Abstract.
The four serotypes of dengue virus (DENV-1, -2, -3, and -4) have had a rapidly expanding geographic range and are now endemic in over 100 tropical and subtropical countries. Sri Lanka has experienced periodic dengue outbreaks since the 1960s, but since 1989 epidemics have become progressively larger and associated with more severe disease. The dominant virus in the 2012 epidemic was DENV-1, but DENV-4 infections were also commonly observed. DENV-4 transmission was first documented in Sri Lanka when it was isolated from a traveler in 1978, but has been comparatively uncommon since dengue surveillance began in the early 1980s. To better understand the molecular epidemiology of DENV-4 infections in Sri Lanka, we conducted whole-genome sequencing on dengue patient samples from two different geographic locations. Phylogenetic analysis indicates that all sequenced DENV-4 strains belong to genotype 1 and are most closely related to DENV-4 viruses previously found in Sri Lanka and those recently found to be circulating in India and Pakistan.
INTRODUCTION
Dengue is the most important arboviral disease afflicting humans in the tropical and subtropical regions of the world.1,2 Over the past 40 years, dengue virus (DENV) 1–4 have dramatically increased their geographic distribution; indeed, much of the tropical and subtropical world is now hyperendemic with multiple serotypes in co-circulation at any given time.3 Currently, an estimated 3.6 billion people live in areas of risk, with an estimated 390 million new infections annually, of which 96 million are symptomatic dengue fever (DF) and 2 million are severe dengue hemorrhagic fever (DHF)/dengue shock syndrome.4 In addition to the substantial public health burden associated with the disease, dengue also has a profoundly negative economic impact on afflicted countries through reductions in worker productivity and tourism.5,6 Furthermore, with increasing global population growth, urbanization and international transportation of people and goods, dengue incidence is likely to continue its explosive growth for the foreseeable future.7
The first serologically confirmed DF case in Sri Lanka was reported in 1962 and the island-country experienced small, periodic DF epidemics thereafter with evidence of all four DENV serotypes in circulation.8,9 In 1989, Sri Lanka experienced its first epidemic of DHF, driven primarily by DENV-3.8,10,11 Since then, the number of DF and DHF cases have increased more than 10-fold,12 contributing to the major socioeconomic burden that dengue now poses in Sri Lanka.13 In the epidemics of the 1990s and early 2000s, DENV-1, -2, and -3 were isolated and DENV-3 was considered to be the dominant serotype.14 The most recent epidemic in 2009 was driven primarily by a new genotype of DENV-1 with 35,508 cases and 346 deaths reported.15 An average of roughly 40,000 dengue cases have since been reported annually, with an incidence rate of 175/100,000 population.16,17 Beginning in 2012, enhanced surveillance identified an increase in DENV-4 infections.18 Full-genome sequencing of DENV-4 strains from two geographically distinct sites in Sri Lanka was conducted to better understand the molecular epidemiology of this emergent serotype.
METHODS
Clinical cohorts.
Samples were selected from two study cohorts in Sri Lanka; one study was based out of Colombo, and the other study was based out of Galle (Table 1). Thirteen samples were selected from the Colombo study, where researchers reported acute dengue in 43.6% and DHF in 11% of their cases.18 DENV-4 was found in 15% of their cases and DENV-1 in 85%.18 Ten samples were selected from the Galle study, where researchers reported acute dengue in 39.8%.19 DENV-4 was found in 7.1% of their cases, DENV-1 in 91.2% and DENV-2 in 1.7%.19 Ethics review and approval was obtained from the participating institutions.
Table 1.
Characteristics of samples
| GenBank accession no. | Country | City | Sex | Age | Days of fever | Time |
|---|---|---|---|---|---|---|
| KX059015 | Sri Lanka | Colombo | F | 12 | 2 | 2012.641 |
| KX059016 | Sri Lanka | Colombo | F | 9 | 2 | 2012.375 |
| KX059017 | Sri Lanka | Colombo | M | 8 | 1 | 2012.773 |
| KX059018 | Sri Lanka | Colombo | F | 51 | 1 | 2012.381 |
| KX059019 | Sri Lanka | Colombo | M | 11 | 4 | 2012.603 |
| KX059020 | Sri Lanka | Colombo | M | 65 | 4 | 2012.822 |
| KX059021 | Sri Lanka | Colombo | F | 25 | 5 | 2012.868 |
| KX059022 | Sri Lanka | Colombo | F | 46 | 5 | 2012.89 |
| KX059023 | Sri Lanka | Colombo | M | 46 | 2 | 2012.975 |
| KX059024 | Sri Lanka | Colombo | F | 42 | 4 | 2013.041 |
| KX059025 | Sri Lanka | Colombo | M | 20 | 3 | 2013.101 |
| KX059026 | Sri Lanka | Colombo | M | 4 | 3 | 2013.142 |
| KX059027 | Sri Lanka | Colombo | M | 23 | 5 | 2013.211 |
| KX059028 | Sri Lanka | Galle | M | 34 | 5 | 2012.567 |
| KX059029 | Sri Lanka | Galle | M | 49 | 5 | 2012.627 |
| KX059030 | Sri Lanka | Galle | M | 43 | 3 | 2012.622 |
| KX059031 | Sri Lanka | Galle | M | 20 | 5 | 2012.663 |
| KX059032 | Sri Lanka | Galle | M | 23 | 3 | 2012.679 |
| KX059033 | Sri Lanka | Galle | M | 26 | 2 | 2012.753 |
| KX059034 | Sri Lanka | Galle | F | 16 | 1 | 2012.764 |
| KX059035 | Sri Lanka | Galle | M | 28 | 3 | 2012.786 |
| KX059036 | Sri Lanka | Galle | M | 31 | 3 | 2012.879 |
| KX059037 | Sri Lanka | Galle | M | 24 | 2 | 2012.921 |
F = female; M = male.
Full-genome sequencing.
Samples for full-genome sequencing were selected to evenly cover a temporal range from May 2012 to March 2013. DENV RNA was extracted primarily from acute sera, but also from culture supernatant when the primary sample was depleted, using the QIAamp® Viral RNA Mini Kit (Qiagen™, Valencia, CA). Complementary DNA synthesis was performed using SuperScript™ III First-Strand Synthesis System for reverse transcription-polymerase chain reaction (PCR) (Invitrogen™, Carlsbad, CA). The DENV full genome was divided into five overlapping amplicons (see Supplemental Figure 1) and sequenced according to the methods previously described.20 PCR cycling parameters for the five amplicons can be found in Supplemental Table 1. PCR products were separated on a 1% agarose TAE gel and amplicons purified using a Qiagen QIAquick® PCR Purification Kit. Purified amplicons were sequenced by Sanger sequencing.21 A complete list of sequencing primers can be found in Supplemental Table 2.
Phylogenetic analyses and molecular dating.
For phylogenetic reconstruction using E-genes, a total of 1,062 full E-gene sequences of DENV-4 with collection date information were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Because of the large number of sequences, a neighbor-joining tree was constructed first and similar sequences with same years of isolation were removed so that a total of 558 unique sequences were combined with the 23 newly sequenced isolates from Sri Lanka. All sequences were confirmed as nonrecombinant by using RDP, GENECONV, and BootScan methods in RDP3.22 Nucleotide sequences were aligned using MUSCLE and a nonbootstrapped maximum likelihood (ML) phylogenetic reconstruction was made with PhyML 3.0.23,24 The best-fit model, the GTR+I+Γ, was selected in jModelTest 2.1.7.25,26 We used Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen/) to evaluate the possibility of having temporal structure on the ML tree. Given the apparent clock-likeness signal on the tree (R2 = 0.4541), a Bayesian approach implemented in the BEAST v1.8.2 was then used to date the time to most recent common ancestor.27 Analysis was carried out with an uncorrelated lognormal distribution (UCLD) relaxed clock model, using a Bayesian skyline plot coalescent tree prior. For UCLD-relaxed clock analysis, we used a GTR+I+Γ substitution model. A total of 400 million Markov chain Monte Carlo states, and used Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer/) to ensure a convergence of parameters was reached (i.e., effective sample sizes > 200). After removal of 10% of the states as burn-in, a maximum clade credibility tree was generated using TreeAnnotator implemented in BEAST software package. The phylogenetic tree was visualized and edited using FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
For phylogenetic reconstruction using full genomes, all complete DENV-4 sequences with collection date information were downloaded from GenBank (160 sequences) and combined with our current 23 sequenced samples, for a total of 183 sequences.
For comparison with other available Sri Lankan strains, all Sri Lankan sequences and representative sequences from genotypes I and II were downloaded from GenBank and combined with our current 23 sequenced samples, for a total of 37 sequences. Because of the variable sequence lengths, the largest overlapping region of 296 bp for positions 787–1,083 was used. Sequences were aligned using MAFFT and identical sequences were removed (20 sequences), leaving a total of 17 sequences for analysis.
RESULTS
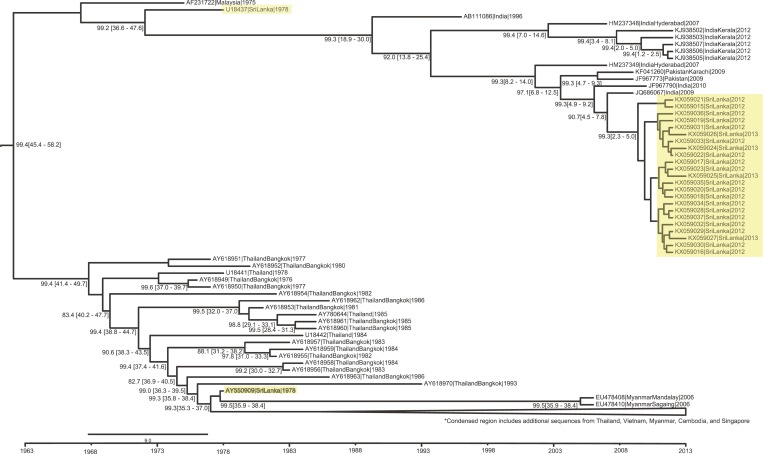
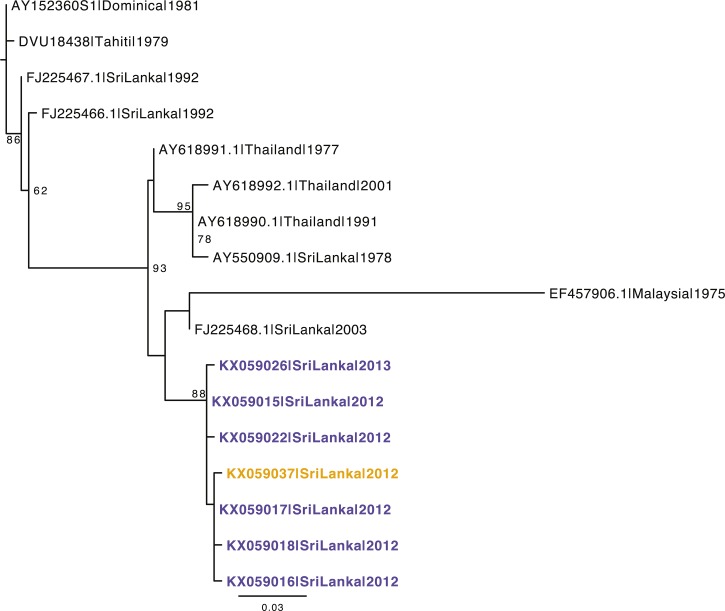
Two major lineages of DENV-4 have been observed in Sri Lanka since collection began, with the earliest examples from 1978 (AY550909 and U18437) (Supplemental Figure 2, Figure 1). In comparison, all 23 DENV-4 from our current study belong to genotype 1 and are most closely related to the 1978 Sri Lankan strain (U18437) and recent strains from India and Pakistan (Figure 1). From the phylogenetic analysis with all available DENV-4 sequences from Sri Lanka (Figure 2), we observe that the viruses isolated in 2003–2004 also belong to the same genotype as the 1978 and 2012–2013 isolates.
Figure 1.
Selection of temporally structured maximum clade credibility phylogenetic tree for dengue virus-4. For selected major nodes, bootstrap values as percentages and time 95% highest posterior density intervals in brackets are shown. Highlighted in gray are strains from Sri Lanka, where blue color represents viruses from Colombo and red from Galle. Condensed region, as noted, include additional sequences from Thailand, Vietnam, Myanmar, Cambodia, and Singapore.
Figure 2.
Maximum likelihood tree of all available, nonredundant dengue virus-4 sequences from Sri Lanka along with selected sequences from neighboring regions. The 296 base sequences common to all strains were used to conduct the analysis. Strains from the current study are in bold with strains from Colombo depicted in blue and those from Galle depicted in yellow.
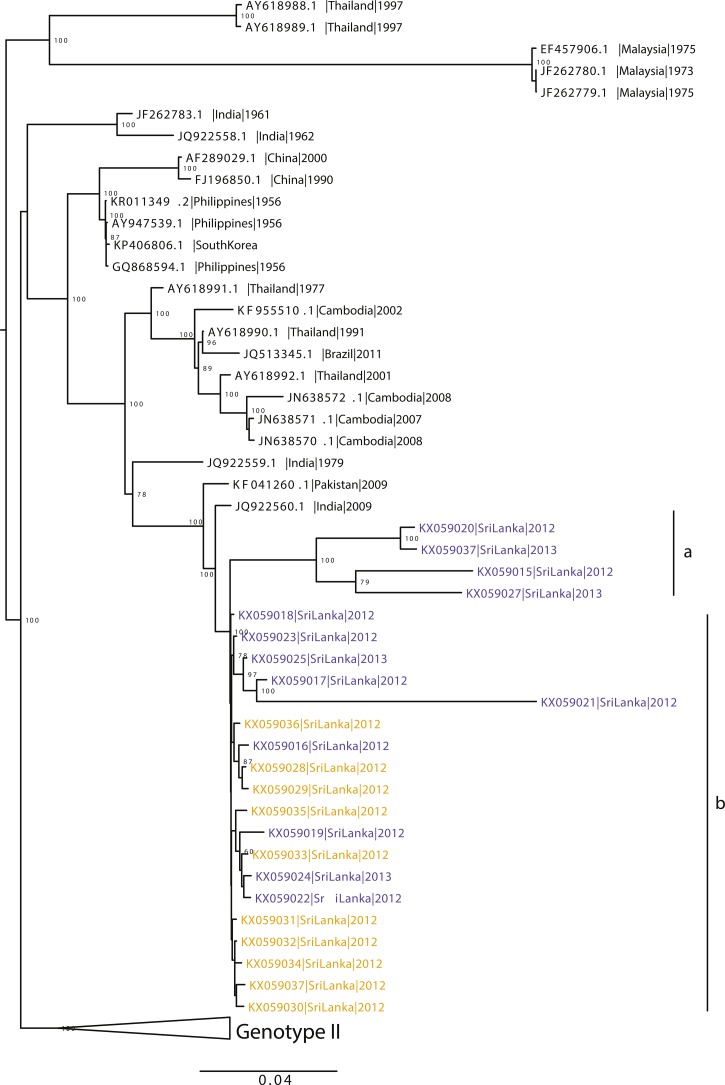
Unlike the results from our complete E-gene-only analysis, which showed that all circulating DENV-4 strains were highly related to one another (Figures 1 and 2), the higher resolution full-genome analysis suggests that there are at least two distinct clades of viruses circulating in the Colombo cohort (Figure 3). One of these clades is closely related to all of the isolates circulating in the monophyletic Galle cohort (Figure 3). In comparison to other full-genome sequences in GenBank, the data largely concur with the E-gene analysis in that the strains from our current study are most closely related to strains from India and Pakistan (Figure 3).
Figure 3.
Maximum likelihood tree of dengue virus-4, genotype I created using all available full genomes on National Center for Biotechnology Information. Strains from the current study are in bold with strains from Colombo depicted in blue and those from Galle depicted in yellow. Clade “a” indicates the clade found only in Colombo and clade “b” indicates the clade found in both Galle and Colombo.
The 23 DENV-4 strains were then compared with the most closely related full-genome strains from India (JQ922560) and Pakistan (KF041260), as well as partial genomes of other Sri Lankan DENV-4 strains available in National Center for Biotechnology Information (AY550909, FJ225466, FJ225467, FJ225468, FJ225469, FJ225470, and FJ225471) (Supplemental Figure 3). The comparison between the full-genome sequences show nucleotide level differences in all parts of the genome except for the 5′ end and the M gene. The greatest number of amino acid changes between the current 23 Sri Lankan strains and the Indian and Pakistani strain was located in the E-gene with an average of 4.35 and 3.91 amino acid changes, respectively.
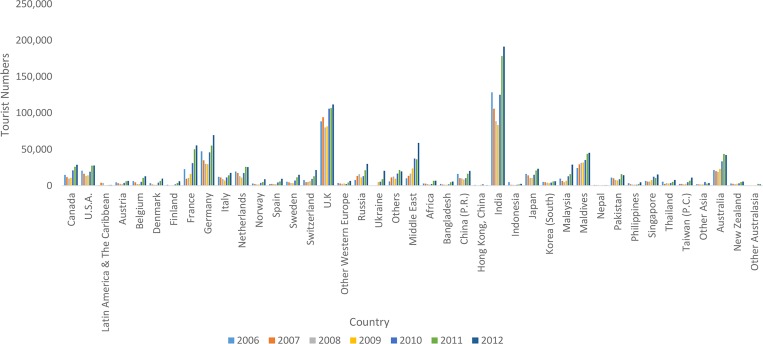
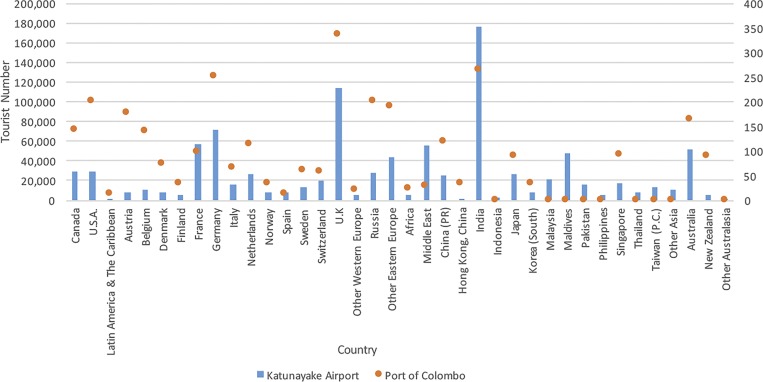
To gain further insight into the relationship between virus circulation in Sri Lanka and other countries around the world, we conducted an analysis of travel information for Sri Lanka between 2006 and 2012 using information from the Sri Lanka Tourism Development Authority. Our analysis indicates that since 2006, the largest number of visitors to Sri Lanka originated from India (Figure 4). The most predominant mode of documented transportation from India to Sri Lanka is by air travel with less than 1% of all documented travelers from India arriving by sea (Figure 5). It should be noted, however, that these figures do not include the undocumented travel between India and Sri Lanka during the 25-year period of political unrest beginning in the 1980s.
Figure 4.
Number of tourists traveling into Sri Lanka from 2006 to 2012 (Sri Lanka Tourism Development Authority).
Figure 5.
Number of tourists by mode of transportation into Sri Lanka in 2012 (Sri Lanka Tourism Development Authority).
DISCUSSION
The 2012–2013 dengue epidemic in Sri Lanka was driven predominantly by DENV-1 with approximately 20% of dengue cases caused by DENV-4.18 In this study, we have sequenced the full genomes of 23 isolates of DENV-4 collected from two cities in Sri Lanka, Colombo, and Galle, to better understand the evolution of these viruses and how they relate to other DENV-4 strains globally. Within the relatively small sampling of our current cohorts, we have found the diversity of DENV-4 to be higher in Colombo compared with Galle. Based on our analysis, there is no clear genetic divergence of our newly isolated viruses from the different locations within the country, suggesting that the virus in circulation most likely originated as a single introduction.
Although we conducted full-genome sequencing for our current isolates, most publicly available information for DENV-4 covers only the sequences coding for one or more of the structural genes. Unfortunately, this also includes the majority of the DENV-4 sequences available from Sri Lanka; the two samples from 1978 include nucleotides 1–2322 of the polyprotein coding sequence, whereas the six samples from 1992, 2003, and 2004 include nucleotides 137–1074 of the polyprotein coding sequence.12 In an effort to fully use these disparate data, we conducted three phylogenetic analyses in parallel.
In accordance with previous findings, our E-gene analysis shows that two major lineages of DENV-4 have been observed in Sri Lanka since surveillance began, with data indicating that the currently circulating strain is a descendent of the Sri Lanka virus from 1978 (U18437).12 Our analysis also suggest that this virus spread to India and/or Pakistan before reemerging in Sri Lanka in 2009 (95% highest posterior density intervals 4.5–7.8 years ago). For increased resolution, we conducted a full-genome phylogenetic analysis with available full DENV-4 genomes. Overall, the full-genome analysis largely concurred with the E-gene analysis indicating that our study strains are most closely related to recent strains from India and Pakistan. However, due to the lack of additional full-genome sequences from Sri Lanka, we are unable to further characterize the relationship between the isolates in our current study to those which have circulated in the past.
There have been fewer DENV-4 outbreaks globally compared with the other three DENV serotypes and the cause of the unusually high number of DENV-4 cases in the ongoing Sri Lankan outbreak is unknown.28 One possibility is that the current Sri Lankan DENV-4 strain may have recently acquired mutations that have increased epidemic potential of the virus. This is not without precedent; the South Pacific islands have observed alternations of silent transmission and explosive epidemic transmission from closely related American genotype II DENV-2 strains in the 1970s.29 Puerto Rico has also observed genetic shifts in DENV-2 since the late 1980s that correlate with increased epidemic potential.30 Interestingly, recent studies have shown that sequences outside of the E-gene in these Puerto Rican viruses can have a large impact on pathogenicity and epidemic potential.31 We are limited in exploring this at the time, however, due to the paucity of other DENV-4 full-genome sequence from Sri Lanka. As such, we would like to emphasize the importance of moving toward the use of full-genome sequencing rather than just the E-gene so that these other important regions can be examined as well. Given the recent technologic improvements and rapidly falling costs of sequencing, we feel this is an achievable goal.32 Combined with good epidemiological data, this knowledge has the potential to transform the current paradigm of retrospective study into one of active prediction and targeted, preventative action.
Comparison at the amino acid level across the current 23 Sri Lankan strain with the most closely related Indian and Pakistani strain showed minor differences. The 5′ end of all strains showed no differences, suggesting the conservative need of this region for viral survival not only in DENV-1, but also in DENV-4.33 The greatest number of differences was observed in the E-gene when compared with both the Indian and Pakistani strains, a region highly studied as the structure of the protein can affect the interaction of the virus with host cells. Among the Sri Lankan strains, the greatest change in the E-gene is observed when comparing the current strains with one isolated from 1978, which illustrates the adaptive changes that the virus has made over time. Unfortunately, the other available Sri Lankan strains do not have a full coverage of the E-gene to conduct an informative comparison of the Sri Lankan strains temporally.
Given the large gaps in the surveillance data for the intervening years between our current isolates and those from previous three decades, we are unable to decisively determine whether the virus from 1978 continued to circulate within Sri Lanka until 2012 or disappeared and was reintroduced from India and/or Pakistan, seeding the outbreak observed in our current study. Further complicating matters, our travel data analysis indicates robust movement of people between these three countries and highlights the importance of better understanding the virus exchange dynamics between these neighboring countries. Surveillance efforts in Sri Lanka at both the local and national levels have greatly improved in recent years, and it is vital that these surveillance efforts continue both for disease management as well as outbreak characterization.15,18,34 Increased surveillance in Sri Lanka and its neighbors, combined with sequence data will strengthen our ability to characterize the evolution and circulation patterns of the four dengue serotypes and it is important that these efforts continue.
Supplementary Material
Supplemental Figure and Tables.
Note: Supplemental figures and table appear at www.ajtmh.org.
REFERENCES
- 1.Gubler DJ, 1997. Epidemic dengue/dengue haemorrhagic fever: a global public health problem in the 21st century. Dengue Bull 21: 1–14. [Google Scholar]
- 2.World Health Organization , 2012. Global Strategy for Dengue Prevention and Control 2012–2020. Geneva, Switzerland: World Health Organization Press. [Google Scholar]
- 3.Wilder-Smith A1, Gubler DJ, 2008. Geographic expansion of dengue: the impact of international travel. Med Clin North Am 92: 1377–1390. [DOI] [PubMed] [Google Scholar]
- 4.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI, 2013. The global distribution and burden of dengue. Nature 496: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beatty ME, Beutels P, Meltzer MI, Shepard DS, Hombach J, Hutubessy R, Dessis D, Coudeville L, Dervaux B, Wichmann O, Margolis HS, Kuritsky JN, 2011. Health economics of dengue: a systematic literature review and expert panel’s assessment. Am J Trop Med Hyg 84: 473–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shepard DS, Undurraga EA, Betancourt-Cravioto M, Guzman MG, Halstead SB, Harris E, Mudin RN, Murray KO, Tapia-Conyer R, Gubler DJ, 2014. Approaches to refining estimates of global burden and economics of dengue. PLoS Negl Trop Dis 8: e3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubler DJ, 2011. Dengue, urbanization and globalization: the Unholy Trinity of the 21(st) century. Trop Med Health 39: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vitarana T, Jayakuru WS, Withane N, 1997. Historical account of dengue haemorrhagic fever in Sri Lanka. Dengue Bull 21: 2. [Google Scholar]
- 9.Messer WB, Vitarana UT, Sivananthan K, Elvtigala J, Preethimala LD, Ramesh R, Withana N, Gubler DJ, De Silva AM, 2002. Epidemiology of dengue in Sri Lanka before and after the emergence of epidemic dengue hemorrhagic fever. Am J Trop Med Hyg 66: 765–773. [DOI] [PubMed] [Google Scholar]
- 10.Lanciotti RS, Gubler DJ, Trent DW, 1997. Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol 78: 2279–2284. [DOI] [PubMed] [Google Scholar]
- 11.Lanciotti RS, Lewis JG, Gubler DJ, Trent DW, 1994. Molecular evolution and epidemiology of dengue-3 viruses. J Gen Virol 75: 65–75. [DOI] [PubMed] [Google Scholar]
- 12.Kanakaratne N, Wahala WMPB, Messer WB, Tissera HA, Shahani A, Abeysinghe N, de Silva AM, Gunasekera M, 2009. Severe dengue epidemics in Sri Lanka, 2003–2006. Emerg Infect Dis 15: 192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thalagala N, Tissera H, Palihawadana P, Amarasinghe A, Ambagahawita A, Wilder-Smith A, Shepard DS, Tozan Y, 2016. Costs of dengue control activities and hospitalizations in the public health sector during an epidemic year in urban Sri Lanka. PLoS Negl Trop Dis 10: e0004466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM, 2003. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis 9: 800–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tissera HA, Ooi EE, Gubler DJ, Tan Y, Logendra B, Wahala WM, de Silva AM, Abeysinghe MR, Palihawadana P, Gunasena S, Tam CC, Amarasinghe A, Letson GW, Margolis HS, De Silva AD, 2011. New dengue virus type 1 genotype in Colombo, Sri Lanka. Emerg Infect Dis 17: 2053–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Health EUoMo , Disease Surveillance. Available at: http://www.epid.gov.lk/web/index.php. Accessed February 16, 2013.
- 17.Initiative DV, 2015. Dengue in Sri Lanka: Burden, Challenges and Prevention Strategies–An Exclusive Interview with Dr. Hasitha Tissera. Available at: http://www.denguevaccines.org/dengue-sri-lanka-burden-challenges-and-prevention-strategies-exclusive-interview-dr-hasitha-tissera. Accessed November 17, 2016.
- 18.Tissera H, Amarasinghe A, Gunasena S, DeSilva AD, Yee LW, Sessions O, Muthukuda C, Palihawadana P, Lohr W, Byass P, Gubler DJ, Wilder-Smith A, 2016. Laboratory-enhanced dengue sentinel surveillance in Colombo District, Sri Lanka: 2012–2014. PLoS Negl Trop Dis 10: e0004477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodinayake CK, Tillekeratne LG, Nagahawatte A, Devasiri V, Kodikara Arachichi W, Strouse JJ, Sessions OM, Kurukulasooriya R, Uehara A, Howe S, Ong XM, Tan S, Chow A, Tummalapalli P, De Silva AD, Østbye T, Woods CW, Gubler DJ, Reller ME, 2016. Emergence of epidemic dengue-1 virus in the southern province of Sri Lanka. PLoS Negl Trop Dis 10: e0004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christenbury JG, Aw PP, Ong SH, Schreiber MJ, Chow A, Gubler DJ, Vasudevan SG, Ooi EE, Hibberd ML, 2010. A method for full genome sequencing of all four serotypes of the dengue virus. J Virol Methods 169: 202–206. [DOI] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson AR, 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P, 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26: 2462–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O, 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 25.Darriba D, Taboada GL, Doallo R, Posada D, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guindon S, Gascuel O, 2003. A Simple, Fast and Accurate Method to Estimate Large Phylogenies by Maximum-Likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 27.Drummond AJ, Suchard MA, Xie D, Rambaut A, 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messina JP, Brady OJ, Scott TW, Zou C, Pigott DM, Duda KA, Bhatt S, Katzelnick L, Howes RE, Battle KE, Simmons CP, Hay SI, 2014. Global spread of dengue virus types: mapping the 70 year history. Trends Microbiol 22: 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steel A, Gubler DJ, Bennett SN, 2010. Natural attenuation of dengue virus type-2 after a series of island outbreaks: a re-trospective phylogenetic study of events in the South Pacific three decades ago. Virology 405: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett SN, Holmes E, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO, 2006. Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J Gen Virol 87: 9. [DOI] [PubMed] [Google Scholar]
- 31.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE, 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aw PP, de Sessions PF, Wilm A, Hoang LT, Nagarajan N, Sessions OM, Hibberd ML, 2014. Next-generation whole genome sequencing of dengue virus. Methods Mol Biol 1138: 175–195. [DOI] [PubMed] [Google Scholar]
- 33.Tang Y, Rodpradit P, Chinnawirotpisan P, Mammen MP, Li T, Lynch JA, Putnak R, Zhang C, 2010. Comparative analysis of full-length genomic sequences of 10 dengue serotype 1 viruses associated with different genotypes, epidemics, and disease severity isolated in Thailand over 22 years. Am J Trop Med Hyg 83: 1156–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tissera H, Amarasinghe A, De Silva AD, Kariyawasam P, Corbett KS, Katzelnick L, Tam C, Letson GW, Margolis HS, de Silva AM, 2014. Burden of dengue infection and disease in a pediatric cohort in urban Sri Lanka. Am J Trop Med Hyg 91: 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure and Tables.