Abstract.
O’nyong-nyong virus (ONNV), an alphavirus closely related to chikungunya virus (CHIKV), has been the documented cause of two large outbreaks in east Africa; however, little is known about the contribution of ONNV to cases of acute febrile illness during interepidemic periods. An ONNV real-time reverse transcription polymerase chain reaction (rRT-PCR) was developed and evaluated using clinical and mosquito pool samples. The ONNV rRT-PCR linear range extended from 8.0 to 2.0 log10 copies/μL, and the lower limit of 95% detection was 22.4 copies/μL. No cases of ONNV infection were identified in serum from 385 Kenyan children who presented with an acute febrile illness. Additionally, ONNV was not detected in 120 mosquito pools collected in coastal and western Kenya. The ONNV rRT-PCR demonstrated good analytical sensitivity when performed in monoplex or as a component of an ONNV–CHIKV duplex assay. This assay should provide a useful diagnostic for the detection of ONNV in surveillance studies.
O’nyong-nyong virus (ONNV) is an arbovirus in the family Togaviridae (genus Alphavirus). ONNV is closely related genetically and antigenically to chikungunya virus (CHIKV),1 and both pathogens cause an acute systemic febrile illness with prominent arthralgia.2,3 However, ONNV is unique among the alphaviruses in that it is transmitted by Anopheles mosquitoes, the vectors of human malaria.4,5 ONNV was identified as the cause of two large, well-documented outbreaks in east Africa. The first, during which the virus was identified, began in northwestern Uganda in 1959 and subsequently spread into Kenya, Tanzania, and Malawi with cases documented as far west as Senegal.6–8 ONNV caused a second outbreak in 1996–1997 that began in southwestern Uganda.9 The last reported cases of ONNV in Kenya occurred in 1961.7,10,11 ONNV was isolated from Anopheles funestus mosquitoes captured on the Kano Plain of Kenya in 1981,7 and in 2013, ONNV was diagnosed in a German traveler who returned from east Africa, including a 2-week stay in the Kisumu region of Kenya immediately preceding symptom onset.12
Little is known about interepidemic ONNV transmission or the contribution of ONNV to acute febrile illnesses in Kenya. Recently, high seroprevalence rates for ONNV and CHIKV were detected in villages of coastal Kenya, including individuals with antibodies to one or both viruses.10 Given the difficulty in distinguishing clinical illnesses caused by ONNV from those caused by pathogens such as malaria or CHIKV, we sought to develop a sensitive and specific real-time reverse transcription polymerase chain reaction (rRT-PCR) for the detection and quantitation of ONNV in patient samples. Only one such assay has been published,13 and the purpose of this study was to design an assay for use as either a monoplex test or in multiplex with real-time assays for CHIKV as well as malaria, previously developed by our group.14,15
To design the ONNV rRT-PCR, all available complete genome sequences of ONNV (N = 3) and Igbo-Ora virus (N = 1) in the Nucleotide database (National Center for Biotechnology Information) were aligned using MegAlign Software (DNAStar, Madison, WI). The resulting majority consensus sequence for ONNV was aligned against a CHIKV consensus sequence generated using 141 complete CHIKV genome sequences. The CHIKV consensus has been described previously.15 A target region was identified that 1) was highly conserved among CHIKV sequences in the CHIKV alignment and 2) contained a sufficient number of mismatches between ONNV and CHIKV to design specific primers and probes. This region included a portion of the 5′ untranslated region and nonstructural protein 1 gene of ONNV. Primers and probes were designed using Primer3 software (Whitehead Institute for Biomedical Research, Cambridge, MA) and selected for evaluation if they 1) had ≥ 4 mismatches to the CHIKV consensus and 2) were compatible in multiplex with assays previously designed by our group (evaluated using http://biocompute.bmi.ac.cn/MPprimer/primer_dimer.html). Primers and probes were evaluated at varying concentrations, from 100 to 400 nM in the final reaction mixture, using genomic RNA from a reference ONNV strain (MP30), and the primer–probe pair with the best analytical sensitivity was selected for use (Table 1).
Table 1.
Primer and probe sequences included in the O’nyong-nyong virus (ONNV) real-time reverse transcription polymerase chain reaction
| Name | Sequence (5′ → 3′) | Concentration* (nM) | Location† |
|---|---|---|---|
| ONNV forward | CGCAGCTTACGGGTTTCATA | 200 | 21–40 |
| ONNV reverse | GCAACGCCTTCAGAAACGC | 200 | 116–134 |
| ONNV probe‡ | TGCTCTACTCTGCATTGCAAGA | 400 | 42–63 |
The concentration of each oligonucleotide in the final reaction mixture is provided.
Genomic locations are provided based on the reference sequence ONNV Gulu strain (Genbank: M20303.1).
The 5′ fluorophore and 3′ quencher on the ONNV probe were FAM and BHQ-1, respectively.
The ONNV rRT-PCR was performed using 5 μL of RNA eluate in 25 μL reactions of the SuperScript III Platinum One-Step qRT-PCR Kit (Thermo Fisher Scientific, Waltham, MA) on a Rotor-Gene Q instrument (Qiagen, Germantown, MD). Final primer and probe concentrations are shown in Table 1. Cycling conditions were the following: 52°C for 15 minutes; 94°C for 2 minutes; and 45 cycles of 94°C for 15 seconds, 55°C for 40 seconds (signal acquired), and 68°C for 20 seconds. Analytical characterization was performed according to published recommendations16 and consistent with previous reports.14,15,17 For assay interpretation, a crossing threshold was set manually, such that the threshold crossed curves at the start of the linear phase of the amplification plot over the entire linear range. Exponential curves that crossed this threshold prior to cycle 45 were considered positive for ONNV. Linear range, precision, and lower limit of 95% detection experiments were performed using synthesized Ultramer Oligos (Integrated DNA Technologies, Coralville, IA) containing the assay target sequence.
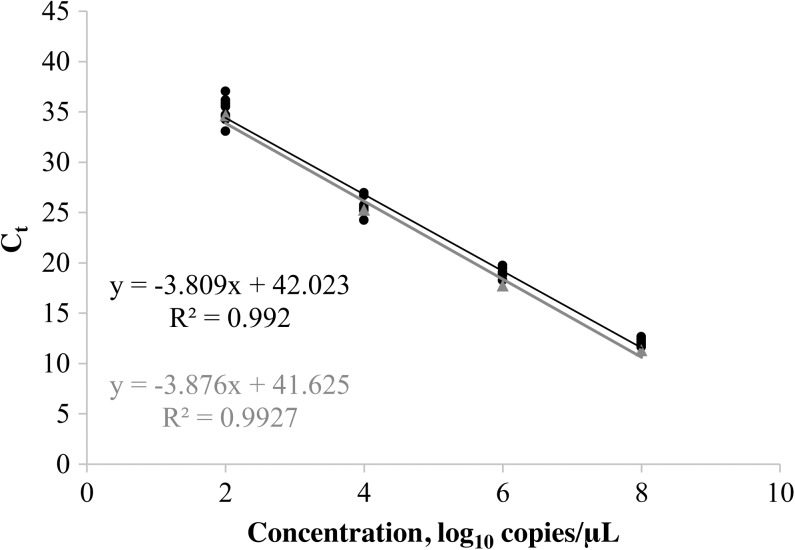
The linear range of the ONNV rRT-PCR extended from 8.0 to 2.0 log10 copies/μL (Figure 1). The lower limit of 95% detection was 22.4 copies/μL (95% confidence interval: 17.3–36.7), as calculated by probit analysis (SPSS software; IBM, Armonk, NY). Precision of the ONNV rRT-PCR at four concentrations is shown in Table 2. Performance of the ONNV rRT-PCR was also evaluated as a component of an ONNV–CHIKV duplex assay, which included CHIKV primers and probes that have been described previously.15 The linearity of ONNV detection in the duplex assay is shown in Figure 1.
Figure 1.
Linear range of the ONNV real-time reverse transcription polymerase chain reaction performed in monoplex (black trend line) and as part of an ONNV–CHIKV duplex assay (gray trend line). Ct values for individual replicates are shown for the ONNV monoplex (•) and ONNV–CHIKV duplex (▴) assays. CHIKV = chikungunya virus; ONNV = O’nyong-nyong virus.
Table 2.
Within-run, between-run, and total imprecision of the O’nyong-nyong virus (ONNV) real-time reverse transcription polymerase chain reaction determined using quantitated ssDNA controls
| Within-run | Between-run | Within-laboratory | |||||
|---|---|---|---|---|---|---|---|
| Concentration* | Mean Ct | SD | %CV | SD | %CV | SD | %CV |
| 8.0 | 12.06 | 0.31 | 2.54 | 0.23 | 1.92 | 0.32 | 2.63 |
| 6.0 | 19.00 | 0.45 | 2.36 | 0.17 | 0.89 | 0.36 | 1.89 |
| 4.0 | 25.60 | 0.89 | 3.48 | 0.24 | 0.92 | 0.67 | 2.63 |
| 2.0 | 35.25 | 1.06 | 2.99 | 0.83 | 2.35 | 1.12 | 3.17 |
%CV = percent coefficient variation; SD = standard deviation.
Values in log10 copies/μL of eluate.
Assay exclusivity was confirmed by testing genomic RNA from the following: dengue virus, serotypes 1–4; two strains of CHIKV; St. Louis encephalitis virus; four strains of West Nile virus, Zika virus; tick-borne encephalitis virus, Japanese encephalitis virus; Semliki Forest virus; Mayaro virus; Ross River virus; Getah virus; Barmah Forest virus; and Una virus. Additionally, extracted RNA from 10 patients with hepatitis C virus infection was tested. No signal was detected from these viruses or samples in the ONNV rRT-PCR.
Following the analytical characterization of the ONNV rRT-PCR, we tested serum from 385 randomly selected children enrolled in an ongoing, acute febrile illness surveillance study in Kenya. Children aged 1–17 years with documented fever (≥ 38°C) were enrolled at two health-care sites on the Indian Ocean coast (Ukunda Health Centre and Msambweni District Hospital) and two sites in Western Kenya (Obama Children’s Hospital in Kisumu and Chulaimbo District Hospital). All participants had demographic and health history information collected and a physical examination and blood draw at enrollment. Mean age was 5.05 years (standard deviation [SD]: 3.46), and 193/383 patients (50.4%) with recorded clinical data were female. Samples were obtained at a mean of 2.46 days after symptom onset (SD: 0.86). Total nucleic acids were extracted from 140 μL serum using an easyMAG instrument (Biomerieux, Durham, NC) and a 60 μL elution volume. In total, 158 patients (41.0%) tested positive for malaria using a real-time PCR that has been described previously.14 Study protocol was reviewed and approved by the Stanford University Institutional Review Board (no. 31488) and the Kenya Medical Research Institute Scientific and Ethical Review Committee (SSC 2611).
Additionally, 120 mosquito pools, including legs from 1,128 individual mosquitoes, were tested for ONNV RNA: 82 pools of Anopheles mosquitoes (An. funestus, N = 32; Anopheles gambiae, N = 40; unspeciated, N = 10) and 38 pools of Aedes aegypti. Mosquitoes were collected at all life stages from both regions using a combination of methods (ovitraps for all Ae. aegypti pools; larval and pupal sampling, 56 pools; backpack aspirators, 24 pools; and BG sentinel traps, Biogents AG, Regensburg, Germany; 2 pools). Immatures were reared to adulthood and then morphologically identified. One leg from each mosquito was dissected and placed into pools of 1–26 mosquito legs (mean: 9.4, SD: 9.0) by species, sex, trap type, and date of collection.18 Pooled legs were homogenized using a Bead Ruptor 24 (Omni International, Kennesaw, GA) and pool RNA was extracted using a MagMAX Express instrument (Thermo Fisher Scientific).
No participants had detectable ONNV RNA. In total, 158 patients tested positive for malaria and 32 patients tested positive for CHIKV using real-time molecular diagnostics. All mosquito pools also tested negative for ONNV. Known positive clinical samples and/or mosquito samples were not available for testing.
Although ONNV was not detected in clinical samples from symptomatic participants or in mosquito pools collected in coastal and western Kenya, these data add to the mystery that surrounds ONNV in east Africa. This virus has resulted in two large outbreaks in the region, both centered in Uganda, and it has sporadically been detected during interepidemic periods. In a seroprevalence study of animal populations in the greater Congo Basin, African forest buffalo (Syncerus caffer nanus), duikers (Cephalophus and Philantomba spp.), and mandrills (Mandrillus sphinx) were found to have antibodies to ONNV, indicating that these may be reservoir populations for the virus, which has not been isolated from naturally infected animals other than humans.19 A seroprevalence study involving 1,848 participants from two village clusters in coastal Kenya found that 56% of individuals had antibodies to ONNV and 38% had high antibody titers to both ONNV and CHIKV.10 Age was significantly associated with antibody detection, though young adults were more likely to be seropositive than older adults.10
Notably, our participants had a high incidence of malaria, as detected by rRT-PCR in serum, indicating exposure to An. gambiae mosquitoes, which are the principal vectors of human malaria. Although ONNV has been shown to infect and disseminate in An. gambiae (as well as Ae. aegypti), An. funestus has been considered the principal vector during outbreaks due to the frequency of its identification in affected areas during outbreak periods.20,21 Interestingly, during an investigation of the 1960 ONNV outbreak in the Masaka District of Uganda, researchers noted an unexpected drop in malaria rates, identified through monthly testing of infants.22 Therefore, the absence of ONNV detection in our study may have resulted from ONNV causing infrequent, large outbreaks, while accounting for few cases of acute febrile illness during interepidemic periods. Alternatively, the high incidence of malaria may indicate that ONNV is unlikely in this population, either due to interactions between these pathogens or exposure of humans to a less-fit mosquito vector for natural ONNV transmission.
In conclusion, we described the design and evaluation of a new rRT-PCR for ONNV that demonstrates good analytical performance when performed in monoplex or as a component of an ONNV–CHIKV duplex assay. This assay should provide a useful diagnostic for the accurate detection of ONNV, which has demonstrated potential to cause large-scale human outbreaks, but remains elusive.
Acknowledgments:
It is a pleasure to record our gratitude for the assistance extended to us, in the work reported here, by the following: Lark Coffey at the University of California, Davis, for provision of the ONNV strain used in these studies; the Clinical Virology Laboratory staff at Stanford for their assistance in coordinating nucleic acid extractions and rRT-PCR performance; and the study participants and their families in Kenya for agreeing to contribute to this research. Quote taken directly from reference number 5.
REFERENCES
- 1.Powers AM, Brault AC, Tesh RB, Weaver SC, 2000. Re-emergence of chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol 81: 471–479. [DOI] [PubMed] [Google Scholar]
- 2.Rwaguma EB, Lutwama JJ, Sempala SD, Kiwanuka N, Kamugisha J, Okware S, Bagambisa G, Lanciotti R, Roehrig JT, Gubler DJ, 1997. Emergence of epidemic O’nyong-nyong fever in southwestern Uganda, after an absence of 35 years. Emerg Infect Dis 3: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shore H, 1961. O’nyong-nyong fever: an epidemic disease in east Africa. Trans R Soc Trop Med Hyg 55: 361–373. [DOI] [PubMed] [Google Scholar]
- 4.Vanlandingham DL, Hong C, Klingler K, Tsetsarkin K, McElroy KL, Powers AM, Lehane MJ, Higgs S, 2005. Differential infectivities of O’nyong-nyong and chikungunya virus isolates in Anopheles gambiae and Aedes aegypti mosquitoes. Am J Trop Med Hyg 72: 616–621. [PubMed] [Google Scholar]
- 5.Williams MC, Woodall JP, Corbet PS, Gillett JD, 1965. O’nyong-nyong fever: an epidemic virus disease in East Africa. 8. Virus isolations from Anopheles mosquitoes. Trans R Soc Trop Med Hyg 59: 300–306. [DOI] [PubMed] [Google Scholar]
- 6.Haddow AJ, Davies CW, Walker AJ, 1960. O’nyong-nyong fever: an epidemic virus disease in east Africa. Trans R Soc Trop Med Hyg 54: 517–522. [DOI] [PubMed] [Google Scholar]
- 7.Johnson BK, Gichogo A, Gitau G, Patel N, Ademba G, Kirui R, Highton RB, Smith DH, 1981. Recovery of O’nyong-nyong virus from Anopheles funestus in western Kenya. Trans R Soc Trop Med Hyg 75: 239–241. [DOI] [PubMed] [Google Scholar]
- 8.Williams MC, Woodall JP, 1961. O’nyong-nyong fever: an epidemic virus disease in east Africa. II. Isolation and some properties of the virus. Trans R Soc Trop Med Hyg 55: 135–141. [DOI] [PubMed] [Google Scholar]
- 9.Lanciotti RS, Ludwig ML, Rwaguma EB, Lutwama JJ, Kram TM, Karabatsos N, Cropp BC, Miller BR, 1998. Emergence of epidemic O’nyong-nyong fever in Uganda after a 35-year absence: genetic characterization of the virus. Virology 252: 258–268. [DOI] [PubMed] [Google Scholar]
- 10.LaBeaud AD, Banda T, Brichard J, Muchiri EM, Mungai PL, Mutuku FM, Borland E, Gildengorin G, Pfeil S, Teng CY, Long K, Heise M, Powers AM, Kitron U, King CH, 2015. High rates of O’nyong nyong and chikungunya virus transmission in coastal Kenya. PLoS Negl Trop Dis 9: e0003436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams MC, Woodall JP, Gillett JD, 1965. O’nyong-nyong fever: an epidemic virus disease in east Africa. Vii. Virus isolations from man and serological studies up to July 1961. Trans R Soc Trop Med Hyg 59: 186–197. [DOI] [PubMed] [Google Scholar]
- 12.Tappe D, Kapaun A, Emmerich P, Campos Rde M, Cadar D, Gunther S, Schmidt-Chanasit J, 2014. O’nyong-nyong virus infection imported to Europe from Kenya by a traveler. Emerg Infect Dis 20: 1766–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DR, Lee JS, Jahrling J, Kulesh DA, Turell MJ, Groebner JL, O’Guinn ML, 2009. Development of field-based real-time reverse transcription-polymerase chain reaction assays for detection of chikungunya and O’nyong-nyong viruses in mosquitoes. Am J Trop Med Hyg 81: 679–684. [DOI] [PubMed] [Google Scholar]
- 14.Waggoner JJ, Abeynayake J, Balassiano I, Lefterova M, Sahoo MK, Liu Y, Vital-Brazil JM, Gresh L, Balmaseda A, Harris E, Banaei N, Pinsky BA, 2014. A multiplex nucleic acid amplification test for the diagnosis of dengue, malaria, and leptospirosis. J Clin Microbiol 52: 2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waggoner JJ, Ballesteros G, Gresh L, Mohamed-Hadley A, Tellez Y, Sahoo MK, Abeynayake J, Balmaseda A, Harris E, Pinsky BA, 2016. Clinical evaluation of a single-reaction real-time RT-PCR for pan-dengue and chikungunya virus detection. J Clin Virol 78: 57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burd EM, 2010. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev 23: 550–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waggoner JJ, Gresh L, Mohamed-Hadley A, Ballesteros G, Davila MJ, Tellez Y, Sahoo MK, Balmaseda A, Harris E, Pinsky BA, 2016. Single-reaction multiplex reverse transcription PCR for detection of Zika, chikungunya, and dengue Viruses. Emerg Infect Dis 22: 1295–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaBeaud AD, Sutherland LJ, Muiruri S, Muchiri EM, Gray LR, Zimmerman PA, Hise AG, King CH, 2011. Arbovirus prevalence in mosquitoes, Kenya. Emerg Infect Dis 17: 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kading RC, Borland EM, Cranfield M, Powers AM, 2013. Prevalence of antibodies to alphaviruses and flaviviruses in free-ranging game animals and nonhuman primates in the greater Congo basin. J Wildl Dis 49: 587–599. [DOI] [PubMed] [Google Scholar]
- 20.Corbet PS, Williams MC, Gillett JD, 1961. O’Nyong-nyong fever: an epidemic virus disease in east Africa. IV. Vector studies at epidemic sites. Trans R Soc Trop Med Hyg 55: 463–480. [DOI] [PubMed] [Google Scholar]
- 21.Lutwama JJ, Kayondo J, Savage HM, Burkot TR, Miller BR, 1999. Epidemic O’nyong-nyong fever in southcentral Uganda, 1996–1997: entomologic studies in Bbaale village, Rakai District. Am J Trop Med Hyg 61: 158–162. [DOI] [PubMed] [Google Scholar]
- 22.De Zulueta J, Woodall JP, Cullen JR, Williams MC, Kafuko GW, Gillett JD, 1962. An observation on the possible effect of O’nyong-nyong fever on malaria. Bull World Health Organ 26: 135–139. [PMC free article] [PubMed] [Google Scholar]