosquito gene expression
Cohorts of 45–60 mosquitoes were dissected 1 or 24 h after provision of a meal of human RBCs supplemented with soluble PfPs (1:100 dilution [12]) in the presence or absence of ILP3 or ILP4 (170 pM). Midguts were collected into TriZOL (Invitrogen, Carlsbad, CA) for extraction of total RNA according to the manufacturer’s protocol. cDNA was synthesized using the QuantiTec Reverse Transcription Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) for analyses of immune gene expression was performed on an ABI 7300 Sequence Detection System (Applied Biosystems, Foster City, CA) using gene-specific primers for defensin, NOS, TEP1, APL1, LRIM1, and S7 with SYBR Green PCR Mastermix as previously described [8,12]. Anopheles stephensi ribosomal protein S7 gene was used to normalize gene target data. Data were analyzed using the ΔΔCt method [74]. These assays were completed with 3–7 separate cohorts of A. stephensi. Data were analyzed by t-test to determine differences between ILP-treated and control groups (PfPs only, set at 0). P-values were deemed significant when P ≤ 0.05.
Western blotting
For analyses of cell signaling in vivo, mosquitoes were fed 170 pM ILP3 or ILP4 in a meal of uninfected human RBCs for 15 min using a Hemotek artificial circulation system. After feeding, 60–90 mosquitoes were dissected and midguts were processed as previously described [14]. After processing, midguts were placed into cell extraction buffer (Invitrogen), incubated at 4°C for 1 h, and periodically vortexed. Cell lysates were then cleared at 16 000 g for 10 min and protein supernatants were mixed with sample loading buffer (125 mM Tris–HCl pH 6.8, 10% glycerol, 10% SDS, 0.006% bromophenol blue, and 130 mM dithiothreitol) and boiled for 5 min. Proteins were separated on 10% SDS–PAGE and transferred onto the nitrocellulose membranes (BioRad, Hercules, CA). Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline/Tween-20 (TBS-T) for 1 h at room temperature. For detection of phosphorylated ERK (pERK), membranes were incubated overnight at 4°C with a 1:10 000 dilution of anti-pERK antibody (Cell Signaling, Danvers, MA) in 5% nonfat dry milk. Membranes were then washed three times with TBS-T for 5 min and incubated with a 1:10 000 dilution of secondary HRP-conjugated rabbit antimouse antibody (Sigma-Aldrich) in 5% nonfat dry milk overnight. Detection of phosphorylated FOXO (pFOXO; Cell Signaling, 1:1000 primary, 1:2000 goat antirabbit secondary), phosphorylated Akt (pAkt; Cell Signaling, 1:1000 primary, 1:3000 goat antirabbit secondary), phosphorylated JNK1/3 (pJNK1/3; Sigma-Aldrich, 1:1000 primary, 1:3000 goat antirabbit secondary), and GAPDH (Cell Signaling, 1:10 000 primary, 1:200 000 goat antirabbit secondary) followed the same protocol. Proteins were visualized by incubating membranes with SuperSignal West Dura chemiluminescent reagent (Pierce, Rockford, IL) for 3 min and exposing on an Image Station 4000 Pro digital imager (Kodak, Rochester, NY) for 1–5 min. Densitometry of pERK and pJNK Western blots was performed using the ImageJ (http://rsbweb.nih.gov/ij/) gel analysis tool. In vivo data were normalized to untreated control groups (RBCs only; set at 1) as well as a Coomassie brilliant blue loading control. These experiments were replicated 3–4 times with independent cohorts of mosquitoes. Data were analyzed by t-test to determine significant differences between ILP-treated and control groups. P-values were deemed significant when P ≤ 0.05.
Metabolomics, hemoglobin content, and fat content
For metabolomic analyses, mosquitoes were fed 170 pM ILP3 or ILP4 in a meal of human RBCs for 15 min using a Hemotek artificial circulation system. Midguts were dissected at 1 and 24 h postfeeding and washed 5–10 times with PBS until clear of blood to eliminate metabolites derived from the blood meal. Samples were processed as described in Supplementary Methods. Fat from mosquitoes (30–200/treatment) was determined using Folch extraction, drying the extracted organic layer under nitrogen, and weighing the final dry pellets as described [75]. Data were normalized per mosquito. For assays of hemoglobin content as a proxy for ingested blood, mosquitoes were allowed to feed on a meal of human RBCs supplemented with or without ILPs for 30 min. At 1 h postfeeding, whole mosquitoes were flash-frozen in liquid nitrogen and lysates were used for determination of hemoglobin with Drabkin’s reagent.
Behavioral assays
For assays of blood-feeding behavior, mosquitoes were maintained on water only for 24 h prior to feeding 170 pM ILP3 or ILP4 in a meal of uninfected human RBCs. Blood-fed mosquitoes were allowed to completely digest the blood meal for 72 h and were then offered a second blood meal of uninfected RBCs. Mosquitoes were given the opportunity to feed for 15 min. After this time, fed and unfed mosquitoes were counted to determine the proportion of engorged mosquitoes in each group. Experiments were independently replicated three times and data were analyzed by χ2 test to determine significant differences between groups. For assays of flight activity, mosquitoes were starved for 24 h (water only) prior to feeding 170 pM ILP3 or ILP4 in a meal of uninfected human RBCs. Blood-fed mosquitoes were allowed to rest for 24 h, transferred to individual pint-sized containers, and their behavior was recorded for 10 min. Time spent flying (seconds) was measured for each mosquito using BORIS observational software (http://penelope.unito.it/boris?page=home). Experiments were replicated with mosquitoes (n = 5) from six separate cohorts (30 mosquitoes total) and data points for each group were pooled for analysis. Data were analyzed using a zero-inflated negative binomial regression model to determine significant differences between groups. P-values were deemed significant when P ≤ 0.05.
Supplementary Material
Acknowledgments
Funding
Funding for these studies was provided by the United States National Institutes of Health National Institute of Allergy and Infectious Diseases (NIH NIAID) [grants AI073745, AI080799, and AI107263] and an F31-NRSA AI100589 ( J.E.P.), as well as National Science Foundation dissertation improvement [grant 1310194] ( J.E.P.).
We thank Caitlin Tiffany for assistance with P. falciparum culture and mosquito infection. We also thank Dr Mark Brown for assistance with the ILPs used in this work.
Abbreviations
- 4H-PG
4-hydroxy-L-phenylglycine
- 5MT
5-methoxytryptamine
- CPT1
carnitine palmitolytransferase-1
- ERK
extracellular signal-regulated kinase
- FFA
free fatty acids
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GHB
γ-hydroxybutyrate
- HA
heptelidic acid
- IIS
insulin/insulin-like growth factor signaling
- ILPs
insulin-like peptides
- JNK
c-Jun N-terminal kinase
- LRIM
leucine-rich repeat immune
- NOS
nitric oxide synthase
- NTs
neurotransmitters
- pERK
phosphorylated ERK
- PfPs
Plasmodium falciparum products
- RBCs
red blood cells
- TEP
thioester-containing protein
- TBS-T
Tris-buffered saline/Tween-20
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Author Contribution
J.E.P., E.E.L., M.A.R., C.G., and S.L. conceived of and designed experiments; J.E.P., N.P., E.N., G.S., E.P., R.P., K.W.C., G.W., and H.S. performed experiments; J.E.P., N.P., E.E.L., C.G., and S.L. analyzed the data; J.E.P., E.E. L., M.A.R., C.G., and S.L. wrote and edited the manuscript.
References
- 1.Luckhart S, Riehle MA. The insulin signaling cascade from nematodes to mammals: insights into innate immunity of Anopheles mosquitoes to malaria parasite infection. Dev Comp Immunol. 2007;31:647–656. doi: 10.1016/j.dci.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nässel DR, Liu Y, Luo J. Insulin/IGF signaling and its regulation in Drosophila. Gen Comp Endocrinol. 2015;221:255–266. doi: 10.1016/j.ygcen.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 3.Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
- 4.Cator LJ, Pietri JE, Murdock CC, Ohm JR, Lewis EE, Read AF, et al. Immune response and insulin signalling alter mosquito feeding behaviour to enhance malaria transmission potential. Sci Rep. 2015;5:11947. doi: 10.1038/srep11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corby-Harris V, Drexler A, Watkins de Jong L, Antonova Y, Pakpour N, Ziegler R, et al. Activation of Akt signaling reduces the prevalence and intensity of malaria parasite infection and lifespan in Anopheles stephensi mosquitoes. PLoS Pathog. 2010;6:e1001003. doi: 10.1371/journal.ppat.1001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drexler A, Nuss A, Hauck E, Glennon E, Cheung K, Brown M, et al. Human IGF1 extends lifespan and enhances resistance to Plasmodium falciparum infection in the malaria vector Anopheles stephensi. J Exp Biol. 2013;216:208–217. doi: 10.1242/jeb.078873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drexler A, Pietri JE, Pakpour N, Hauck E, Wang B, Glennon EK, et al. Human IGF1 regulates midgut oxidative stress and epithelial homeostasis to balance lifespan and Plasmodium falciparum resistance in Anopheles stephensi. PLoS Pathog. 2014;10:e1004231. doi: 10.1371/journal.ppat.1004231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauck ES, Antonova-Koch Y, Drexler A, Pietri J, Pakpour N, Liu D, et al. Overexpression of phosphatase and tensin homolog improves fitness and decreases Plasmodium falciparum development in Anopheles stephensi. Microb Infect. 2013;15:775–787. doi: 10.1016/j.micinf.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton AA, Lee Y, Coulibaly CA, Rashbrook VK, Cornel AJ, Lanzaro GC, et al. Identification of three single nucleotide polymorphisms in Anopheles gambiae immune signaling genes that are associated with natural Plasmodium falciparum infection. Malaria J. 2010;9:160. doi: 10.1186/1475-2875-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang M-A, Mott TM, Tapley EC, Lewis EE, Luckhart S. Insulin regulates aging and oxidative stress in Anopheles stephensi. J Exp Biol. 2008;211:741–748. doi: 10.1242/jeb.012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luckhart S, Giulivi C, Drexler AL, Antonova-Koch Y, Sakaguchi D, Napoli E, et al. Sustained activation of Akt elicits mitochondrial dysfunction to block Plasmodium falciparum infection in the mosquito host. PLoS Pathog. 2013;9:e1003180. doi: 10.1371/journal.ppat.1003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pakpour N, Corby-Harris V, Green GP, Smithers HM, Cheung KW, Riehle MA, et al. Ingested human insulin inhibits the mosquito NF-κB-dependent immune response to Plasmodium falciparum. Infect Immun. 2012;80:2141–2149. doi: 10.1128/IAI.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietri JE, Pietri EJ, Potts R, Riehle MA, Luckhart S. Plasmodium falciparum suppresses the host immune response by inducing the synthesis of insulin-like peptides (ILPs) in the mosquito Anopheles stephensi. Dev Comp Immunol. 2015;53:134–144. doi: 10.1016/j.dci.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surachetpong W, Singh N, Cheung KW, Luckhart S. MAPK ERK signaling regulates the TGF-β1-dependent mosquito response to Plasmodium falciparum. PLoS Pathog. 2009;5:e1000366. doi: 10.1371/journal.ppat.1000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surachetpong W, Pakpour N, Cheung KW, Luckhart S. Reactive oxygen species-dependent cell signaling regulates the mosquito immune response to Plasmodium falciparum. Antioxid Redox Signal. 2011;14:943–955. doi: 10.1089/ars.2010.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broughton S, Alic N, Slack C, Bass T, Ikeya T, Vinti G, et al. Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS ONE. 2008;3:e3721. doi: 10.1371/journal.pone.0003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown MR, Clark KD, Gulia M, Zhao Z, Garczynski SF, Crim JW, et al. An insulin-like peptide regulates egg maturation and metabolism in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2008;105:5716–5721. doi: 10.1073/pnas.0800478105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulia-Nuss M, Robertson AE, Brown MR, Strand MR. Insulin-like peptides and the target of rapamycin pathway coordinately regulate blood digestion and egg maturation in the mosquito Aedes aegypti. PLoS ONE. 2011;6:e20401. doi: 10.1371/journal.pone.0020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sim C, Denlinger DL. A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquito Culex pipiens. Insect Mol Biol. 2009;18:325–332. doi: 10.1111/j.1365-2583.2009.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Liu J, Li CR, Momen B, Kohanski RA, Pick L. Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proc Natl Acad Sci USA. 2009;106:19617–19622. doi: 10.1073/pnas.0905083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marquez AG, Pietri JE, Smithers HM, Nuss A, Antonova Y, Drexler AL, et al. Insulin-like peptides in the mosquito Anopheles stephensi: identifigcation and expression in response to diet and infection with Plasmodium falciparum. Gen Comp Endocrinol. 2011;173:303–312. doi: 10.1016/j.ygcen.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3:711–722. doi: 10.1016/S1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 23.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker T, Loch G, Beyer M, Zinke I, Aschenbrenner AC, Carrera P, et al. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463:369–373. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- 25.Delaney JR, Stöven S, Uvell H, Anderson KV, Engström Y, Mlodzik M. Cooperative control of Drosophila immune responses by the JNK and NF-κB signaling pathways. EMBO J. 2006;25:3068–3077. doi: 10.1038/sj.emboj.7601182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kallio J, Leinonen A, Ulvila J, Valanne S, Ezekowitz RA, Rämet M. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microb Infect. 2005;7:811–819. doi: 10.1016/j.micinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Nemoto S, DiDonato JA, Lin A. Coordinate regulation of IκB kinases by mitogen-activated protein kinase kinase kinase 1 and NF-κB-inducing kinase. Mol Cell Biol. 1998;18:7336–7343. doi: 10.1128/MCB.18.12.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park M, Brady H, Ruocco MG, Sun H, Williams D, Lee SJ, et al. Targeting of TAK1 by the NF-κB protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 2004;18:584–594. doi: 10.1101/gad.1168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Q, Lee FS. Mitogen-activated protein kinase/ERK kinase kinases 2 and 3 activate nuclear factor-κB through IκB kinase-α and IκB kinase-β. J Biol Chem. 1999;274:8355–8358. doi: 10.1074/jbc.274.13.8355. [DOI] [PubMed] [Google Scholar]
- 30.Buchon N, Broderick NA, Kuraishi T, Lemaitre B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 2010;8:152. doi: 10.1186/1741-7007-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuervo AM. Autophagy and aging: keeping that old broom working. Trend Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamin JL, Sumpter R, Jr, Levine B, Hooper LV. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling regulated pathogen resistance. Proc Natl Acad Sci USA. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randow F, Münz C. Autophagy in the regulation of pathogen replication and adaptive immunity. Trends Immunol. 2012;33:475–487. doi: 10.1016/j.it.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren C, Finkel SE, Tower J. Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity. Exp Gerontol. 2009;44:228–235. doi: 10.1016/j.exger.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Hang S, Purdy AE, Watnick PI. Mutations in the IMD pathway and mustard counter Vibrio cholerae suppression of intestinal stem cell division in Drosophila. mBio. 2013;4:e00337–13. doi: 10.1128/mBio.00337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, et al. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato-Miyata Y, Muramatsu K, Funakoshi M, Tsuda M, Aigaki T. Overexpression of dilp2 causes nutrient-dependent semi-lethality in Drosophila. Front Physiol. 2014;5:3312. doi: 10.3389/fphys.2014.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiAngelo JR, Bland ML, Bambina S, Cherry S, Birnbaum MJ. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc Natl Acad Sci USA. 2009;106:20853–20858. doi: 10.1073/pnas.0906749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers MC, Song KH, Schneider DS. Listeria monocytogenes infection causes metabolic shifts in Drosophila melanogaster. PLoS ONE. 2012;7:e50679. doi: 10.1371/journal.pone.0050679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila. Curr Biol. 2006;16:1977–1985. doi: 10.1016/j.cub.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 42.Hang S, Purdy AE, Robins WP, Wang Z, Mandal M, Chang S, et al. The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe. 2014;16:592–604. doi: 10.1016/j.chom.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng S-C, Joosten LAB, Netea MG. The interplay between central metabolism and innate immune responses. Cytokine Growth Factor Rev. 2014;25:707–713. doi: 10.1016/j.cytogfr.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Liu TF, Brown CM, El Gazzar M, McPhail L, Millet P, Rao A, et al. Fueling the flame: bioenergy couples metabolism and inflammation. J Leukoc Biol. 2012;92:499–507. doi: 10.1189/jlb.0212078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Ye L, Araki K, Ahmed R. mTOR, linking metabolism and immunity. Semin Immunol. 2012;24:429–435. doi: 10.1016/j.smim.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Y, Das S, Cirimotich C, Souza-Neto JA, Mclean KJ, Dimopoulos G. Engineered Anopheles immunity to Plasmodium infection. PLoS Pathog. 2011;7:e1002458. doi: 10.1371/journal.ppat.1002458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biteau B, Hochmuth CE, Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kux K, Pitsouli C. Tissue communication in regenerative inflammatory signaling: lessons from the Fly gut. Front Cell Infect Microbiol. 2014;4:695. doi: 10.3389/fcimb.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brandon M, Pennington JE, Isoe J, Zamora J, Schillinger A-S, Miesfeld RL. TOR signaling is required for amino acid stimulation of early trypsin protein synthesis in the midgut of Aedes aegypti mosquitoes. Insect Biochem Mol Biol. 2008;38:916–922. doi: 10.1016/j.ibmb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rottenberg H, Hashimoto K. Fatty acid uncoupling of oxidative phosphorylation in rat liver mitochondria. Biochemistry. 1986;25:1747–1755. doi: 10.1021/bi00355a045. [DOI] [PubMed] [Google Scholar]
- 52.Rottenberg H, Steiner-Mordoch S. Free fatty acids decouple oxidative phosphorylation by dissipating intramembranal protons without inhibiting ATP synthesis driven by the proton electrochemical gradient. FEBS Lett. 1986;202:314–318. doi: 10.1016/0014-5793(86)80708-6. [DOI] [PubMed] [Google Scholar]
- 53.Skulachev V. Fatty acid circuit as a physiological mechanism of uncoupling of oxidative phosphorylation. FEBS Lett. 1991;294:158–162. doi: 10.1016/0014-5793(91)80658-P. [DOI] [PubMed] [Google Scholar]
- 54.Coates PM, Hale DE, Stanley CA, Corkey BE, Cortner JA. Genetic deficiency of medium-chain acyl coenzyme A dehydrogenase: studies in cultured skin fibroblasts and peripheral mononuclear leukocytes. Pediatr Res. 1985;19:671–676. doi: 10.1203/00006450-198507000-00007. [DOI] [PubMed] [Google Scholar]
- 55.Sweetman L, Weyler W, Nyhan WL, de Céspedes C, Loria AR, Estrada Y. Abnormal metabolites of isoleucine in a patient with propionyl-CoA carboxylase deficiency. Biomed Mass Spectrom. 1978;5:198–207. doi: 10.1002/bms.1200050307. [DOI] [PubMed] [Google Scholar]
- 56.Matthews BJ, McBride CS, DeGennaro M, Despo O, Vosshall LB. The neurotranscriptome of the Aedes aegypti mosquito. BMC Genomics. 2016;17:32. doi: 10.1186/s12864-015-2239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Besson MT, Re DB, Moulin M, Birman S. High affinity transport of taurine by the Drosophila aspartate transporter dEAAT2. J Biol Chem. 2005;280:6621–6626. doi: 10.1074/jbc.M412440200. [DOI] [PubMed] [Google Scholar]
- 58.El Idrissi A, Trenkner E. Taurine as a modulator of excitatory and inhibitory neurotransmission. Neurochem Res. 2004;29:189–197. doi: 10.1023/B:NERE.0000010448.17740.6e. [DOI] [PubMed] [Google Scholar]
- 59.Oja S, Saransaari P. Kinetic analysis of taurine influx into cerebral cortical slices from adult and developing mice in different incubation conditions. Neurochem Res. 1996;21:161–166. doi: 10.1007/BF02529133. [DOI] [PubMed] [Google Scholar]
- 60.Trenkner E. The role of taurine and glutamate during early postnatal cerebellar development of normal and Weaver mutant mice. Advanc Experiment Med Biol. 1990;268:239–244. doi: 10.1007/978-1-4684-5769-8_27. [DOI] [PubMed] [Google Scholar]
- 61.Stathopoulos S, Neafsey DE, Lawniczak MKN, Muskavitch MAT, Christophides GK. Genetic dissection of Anopheles gambiae gut epithelial responses to Serratia marcescens. PLoS Pathog. 2014;10:e1003897. doi: 10.1371/journal.ppat.1003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dzitoyeva S, Dimitrijevic N, Manev H. γ-Aminobutyric acid B receptor 1 mediates behavior-impairing actions of alcohol in Drosophila: adult RNA interference and pharmacological evidence. Proc Natl Acad Sci USA. 2003;100:5485–5490. doi: 10.1073/pnas.0830111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci USA. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shohat-Ophir G, Kaun KR, Azanchi R, Mohammed H, Heberlein U. Sexual deprivation increases ethanol intake in Drosophila. Science. 2012;335:1351–1355. doi: 10.1126/science.1215932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kato M, Sakai K, Endo A. Koningic acid (heptelidic acid) inhibition of glyceraldehyde 3-phosphate dehydrogenases from various sources. Biochem Biophys Acta. 1992;1120:113–116. doi: 10.1016/0167-4838(92)90431-c. [DOI] [PubMed] [Google Scholar]
- 66.Obici S, Feng Z, Arduini A, Conti R, Rossetti L. Inhibition of hypothalamic carnitine palmitoyltransferase-1 decreases food intake and glucose production. Nat Med. 2003;9:756–761. doi: 10.1038/nm873. [DOI] [PubMed] [Google Scholar]
- 67.Pocai A, Lam TK, Obici S, Gutierrez-Juarez R, Muse ED, Arduini A, et al. Restoration of hypothalamic lipid sensing normalizes energy and glucose homeostasis in overfed rats. J Clin Invest. 2006;116:1081–1091. doi: 10.1172/JCI26640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loftus TM, Jaworsky DE, Frehywot GL, Townsend CA, Ronnett GV, Lane MD, et al. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science. 2000;288:2379–2381. doi: 10.1126/science.288.5475.2379. [DOI] [PubMed] [Google Scholar]
- 69.Marty N, Dallaporta M, Thorens B. Brain glucose sensing, counterregulation, and energy homeostasis. Physiology. 2007;22:241–251. doi: 10.1152/physiol.00010.2007. [DOI] [PubMed] [Google Scholar]
- 70.Wen Z, Gulia M, Clark KD, Dhara A, Crim JW, Strand MR, et al. Two insulin-like peptide family members from the mosquito Aedes aegypti exhibit different biological and receptor binding activities. Mol Cell Endocrinol. 2010;328:47–55. doi: 10.1016/j.mce.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giulivi C, Ross-Inta C, Horton AA, Luckhart S. Metabolic pathways in Anopheles stephensi mitochondira. Biochem J. 2008;415:309–316. doi: 10.1042/BJ20080973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Birse RT, Choi J, Reardon K, Rodriguez J, Graham S, Diop S, et al. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 2010;12:533–544. doi: 10.1016/j.cmet.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.You Y-j, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-β signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 75.Ross-Inta CM, Zhang YF, Almendares A, Giulivi C. Threonine-deficient diets induced changes in hepatic bioenergetics. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1130–G1139. doi: 10.1152/ajpgi.90545.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.