Abstract
Currently, major concerns about the safety and efficacy of RNA interference (RNAi)-based bone anabolic strategies still exist because of the lack of direct osteoblast-specific delivery systems for osteogenic siRNAs. Here we screened the aptamer CH6 by cell-SELEX, specifically targeting both rat and human osteoblasts, and then we developed CH6 aptamer–functionalized lipid nanoparticles (LNPs) encapsulating osteogenic pleckstrin homology domain-containing family O member 1 (Plekho1) siRNA (CH6-LNPs-siRNA). Our results showed that CH6 facilitated in vitro osteoblast-selective uptake of Plekho1 siRNA, mainly via macropinocytosis, and boosted in vivo osteoblast-specific Plekho1 gene silencing, which promoted bone formation, improved bone microarchitecture, increased bone mass and enhanced mechanical properties in both osteopenic and healthy rodents. These results indicate that osteoblast-specific aptamer-functionalized LNPs could act as a new RNAi-based bone anabolic strategy, advancing the targeted delivery selectivity of osteogenic siRNAs from the tissue level to the cellular level.
Metabolic skeletal disorders associated with impaired bone formation (for example, osteoporosis) remain major clinical challenges. RNA interference (RNAi)-based approaches aimed at promoting osteo-blastic bone formation may hold therapeutic potential1,2. However, a major bottleneck for translating RNAi-based approaches into clinical application is the lack of osteoblast-specific osteogenic siRNA delivery systems3.
Plekho1 (also known as casein kinase-2 interacting protein-1 (CKIP-1)) has been identified as an intracellular negative regulator of bone formation that does not affect bone resorption4. Previously we developed a targeting system involving dioleoyl trimethylammonium propane (DOTAP)-based cationic liposomes attached to six repetitive sequences of aspartate, serine and serine ((AspSerSer)6), which had good affinity for the physiochemical features of the bone-formation surface when compared to the bone-resorption surface. By using this system, osteogenic Plekho1 siRNA was specifically delivered to the bone-formation surface to promote bone formation5. However, as the system was not specific to osteoblasts at the cellular level, other non-osteoblasts near the bone-formation surface, including endothelial cells and lymphocytes, may also be targeted, which arouses concerns about efficacy and potential toxic side effects5–7. In addition, other potential concerns, including mononuclear phagocyte system (MPS)-induced dose reduction8,9, inefficient nanoparticle extravasation caused by large particle size (larger than bony sinusoids (80–100 nm))5,10 and detrimental hepatocyte accumulation of siRNA11, should also be considered by drug developers.
Here we sought to develop an osteoblast-specific delivery system for osteogenic siRNAs. First we used cell-based systematic evolution of ligands by exponential enrichment (cell-SELEX) to select osteoblast-specific aptamers, which are single-stranded oligonucleotides that use distinct tertiary structures to specifically bind to target cells12,13. By performing positive selection with osteoblasts and negative selection with hepatocytes and peripheral blood mononuclear cells (PBMCs), we aimed to screen an aptamer that could achieve direct osteoblast-specific delivery of osteogenic siRNAs and minimal hepatocyte and PBMC accumulation of osteogenic siRNAs. Second, using lipid nanoparticles (LNPs), which are <90 nm (ref. 14) and have high polyethylene glycol (PEG) shielding, as the siRNA carriers, we aimed to realize efficient siRNA extravasation and reduce MPS uptake15,16.
The osteoblast-specific aptamer CH6 was selected by cell-SELEX. When rendered with nuclease-resistance and conjugated to LNPs encapsulating osteogenic Plekho1 siRNA, CH6 achieved osteoblast-specific delivery of siRNAs and facilitated bone formation in osteopenic and healthy rodents. This demonstrates a real osteoblast-specific delivery system at the cellular level.
RESULTS
Selection of osteoblast-specific aptamers
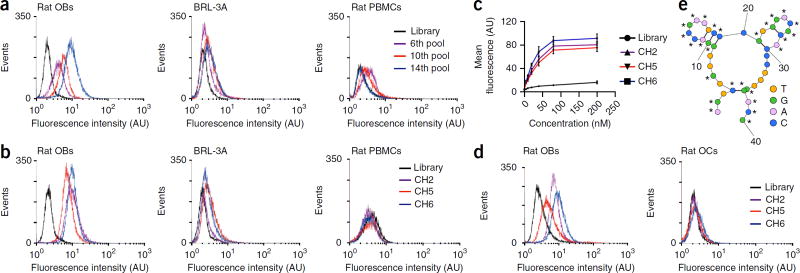
We used rat primary osteoblasts to select aptamers by cell-SELEX from a library composed of 1015 different ssDNA sequences. With increasing rounds of selection we observed progressive enhancement of fluorescence intensity for fluorescein amidite (FAM)-labeled ssDNA pools in target cells (rat primary osteoblasts) by flow cytometry analysis, whereas we observed no obvious change of fluorescence intensity in non-target cells (rat liver cell line BRL-3A and rat PBMCs) (Fig. 1a). After 14 rounds of selection, we sequenced the highly enriched ssDNA pool and chose 20 representative sequences (from 200 clones), on the basis of their predicted secondary structures, for truncation to remove the non-critical bases17 (Supplementary Table 1). Flow cytometry analysis showed that three aptamer candidates (CH2, CH5 and CH6) had good binding ability to osteoblasts (Fig. 1b) with equilibrium dissociation constants (Kd) in the nanomolar-to-picomolar range (Fig. 1c).
Figure 1.
Cell-SELEX for the identification of osteoblast-specific aptamers. (a) Binding ability of the enriched pools to target cells (rat primary osteoblasts) and non-target cells (rat liver cell line BRL-3A and PBMCs) as determined by flow cytometry. The concentration of the enriched pools in the binding buffer was 250 nM. Nonspecific binding was measured using an FAM-labeled unselected library. AU, arbitrary units; events, number of cells with the given fluorescence intensity. (b) Binding ability of the FAM-labeled aptamer candidates (CH2, CH5 and CH6) to target cells and non-target cells as determined by flow cytometry. (c) Flow cytometry to determine the binding affinity of the aptamer candidates for the target cells. Kd (CH6) = 25.90 ± 3.66 nM; Kd (CH2) = 31.02 ± 4.76 nM; Kd (CH5) = 34.87 ± 5.41 nM. The concentrations of the aptamer candidates ranged from 5 nM to 200 nM. The data are presented as the mean ± sd; n = 3. (d) The binding ability of the FAM-labeled, 2′-O-methyl-nucleotide substitutions–modified aptamer candidates (CH2, CH5 and CH6) to rat primary osteoblasts (left) and osteoclasts (right) determined by flow cytometry. (e) Proposed secondary structure of CH6. 2′-O-methoxy nucleotides are marked with the asterisk symbol (*). Thick lines, hydrogen bonds between base pairs; thin lines, main phosphodiester bond; Rat OBs, rat primary osteoblasts; Rat OCs, rat osteoclasts; Rat PBMCs, rat peripheral blood mononuclear cells; BRL-3A, rat liver cell line.
To minimize nuclease degradation, CH2, CH5 and CH6 were modified with 2′-O-methyl-nucleotide substitutions18,19 and flow cytometry analysis revealed that they still bound with high affinity to rat osteoblasts but not rat osteoclasts (Fig. 1d). CH2 and CH6 aptamers also bound to human primary osteoblasts but not human osteoclasts or liver cells (THLE-3) (Supplementary Fig. 1). We chose the CH6 aptamer because it had satisfactory secondary structure and shorter nucleotide sequences (Fig. 1e), making it easier to synthesize and conjugate to LNPs-siRNA than CH2.
In vitro analysis of cellular uptake
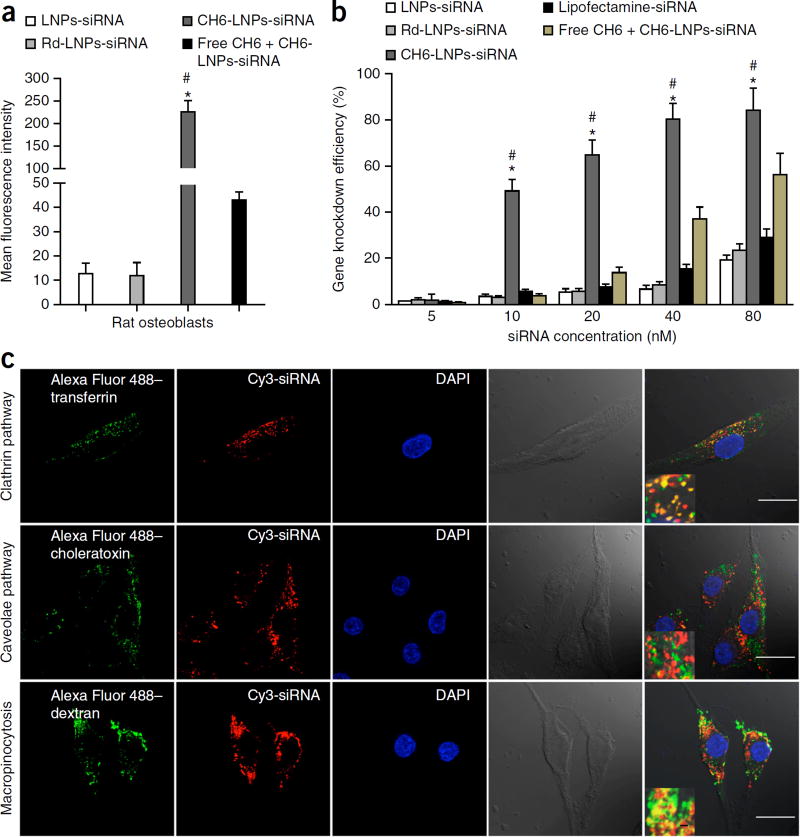
Next, we prepared and characterized CH6 aptamer–functionalized LNPs encapsulating osteogenic Plekho1 siRNA. CH6-LNPs-siRNA had a diameter of 84.0 ± 5.3 nm, encapsulation efficiency above 80%, uniform particle shape, good serum stability, no detectable cytotoxicity and high in vitro osteoblast selectivity (Supplementary Fig. 2; Supplementary Table 2). A random sequence (Rd) served as the negative control aptamer. We labeled Plekho1 siRNA with Cy3 and investigated the effect of the CH6 aptamer versus the Rd aptamer on the cellular uptake of siRNA. The most intense fluorescence signals were detected in osteoblasts incubated with CH6-LNPs-siRNA, and fluorescence signals were rarely observed in osteoblasts treated with no aptamer–conjugated LNPs-siRNA or Rd-LNPs-siRNA (Fig. 2a). Pretreatment of osteoblasts with free CH6 aptamer resulted in a significantly decreased fluorescence signal of CH6-LNPs-siRNA in osteoblasts (P < 0.05; Fig. 2a). We also examined the Plekho1 gene knockdown efficiency of different siRNA formulations in osteoblasts. The gene knockdown efficiency for CH6-LNPs-siRNA was significantly higher than other siRNA formulations (P < 0.05) (Fig. 2b).
Figure 2.
Cellular selectivity, gene knockdown efficiency and mechanism of cellular uptake of various siRNA formulations. (a) Cellular selectivity: fluorescence intensity of FAM-labeled siRNA in rat primary osteoblasts incubated with LNPs-siRNA, Rd-LNPs-siRNA, CH6-LNPs-siRNA and free CH6 + CH6-LNPs-siRNA. (b) Gene knockdown efficiency: percentage changes of gene expression were quantified after the incubation of the siRNA formulations as described in a as well as the positive control Lipofectamine 2000–siRNA with rat primary osteoblasts. The data are presented as the mean ± sd; n = 3 per group. One-way analysis of variance (ANOVA) with LSD’s post hoc test was performed to determine inter-group differences. (c) Pathway for the cellular uptake of CH6-LNPs-siRNA: intracellular co-localization of Cy3-labeled Plekho1 siRNA (red) and Alexa Fluor 488–labeled endocytic markers (transferrin, choleratoxin and dextran; green) in rat primary osteoblasts. The nucleus was counterstained with DAPI (blue). Scale bars, 25 µm, large panels and 1 µm, insets. Fourth column, bright-field images of osteoblasts; last column, merged images from all columns. *P < 0.05 for a comparison of CH6-LNPs-siRNA with either LNPs-siRNA or Rd-LNPs-siRNA; #P < 0.05 for a comparison of CH6-LNPs-siRNA with free CH6 + CH6-LNPs-siRNA.
To investigate the cellular mechanism through which CH6-LNPs-siRNA is taken up by osteoblasts, we labeled Plekho1 siRNA with Cy3 and incubated osteoblasts with different siRNA formulations in the presence of Alexa Fluor 488–labeled endocytic markers20. We detected by confocal microscopy numerous instances of co-localization of siRNA with transferrin (a marker for clathrin-mediated endocytosis) and dextran (a marker for macropinocytosis), but not with choleratoxin (a marker for caveolae-mediated endocytosis) in osteoblasts incubated with CH6-LNPs-siRNA (Fig. 2c), indicating that CH6-LNPs-siRNA can be taken up via clathrin-mediated endocytosis as well as by macropinocytosis. In contrast, Rd-LNPs-siRNA (Supplementary Fig. 3a) and LNPs-siRNA (data not shown) were found to enter osteoblasts only via clathrin-mediated endocytosis. Chemical inhibition experiments21 confirmed these findings from the confocal imaging (Supplementary Fig. 3b,c). Additionally, confocal images showed that CH6-LNPs-siRNA induced membrane ruffling and lysosome escape of siRNA (attributes of macropinocytosis22) in osteoblasts, but we did not observe this for either LNP-siRNA or Rd-LNP-siRNA (Supplementary Fig. 3d,e).
Tissue distribution and cell-specific delivery in vivo
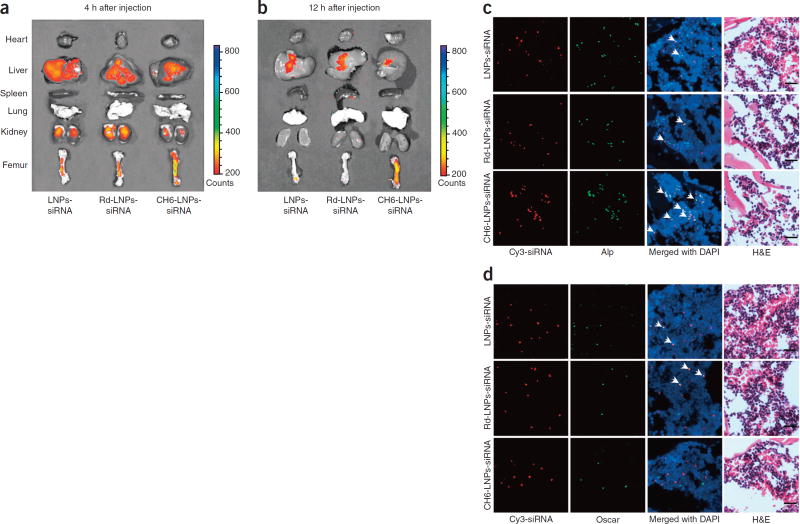
We used biophotonic imaging technology to examine the tissue distribution of Cy3-labeled siRNA delivered by different carriers. At each of the time points examined, fluorescence signal intensity of CH6-LNPs-siRNA in bone was significantly higher, whereas the fluorescence signal intensities in liver and kidney were significantly lower when compared to LNPs-siRNA or Rd-LNPs-siRNA, which was consistent with quantitative analysis by a fluorescence microplate reader system (P < 0.05) (Fig. 3a,b; Supplementary Fig. 4a). Very low fluorescence signals in the heart, spleen and lungs were detected in all administration groups (Fig. 3a,b).
Figure 3.
Tissue distribution and cell-selective delivery of various siRNA formulations in vivo. (a,b) Tissue distribution: localization of Cy3-labeled siRNA in rats administered LNPs-siRNA, Rd-LNPs-siRNA or CH6-LNPs-siRNA, visualized by biophotonic imaging at 4 h (a) and 12 h (b). The intensities of the fluorescence signals in the isolated hearts, livers, spleens, lungs, kidneys and femurs are presented. Green, highest siRNA fluorescence intensity. (c,d) Cellular selectivity of siRNA in bone: fluorescence micrographs of cryosections from distal femur after injection of LNPs-siRNA (top row), Rd-LNPs-siRNA (middle row) or CH6-LNPs-siRNA (bottom row). The siRNA was labeled with Cy3 (red; left column). Immunofluorescence staining was performed to detect Alp+ osteoblasts (green; middle left column in c) and Oscar+ pre-osteoclasts/osteoclasts (green; middle left column in d). Merged images with DAPI staining show co-localization of siRNA with osteoblasts (Alp+ cells, arrows and yellow color, middle right column in c) or pre-osteoclasts/osteoclasts (Oscar+ cells, arrows and yellow color, middle right column in d). H&E staining of the same sections is included at right. Scale bars, 20 µm.
To investigate whether CH6 could facilitate delivery of siRNA in an osteoblast-selective manner in vivo, we examined the co-localization of Cy3-labeled siRNA and markers of osteoblasts (alkaline phosphatase (Alp)23 and osteocalcin (OCN)24) and osteoclasts (osteoclast-associated receptor (Oscar)25,26 and cathepsin K27) in cryosections from the distal femurs of rats injected with different siRNA formulations. We found numerous instances of co-localization of siRNA with Alp+ or OCN+ cells in rats when CH6-LNPs-siRNA were administered, whereas there were few instances of such overlapping staining in rats when LNPs-siRNA or Rd-LNPs-siRNA were administered (Fig. 3c; Supplementary Fig. 4b). Furthermore, we found no obvious overlapping staining for siRNA and Oscar+ or cathepsin K+ cells in rats treated with CH6-LNPs-siRNA. There were, however several instances of overlapping staining for siRNA and Oscar+ or cathepsin K+ cells in rats treated with LNPs-siRNA or Rd-LNPs-siRNA (Fig. 3d; Supplementary Fig. 4c). We also examined the cellular distribution of siRNA in hepatocytes and the MPS (PBMCs and Kupffer cells); CH6 reduced the uptake of siRNA by hepatocytes, Kupffer cells and PBMCs (Supplementary Fig. 4d,e). Compared to (AspSerSer)6-liposome-siRNA, all LNP-based siRNA delivery systems exhibited weaker fluorescence intensities in the liver (Supplementary Fig. 4d).
In vivo gene knockdown and dose-response pattern
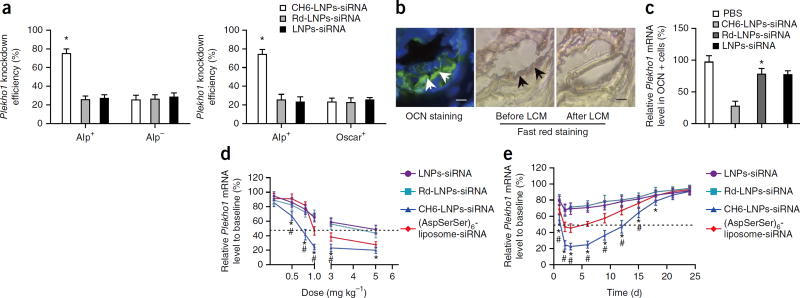
We next examined the gene knockdown efficiency of Plekho1 siRNA in sorted cells from the bone marrow of rats injected with different siRNA formulations by fluorescence-activated cell sorting (FACS) in combination with real-time PCR. Compared to LNPs-siRNA or Rd-LNPs-siRNA, the gene knockdown efficiency of CH6-LNPs-siRNA was significantly higher in Alp+ or OCN+ cells than in Alp−, OCN− or Oscar+ cells (P < 0.05) (Fig. 4a; Supplementary Fig. 5a). Results from laser-capture microdissection (LCM) in combination with real-time PCR also showed that CH6 could facilitate Plekho1 gene knockdown in OCN+ or Alp+ cells (Fig. 4b,c; Supplementary Fig. 5b).
Figure 4.
Cell-selective gene knockdown efficiency, dose-response pattern and persistence of gene silencing in vivo. (a) The knockdown efficiencies of Plekho1 mRNA in Alp+, Alp−, and Oscar+ cells after the administration of LNPs-siRNA, Rd-LNPs-siRNA, or CH6-LNPs-siRNA, as detected by FACS in combination with real-time PCR. (b) Osteoblasts are identified by OCN staining (arrows) in cryosections from the distal femurs (left). The adjacent sections are indicated by fast red staining before (middle) and after (right) LCM. Scale bars, 50 µm. (c) Real-time PCR was used to analyze the Plekho1 mRNA expression (relative to baseline) in OCN+ cells of rats injected with the specified siRNA formulation. PBS, control. (d) Plekho1 mRNA level (measured by real-time PCR) after a tail vein injection of LNPs-siRNA or Rd-LNPs-siRNA or CH6-LNPs-siRNA or (AspSerSer)6-liposome-siRNA at doses ranging from 0.1 to 5.0 mg kg−1. (e) Plekho1 mRNA level measured by real-time PCR after a single injection of the indicated siRNA formulations at a single dose of 1.0 mg kg−1. Dashed line, 50% Plekho1 mRNA level relative to baseline. Data in a,c–e are presented as means ± sd; n = 6 per group. *P < 0.05 for a comparison of CH6-LNPs-siRNA with LNPs-siRNA or Rd-LNPs-siRNA; #P < 0.05 for a comparison of CH6-LNPs-siRNA with (AspSerSer)6-liposome-siRNA. One-way ANOVA with LSD’s post hoc test was performed to determine inter-group differences.
We also determined the dose-response pattern and persistence of Plekho1 gene silencing in OCN+ cells sorted from rats administered with CH6-LNPs-siRNA, LNPs-siRNA, Rd-LNPs-siRNA or (AspSerSer)6-liposome-siRNA. The Plekho1 mRNA level decreased in a dose-dependent manner and was lowest (~20% of baseline) in rats administered with CH6-LNPs-siRNA at an siRNA dose of 1.0 mg kg−1 when compared to the other LNPs-based siRNA formulations or (AspSerSer)6-liposome-siRNA (Fig. 4d). After a single injection of siRNA formulations at an siRNA dose of 1.0 mg kg−1, our results showed that CH6-LNPs-siRNA maintained a low Plekho1 mRNA level (<50% of the Plekho1 mRNA level at baseline) for a longer time (~12 d) when compared to the other LNPs-based siRNA formulations or to (AspSerSer)6-liposome-siRNA (~7 d) (Fig. 4e).
Bone anabolic action
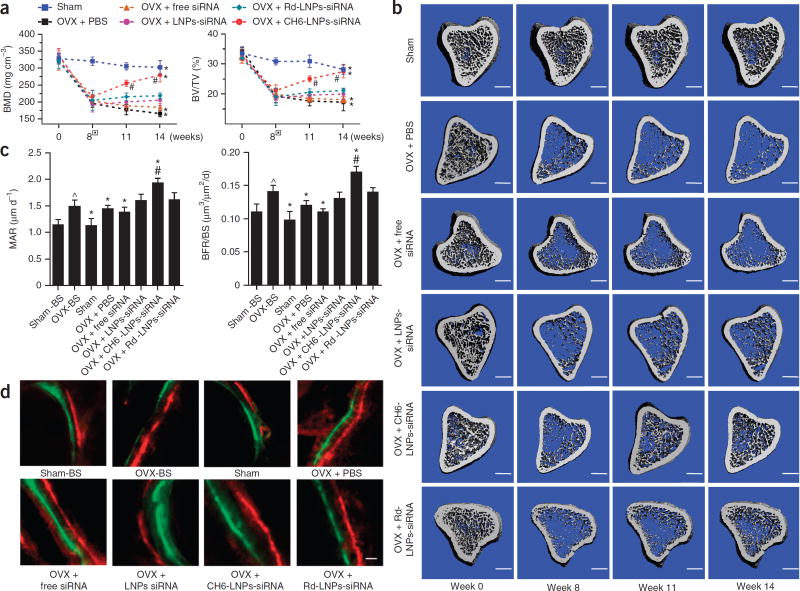
We administered six periodic injections of Plekho1 siRNA (1.0 mg kg−1) encapsulated in LNPs-based carriers to ovariectomized (OVX) rats at an interval of once per week and performed in vivo microcomputed tomography (microCT) scanning every 3 weeks during the treatment period (Supplementary Fig. 6a). A statistical analysis of the in vivo microCT data by repeated-measures ANOVA showed that both the ‘time effect’ and the ‘time by group interaction effect’ were substantial for all of the examined variables, indicating both considerable changes in the examined variables over time and remarkably different change patterns over time among the examined groups after the administration. After treatment, all the in vivo microCT parameters, including bone mineral density (BMD), relative bone volume (BV/TV), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), trabecular number (Tb.N) and structural model index (SMI), were obviously improved and were nearly restored to the pre-surgery levels in the OVX + CH6-LNPs-siRNA group when compared to the OVX + Rd-LNPs-siRNA group (P < 0.05) (Fig. 5a; Supplementary Fig. 6b). Better-organized microarchitecture and higher bone mass were consistently found in trabecular bone from the OVX rats treated with CH6-LNPs-siRNA when compared to Rd-LNPs-siRNA (Fig. 5b).
Figure 5.
MicroCT examinations of proximal tibia and histomorphometric analysis of trabecular bone at the distal metaphysis of femur in osteopenic rats. (a) BMD and BV/TV monitoring over time by in vivo microCT examination of the rats in sham, OVX + PBS, OVX + free siRNA, OVX + CH6-LNPs-siRNA, OVX + Rd-LNPs-siRNA and OVX + LNPs-siRNA groups. The boxes indicate the initiation of siRNA treatment at week 8 after surgery. Repeated-measures ANOVA was conducted to determine whether (i) there was a time effect and (ii) if there was a time by group interaction effect. (b) Representative 3D microarchitecture of the proximal tibia in each group (n = 9), obtained by in vivo microCT examination. Scale bars, 1.0 mm. (c) Analysis of bone histomorphometric parameters (MAR and BFR/BS) from rats in each group. One-way ANOVA with LSD’s post hoc test was performed to determine inter-group differences. (d) Bone formation examined by sequential labeling with fluorescent dye in non-decalcified bone sections. Representative fluorescent micrographs of the trabecular bone sections showing xylenol (red) and calcein (green) labeling in each group (n = 9). Scale bar, 10 µm. BMD, bone mineral density; BV/TV, relative bone volume; MAR, mineral apposition rate; BFR/BS, bone formation rate; sham-BS, sham baseline; OVX-BS, OVX baseline before treatment initiation. The data are presented as the mean ± sd; n = 9 per group. *P < 0.05 for a comparison of OVX + CH6-LNPs-siRNA or OVX + PBS or OVX + free siRNA or sham group with OVX + Rd-LNPs-siRNA group; #P < 0.05 for a comparison of OVX + CH6-LNPs-siRNA with OVX-BS group; ^P < 0.05 for a comparison of OVX-BS with sham-BS.
Histomorphometric analysis further demonstrated that the mineral apposition rate (MAR), bone formation rate (BFR/BS), osteoblast surface (Ob.S/BS) and osteoblast number (Ob.N/B.Pm) were significantly increased from baseline and were obviously higher in the OVX + CH6-LNPs-siRNA group when compared to the OVX + Rd-LNPs-siRNA group after 6 weeks of siRNA treatment (P < 0.05) (Fig. 5c; Supplementary Fig. 6c). A larger width between the xylenol and the calcein labeling bands was consistently found in the OVX rats treated with CH6-LNPs-siRNA compared to Rd-LNPs-siRNA (Fig. 5d). No significant difference between Oc.S/BS and Oc.N/B.Pm was found among the siRNA groups (P > 0.05; Supplementary Fig. 6c). Results from mechanical testing, bone turnover marker assays and microCT examination of cortical bone and fifth vertebrae also evidenced that CH6-LNPs-siRNA achieved a better therapeutic effect when compared to the other siRNA groups (Supplementary Fig. 6d–g).
We administered six periodic injections of Plekho1 siRNA (1.0 mg kg−1) encapsulated in LNPs, Rd-LNPs, CH6-LNPs or (AspSerSer)6-liposome to 6-month-old female healthy Sprague-Dawley rats at an interval of once per week. Results from microCT examination and bone turnover marker assays showed that CH6-LNPs-siRNA significantly improved bone formation and bone mass when compared to other LNPs-based siRNA formulations or (AspSerSer)6-liposome-siRNA (P < 0.05) (Supplementary Fig. 7). No obvious toxicity and immune response were found in rats after either a single dose or multiple doses of CH6-LNPs-siRNA (Supplementary Table 4; Supplementary Fig. 8).
DISCUSSION
To our knowledge, this is the first report of aptamer-functionalized lipid nanoparticles that can directly target osteoblasts for osteogenic siRNA delivery, and as such these findings may help enable the development of RNAi-based bone anabolic therapies. Relevant to its translational potential, the aptamer CH6 specifically targets both rat and human osteoblasts.
CH6-LNPs-siRNA achieved better gene silencing and bone anabolic action when compared to (AspSerSer)6-liposome-siRNA (Supplementary Fig. 7). This difference could be partly explained by the distinct mechanisms responsible for targeted delivery. The (AspSerSer)6 moiety was designed to target the physiochemical features of the bone formation surface, and as such it targeted osteo-blasts at a tissue level. In contrast, the CH6 aptamer was selected to directly target osteoblasts at cellular level, which is more specific and efficient.
Mechanistically, we found that the CH6 aptamer specifically induced macropinocytosis and facilitated siRNA entry in osteoblasts, which was evidenced by the co-localization of the encapsulated siRNA with a macropinocytosis marker, and by the fact that macropinocytosis inhibitors mediated blockade of siRNA cellular uptake and membrane ruffling appearance. Confocal imaging data suggested that the CH6 aptamer facilitated lysosome escape of siRNA, which reflects another feature of macropinocytosis22. Taken together, macropinocytosis induced by the CH6 aptamer can also partially explain the high gene knockdown efficiency and enhanced bone anabolic action of CH6-LNPs-siRNA.
In summary, CH6 aptamer-functionalized LNPs update targeted delivery systems from a tissue level to a cellular level, which facilitates the clinical translation of an RNAi-based bone anabolic strategy.
ONLINE METHODS
Cell culture
BRL-3A (rat liver; ATCC CRL-1442) and THLE-3 (human liver; ATCC CRL-11233) cells were cultured in DMEM supplemented with 10% FBS and 100 units ml−1 penicillin-streptomycin. Rat PBMCs were obtained by density gradient centrifugation with Ficoll-Paque PREMIUM (GE Healthcare). Rat primary osteoblasts (CHI Scientific) and human primary osteoblasts (PriCells) were cultured in DMEM/F12 supplemented with 10% FBS and 100 units ml−1 penicillin-streptomycin. Rat pre-osteoclasts (B-Bridge International) and human osteoclasts (PriCells) were cultured in DMEM containing 10% FBS, 100 units ml−1 penicillin-streptomycin, macrophage colony-stimulating factor (M-CSF) and receptor for activation of nuclear factor kappa B ligand (RANKL). Phenotypes of osteoblasts and osteoclasts were confirmed by real-time PCR as described previously28–31. The primers are listed in Supplementary Table 3. We found no mycoplasma infection for the above cell lines; we also validated their phenotypes.
Random library and primers
The ssDNA library contained a central randomized sequence of 40 nucleotides: 5′-ATCCAGAGTGACGCAGCA-40nt-TGGACACGGTGGCTTAGT-3′. An FAM-labeled forward primer (5′-FAM-ATCCAGAGTGACGCAGCA-3′) and a biotin-labeled reverse primer (5′-biotin-ACTAAGCCACCGTGTCCA-3′) were used in the PCR for the synthesis of dsDNA. After alkaline denaturing (0.2 M NaOH), the FAM-labeled ssDNA was separated from the biotin-labeled ssDNA by streptavidin-coated magnetic beads (Promega). The library, primers and random sequence (Rd; 5′-CTACTGGACTTCATCGGAGCTAGGTCATCGCTTGCATGCA-3′) were synthesized by Sangon, Shanghai, China.
Cell-SELEX
Briefly, ssDNA library was incubated with 1–2 × 106 target cells at 37 °C for 0.5–1 h. After washing, the bound DNAs were eluted and then incubated with non-target cells at 37 °C for 1 h. The supernatant was desalted and amplified by PCR. The desired ssDNA was separated by streptavidin-coated magnetic beads (Promega). After multiple rounds of selection, the enriched ssDNA pool was cloned into TOP10 chemically competent Escherichia coli with the TA cloning kit (Invitrogen) and sequenced by Sangon, Shanghai, China. Binding assays of enriched ssDNA with the target cells and non-target cells were performed by a FACScan cytometer (BD Immunocytometry Systems). Secondary structures were predicted by RNAstructure 5.6 software. The equilibrium dissociation constants (Kd) were calculated by the equation Y = BmaxX/(Kd + X) (refs. 13, 32).
Preparation and evaluation of siRNA formulations in vitro
siRNA formulations were prepared using the established protocol33 and evaluated by PAGE (PAGE). The in vitro characteristics were examined in the following respects: (1) Hydrodynamic diameters and zeta potentials were measured by laser light scattering using a DELSA 440 SX Zeta Potential Analyzer (Beckman Coulter Corporation) at 25 °C following dilution with distilled water. (2) Aptamer loading efficiency was expressed by the amount of aptamer on LNPs divided by the amount of aptamer on PEG 2000–DSPE. (3) Morphology: 10 µl of siRNA formulation dispersion was placed on a copper grid covered by a holey carbon film. Excess sample was blotted with filter paper, and the grid was rapidly plunged into liquid ethane (cooled to approximately −180 °C with liquid nitrogen) in a cryo-box (Carl Zeiss NTS GmbH, D-Oberkochen). The frozen specimen was then transferred via a cryo-transfer unit (Gatan 626-DH) to a transmission electron microscope (FEI/Philips Tecnai 12 BioTWIN) with a pre-cooled cryo-specimen holder. (4) The encapsulation efficiency of the siRNA was assessed by a Quant-iT RiboGreen RNA assay (Invitrogen). (5) Serum stability of siRNA and CH6 was assessed by electrophoresis. (6) Cytotoxicity was determined using MTT assays (Sigma). (7) The effect of LNPs-siRNA on the osteoblast-binding ability of Cy3-labeled aptamer was examined by a FACScan cytometer (BD Immunocytometry Systems). (8) Cellular uptake of Cy3-labeled siRNA in vitro: LNPs-siRNA or Rd-LNPs-siRNA or CH6-LNPs-siRNA or free CH6 (pre-incubation for 1 h) + CH6-LNPs-siRNA was incubated with rat osteoblasts at 37 °C for 8 h. Fluorescence was determined with a FACScan cytometer (BD Immunocytometry Systems). (9) In vitro knockdown efficiency: LNPs-siRNA, Rd-LNPs-siRNA, CH6-LNPs-siRNA, free CH6 + CH6-LNPs-siRNA and positive control Lipofectamine 2000–siRNA were incubated with rat osteoblasts for 8 h in serum-free medium. The medium was replaced with fresh medium supplemented with 10% FBS and maintained at 37 °C for 48 h. The Plekho1 mRNA expression levels were quantified by real-time PCR5. The primers are listed in Supplementary Table 3.
Confocal imaging for endocytosis pathways
Rat primary osteoblasts were incubated with siRNA formulations with Plekho1 siRNA concentration of 250 nM at 37 °C for 4 h in serum-free medium. Alexa Fluor 488–labeled endocytic markers (500 µg ml−1 dextran, 50 µg ml−1 transferrin or 5 µg ml−1 choleratoxin) were added during the final 30 min of the incubation. The cells were fixed, counterstained with DAPI and observed by confocal microscopy. To examine membrane ruffling, osteoblasts were serum starved for 1 h, followed by the addition of siRNA formulations for 4 h. The cells were fixed and stained with FITC-labeled phalloidin (Sigma) for visualization by confocal microscopy. To investigate the sub-cellular trafficking, osteoblasts were incubated with siRNA formulations for 4 h. 75 nM LysoTracker Green DND-26 (Invitrogen) was added to the cells during the final 1 h of the incubation. The cells were fixed and prepared for visualization by confocal microscopy.
Chemical inhibition of endocytosis pathways
Rat primary osteoblasts were seeded in flasks with 1 × 105 cells and incubated overnight until 70–80% confluence. Inhibitors of endocytosis pathways were pre-incubated with osteo-blasts in serum-free medium at 37 °C for 0.5 h before addition of siRNA formulations. After 4 h, fluorescence was determined with a FACScan cytometer (BD Immunocytometry Systems).
Tissue distribution of siRNA
According to previously established protocols5, fluorescence imaging of the Cy3-labeled siRNA distribution in tissues was performed using an IVIS Lumina XR imaging system and fluorescence intensity in different organ samples was measured by a microplate reader system (Bioscan, Washington, DC).
Animal handling
All animals were maintained under a 12-h light–12-h dark cycle in a temperature-controlled room with ad libitum access to water and food. Six-month-old female Sprague-Dawley rats were used for the in vivo experiments. To evaluate the therapeutic effects of siRNA formulations, 6-month-old female Sprague-Dawley rats were ovariectomized or sham-operated and then left untreated for 8 weeks until siRNA administration.
In vivo localization of Cy3-labeled siRNA in livers
Portions of the livers were placed in 4% paraformaldehyde overnight. Before embedding and sectioning, the liver pieces were incubated in 30% sucrose for 6 h. The tissues were embedded in optimal cutting temperature medium (OCT) and cryosectioned. The sections were washed with PBS, permeabilized with 0.1% Triton X-100 in PBS, stained with Alexa Fluor 488–phalloidin (Invitrogen) and mounted with DAPI. Images were acquired with a confocal microscopy.
In vivo Cy3-labeled siRNA cellular uptake in PBMCs
Blood samples of rats injected with siRNA formulations were collected and PBMCs were obtained by density gradient centrifugation with Ficoll-Paque PREMIUM. Fluorescence was determined with a FACScan cytometer (BD Immunocytometry Systems).
In vivo cell selectivity and gene silencing
According to previously established protocols5,7, in vivo cell-selectivity was examined by immunohistochemistry. In vivo gene silencing was tested by fluorescence-activated cell sorting (FACS) or Laser capture microdissection (LCM) combined with real-time PCR.
Analysis of bone anabolic action
According to previously established protocols5,7,28,34, bone mass and bone microarchitecture were evaluated by a microCT system (vivaCT 40, SCANCO MEDICAL, Switzerland). Bone formation was tested by an EXAKT Cut/grinding System (EXAKT Technologies, Inc, Germany) combined with professional image analysis software (ImageJ, NIH, USA and BIOQUANT OSTEO, Version 13.2.6, Nashville, TN, USA). Mechanical properties were examined by a material test machine (H25KS; Hounsfield Test Equipment Ltd., UK) combined with built-in software (QMAT Professional; Tinius Olsen, Inc., Horsham, PA, USA).
Assays of biochemical and hematological parameters, immune factors and bone turnover markers
Hematological parameters (TP, RBCs, hemoglobin, WBCs and PLTs) were analyzed using an Auto Hematology Analyzer (Mindray International Ltd., China)35. Clinical biochemical parameters (ALT, AST and BUN) were analyzed using a clinical chemistry analyzer (Cruinn Diagnostics Ltd., Ireland)35. Immune factors (IL-6, TNF-α, IFN-γ and IL-2) were quantified using commercially available kits (Life Technologies). Bone turnover markers (PINP and TRACP 5b) were determined using commercially available kits (Immunodiagnostic Systems). All the animal study procedures were approved by the Animal Experimentation Ethics Committee of the Hong Kong Baptist University (ref. no.: HASC/12–13/0032).
Statistical analysis
All the variables were expressed as mean ± sd. For the repeated measurement data from longitudinal in vivo microCT examination, repeated-measures analysis of variance (ANOVA) was conducted to determine whether (a) there was a time effect and (b) if there was a time by group interaction effect. For the non-repeated measurement data, one-way ANOVA with LSD’s post hoc test was performed to determine inter-group differences in the study variables. All the statistical data were analyzed by a contract service from Bioinformedicine (San Diego, California, USA, http://www.bioinformedicine.com/index.php). A statistical software SPSS (version 22.0) was used and P < 0.05 was considered to be statistically significant. For the in vitro experiments, sample size was pre-determined based on preliminary results. For the in vivo experiments, sample size was pre-determined by a power calculation and our previously published paper5. The rats were grouped randomly and blindly to researchers. The rats in poor body condition were excluded.
Supplementary Material
Acknowledgments
We thank the academic staff (L. Qin) from the Chinese University of Hong Kong and H.Y.S. Cheung from the Institute for Advancing Translational Medicine in Bone & Joint Diseases, Hong Kong Baptist University for providing critical comments and technical support. This study was supported by the Ministry of Science and Technology of China (2013ZX09301307 to A.L.), the Hong Kong General Research Fund (HKBU479111 to G.Z., HKBU478312 to G.Z., HKBU12102914 to G.Z. and HKBU261113 to A.L.), the Natural Science Foundation Council of China (81272045 to G.Z., 81228013 to J.X. and 21221003 to W.T.), the Research Grants Council & Natural Science Foundation Council of China (N_HKBU435/12 to G.Z.), the Interdisciplinary Research Matching Scheme (IRMS) of Hong Kong Baptist University (RC-IRMS/12-13/02 to A.L. and RC-IRMS/13-14/02 to G.Z.), the Hong Kong Baptist University Strategic Development Fund (SDF) (SDF13-1209-P01 to A.L.), the Hong Kong Research Grants Council (RGC) Early Career Scheme (ECS) (489213 to B.-T.Z.), the Faculty Research Grant of Hong Kong Baptist University (FRG1/13-14/024 and FRG2/12-13/027 to G.Z.), the China Academy of Chinese Medical Sciences (Z0252 and Z0293 to A.L.), the Chinese National High-Tech Research and Development Programme (2012AA022501 to N.S.), the National Key Technologies R&D Programs for New Drugs of China (2012ZX09301003-001-001 and 2014ZX09J14106-04C to L.Z.), the Collaborative Research Programme (CRP)-International Centre for Genetic Engineering and Biotechnology (ICGEB) grant (CRP/CHN13-02 to L.Z.), the Chinese National Natural Science Foundation Project (81261160503 to L.Z.), the National Key Scientific Program of China (2011CB911000 to W.T.), the National Institutes of Health (GM079359 to W.T.) and the Beijing Natural Science Foundation (7131012 to L.Z.). The statistical analysis was performed by a contract service from Bioinformedicine (http://www.bioinformedicine.com/index.php).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
G.Z., A.L. and L.Z. supervised the whole project. C. Liang, B.G. and H.W. performed the major research and wrote the manuscript in equal contribution. D.L., J.L., C.W., W.K.L., Changwei Lu, Y.C. and L.D. provided the technical support. X.H., D.W.T.A., Cheng Lu, H.L., S.L., B.-T.Z., N.S., Z.Y., X.P., H.Z., K.Y., A.Q., P.S., J.X., L.X., Z.L., Z.B., F.H. and W.T. provided their professional expertise.
COMPETING FINANCIAL INTERSTS
The authors declare no competing financial interests
References
- 1.López-Fraga M, Martinez T, Jimenez A. RNA interference technologies and therapeutics: from basic research to products. BioDrugs. 2009;23:305–332. doi: 10.2165/11318190-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Grainger DW. RNA therapeutics targeting osteoclast-mediated excessive bone resorption. Adv. Drug Deliv. Rev. 2012;64:1341–1357. doi: 10.1016/j.addr.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu K, et al. Targeting WW domains linker of HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nat. Cell Biol. 2008;10:994–1002. doi: 10.1038/ncb1760. [DOI] [PubMed] [Google Scholar]
- 5.Zhang G, et al. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat. Med. 2012;18:307–314. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, Miller SC, Kopeckova P, Kopecek J. Bone-targeting macromolecular therapeutics. Adv. Drug Deliv. Rev. 2005;57:1049–1076. doi: 10.1016/j.addr.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, et al. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 8.Li SD, Huang L. Pharmacokinetics and biodistribution of nanoparticles. Mol. Pharm. 2008;5:496–504. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- 9.Zamboni WC. Liposomal, nanoparticle, and conjugated formulations of anticancer agents. Clin. Cancer Res. 2005;11:8230–8234. doi: 10.1158/1078-0432.CCR-05-1895. [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Kucharski C, Lin X, Uludag H. Bisphosphonate-coated BSA nanoparticles lack bone targeting after systemic administration. J. Drug Target. 2010;18:611–626. doi: 10.3109/10611861003622560. [DOI] [PubMed] [Google Scholar]
- 11.Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J. Clin. Invest. 2007;117:3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye M, et al. Generating aptamers by cell-SELEX for applications in molecular medicine. Int. J. Mol. Sci. 2012;13:3341–3353. doi: 10.3390/ijms13033341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W. Development of DNA aptamers using cell-SELEX. Nat. Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 14.Tam YY, Chen S, Cullis PR. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics. 2013;5:498–507. doi: 10.3390/pharmaceutics5030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auguste DT, et al. Triggered release of siRNA from poly(ethylene glycol)-protected, pH-dependent liposomes. J. Control. Release. 2008;130:266–274. doi: 10.1016/j.jconrel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mui BL, et al. Infuence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. Nucleic Acids. 2013;2:e139. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rockey WM, et al. Rational truncation of an RNA aptamer to prostate-specifc membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011;21:299–314. doi: 10.1089/nat.2011.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert JC, et al. First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation. 2007;116:2678–2686. doi: 10.1161/CIRCULATIONAHA.107.724864. [DOI] [PubMed] [Google Scholar]
- 20.Cao H, Chen J, Awoniyi M, Henley JR, McNiven MA. Dynamin 2 mediates fuidphase micropinocytosis in epithelial cells. J. Cell Sci. 2007;120:4167–4177. doi: 10.1242/jcs.010686. [DOI] [PubMed] [Google Scholar]
- 21.Vercauteren D, et al. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol. Ther. 2010;18:561–569. doi: 10.1038/mt.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim JP, Gleeson PA. Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol. Cell Biol. 2011;89:836–843. doi: 10.1038/icb.2011.20. [DOI] [PubMed] [Google Scholar]
- 23.Zernik J, Twarog K, Upholt WB. Regulation of alkaline phosphatase and alpha 2(I) procollagen synthesis during early intramembranous bone formation in the rat mandible. Differentiation. 1990;44:207–215. doi: 10.1111/j.1432-0436.1990.tb00619.x. [DOI] [PubMed] [Google Scholar]
- 24.Fukushima N, Hiraoka K, Shirachi I, Kojima M, Nagata K. Isolation and characterization of a novel peptide, osteoblast activating peptide (OBAP), associated with osteoblast differentiation and bone formation. Biochem. Biophys. Res. Commun. 2010;400:157–163. doi: 10.1016/j.bbrc.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa S, et al. Involvement of FcRgamma in signal transduction of osteoclast-associated receptor (OSCAR) Int. Immunol. 2004;16:1019–1025. doi: 10.1093/intimm/dxh103. [DOI] [PubMed] [Google Scholar]
- 26.Herman S, et al. Induction of osteoclast-associated receptor, a key osteoclast costimulation molecule, in rheumatoid arthritis. Arthritis Rheum. 2008;58:3041–3050. doi: 10.1002/art.23943. [DOI] [PubMed] [Google Scholar]
- 27.Lotinun S, et al. Osteoclast-specifc cathepsin K deletion stimulates S1P–dependent bone formation. J. Clin. Invest. 2013;123:666–681. doi: 10.1172/JCI64840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo B, et al. Therapeutic RNA interference targeting CKIP-1 with a cross-species sequence to stimulate bone formation. Bone. 2014;59:76–88. doi: 10.1016/j.bone.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Sanguineti R, Storace D, Monacelli F, Federici A, Odetti P. Pentosidine effects on human osteoblasts in vitro. Ann. NY Acad. Sci. 2008;1126:166–172. doi: 10.1196/annals.1433.044. [DOI] [PubMed] [Google Scholar]
- 30.Kishimoto Y, et al. Gene expression relevant to osteoclastogenesis in the synovium and bone marrow of mature rats with collagen-induced arthritis. Rheumatology (Oxford) 2004;43:1496–1503. doi: 10.1093/rheumatology/keh395. [DOI] [PubMed] [Google Scholar]
- 31.Susa M, Luong-Nguyen NH, Cappellen D, Zamurovic N, Gamse R. Human primary osteoclasts: in vitro generation and applications as pharmacological and clinical assay. J. Transl. Med. 2004;2:6. doi: 10.1186/1479-5876-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang M, et al. Developing aptamer probes for acute myelogenous leukemia detection and surface protein biomarker discovery. J. Hematol. Oncol. 2014;7:5. doi: 10.1186/1756-8722-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H, et al. Preparation of aptamer-functionalized lipid nanoparticles (LNPs) encapsulating siRNAs. Protocol Exchange. 2014 [Google Scholar]
- 34.Hao YJ, et al. Changes of microstructure and mineralized tissue in the middle and late phase of osteoporotic fracture healing in rats. Bone. 2007;41:631–638. doi: 10.1016/j.bone.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.