Abstract.
Nontuberculous mycobacteria (NTM) cause significant pulmonary infections in humans. Researchers have reported an association between interferon-gamma receptor-1 (IFN-γR1 or IFNGR1) deficiency and susceptibility to NTM, but the relevance of polymorphism within these genes is not yet clear. In this study, a single nucleotide polymorphism (SNP), T to C, at position-56 in NTM patients with pulmonary disease was investigated. Molecular identification of Mycobacterium isolates was performed with hsp65 genes using polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). Then, the host genomic DNA from confirmed NTM patients (N = 80) and control subjects (N = 80) were screened for SNPs of IFNGR1 (T-56C) by PCR-RFLP. The results indicated that NTM patients had higher TC (26/80; 32.5%) or CC (46/80; 57.5%) genotypes in comparison with control groups (TC genotypes [22/80, 27.5%]; CC genotypes [6/80, 7.5%]) (P < 0.05). In this regard, all the patients infected with rapid-growing Mycobacterium (RGM, i.e., Mycobacterium chelonae and Mycobacterium fortuitum) had CC genotypes (100%). In contrary, only 50.7% (35/69) of infected patients with slow-growing Mycobacterium (i.e., Mycobacterium simiae, Mycobacterium kansasii, and Mycobacterium avium-intracellulare) had CC genotypes. Thus, patients with CC mutation in IFNGR1 at position-56 are more likely to develop RGM infection. In overall, there is a significant association between SNP of IFNGR1 at position-56 and susceptibility to NTM infection. Based on these data, we propose SNP of IFNGR1 at position-56 as a suitable “biomarker” for identifying populations at higher risk of infection.
INTRODUCTION
Nontuberculous mycobacteria (NTM) are free-living organisms that commonly occur in different environmental reservoirs, including water, soil, and biofilms.1–3 They cause a variety of diseases in humans, notably severe, protracted lung disease in patients with underlying lung disorders.4 Conditions such as human immunodeficiency virus (HIV) infection, acquired immune deficiency syndrome (AIDS), immunosuppressive therapy, cancer, autoimmune disease, and diabetes mellitus can predispose a person to NTM infection.5–10 However, it is not clear why some infected people become ill or why certain individuals have a particular presentation of NTM diseases.6 Normally, the occurrence of NTM infections in a “well-defined population” suggests an immune deficiency in the host genetic system.6 During the recent years, a considerable number of candidate-based association studies have been performed to demonstrate the association of predicted functional DNA variations with NTM infection.6 For example, polymorphisms of tumor necrosis factor (TNF-α) gene, interleukin-12-receptor-1 (IL-12R1), and interferon-gamma (IFN-γ) signaling were shown to be associated with susceptibility to Mycobacterium.7–15 Among host genetic factors, IFN-γ plays a key role in protective immune response against mycobacterial infection.16,17 IFN-γ executes its antimycobacterial effector mechanism by the activation of macrophages and dendritic cells via interaction with its receptor complex.12,17,18 Generally, IFN-γ mediates its action through a receptor complex (IFN-γ R or IFNGR) composed of two heterologous subunits, a ligand-binding subunit (IFNGR1), and an accessory subunit (IFNGR2) on the cell surface.17–19 The gene coding for IFNGR1 is located on the long arm of chromosome 6 at position 6q23.3, which consists of seven exons.12,19,20 Patients with complete or partial IFNGR1 receptor deficiency are highly susceptible to infection by NTM, Mycobacterium tuberculosis, and some Salmonella species.10,21 In 2006, Sampaio and others provided detailed evidences on the genetic mechanisms underlying polymorphisms of IFNGR1 and susceptibility to pulmonary tuberculosis (PTB).22 They showed that the substitution of wild type (T) to variant (C) forms in IFNGR1 at position-56 caused 3-fold lower expression of a receptor gene on the cell surface.10,23 This will consequently lead to decrease in the binding site for the transcription factor of activator protein 2 and 4 (AP-2 and AP-4) on both strands.10,23 His findings support the hypothesis that variation in IFN-γ gene might increase the risk of TB development.10 In a study performed by Varahram and others, a correlation between single nucleotide polymorphisms (SNPs) of IFNGR1 and susceptibility to Iranian PTB patients were demonstrated.15 In a convincing report, Velayati and others showed an association between polymorphisms of IFNGR1 at position-56 with susceptibility to leprosy in children of the same family.24 Even though most of the available data outlines the importance of polymorphisms and susceptibility to M. tuberculosis species, but it is expected that polymorphisms in IFNGR1 at position-56 might possibly affect the NTM development and outcomes, which need to be investigated.15,20,24 In the present study, we aimed to study the IFNGR1 T-56C SNP in patients with pulmonary disease that were infected with NTM. Our results showed a significant correlation between IFNGR1 T-56C SNP and NTM infection among studied populations.
MATERIALS AND METHODS
Study group.
The investigation was conducted from December 2012 to June 2016 at the National Research Institute of Tuberculosis and Lung Diseases (NRITLD). NRITLD is the only World Health Organization–approved center for the detection and diagnosis of Mycobacterium infection in Iran.15 A total of 80 patients with pulmonary disease with confirmed culture-positive NTM results and 80 healthy volunteers (referred to as normal controls) were included in the study. Patient’s confirmation with NTM infection was based on the American Thoracic Society and the Infectious Disease Society of America guidelines.25 In brief, the diagnosis was based on the presence of clinical symptoms, radiologic abnormalities, and microbiologic cultures in conjunction with the exclusion of other potential etiologies. Patients and control subjects were matched for age, sex, and nationality.
Ethics.
The Institutional Review Board at the NRITLD approved the study and all the patients signed informed consent (MRC-2013/028a/sbmu).
Mycobacterial isolation.
Collected sputum samples from each patient were digested and decontaminated by Petroff’s method.15 Lowenstein–Jensen media were used for bacterial growth. The extracted DNA from culture-positive samples was used for identification.26
Mycobacterial identification.
The isolates were characterized using hsp65 genes spacer polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). For hsp65 gene, a PCR reaction was amplified in 50-mL mixtures containing 4 pmol of specific primers (TB15 [59-CGT AYG ACG AAG AGG CCC GT-39] and TB17 [59-WAS GGR TCC TCS AGG ACS GC-39], 1 mL deoxynucleotide triphosphates (dNTPs), 1.5 mL MgCl, 0.25 mL (1 U) Taq polymerase, 2.5 mL (1%) dimethyl sulfoxide, 5 mL PCR buffer, and 5 mL (20 ng) of extracting DNA.27,28 The reaction mixture was subjected to 30 amplification cycles (20 seconds at 95°C, 1 minute at 60°C, 40 seconds at 72°C) followed by a 5-minute extension at 95°C.27,28 The PCR product of the first step (470 base pairs [bp]) was used for the second amplification using primers TB11 (59-ACC AAC GAT GGT GTG TCC AT-39) and TB12 (59-CTT GTC GAA CCG CAT ACC CT), which produced a segment of 439 bp (26). PCR products were digested with 5 U of restriction enzyme HaeIII and BstEII for 24 hours at 37°C. The pattern of digested products was analyzed using 8% polyacrylamide gel.27,28
IFN-γ genotyping.
SNPs in IFN-γ receptor-1 (T-56 C) was performed using PCR-RFLP.10,24 The target DNA was amplified in a PCR reaction mixture containing 1× reaction buffer (50 mM KCl, 2 mM MgCl2), 67 mM Tris-HCl (pH 9.0), 0.2 U of Taq DNA-polymerase (Roche, Basel, Switzerland), 2 mM of dNTPs, and 20 pmol of each primer. For IFN-γ receptor-1 (T-56 C), the following primers were used to amplify a 455-bp product: 5′-GGTGACGGAAGTGACGTAAGG, reverse 5′-GGTCGCTGGCTCCAA. The PCR products of IFN-γ receptor-1 (T-56 C) were digested with 2 U enzymes of Bst110.22,29,30 Digested products were run on 8% polyacrylamide gel and were stained with silver nitrate.24
Statistical analyses.
Statistical analysis was performed using χ2 test to determine the statistical difference between studied groups. A P value less than 0.05 was considered statistically significant. Data were analyzed using SPSS version 18 software (IBM, Chicago, IL).15
RESULTS
Participants and demographics.
During the study period, 100 patients with confirmed NTM infection were hospitalized to NRITLD for diagnosis and treatment. Twenty patients were excluded because they either had NTM and TB (6/100; 6%) or NTM and lung cancer (3/100; 3%) together. Few patients (11/100; 11%) were not ready to donate their blood for research purposes. Thereby, in total, 80 cases and 80 controls enrolled in this study. The mean ages of patients and control were 48.2 ± 21.2 and 45.5 ± 20.16 years, respectively. Majority of patients (51; 63.7%) were over 45 years of age, and five were more than 70 years of age (5; 6.25%). Forty-eight (60%) patients were male and thirty-two (40%) were female. Fisher’s exact test showed a correlation between old age and susceptibility to NTM (P value < 0.001).
NTM isolation and IFN-γ receptor genotyping.
Majority of patients with pulmonary disease (69/80; 86.2%) were infected with slow-growing Mycobacterium (SGM) (Figure 1). Among identified SGM, M. simiae (62/80; 77.5%) and Mycobacterium kansasii (4/80; 5%) were the most frequent isolates (Table 1). Eleven patients (11/80; 13.75%) had been infected with rapid-growing Mycobacterium (RGM) (Table 1). For IFN-γ receptor genotyping, one type of T to C substitution at position-56 occurred which resulted in three types of genotypes (TT, TG, and GG) (Figure 2). As shown in Table 2, the frequency of the C allele (112 [70.0%]) was significantly more common in the NTM group (odds ratio = 0.6; 95% confidence interval = 0.4–0.9; P < 0.05), whereas the frequency of the C allele in the control group was as low as 27. 5%. In overall, we found only eight patients (10%) with TT genotypes; 72 patients (90%) had either heterozygous (TC: 32/80; 40%) or homozygous (CC: 40/80; 50%) mutations. Comparison of IFN-γ receptor genotyping among patients revealed that all cases with RGM infection had CC genotypes (11/11; 100%), whereas 50.7% (35/69) of patients with SGM, showed CC genotypes. In control subjects, TT genotypes was more frequent (56/80; 65%) than TC and CC genotypes (Table 2).
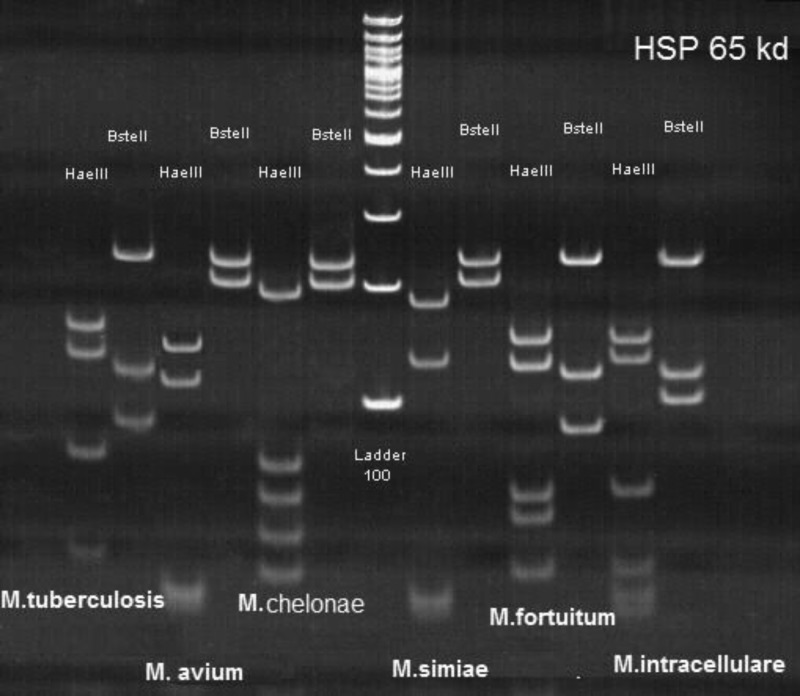
Figure 1.
The pattern of Mycobacterium isolates that were characterized using hsp65 genes spacer polymerase chain reaction restriction fragment length polymorphism.
Table 1.
The allele and genotyping frequencies of IFN-γ receptor-1 (T-56 C) among patients with different NTM species
| Isolated Mycobacterium | Total = 80 | IFN-γ receptor-1 (T-56 C) |
|---|---|---|
| Mycobacterium simiae | 62 | TT = 8 (12.9%) |
| TC = 25 (40.3%) | ||
| CC = 29 (46.7%) | ||
| Mycobacterium kansasii | 4 | TT = 0 |
| TC = 1 (25 | ||
| CC = 3 (75%) | ||
| Mycobacterium avium-intracellulare | 3 | CC = 3 (100%) |
| Mycobacterium chelonae | 6 | TT = 0 |
| TC = 0 | ||
| CC = 6 (100%) | ||
| Mycobacterium fortuitum | 5 | TT = 0 |
| TC = 0 | ||
| CC = 5 (100%) |
IFN = interferon; NTM = nontuberculous mycobacteria.
Figure 2.
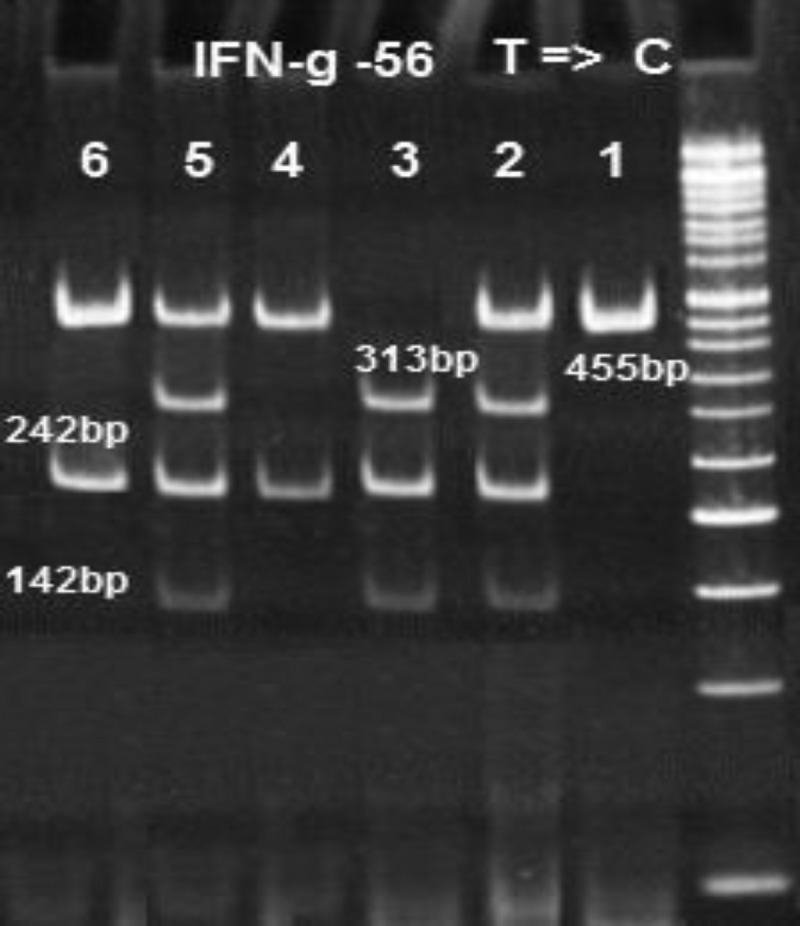
The electrophoresis pattern of polymerase chain reaction (PCR) restriction fragment length polymorphism of single nucleotide polymorphisms of interferon-gamma receptor-1 (IFNGR1) at position-56. The PCR product is 455 base pairs (bp) (line 1). After digestion with Bst1, three genotypes were obtained: TT (line 3 = 313 + 215 + 142 bp), TC (lines 2 and 5 = 455 + 313 + 216 + 142 bp), and CC (lines 4 and 6 = 455 + 215 bp).
Table 2.
The allele and genotyping frequencies of IFN-γ receptor-1 (T-56 C)
| Genotypes | Control (80) | NTM cases (80) | OR (95% CI) | P value |
|---|---|---|---|---|
| IFN-γ receptor-1 allele | ||||
| T | 126 (72.5%) | 48 (30.0%) | 0.05 | |
| C | 44 (27.5%) | 112 (70.0%) | 0.6 (0.4–0.9) | |
| Genotype | ||||
| TT | 52 (65%) | 8 (10.0%) | 0.6 (0.3–1.2) | S |
| TC | 22 (27.5%) | 32 (40.0%) | 1.3 (0.5–1.8) | S |
| CC | 6 (7.5%) | 40 (50.0%) | 1.5 (1.4–1.7) | S |
CI = confidence interval; IFN = interferon; NTM = nontuberculous mycobacteria; OR = odds ratio.
DISCUSSION
To our knowledge, this is the first case-control study which demonstrates association of IFN-γ receptor-1 (-56 CC) with susceptibility to NTM infection among Iranian patients with pulmonary disease.
As shown in Table 2, majority of patients (N = 72) had either TC (40%) or CC genotyping (50%). Normally, a switch from T to C at position-56 in the IFNGR1 promoter leads to a lower expression of luciferase in a standard test system.10 Therefore, lower levels of IFNGR1 expression would lead to reduced IFN-γ-mediated immunity against intracellular microbes by at least partially blocking the pathways which activate macrophages or process antigens.10,11,19,29,31 In this regard, Velayati and others showed an association between autosomal recessive disorders of IFN-γ receptor-1 (T-56 C) with susceptibility to leprosy in children of the same family.22 He investigated an influence of SNPs of various candidate genes, that is, vitamin D receptor, natural resistance-associated macrophage protein 1 gene, TNF-α, IL-10, IL-12 receptor (IL_12R1). However, polymorphism of IFN-γ receptor-1 (T-56 C) was shown to be the most effective SNP.24 In the present study, all patients (N = 11) with RGM infection (i.e., Mycobacterium chelonae and Mycobacterium fortuitum) had CC alleles. This might explain the correlation of IFN-γ receptor-1 (T-56 C) polymorphisms with RGM infection. Indeed, the increased susceptibility to NTM in a host with defective immunity was first reported in four children of the same family in 1995. These children were infected with M. chelonae, M. fortuitum, and two different subspecies of Mycobacterium avium-intracellulare.32 A whole genome search for homozygosity in three of the affected children showed a mutation in the coding region of IFNGR1.31 This mutation then resulted in the complete absence of IFN-γR1 expression at the cell surface and was subsequently identified as the cause of the defect in these children. Thereafter, other investigators outlined the importance of molecular defects in IFN-γ-dependent signaling with susceptibility to disseminated NTM infection.32–37 They showed that mutations in each of the IFN-γ signaling genes have either complete or partial deficiency of the encoded protein.32,35,36 The partial gene deficiency negatively affects the expression of aberrant proteins and as a result IFN-γ cannot be mediated.32 Complete deficiency of IFNγR1 has a much worse prognosis than partial deficiency. Hence, mutated genes in IFNγR1 can play a critical role in immunity against mycobacterial infection.38 In most previous studies,32–36 the NTM-infected patients were either children or had underlying diseases (e.g., HIV/AIDS). In the present study, the majority of patients were above 45 years of age, without any underlying disorder. These data might suggest a high risk of NTM infection in elderly people. A recent study by Mirsaeidi and others has highlighted the risk factors that are associated with NTM infection in North America.39 They demonstrated that while the U.S. population is aging, NTM diseases are increasing in an elderly populations.39 Therefore, they proposed that the elderly population in any community may be more vulnerable to NTM infection. Addressing the importance of polymorphisms of IFNγR1 at position-56 with susceptibility to mycobacterial infection, Wang and others performed an extensive meta-analysis of 56 CC in M. tuberculosis.20 They analyzed 1,497 TB cases and 1,742 healthy controls from six case-control studies that were reported from African, Asian, and Caucasian populations. They showed that IFNGR1 T-56C polymorphism is possibly associated with increased TB risk in Africans, but not in Asian or Caucasians.20 Wang and others had examined the Korean and Chinese populations as representatives of Asian communities,20 whereas the Iranian ethnicity is different from other Asians and the results may be variable. For example, Biranvand and others reported an association between SNPs in IFNGR1 T-56C polymorphisms and susceptibility to TB.40 Today it is known that reported discrepancies between studies regarding the impact of IFNGR1 polymorphisms on susceptibility to Mycobacterium infection may be due to population genetic and/or environmental factors. In this study, Mycobacterium simiae was the most prominent NTM among PTB-infected patients (62/80; 77.5%). Previous reports also documented M. simiae as the most frequent SGM species in Iran and in the Middle East region.41,42 Of 62 infected patients with M. simiae, 54 (87.0%) had SNP in IFNGR1 T-56C position. We also found eight M. simiae-infected patients with no polymorphisms in IFNGR1 at position-56 (TT genotyping), five of which were more than 60 years of age and three had diabetes mellitus. Therefore, we propose a further investigation in determining the associated risk factors with NTM infection.39
In conclusion, the 56 CC genotypes of IFN-γ receptor-1 were significantly associated with NTM infection. Although the number of studied population was low, patients belonged to a different geographical region of Iran that were referred to NRITLD for diagnosis and treatment. The high prevalence of CC genotypes among randomly selected patients with pulmonary disease outlines the correlation of this mutation with susceptibility to NTM infection.
REFERENCES
- 1.Cassidy PM, Hedberg K, Saulson A, McNelly E, Winthrop KL, 2009. Nontuberculous mycobacterial disease prevalence and risk factors: a changing epidemiology. Clin Infect Dis 49: 124–129. [DOI] [PubMed] [Google Scholar]
- 2.Falkinham JO, 2009. Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J Appl Microbiol 107: 356–367. [DOI] [PubMed] [Google Scholar]
- 3.Hruska K, Kaevska M, 2012. Mycobacteria in water, soil, plants and air: a review. Vet Med (Praha) 57: 623–679. [Google Scholar]
- 4.Winthrop KL, Chang E, Yamashita S, Iademarco MF, LoBue PA, 2009. Nontuberculous mycobacteria infections and anti-tumor necrosis factor-alpha therapy. Emerg Infect Dis 15: 1556–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glassroth J, 2008. Pulmonary disease due to nontuberculous mycobacteria. Chest 133: 243–251. [DOI] [PubMed] [Google Scholar]
- 6.Johnson MM, Odell JA, 2014. Nontuberculous mycobacterial pulmonary infections. J Thorac Dis 6: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benator DA, Gordin FM, 1996. Nontuberculous mycobacteria in patients with human immunodeficiency virus infection. Semin Respir Infect 11: 285–300. [PubMed] [Google Scholar]
- 8.Keating MR, Daly JS; AST Infectious Diseases Community of Practice , 2013. Nontuberculous mycobacterial infections in solid organ transplantation. Am J Transplant 13: 77–82. [DOI] [PubMed] [Google Scholar]
- 9.Velayati AA, Bakayev V, Bahadori M, Tabatabaei SJ, Alaei A, Farahbood A, Masjedi MR, 2007. Religious and cultural traits in HIV/AIDS epidemics in sub-Saharan Africa. Arch Iran Med 10: 486–497. [PubMed] [Google Scholar]
- 10.Cooke GS, Campbell SJ, Sillah J, Gustafson P, Bah B, Sirugo G, Bennett S, McAdam KP, Sow O, Lienhardt C, Hill AV, 2006. Polymorphism within the interferon-gamma/receptor complex is associated with pulmonary tuberculosis. Am J Respir Crit Care Med 174: 339–343. [DOI] [PubMed] [Google Scholar]
- 11.Park HY, Kwon YS, Ki CS, Suh GY, Chung MP, Kim H, Kwon OJ, Koh WJ, 2008. Interleukin-12 receptor beta1 polymorphisms and nontuberculous mycobacterial lung diseases. Lung 186: 241–245. [DOI] [PubMed] [Google Scholar]
- 12.Hwang JH, Kim EJ, Koh WJ, Kim SY, Lee SH, Suh GY, Kwon OJ, Ki CS, Ji Y, Kang M, Kim DH, 2007. Polymorphisms of interferon-gamma and interferon-gamma receptor 1 genes and non-tuberculous mycobacterial lung diseases. Tuberculosis (Edinb) 87: 166–171. [DOI] [PubMed] [Google Scholar]
- 13.Mansouri D, Mahdaviani SA, Khalilzadeh S, Mohajerani SA, Hasanzad M, Sadr S, Nadji SA, Karimi S, Droodinia A, Rezaei N, Linka RM, Bienemann K, Borkhardt A, Masjedi MR, Velayati AA, 2012. IL-2-inducible T-cell kinase deficiency with pulmonary manifestations due to disseminated Epstein-Barr virus infection. Int Arch Allergy Immunol 158: 418–422. [DOI] [PubMed] [Google Scholar]
- 14.Bakayev VV, Mohammadi F, Bahadori M, Sheikholslami M, Javeri A, Masjedi MR, Velayati AA, 2004. Arylamine N-acetyltransferase 2 slow acetylator polymorphisms in unrelated Iranian individuals. Eur J Clin Pharmacol 60: 467–471. [DOI] [PubMed] [Google Scholar]
- 15.Varahram M, Farnia P, Nasiri MJ, Karahrudi MA, Kazempour Dizagie M, Velayati AA, 2014. Association of Mycobacterium tuberculosis lineages with IFN-γ and TNF-α gene polymorphisms among pulmonary tuberculosis patient. Mediterr J Hematol Infect Dis 6: e2014015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper AM, Khader SA, 2008. The role of cytokines in the initiation, expansion, and control of cellular immunity to tuberculosis. Immunol Rev 226: 191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shtrichman R, Samuel CE, 2001. The role of gamma interferon in antimicrobial immunity. Curr Opin Microbiol 4: 251–259. [DOI] [PubMed] [Google Scholar]
- 18.Merlin G, van der Leede BJ, McKune K, Knezevic N, Bannwarth W, Romquin N, Viegas-Pequignot E, Kiefer H, Aguet M, Dembic Z, 1997. The gene for the ligand binding chain of the human interferon gamma receptor. Immunogenetics 45: 413–421. [DOI] [PubMed] [Google Scholar]
- 19.Le Coniat M, Alcaide-Loridan C, Fellous M, Berger R, 1989. Human interferon gamma receptor 1 (IFNGR1) gene maps to chromosome region 6q23-6q24. Hum Genet 84: 92–94. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Ren W, Zhang X, Liu Y, Li C, 2014. Association between interferon gamma receptor 1-56C/T gene polymorphism and tuberculosis susceptibility: a meta-analysis. Chin Med J (Engl) 127: 3782–3788. [PubMed] [Google Scholar]
- 21.Jouanguy E, Lamhamedi-Cherradi S, Altare F, Fondanèche MC, Tuerlinckx D, Blanche S, Emile JF, Gaillard JL, Schreiber R, Levin M, Fischer A, Hivroz C, Casanova JL, 1997. Partial interferon-gamma receptor 1 deficiency in a child with tuberculoid bacillus Calmette-Guérin infection and a sibling with clinical tuberculosis. J Clin Invest 100: 2658–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sampaio MC, Coutinho A, 2007. Immunity to microbes: lessons from primary immunodeficiencies. Infect Immun 75: 1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jüliger S, Bongartz M, Luty AJ, Kremsner PG, Kun JF, 2003. Functional analysis of a promoter variant of the gene encoding the interferon-gamma receptor chain I. Immunogenetics 54: 675–680. [DOI] [PubMed] [Google Scholar]
- 24.Velayati AA, Farnia P, Khalizadeh S, Farahbod AM, Hasanzadh M, Sheikolslam MF, 2011. Interferon-gamma receptor-1 gene promoter polymorphisms and susceptibility to leprosy in children of a single family. Am J Trop Med Hyg 84: 627–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K, ATS Mycobacterial Diseases Subcommittee; American Thoracic Society; Infectious Disease Society of America , 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175: 367–416. [DOI] [PubMed] [Google Scholar]
- 26.Miller SA, Dykes DD, Polesky HF, 1989. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T, 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol 31: 175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velayati AA, Farnia P, Mozafari M, Malekshahian D, Seif S, Rahideh S, Mirsaeidi M, 2014. Molecular epidemiology of nontuberculous mycobacteria isolates from clinical and environmental sources of a metropolitan city. PLoS One 9: e114428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thye T, Burchard GD, Nilius M, Müller-Myhsok B, Horstmann RD, 2003. Genome wide linkage analysis identifies polymorphism in the human interferon-gamma receptor affecting Helicobacter pylori infection. Am J Hum Genet 72: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kardum LB, Etokebe GE, Knezevic J, 2005. Interferon-γ receptor-1 gene promoter polymorphisms (G 611A; T-56C) and susceptibility to tuberculosis. Scand J Immunol 63: 142–150. [DOI] [PubMed] [Google Scholar]
- 31.Newport MJ, Huxley CM, Huston S, Hawrylowicz CM, Oostra BA, Williamson R, Levin M, 1996. A mutation in the interferon-γ receptor gene and susceptibility to mycobacterial infection. N Engl J Med 335: 1941–1948. [DOI] [PubMed] [Google Scholar]
- 32., D’Souza S, Levin M, Newport MJ, Kalabalikis P, Brown IN, Lenicker HM, Agius PV, Davies EG, Thrasher A, Klein N, Blackwell JM, 1995. Familial disseminated atypical mycobacterial infection in childhood: a human mycobacterial susceptibility gene? Lancet 345: 79–83. [DOI] [PubMed] [Google Scholar]
- 33.Bellamy R, 2003. Susceptibility to mycobacterial infections: the importance of host genetics. Genes Immun 23: 4–11. [DOI] [PubMed] [Google Scholar]
- 34.Guide SV, Holland SM, 2002. Host susceptibility factors in mycobacterial infection. Genetics and body morphotype. Infect Dis Clin North Am 16: 163–186. [DOI] [PubMed] [Google Scholar]
- 35.Dorman SE, Holland SM, 1998. Mutation in the signal-transducing chain of the interferon-gamma receptor and susceptibility to mycobacterial infection. J Clin Invest 101: 2364–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altare F, Jouanguy E, Lamhamedi-Cherradi S, Fondanéche MC, Fizame C, Ribiérre F, Merlin G, Dembic Z, Schreiber R, Lisowska-Grospierre B, Fischer A, Seboun E, Casanova JL, 1998. A causative relationship between mutant IFNgR1 alleles and impaired cellular response to IFNγ in a compound heterozygous child. Am J Hum Genet 62: 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dupuis S, Döffinger R, Picard C, Fieschi C, Altare F, Jouanguy E, Abel L, Casanova JL, 2000. Human interferon-γ-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol Rev 178: 129–137. [DOI] [PubMed] [Google Scholar]
- 38.Lake MA, Ambrose LR, Lipman M C I, Lowe DM, 2016. “Why me, why now?” Using clinical immunology and epidemiology to explain who gets nontuberculous mycobacterial infection. BMC Med 14: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirsaeidi M, Farshidpour M, Allen MB, Ebrahimi G, Falkinham JO, 2014. Highlight on advances in nontuberculous mycobacterial disease in North America. Biomed Res Int 2014: 919474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biranvand E, Abedian Kenary S, Ghaheri A, Rezaei MS, Hasannia H, Nasrolahi M, Parsaee MR, Ahanjan M, Biranvand B, Ahmadi Basiri E, Jivad F, 2011. Interferon-gamma gene polymorphism in patients with tuberculosis. Med Lab J 5: 18–23. [Google Scholar]
- 41.Baghaei P, Tabarsi P, Farnia P, Marjani M, Sheikholeslami FM, Chitsaz M, Gorji Bayani P, Shamaei M, Mansouri D, Masjedi MR, Velayati AA, 2012. Pulmonary disease caused by Mycobacterium simiae in Iran’s national referral center for tuberculosis. J Infect Dev Ctries 6: 23–28. [DOI] [PubMed] [Google Scholar]
- 42.Velayati AA, Farnia P, Mozafari M, Mirsaeidi M, 2015. Nontuberculous mycobacteria isolation from clinical and environmental samples in Iran: twenty years of surveillance. BioMed Res Int 2015: 254285. [DOI] [PMC free article] [PubMed] [Google Scholar]