Abstract
Melanoma is one of the most immunogenic tumors, and extensive lists of potential tumor rejection antigens have been collected during the last decades. By isolating human leukocyte antigen (HLA) class I complexes from five melanoma cell lines (FM-82, FM-93/2, Mel-624, MeWo and SK-Mel-5) and sequencing HLA-eluted peptides by mass spectrometry, we identified over 10,000 unique peptides with high confidence. The majority of the peptides were 8–11 amino acids in length and were predicted to bind to the respective HLA alleles. Over 250 epitopes, corresponding to previously described tumor-associated antigens, were identified, suggesting that HLA peptidome analysis may facilitate the characterization of putative tumor rejection antigens. MeWo and SK-Mel-5 cell lines were further interrogated for neo-epitopes, revealing one peptide from MeWo cells carrying an amino acid mutation. We also observed a remarkable overlap between A*03:01 peptides eluted from Mel-624 cells and A*03:01 peptides recovered from soluble HLA complexes purified from two melanoma patients, shedding light on the similarity of the HLA peptidome in cell lines and in patient-derived material. The reliable characterization of the HLA class I peptidome in melanoma promises to facilitate the identification of tumor rejection antigens and the development of immunotherapeutic strategies.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-016-1897-3) contains supplementary material, which is available to authorized users.
Keywords: HLA, Melanoma, Immunopeptidome, Tumor-associated antigen, Mass spectrometry, Immunocapture
Introduction
Immunotherapeutic strategies are gaining considerable importance for the treatment of various types of malignancies. Therapeutic approaches include the development of antibodies against immunological checkpoints [1, 2], antibody–cytokine fusion proteins [3], bispecific antibodies [4], lymphokine-activated adoptive cell therapies [5, 6], as well as cancer vaccines [7].
Over the last decades, substantial evidence was collected, indicating that melanoma cells can be recognized by cytotoxic T cells through the presentation of tumor rejection antigens and neo-epitopes onto HLA class I molecules [8–10]. Thus, boosting a cancer patient’s immune system represents a promising strategy for the treatment of melanoma, especially considering the high mutation rate of this cancer type [11, 12]. To understand the molecular basis for the tumor rejection process, a detailed knowledge of HLA class I peptides presented by malignant cells is essential. A detailed knowledge of the HLA peptidome facilitates the study of tumor-reactive T cell specificities, either by multiplex tetramer analysis [13, 14] or by peptide stimulation assays [15]. In principle, these investigations could be applied to cancer patients, helping profile their response to therapy. Indeed, recent reports have shown that the breadth and frequency of anti-melanoma T cell specificities increase, after therapeutic intervention with the anti-CTLA-4 antibody ipilimumab [16].
Since the first direct identification of a peptide, recognized by melanoma-specific T cells, by mass spectrometry in 1994 [17], several studies investigated the HLA peptidome of melanoma cells [18, 19]. Both studies, however, purified HLA complexes from >109 cells per analysis, which was necessary due to substantial sample loss during the purification process, estimated to be 95 % [20]. Recent advances in mass spectrometry and sample preparation protocols have made it possible to identify thousands of HLA class I-bound peptides from 108 cells [21, 22]. These technological improvements facilitate the investigation of tumor-associated antigens by mass spectrometry, allowing a comparison with other epitopes (e.g., peptides derived from housekeeping proteins) presented on HLA class I molecules.
In this work, we report on the confident identification of over 10,000 HLA class I-bound peptides from five human melanoma cell lines (FM-82, FM-93/2, Mel-624, MeWo and SK-Mel-5). Analysis of the identified sequences revealed the presence of more than 250 peptides from previously described tumor-associated antigens. Furthermore, we present for the first time the direct mass spectrometry-based identification of a neo-epitope purified from a human melanoma cell line. The amino acid substitution in this peptide led to an increased binding affinity to the cognate HLA allele. Finally, a comparison between peptides isolated from melanoma cell lines and from the serum of two HLA-matched melanoma patients and two healthy donors revealed similarities on the level of presented peptides.
Materials and methods
Cell lines and antibodies
Cell lines FM-82, FM-93/2, Mel-624 and MeWo were obtained from European Searchable Tumour Line Database (ESTDAB) [23], SK-Mel-5 was obtained from CLS Cell Lines Services, and A375 was obtained from American Type Culture Collection (ATCC). Cells were grown in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10 % FBS at 37 °C and 5 % CO2. HB-95 hybridoma cells were cultivated in CD Hybridoma medium supplemented with 2 mM glutamine in a shaking incubator at 37 °C and 5 % CO2. The W6/32 antibody was purified from HB-95 supernatant using protein A Sepharose and subsequently coupled to AminoLink Plus Coupling Resin (Thermo Fisher Scientific) following the manufacturer`s instructions.
Cell lysis and affinity purification of HLA class I molecules and enrichment of HLA-bound peptides
Cells were washed with PBS, harvested using a cell scraper, washed twice again with PBS and lysed on ice at a density of 2 × 107–5 × 107 cells/ml lysis buffer (0.5 % IGEPAL CA-630, 0.25 % sodium deoxycholate, 1 mM EDTA, 0.2 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride (PMSF), Roche Complete Protease Inhibitor Cocktail in PBS) for 1 h. Lysates were cleared by centrifugation at 21,000g for 30 min at 4 °C and snap frozen for storage at −80 °C. HLA class I complexes were purified from cleared lysate using W6/32 antibody-coupled resin by incubation for 2 h at 4 °C. The resin was washed once with lysis buffer, then buffer A (150 mM NaCl, 20 mM Tris, pH 7.4), buffer B (400 mM NaCl, 20 mM Tris, pH 7.4), buffer A again and buffer C (20 mM Tris, pH 8.0) at 4 °C. Peptide–HLA complexes were eluted using 0.1 M acetic acid. Peptides were separated from protein by elution from C18 Macro SpinColums (Harvard Apparatus) with 30 % acetonitrile (ACN), and subsequently dried and stored at −20 °C.
cDNA analysis of NY-ESO-1 and GAPDH
cDNA of the cell lines A375, FM-82, FM-93/2, Mel-624, MeWo and SK-Mel-5 was generated using the “SuperScript® III CellsDirect™ cDNA Synthesis System” (Thermo Scientific) following the manufacturer’s instructions. mRNA expression of NY-ESO-1 and GAPDH was analyzed via PCR using primers 5′-TGCTTGAGTTCTACCTCGCCA-3′ (forward) and 5′-TATGTTGCCGGACACAGTGAA-3′ (reverse) for NY-ESO-1 and 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and 5′-GAAGATGGTGATGGGATTTC-3′ (reverse) for GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) and Phire polymerase (Thermo Scientific). NY-ESO-1+ A375 cells were used as positive control [24].
Refolding of MHC class I monomers
Extracellular domains of HLA-A*02:01 and HLA-A*26:01 were refolded in the presence of β2-microglobulin and peptides (Biomatik) as previously described [25]. The experimental setup was scaled down to 20 ml and performed without the addition of PMSF. In brief, 6 µM denatured β2-microglobulin and 3 µM denatured HLA alpha chain (both from bacterial inclusion body production) were refolded with 60 µM of peptide for four days at 4 °C in a buffer containing 100 mM Tris HCl pH 8, 400 mM l-arginine hydrochloride, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 2 mM EDTA and protease inhibitor cocktail (Roche). Refolded monomers were concentrated to 1 ml using “Vivaspin 20 (molecular weight cutoff 30,000)” (Sartorius) and subsequently 100 µl were analyzed on a “ÄKTA Purifier” fast protein liquid chromatography (FPLC) with a “Superdex™ 200 10/300 GL” column (both GE Healthcare). Fractions (0.25 ml) of the HLA monomer peaks (peak at 15 ml) were collected and analyzed via SDS-PAGE.
Analysis of HLA class I peptides by ultra high performance liquid chromatography (UHPLC) and mass spectrometry
Liquid chromatography–mass spectrometry (LC-MS) analysis of HLA class I peptides was essentially carried out as described in Ritz et al. [22]. Peptides were analyzed by LC-MS using a Q Exactive Mass Spectrometer connected to an EASY-nLC 1000 (both Thermo Scientific). Peptides were separated by reversed-phase chromatography using an Acclaim PepMap RSLC C18, 50 μm × 150 mm, 2 μm analytical column (Thermo Fisher) at a flow rate of 0.3 μl/min, with linear gradients from 0 to 24 % ACN in 90 min. All buffers contained 0.1 % formic acid. MS spectra were recorded in full ion scan mode from 250 to 2000 m/z, with a resolution of 70,000 and a maximum injection time of 80 ms. Fragment mass spectra (MS/MS) were recorded at a resolution of 17,500 and an injection time of 240 ms. The most intense ten doubly or triply charged ions were selected for higher-energy collisional dissociation (HCD) fragmentation. Dynamic exclusion was set to 15 s.
The datasets were searched using the MaxQuant software (version 1.4.1.2 [26]) against the human reference proteome databases containing 89,649 human proteins downloaded from the UniProt homepage on January 20, 2015. MaxQuant parameters were essentially set as described [21]: (1) digestion mode: unspecific, (2) first search ppm 20; main search ppm 4.5, (3) fragment mass tolerance ppm 20, (4) one variable modification (oxidation of methionine), (5) no specific amino acids for the generation of the decoy databases, (6) peptide false discovery rate (FDR) 1 %, protein FDR 100 %, (7) peptide length allowed: 7–20 amino acids. Reverse hits and contaminants were removed from the “peptide.txt” output file.
To identify melanoma-associated antigens in the HLA analysis, a list of melanoma-associated antigens [10] was retrieved and compared to the identified peptides on the gene level.
HLA binding prediction of identified peptides
Peptide identifications based on the MS/MS data were combined into a final report for each cell line. Identical sequences identified multiple times due to modifications were collapsed into one identification. Peptides with a length between 8 and 11 amino acids were subjected to HLA class I binding prediction analysis using NetMHCpan 2.8 [27], taking into account the respective HLA alleles of the cell lines reported in the literature [23, 28]. The analysis was performed on a cell line by cell line basis. Each peptide was assigned the minimal predicted half maximal inhibitory concentration (IC50).
Graphical representation of HLA-specific motifs
HLA-specific motifs were generated using the Seq2Logo 2.0 server using default settings [29]. Based on the binding prediction analysis, all nine mers were assigned to the allele with the lowest predicted IC50-value and subsequently used for the visualization of respective HLA-specific motifs.
Identification of a neo-epitope and validation of the fragment mass spectrum
The mutational information for MeWo and SK-Mel-5 cell lines, respectively, were downloaded from the “Catalogue of Somatic Mutations in Cancer” (COSMIC) database [30] on September 3, 2015. The positional information for all protein code-changing single nucleotide polymorphisms was used to create 24 mers with the mutated amino acid at position 12. Mass spectrometric data from MeWo or SK-Mel-5 was interrogated with MaxQuant for the presence of mutated sequences using the same search parameters as for other analyses, but taking into account all protein sequences of the human reference proteome and the sequences carrying mutations. The synthetic peptide corresponding to IFRD2437–446H446Y (Biomatik) was analyzed by LC-MS using the same HPLC and MS conditions as for the HLA peptidomes. This allowed for comparison of the retention time and to identify a representative fragment mass spectrum. The fragment mass spectra of the synthetic and the endogenous peptides were extracted with Thermo Xcalibur Qual Browser 2.2 SP1 (Thermo Scientific) and plotted with Excel (Microsoft). Peptide fragments (a, b and y ions) were annotated with the MaxQuant Viewer 1.4.1.2 [26].
Results
Identification of HLA class I-bound peptides from melanoma cell lines
HLA class I peptides were purified from melanoma cell lysates, using an immunocapture procedure depicted in Fig. 1a, based on the pan-HLA class I monoclonal antibody W6/32 [31]. Peptides were subsequently eluted with acid, purified and desalted using C18 spin columns and analyzed by LC-MS. The lysates of five melanoma cell lines (FM-82, FM-93/2, Mel-624, MeWo and SK-Mel-5) were analyzed in independent quadruplicate experiments (quintuplicates for MeWo), leading to the identification of 3566, 3290, 2403, 1679 and 3321 unique peptides with a false discovery rate of 1 %, respectively (Supplementary Tables 1–5 list all identifications). Between 84 and 95 % of the identified peptides ranged between 8 and 11 amino acids in length (Table 1; Fig. 1b–f) demonstrating compatibility with the typical length of HLA-bound peptides. HLA class I binding prediction analysis using NetMHCpan 2.8 revealed that between 79 and 91 % of these 8–11 mers were predicted to bind (IC50 < 500 nM) to the cognate HLA class I molecule, with 45–66 % predicted to be strong binders (IC50 < 50 nM) (Table 1).
Fig. 1.
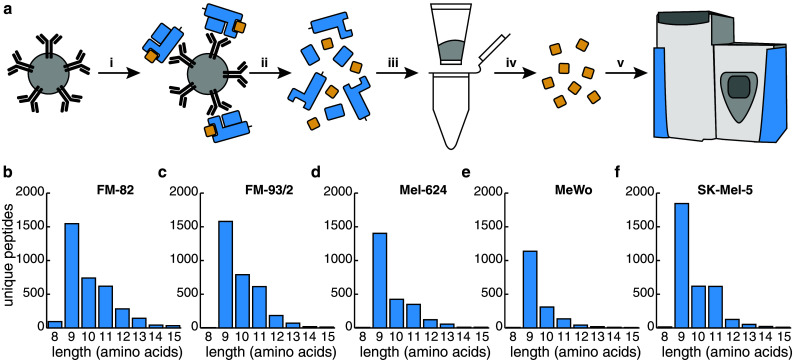
Purification of HLA class I complexes and length distribution of peptides identified from melanoma cell lines. a Schematic representation of HLA class I peptide purification. i HLA complexes are captured using the pan-HLA antibody W6/32 coupled to resin. ii HLA complexes are eluted and denatured under acidic conditions. iii, iv HLA peptides are purified by stepwise elution from C18 resin. v Analysis of purified peptides by LC-MS. b–f Length distribution of peptides identified from five melanoma cell lines. The number of identified peptides between 8 and 15 amino acids is plotted in relation to their length for b FM-82, c FM-93/2, d Mel-624, e MeWo, f SK-Mel-5
Table 1.
HLA-bound peptides identified from melanoma cell lines, their length and binding prediction to respective HLA alleles
| Peptides identified | 8–11 mers | % 8–11 mers | Strong bindersa | % Strong binders of 8–11 mers | Bindersb | % Binders of 8–11 mers | Tumor-associated antigens | |
|---|---|---|---|---|---|---|---|---|
| FM-82 | 3566 | 3001 | 84.2 | 1545 | 51.5 | 2525 | 84.1 | 106 |
| FM-93/2 | 3290 | 2987 | 90.8 | 1680 | 56.2 | 2610 | 87.4 | 110 |
| Mel-624 | 2403 | 2186 | 91.0 | 980 | 44.8 | 1848 | 84.5 | 78 |
| MeWo | 1679 | 1589 | 94.6 | 741 | 46.6 | 1258 | 79.2 | 50 |
| SK-Mel-5 | 3321 | 3094 | 93.2 | 2041 | 66.0 | 2824 | 91.3 | 80 |
aNetMHCpan IC50 < 50 nM
bNetMHCpan IC50 < 500 nM
Based on the binding prediction analysis of identified nine mers, individual HLA-specific motifs for the alleles of each cell line were plotted using the Seq2Logo 2.0 server [29] (Fig. 2). All cell lines shared the frequent A*02:01 allele and corresponding motifs were in excellent agreement with each other and with motifs presented in the literature [21, 32]. Distinct motifs could also be generated for all other alleles expressed by the five cell lines in agreement with the literature motifs [32, 33]. The majority of the peptides were recovered from HLA-A alleles; however, 842 and 874 nine mers were predicted to bind to HLA-B and HLA-C alleles, respectively, allowing the identification of more elusive HLA-C-specific motifs [21, 22, 33].
Fig. 2.
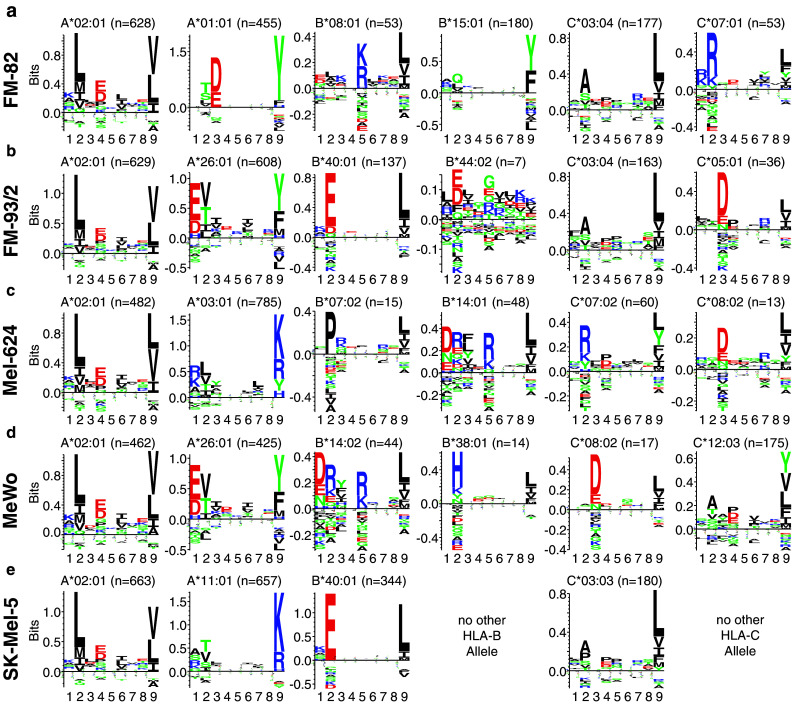
Definition of HLA-specific motifs from the HLA peptidome of melanoma cell lines. IC50 values of all nine mers were predicted for HLA alleles of each cell line [23, 28] using the NetMHCpan 2.8 software [27]. Each peptide was assigned to the HLA allele with the lowest IC50 value for the generation of HLA-specific motifs using Seq2Logo 2.0 [29]. HLA-specific motifs are presented for a FM-82, b FM-93/2, c Mel-624, d MeWo and e SK-Mel-5
Expansion of the catalogue of putative tumor rejection epitopes
The list of HLA-eluted peptides was interrogated for the presence of tumor- and melanoma-associated antigens, using the detailed list described by Andersen et al. [10]. Epitopes were denoted putative tumor-associated antigens if their sequence was identical to one of the genes presented in the above mentioned publication, adding up to a total of 262 epitopes from 72 antigens (Table 2). Of these, 31 demonstrated the exact same sequence as the tumor rejection antigens compiled by Andersen. Interestingly, for gp100/PMEL a total of 32 unique epitopes were identified. While many of the epitopes have been patented and/or characterized in great detail, we also identified previously undescribed epitopes, including predicted strong binders to HLA-A*02:01 originating from prominent melanoma-associated antigens [e.g., KTWDQVPFSV (gp100/PMEL) and KIWEELSVLEV (MAGE-A3/-A6)]. We also detected two length variants of gp100/PMEL epitopes (i.e., SLADTNSLAVV and AMLGTHTMEVTV; the underlined amino acids were either present or absent in the variants). A total of 85 epitopes were predicted to bind to A*02:01, which is probably a result of the high number of A*02:01-eluted peptides identified. The list further contained 22 epitopes originating from B*40:01 and 16 epitopes from C*03:04 or C*03:03, for which only one and two epitopes were presented by Andersen et al. [10], respectively. In contrast to the collection of tumor rejection antigens presented previously, our list of putative tumor rejection antigens is not biased by the frequency of a specific allele in the western population, but shows a strong linear correlation with the total number of peptides identified for a given allele (R 2 = 0.78, Supplementary Fig. 1).
Table 2.
List of peptides identified from tumor-associated antigens
| Gene name (protein name) | Peptide sequence | FM-82 | FM-93/2 | Mel-624 | MeWo | SK-Mel-5 | HLA Alleles (IC50 in nM by NetMHCpan 2.8) |
|---|---|---|---|---|---|---|---|
| SEPT2 (septin-2) | KVNIVPVIAK | X | A*03:01 (46 nM) | ||||
| RLYPWGVVEV | X | X | X | A*02:01 (5 nM) | |||
| VEIEERGVKL | X | X | B*40:01 (47 nM) | ||||
| YIDEQFERY | X | A*01:01 (12 nM) | |||||
| ALDH1A1 (retinal dehydrogenase 1) | ASERGRLLY | X | A*01:01 (35 nM) | ||||
| TMESMNGGKLY | X | No binding predicted | |||||
| ARL4D (ADP-ribosylation factor-like protein 4D) | RLYEMILKR | X | A*03:01 (51 nM) | ||||
| ATIC (IMP cyclohydrolase) | NLYPFVKTV | X | X | A*02:01 (41 nM) | |||
| BCAP31 (B-cell receptor-associated protein 31) | KLDVGNAEV | X | X | X | X | X | A*02:01 (27 nM) |
| CCNB1 (G2/mitotic-specific cyclin-B1) | VMVNQGLTKHMTV | X | A*02:01 (93 nM) | ||||
| VVMVNQGLTK | X | X | A*11:01 (27 nM), A*03:01 (95 nM) | ||||
| GEVDVEQHTL | X | X | B*40:01 (10 nM) | ||||
| TLAKYLMEL | X | X | X | X | A*02:01 (7 nM) | ||
| CCND1 (G1/S-specific cyclin-D1) | ALLESSLRQA | X | X | X | A*02:01 (90 nM) | ||
| EVFPLAMNY | X | X | A*26:01 (22 nM) | ||||
| EVFPLAMNYL | X | A*26:01 (221 nM), C*12:03 (176 nM) | |||||
| CCNI (cyclin-I) | AVLTKQLLH | X | No binding predicted | ||||
| EVIQWLAKL | X | A*26:01 (11 nM) | |||||
| LLDRFLATV | X | X | X | X | A*02:01 (8 nM) | ||
| SLLDRFLATV | X | A*02:01 (4 nM) | |||||
| CDR2 (cerebellar degeneration-related protein 2) | NSELEQQLGATGAY | X | A*01:01 (67 nM) | ||||
| RELEETNQKL | X | X | B*40:01 (19 nM) | ||||
| CEP55 (centrosomal protein of 55 kDa) | AELESKTNTL | X | B*40:01 (34 nM) | ||||
| EVHNLNQLLY | X | X | A*26:01 (22 nM) | ||||
| CPSF1 (cleavage and polyadenylation specificity factor subunit 1) | ALDEKLLNI | X | A*02:01 (47 nM) | ||||
| RLGNSLLLK | X | A*03:01 (17 nM) | |||||
| TLITDGMRSV | X | X | A*02:01 (76 nM) | ||||
| VSDRDRNLMVY | X | A*01:01 (13 nM) | |||||
| CSPG4 (chondroitin sulfate proteoglycan 4) | ALTDIDLQL | X | A*02:01 (62 nM) | ||||
| ASSSAGTDPQLLLY | X | A*01:01 (39 nM) | |||||
| SSSAGTDPQLLLY | X | A*01:01 (14 nM) | |||||
| STDPQHHAY | X | A*01:01 (5 nM) | |||||
| TMLARLASA | X | X | X | A*02:01 (44 nM), B*08:01 (40 nM) | |||
| TPNPALKNGQYW | X | No binding predicted | |||||
| DCT (l-dopachrome tautomerase) | ELAPIGHNRMY | X | A*26:01 (50 nM) | ||||
| ETHLSSKRY | X | X | A*26:01 (207 nM) | ||||
| GTYEGLLRR | X | X | A*03:01 (421 nM), A*11:01 (56 nM) | ||||
| KELPSLHVL | X | B*40:01 (18 nM) | |||||
| LMETHLSSK | X | A*03:01 (122 nM) | |||||
| MTVDSLVNK | X | A*11:01 (9 nM) | |||||
| REQFLGALDL | X | X | B*40:01 (13 nM) | ||||
| SLDDYNHLV | X | A*02:01 (4 nM) | |||||
| SLDDYNHLVTL | X | X | X | A*02:01 (10 nM) | |||
| TRPWSGPYIL | X | C*07:02 (182 nM) | |||||
| TSDQLGYSY | X | A*01:01 (8 nM) | |||||
| VEETPGWPTTL | X | X | B*40:01 (61 nM) | ||||
| YYSVRDTLL | X | C*07:02 (57 nM) | |||||
| EHD2 (EH domain-containing protein 2) | ALASHLIEA | X | X | X | A*02:01 (6 nM) | ||
| GQYSTGKTSFIQY | X | B*15:01 (40 nM) | |||||
| KLPNSVLGR | X | No binding predicted | |||||
| RVHAYIISY | X | A*03:01 (59 nM) | |||||
| ENAH (protein enabled homolog) | ERLERQERL | X | No binding predicted | ||||
| GAGE12I (G antigen 12I) | YYWPRPRRY | X | No binding predicted | ||||
| GPR143 (G-protein-coupled receptor 143) | ATSPPASVR | X | X | A*11:01 (103 nM) | |||
| AVASLLKGR | X | No binding predicted | |||||
| GLSTILLYH | X | A*03:01 (390 nM) | |||||
| HAIPHYVTM | X | X | X | C*03:03 (5 nM), C*03:04 (5 nM) | |||
| MLYYPSVSR | X | A*03:01 (29 nM) | |||||
| HMMR (hyaluronan mediated motility receptor) | KLLEYIEEI | X | X | A*02:01 (2 nM) | |||
| KLQEELNKV | X | X | A*02:01 (12 nM) | ||||
| RLNAALREK | X | A*03:01 (62 nM) | |||||
| HMOX1 (heme oxygenase 1) | EVIPYTPAM | X | X | A*26:01 (5 nM) | |||
| EVIPYTPAMQR | X | No binding predicted | |||||
| EVIPYTPAMQRY | X | A*26:01 (4 nM) | |||||
| HNRNPL (heterogeneous nuclear ribonucleoprotein L) | AAAGGGGGGGRY | X | X | No binding predicted | |||
| AAAGGGGGGGRYY | X | B*15:01 (159 nM) | |||||
| ETLGFLNHY | X | A*26:01 (14 nM) | |||||
| KQPAIMPGQSY | X | B*15:01 (98 nM) | |||||
| KSDALETLGFLNHY | X | No binding predicted | |||||
| MAAAGGGGGGGRY | X | X | X | A*26:01 (170 nM), B*15:01 (59 nM) | |||
| YAVDRAITH | X | X | C*12:03 (59 nM), C*03:04 (273 nM) | ||||
| HSPA1L (heat-shock 70 kDa protein 1-like) | LIFDLGGGTFD | X | No binding predicted | ||||
| TVFDAKRLIGR | X | X | A*11:01 (109 nM) | ||||
| DTERLIGDAAKNQV | X | No binding predicted | |||||
| RLIGDAAKNQV | X | X | X | X | X | A*02:01 (84 nM) | |
| AMTKDNNLL | X | X | No binding predicted | ||||
| IGF2BP3 (insulin-like growth factor 2 mRNA-binding protein 3) | EVLDSLLVQY | X | A*26:01 (47 nM) | ||||
| KIQEILTQV | X | X | X | X | A*02:01 (21 nM) | ||
| TLYNPERTITV | X | X | A*02:01 (80 nM) | ||||
| IL13RA2 (interleukin-13 receptor subunit alpha-2) | LLDTNYNLF | X | No binding predicted | ||||
| LLDTNYNLFY | X | A*01:01 (10 nM) | |||||
| KDM5B (lysine-specific demethylase 5B) | ALKDSVQRA | X | X | No binding predicted | |||
| RLIDLGVGL | X | A*02:01 (20 nM) | |||||
| RQFVTQLY | X | B*15:01 (30 nM) | |||||
| SVAQQLLNGK | X | A*03:01 (20 nM) | |||||
| KIF20A (kinesin-like protein KIF20A) | KEAGNINTSL | X | B*40:01 (14 nM) | ||||
| RVFQGFFTGR | X | X | A*11:01 (22 nM), A*03:01 (72 nM) | ||||
| SEHLDTQKELL | X | B*40:01 (80 nM) | |||||
| LGALS3BP (galectin-3-binding protein) | ALWKEPGSNV | X | A*02:01 (42 nM) | ||||
| NLWDLTDASVV | X | A*02:01 (19 nM) | |||||
| RIDITLSSV | X | A*02:01 (179 nM) | |||||
| MAGEA1 (melanoma-associated antigen 1) | ALREEEEGV | X | No binding predicted | ||||
| EADPTGHSY | X | A*01:01 (79 nM) | |||||
| EVYDGREHSA | X | No binding predicted | |||||
| KEADPTGHSY | X | No binding predicted | |||||
| KVLEYVIKV | X | A*02:01 (5 nM) | |||||
| LTQDLVQEKYLEY | X | A*01:01 (42 nM) | |||||
| SAYGEPRKL | X | C*03:04 (184 nM) | |||||
| MAGEA12 (melanoma-associated antigen 12) | EVVRIGHLY | X | A*26:01 (7 nM) | ||||
| KMAELVHFL | X | X | X | A*02:01 (2 nM) | |||
| MAGEA12; MAGEA6; MAGEA3 (melanoma-associated antigen 12; 6; 3) | KIWEELSVLE | X | No binding predicted | ||||
| MAGEA2 (melanoma-associated antigen 2) | EVFEGREDSVF | X | X | A*26:01 (72 nM) | |||
| MAGEA2; MAGEA2B (melanoma-associated antigen 2; 2B) | KMVELVHFL | X | A*02:01 (2 nM) | ||||
| MAGEA1; MAGEA2; MAGEA2B; MAGEA3; MAGEA6 (melanoma-associated antigen 1; 2; 2B; 3; 6) | REPVTKAEML | X | B*40:01 (255 nM) | ||||
| MAGEA3 (melanoma-associated antigen 3) | EVDPIGHLYIF | X | No binding predicted | ||||
| MAGEA4 (melanoma-associated antigen 4) | GVYDGREHTV | X | A*02:01 (225 nM) | ||||
| MAGEA4; MAGEA8 (melanoma-associated antigen 4; 8) | KVLEHVVRV | X | A*02:01 (9 nM) | ||||
| MAGEA3; MAGE6 (melanoma-associated antigen 3; 6) | KIWEELSVLEV | X | X | A*02:01 (7 nM) | |||
| MAGEC2 (melanoma-associated antigen C2) | ALIEVGPDHFC | X | No binding predicted | ||||
| MDM2 (E3 ubiquitin–protein ligase Mdm2) | RLYDEKQQHI | X | A*02:01 (72 nM) | ||||
| RLYDEKQQHIVY | X | B*15:01 (54 nM) | |||||
| MET (hepatocyte growth factor receptor) | EVIGRGHFGCVY | X | A*26:01 (6 nM) | ||||
| KSEMNVNMKY | X | A*01:01 (193 nM) | |||||
| MFGE8 (lactadherin) | EVRGDVFPSY | X | A*26:01 (15 nM) | ||||
| MLANA (melanoma antigen recognized by T cells 1) | AEEAAGIGIL | X | B*40:01 (9 nM) | ||||
| ALMDKSLHV | X | A*02:01 (4 nM) | |||||
| MMP2 (72 kDa type IV collagenase; PEX) | AVVDLQGGGHSY | X | X | A*26:01 (232 nM) | |||
| NELFA (negative elongation factor A) | KLLDISELDMV | X | X | X | A*02:01 (4 nM) | ||
| NOB1 (RNA-binding protein NOB1) | ALQDIGKNIYTI | X | A*02:01 (59 nM) | ||||
| NPM1 (nucleophosmin) | DIKAKMQAS | X | No binding predicted | ||||
| FEITPPVVL | X | X | B*40:01 (10 nM) | ||||
| KMQASIEKA | X | X | A*02:01 (472 nM) | ||||
| RMTDQEAIQ | X | X | No binding predicted | ||||
| RMTDQEAIQDL | X | X | X | X | A*02:01 (347 nM) | ||
| YEGSPIKVTL | X | X | B*40:01 (24 nM) | ||||
| NUDCD1 (NudC domain-containing protein 1) | AEEHSIATL | X | B*40:01 (13 nM) | ||||
| NUF2 (kinetochore protein Nuf2) | ATAQFKINK | X | A*11:01 (11 nM) | ||||
| EVVEKYEIY | X | A*26:01 (9 nM) | |||||
| KIDEKTAELK | X | No binding predicted | |||||
| OCA2 (P protein) | HVDSTLLQ | X | No binding predicted | ||||
| HVDSTLLQV | X | X | X | X | A*02:01 (170 nM) | ||
| KEPSEIVEL | X | B*40:01 (104 nM) | |||||
| PASD1 (PAS domain-containing protein 1) | QLEERTWLL | X | A*02:01 (79 nM) | ||||
| PGK1 (phosphoglycerate kinase 1) | DNGAKSVVL | X | No binding predicted | ||||
| EGKVLPGVDALSNI | X | No binding predicted | |||||
| PLIN2 (perilipin-2) | RAYQQALSR | X | A*03:01 (140 nM) | ||||
| SLLTSSKGQLQK | X | A*03:01 (53 nM) | |||||
| SVASTITGV | X | A*02:01 (55 nM) | |||||
| TLLSNIQGV | X | A*02:01 (9 nM) | |||||
| TSALPIIQK | X | A*11:01 (24 nM) | |||||
| PMEL (melanocyte protein PMEL; M-alpha; M-beta) | ALDGGNKHFL | X | X | X | X | X | A*02:01 (168 nM) |
| ALLAVGATK | X | A*03:01 (45 nM) | |||||
| ALNFPGSQK | X | A*03:01 (21 nM) | |||||
| AMLGTHTMEV | X | X | X | X | X | A*02:01 (8 nM) | |
| AMLGTHTMEVTV | X | X | A*02:01 (20 nM) | ||||
| ASLIYRRRLMK | X | A*11:01 (61 nM) | |||||
| FALQLHDPSGY | X | C*03:04 (250 nM) | |||||
| FTITDQVPFSVSVSQLR | X | No binding predicted | |||||
| GTATLRLVK | X | X | A*11:01 (22 nM), A*03:01 (121 nM) | ||||
| HQILKGGSGTY | X | B*15:01 (25 nM) | |||||
| HSSSHWLRLP | X | X | No binding predicted | ||||
| HTYLEPGPVTAQ | X | No binding predicted | |||||
| ITDQVPFSV | X | C*05:01 (37 nM), A*02:01 (188 nM) | |||||
| ITDQVPFSVSVSQLR | X | No binding predicted | |||||
| KQDFSVPQL | X | A*02:01 (455 nM) | |||||
| KTWDQVPFS | X | No binding predicted | |||||
| KTWDQVPFSV | X | X | X | X | X | A*02:01 (11 nM) | |
| KTWDQVPFSVSV | X | A*02:01 (13 nM) | |||||
| KTWGQYWQ | X | No binding predicted | |||||
| KTWGQYWQV | X | X | X | X | X | A*02:01 (10 nM) | |
| LSIGTGRAM | X | X | C*03:03 (39 nM), C*03:04 (39 nM) | ||||
| LVLKRCLLH | X | No binding predicted | |||||
| MLGTHTMEV | X | X | X | X | A*02:01 (8 nM) | ||
| RNQPLTFALQLHDPSGY | X | No binding predicted | |||||
| RYGSFSVTL | X | C*07:02 (362 nM) | |||||
| SLADTNSLAV | X | A*02:01 (13 nM) | |||||
| SLADTNSLAVV | X | X | X | X | X | A*02:01 (12 nM) | |
| SVPQLPHSSSHW | X | No binding predicted | |||||
| TITDQVPFSVSVSQLR | X | No binding predicted | |||||
| VLYRYGSFSV | X | X | A*02:01 (6 nM) | ||||
| VLYRYGSFSVTL | X | A*02:01 (41 nM) | |||||
| YRYGSFSVTL | X | X | C*07:01 (10 nM), C*03:03 126 (nM) | ||||
| PPIB (peptidyl-prolyl cis–trans isomerase B) | LLLPGPSAA | X | X | X | X | A*02:01 (75 nM) | |
| PRAME (MELANOMA antigen preferentially expressed in tumors) | AAFDGRHSQTL | X | X | X | X | C*03:03 (7 nM), C*03:04 (7 nM), C*12:03 (16 nM) | |
| ALLERASATL | X | A*02:01 (25 nM) | |||||
| ALLPSLSHC | X | X | No binding predicted | ||||
| GQHLHLETF | X | B*15:01 (96 nM) | |||||
| QLLALLPSL | X | A*02:01 (9 nM) | |||||
| RLDQLLRHV | X | X | X | X | A*02:01 (217 nM) | ||
| RLVELAGQSLLK | X | A*03:01 (35 nM) | |||||
| SLLQHLIGL | X | X | A*02:01 (12 nM) | ||||
| YEDIHGTLHL | X | B*40:01 (15 nM) | |||||
| PRDX5 (peroxiredoxin-5, mitochondrial) | LLDDSLVSI | X | A*02:01 (8 nM) | ||||
| RAN (GTP-binding nuclear protein Ran) | AQYEHDLEVA | X | A*02:01 (303 nM) | ||||
| KLIGDPNLEFV | X | X | A*02:01 (5 nM) | ||||
| RHOC (Rho-related GTP-binding protein RhoC) | FSIDSPDSL | X | C*03:04 (6 nM) | ||||
| IEVDGKQVEL | X | X | B*40:01 (29 nM) | ||||
| RPA1 (replication protein A 70 kDa DNA-binding subunit) | DTEFPNFKY | X | A*01:01 (410 nM) | ||||
| EVFQNANFRSF | X | A*26:01 (43 nM) | |||||
| GAIAAIMQK | X | A*11:01 (32 nM) | |||||
| KAYGASKTFGK | X | X | A*03:01 (32 nM), A*11:01 (35 nM) | ||||
| KVIDQQNGL | X | No binding predicted | |||||
| SGMGSTVSK | X | A*11:01 (31 nM) | |||||
| VSDFGGRSL | X | X | X | X | X | C*03:03 (438 nM), C*03:04 (438 nM), C*05:01 (89 nM) | |
| RPL10A (replication protein A 70 kDa DNA-binding subunit) | FLASESLIKQI | X | X | X | X | A*02:01 (22 nM) | |
| NMVAKVDEV | X | X | X | X | A*02:01 (51 nM) | ||
| RILGPGLNK | X | A*03:01 (21 nM) | |||||
| SLIKQIPRI | X | X | X | A*02:01 (36 nM) | |||
| STIKFQMKK | X | A*11:01 (5 nM) | |||||
| TLYEAVREV | X | X | A*02:01 (9 nM) | ||||
| RPL19 (60S ribosomal protein L19) | HMYHSLYLK | X | A*03:01 (7 nM) | ||||
| ILMEHIHKL | X | X | X | X | X | A*02:01 (2 nM) | |
| ILMEHIHKLK | X | A*03:01 37 (nM) | |||||
| ILMEHIHKLKA | X | A*02:01 (19 nM) | |||||
| KLIKDGLIIRK | X | A*03:01 (49 nM) | |||||
| RILMEHIHKLK | X | A*03:01 (90 nM) | |||||
| RLASSVLRCGK | X | A*03:01 (23 nM) | |||||
| WLDPNETNEI | X | A*02:01 (79 nM) | |||||
| RPS2 (40S ribosomal protein S2) | KEWMPVTKL | X | B*40:01 (28 nM) | ||||
| RPSA (40S ribosomal protein SA) | NTDSPLRY | X | A*01:01 (7 nM) | ||||
| SART1 (U4/U6.U5 tri-snRNP-associated protein 1) | GLLETTVQKV | X | X | A*02:01 (9 nM) | |||
| KSMNANTITK | X | X | A*03:01 (61 nM), A*11:01 (12 nM) | ||||
| RVKAPNKSL | X | No binding predicted | |||||
| SFMBT1 (Scm-like with four MBT domains protein 1) | ESVMINGKY | X | A*26:01 (41 nM) | ||||
| SSDNYEHWLY | X | A*01:01 (5 nM) | |||||
| SH3GLB2 (endophilin-B2) | EVAPPASGTR | X | No binding predicted | ||||
| SOX10 (transcription factor SOX-10) | ETAGPQGPPHY | X | X | A*26:01 (13 nM) | |||
| EVMSNMETF | X | A*26:01 (14 nM) | |||||
| FSYMGPSQRPL | X | X | X | C*03:03 (3 nM), C*03:04 (3 nM) | |||
| GTAAIQAHY | X | X | No binding predicted | ||||
| ISKPPGVAL | X | X | C*03:04 (228 nM), C*12:03 (152 nM) | ||||
| YGHSGQASGLY | X | No binding predicted | |||||
| YSDHQPSGPYY | X | A*01:01 (5 nM) | |||||
| YTDQPSTSQIAY | X | A*01:01 (4 nM) | |||||
| KLADQYPHL | X | X | A*02:01 (6 nM) | ||||
| KTLGKLWRL | X | X | X | A*02:01 (74 nM) | |||
| STAT1 (signal transducer and activator of transcription 1-alpha/beta) | ALLKDQQPGTFLL | X | A*02:01 (22 nM) | ||||
| DSFPMEIRQY | X | X | No binding predicted | ||||
| DSFPMEIRQYL | X | No binding predicted | |||||
| EVVHKIIEL | X | A*26:01 (211 nM) | |||||
| KLQELNYNL | X | X | X | A*02:01 (6 nM) | |||
| QQLDSKFLEQV | X | X | A*02:01 (176 nM) | ||||
| VLWDRTFSL | X | X | X | X | A*02:01 (3 nM) | ||
| VMLDKQKEL | X | A*02:01 (201 nM) | |||||
| SUGT1 (suppressor of G2 allele of SKP1 homolog) | ETFTEGQKL | X | No binding predicted | ||||
| TOP2A (DNA topoisomerase 2-alpha) | KLAEAERVGLHK | X | A*03:01 (59 nM) | ||||
| KLDETGNSL | X | X | X | A*02:01 (68 nM) | |||
| KLDETGNSLK | X | A*03:01 (153 nM) | |||||
| KLDETGNSLKV | X | A*02:01 (29 nM) | |||||
| SLDKDIVAL | X | A*02:01 (59 nM) | |||||
| THFPDETEI | X | B*38:01 (175 nM) | |||||
| AEINNIIKI | X | X | B*40:01 (58 nM) | ||||
| EVTFVPGLY | X | A*26:01 (12 nM) | |||||
| FLYDDNQRV | X | X | X | X | A*02:01 (4 nM) | ||
| KIFDEILVNA | X | A*02:01 (26 nM) | |||||
| LTYNDFINK | X | A*11:01 (8 nM) | |||||
| TP53 (cellular tumor antigen p53) | GLAPPQHLIRV | X | A*02:01 (18 nM) | ||||
| TTK (dual specificity protein kinase TTK) | ESFSGSLGHL | X | No binding predicted | ||||
| TYMS (thymidylate synthase) | NSELSCQ | X | No binding predicted | ||||
| NSELSCQLY | X | A*01:01 (36 nM) | |||||
| YMIAHITGL | X | X | A*02:01 (3 nM) | ||||
| TYR (tyrosinase) | EEYNSHQSL | X | X | B*40:01 (42 nM) | |||
| EVYPEANAPIGHN | X | No binding predicted | |||||
| HTISSDYVIPIGTY | X | A*26:01 (18 nM) | |||||
| KQLPEEKQPLL | X | X | No binding predicted | ||||
| LEEYNSHQSL | X | X | B*40:01 (47 nM) | ||||
| LLMEKEDYHSL | X | X | X | X | X | A*02:01 (6 nM) | |
| MEKEDYHSL | X | B*40:01 (69 nM) | |||||
| NIFDLSAPEKDKFFA | X | A*02:01 (71 nM) | |||||
| SSDYVIPIGTY | X | A*01:01 (23 nM) | |||||
| YVIPIGTYGQM | X | A*26:01 (37 nM) | |||||
| WNK2 (serine/threonine protein kinase WNK2) | HTRTPPIIHR | X | No binding predicted |
Since some of the identified peptides were not predicted to bind to any of the cognate HLA alleles, the possibility of such peptides to form a stable complex with the HLA class I alpha chain and β2-microglobulin was investigated. Therefore, the putative tumor-associated antigens simultaneously identified from MeWo and FM-93/2 cells (AAAGGGGGGGRY, AMTKDNNLL, GTAAIQAHY and DSFPMEIRQY) were obtained as synthetic peptides (see Supplementary Figs. 2–5 for a comparison of synthetic and observed spectra) and added to HLA class I alpha chain and β2-microglobulin during refolding. FPLC profiles of the HLA complexes after refolding (Supplementary Fig. 6) and SDS-PAGE analysis (Supplementary Fig. 7) demonstrated that peptide AMTKDNNLL was able to form a stable complex with HLA-A*02:01, while peptides AAAGGGGGGGRY, GTAAIQAHY and DSFPMEIRQY facilitated the formation of a stable complex with HLA-A*26:01.
Although Table 2 contains peptides from over 70 known tumor-associated antigens, no peptide from the well-described antigen NY-ESO-1 was identified, even though NY-ESO-1 mRNA could be detected in three of the investigated cell lines (Supplementary Fig. 8).
Identification of a neo-epitope presented on the surface of MeWo cells
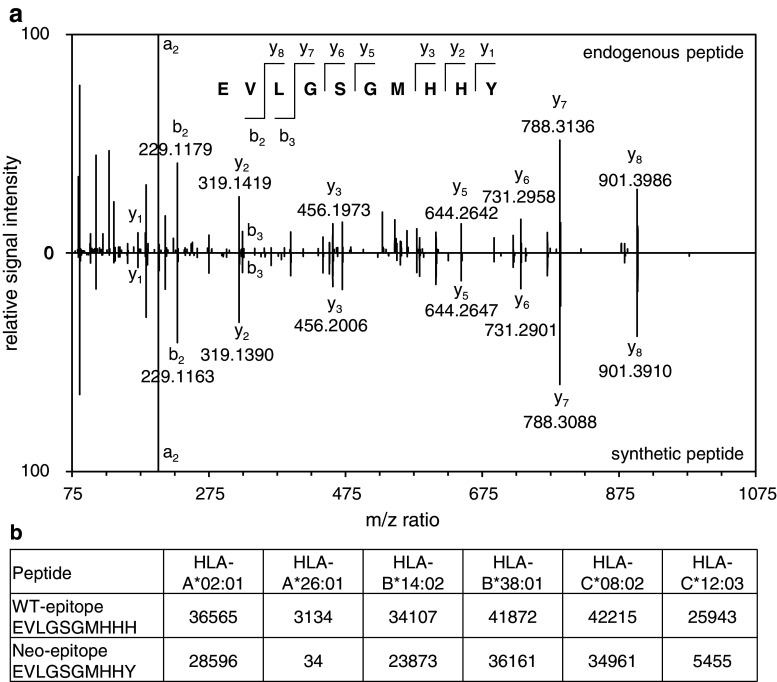
In order to examine whether neo-epitopes could be observed among the purified peptides, we searched the mass spectrometric raw data of the SK-Mel-5 and MeWo cell lines against publicly available catalogs of somatic mutations for these two cell lines downloaded from the COSMIC database [30] concatenated with the human reference proteome database. Out of 3911 mutations for MeWo and 128 mutations for SK-Mel-5, one mutant peptide was identified with high confidence (<1 % false discovery rate) from MeWo cells, corresponding to IFRD2437–446H446Y (interferon-related developmental regulator 2). The mutant peptide sequence could be confirmed by spectral comparison with a synthetic peptide (Fig. 3a). It is interesting to note that the mutated peptide was predicted to be a strong binder of HLA-A*26:01 (IC50 = 34 nM), while the wild type sequence was not predicted to bind to any of the MeWo alleles (IC50 > 3000 nM for all other alleles) (Fig. 3b).
Fig. 3.
Characterization of a neo-epitope identified from MeWo cells. a Sequence confirmation of identified neo-epitope with a synthetic peptide. (a, upper panel) Representative spectrum of endogenous EVLGSGMHHY peptide from the HLA peptidome analysis of MeWo cells. (a, lower panel) Representative spectrum of the synthetic EVLGSGMHHY peptide. b Binding prediction (IC50 in nM) for the wild type (WT) peptide and the neo-epitope to the alleles of the MeWo cell line as predicted by NetMHCpan 2.8 [27]
Comparison of HLA-A*03:01-eluted peptides from melanoma cell lines and patients’ sera
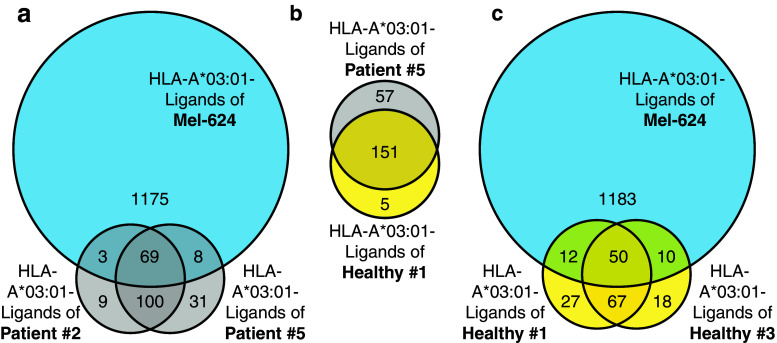
In a previous study [22], we had purified soluble HLA class I complexes (sHLA) from the blood of melanoma patients and healthy donors. Three of the melanoma patients possessed the HLA-A*03:01 haplotype and a significant number of peptides from two of these patients were predicted to bind to this allele. To identify whether the HLA peptidome of melanoma cell lines is comparable to the one of the melanoma patients, we investigated whether the same peptide sequences could be found both in the sHLA peptidome of patients and in the HLA peptidome of the HLA-A*03:01 positive Mel-624 cell line (Fig. 4). We observed a substantial overlap of A*03:01 peptides between the melanoma patients and the cell line (Fig. 4a), between patient and healthy donor (Fig. 4b), and between the healthy donors and the cell line (Fig. 4c). The 80 peptides shared between Mel-624 cells and one or both of the melanoma patients are reported in Supplementary Table 6. One epitope of a tumor-associated antigen was identified in the cell line Mel-624 as well as in the melanoma patient P5, but was not identified in the healthy donors (SLLTSSKGQLQK of perilipin-2 (PLIN2), also reported in Table 2).
Fig. 4.
Comparative analysis of HLA-A*03:01 peptides isolated from Mel-624 cells and sHLA-A*03:01 peptides isolated from the serum of melanoma patients or healthy donors. Peptides between 8 and 12 amino acids in length with a NetMHCpan 2.8 binding prediction of <500 nM to HLA-A*03:01 isolated from Mel-624 cells were compared to peptides with the same characteristics isolated from the serum of melanoma patients or healthy donors [22]. a Venn diagram depicting identifications of HLA-A*03:01 peptides isolated from Mel-624 and/or the sera of two melanoma patients. b Venn diagram depicting identifications of HLA-A*03:01 peptides from the serum of a healthy donor and a melanoma patient. c Venn diagram depicting identifications of HLA-A*03:01 peptides isolated from Mel-624 and/or the sera of two healthy donors
Discussion
In this study, we present an extensive mass spectrometric characterization of the HLA peptidome of five melanoma cell lines, which led to the identification of 10,399 unique peptides with high confidence. Comparison of these peptides with a list of tumor-associated antigens compiled by Andersen et al. revealed that 262 peptides corresponded to antigens, which had previously been described as tumor associated [10]. A peptide containing a mutation was identified when interrogating the MS data for a catalog of somatic mutations. Furthermore, we observed that sHLA peptides isolated from the serum of melanoma patients and from an HLA-matched melanoma cell line display a substantial overlap.
While the experimental atlas included over 10,000 HLA class I-bound peptides, identified with high confidence, it is of note that the majority of HLA ligands originated from HLA-A. A general underrepresentation of HLA-C has been described before, on the level of both surface expressed protein [34] and identified ligands in mass spectrometric experiments [21, 22]. HLA-B ligands, however, have occasionally been found at similar or even higher numbers applying the same procedure as in this paper (see MHC-specific motifs of MAVER-1 and THP-1 cells [22]), indicating that the low number of HLA-B ligands originates from a low expression of HLA-B in the investigated melanoma cell lines.
The HLA peptide atlas, further, included 262 peptides corresponding to previously described tumor-associated antigens and 31 epitopes, which had previously been reported to be potentially immunogenic (Table 2). Interestingly, some well-known tumor-associated antigens were identified with up to 19 different peptides from a single cell line (i.e., PMEL in cell line FM-82). Other antigens (i.e., NY-ESO-1, (E)AAGIGILTV of MLANA/MART-1, or YM(N/D)GTMSQV of tyrosinase) were not present in our analysis. The advent of multiplex tetramer technology, which allows the high-throughput screening of T cell specificities [13], crucially relies on the availability of the exact sequence of presented peptides. Currently, selection of epitopes is often guided by binding predictions. In this study, we demonstrate that the majority of identified peptides were predicted to bind to cognate HLA molecules. However, a substantial portion of the eluted peptides (9–21 %, depending on the cell line) were not predicted to be HLA binders, which may reflect limitations of the prediction algorithms as demonstrated by the ability of four peptides identified from both MeWo and FM-93/2 cells to form stable complexes with the HLA alpha chain and β2-microglobulin (Supplementary Figs. 6 & 7). We therefore believe that it is essential to complement the selection of epitopes with sequences identified by mass spectrometric characterization of HLA peptidomes. The peptidome described in this work could in principle readily be used to complement recently described studies in melanoma patients (e.g., those describing the evolution of T cells in patients, after treatment with ipilimumab, using multiplex tetramer technology [16]).
Mutational information for MeWo and SK-Mel-5 cell lines, which was available from public repositories [30], facilitated the confident identification of one neo-epitope presented by MeWo cells. The identification was confirmed by spectral comparison with the corresponding synthetic peptide. The observation that only a small fraction of possible neo-epitopes are presented to a degree which allows identification by mass spectrometry is in line with a recent report analyzing patient-derived melanoma cell lines [19]. In that study, the authors did not observe any neo-epitope, in spite of having confidently characterized between 2701 and 6397 HLA-bound peptides from three cell lines, whose mutations had previously been identified by whole exome sequencing. This finding is intriguing, especially since melanoma represents one of the most mutated human cancers [11, 12], and since neo-epitopes have recently been proposed as a major factor in the activity of immunotherapy [35, 36]. Mass spectrometry-based methodologies may allow to experimentally measure the relative frequency of different types of peptide display (e.g., those derived from housekeeping proteins, over-expressed antigens, tumor testis antigens and mutant peptides) on HLA class I molecules. This information is likely to be crucially important for a molecular characterization of T cell-based anti-tumor immunity.
A comparison of melanoma patient-derived sHLA peptidomes and data derived from cell lines indicated the presence of shared peptides. Since tumor cell lines do not fully resemble the nature of corresponding solid tumors, as they may change their HLA peptidome under cell culture conditions, it is not surprising to observe differences between the ligandomes. Nevertheless, observation of a significant overlap between cell lines and unrelated patients sera suggests that, at least in part, the sHLA peptidome may represent peptides derived from the tumor. In the future, analysis of matched primary tumor and sHLA peptidomes will be important to understand if this methodology might indeed be useful to probe for tumor-derived antigens. If this is the case, sHLA peptidomics could represent a promising noninvasive strategy for the monitoring of patients during immunotherapeutic intervention.
Immunotherapy is gaining importance for the treatment of cancer and, in particular, for the treatment of melanoma. It is very well established that tumor-associated antigen-specific T cells are found in melanoma patients [10]. Recent advances in multiplex tetramer technology have provided unambiguous evidence that treatment with anti-CTLA-4 therapy broadens the profile and alters the frequency of the melanoma-reactive CD8+ T cell response [16]. In addition, survival of melanoma patients with tumor-associated antigen-specific T cells against NY-ESO-1 and Melan-A was found to be significantly prolonged [37]. These studies reinforce the concept that immunotherapy can be used to boost the patient’s immune system against melanoma malignancies. A detailed characterization of the cancer peptidome, as presented in this article, should facilitate mechanistic studies on the quality and quantity of tumor rejection antigens in melanoma and other cancer types. A molecular characterization of T cell specificities in patients (e.g., before and after pharmacological intervention) may shed light on clinically important issues, such as why only certain individuals respond to immunotherapy. In addition, peptides bound to HLA class I may be considered for vaccination strategies [38] and for the development of T cell receptor-engineered adoptive cell therapies [6].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Camilla Bacci (Philogen SpA) for providing W6/32 antibody.
Funding
This work was supported financially by ETH Zürich, the Swiss National Science Foundation, the European Union’s Seventh Framework Programme (FP7/2007-2013) under Grant Agreement Nos. 305309 (PRIAT) and 305608 (EURenOmics), the Bonveda Foundation and the European Research Council (ERC advanced Grant “ZAUBERKUGEL”).
Abbreviations
- ACN
Acetonitrile
- ATCC
American Type Culture Collection
- COSMIC
Catalogue of Somatic Mutations in Cancer
- ESTDAB
European Searchable Tumour Line Database
- ETH Zurich
Eidgenössische Technische Hochschule Zürich
- FDR
False discovery rate
- FPLC
Fast protein liquid chromatography
- HCD
Higher-energy collisional dissociation
- HLA
Human leukocyte antigen
- IC50
Half maximal inhibitory concentration
- LC-MS
Liquid chromatography–mass spectrometry
- MS
Mass spectrometry
- MS/MS
Fragment mass spectra
- PMSF
Phenylmethylsulfonyl fluoride
- sHLA
Soluble human leukocyte antigen
- UHPLC
Ultra high performance liquid chromatography
- WT
Wild type
Compliance with ethical standards
Conflict of interest
Dario Neri is co-founder of Philogen, shareholder and member of the board. Tim Fugmann and Danilo Ritz are employees of Philochem AG. The authors declare no additional conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Contributor Information
Tim Fugmann, Phone: +41 43 544 88 00, Email: tim.fugmann@philochem.ch.
Dario Neri, Phone: +41 44 633 74 01, Email: neri@pharma.ethz.ch.
References
- 1.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 3.Probst P, Neri D. Immunocytokines for cancer therapy. Forum Immunopathol Dis Therap. 2014;5(1):83–99. doi: 10.1615/ForumImmunDisTher.2015014013. [DOI] [Google Scholar]
- 4.Kontermann RE, Brinkmann U. Bispecific antibodies. Drug Discov Today. 2015;20(7):838–847. doi: 10.1016/j.drudis.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348(6230):62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.June CH, Riddell SR, Schumacher TN. Adoptive cellular therapy: a race to the finish line. Sci Transl Med. 2015;7(280):280ps7. doi: 10.1126/scitranslmed.aaa3643. [DOI] [PubMed] [Google Scholar]
- 7.Melero I, Gaudernack G, Gerritsen W, Huber C, Parmiani G, Scholl S, Thatcher N, Wagstaff J, Zielinski C, Faulkner I, Mellstedt H. Therapeutic vaccines for cancer: an overview of clinical trials. Nat Rev Clin Oncol. 2014;11(9):509–524. doi: 10.1038/nrclinonc.2014.111. [DOI] [PubMed] [Google Scholar]
- 8.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, Samuels Y, Rosenberg SA. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheibenbogen C, Sun Y, Keilholz U, Song M, Stevanovic S, Asemissen AM, Nagorsen D, Thiel E, Rammensee HG, Schadendorf D. Identification of known and novel immunogenic T-cell epitopes from tumor antigens recognized by peripheral blood T cells from patients responding to IL-2-based treatment. Int J Cancer. 2002;98(3):409–414. doi: 10.1002/ijc.10205. [DOI] [PubMed] [Google Scholar]
- 10.Andersen RS, Thrue CA, Junker N, Lyngaa R, Donia M, Ellebaek E, Svane IM, Schumacher TN, Thor Straten P, Hadrup SR. Dissection of T-cell antigen specificity in human melanoma. Cancer Res. 2012;72(7):1642–1650. doi: 10.1158/0008-5472.CAN-11-2614. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, PedBrain I, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR, Australian Pancreatic Cancer Genome I, Consortium IBC, Consortium IM-S Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadrup SR, Schumacher TN. MHC-based detection of antigen-specific CD8+ T cell responses. Cancer Immunol Immunother. 2010;59(9):1425–1433. doi: 10.1007/s00262-010-0824-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 15.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, Wunderlich JR, Somerville RP, Rosenberg SA. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kvistborg P, Philips D, Kelderman S, Hageman L, Ottensmeier C, Joseph-Pietras D, Welters MJ, van der Burg S, Kapiteijn E, Michielin O, Romano E, Linnemann C, Speiser D, Blank C, Haanen JB, Schumacher TN. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med. 2014;6(254):254ra128. doi: 10.1126/scitranslmed.3008918. [DOI] [PubMed] [Google Scholar]
- 17.Cox AL, Skipper J, Chen Y, Henderson RA, Darrow TL, Shabanowitz J, Engelhard VH, Hunt DF, Slingluff CL., Jr Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264(5159):716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 18.Jarmalavicius S, Welte Y, Walden P. High immunogenicity of the human leukocyte antigen peptidomes of melanoma tumor cells. J Biol Chem. 2012;287(40):33401–33411. doi: 10.1074/jbc.M112.358903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritchard AL, Hastie ML, Neller M, Gorman JJ, Schmidt CW, Hayward NK. Exploration of peptides bound to MHC class I molecules in melanoma. Pigment Cell Melanoma Res. 2015;28(3):281–294. doi: 10.1111/pcmr.12357. [DOI] [PubMed] [Google Scholar]
- 20.Hassan C, Kester MG, de Ru AH, Hombrink P, Drijfhout JW, Nijveen H, Leunissen JA, Heemskerk MH, Falkenburg JH, van Veelen PA. The human leukocyte antigen-presented ligandome of B lymphocytes. Mol Cell Proteom. 2013;12(7):1829–1843. doi: 10.1074/mcp.M112.024810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteom. 2015;14(3):658–673. doi: 10.1074/mcp.M114.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritz D, Gloger A, Weide B, Garbe C, Neri D, Fugmann T. High-sensitivity HLA class I peptidome analysis enables a precise definition of peptide motifs and the identification of peptides from cell lines and patients’ sera. Proteomics. 2016;16(10):1570–1580. doi: 10.1002/pmic.201500445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawelec G, Marsh SG. ESTDAB: a collection of immunologically characterised melanoma cell lines and searchable databank. Cancer Immunol Immunother. 2006;55(6):623–627. doi: 10.1007/s00262-005-0117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou J, Voong LN, Mortales CL, Towlerton AM, Pollack SM, Chen X, Yee C, Robbins PF, Warren EH. Epigenetic modulation to enable antigen-specific T-cell therapy of colorectal cancer. J Immunother. 2012;35(2):131–141. doi: 10.1097/CJI.0b013e31824300c7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toebes M, Rodenko B, Ovaa H, Schumacher TN. Generation of peptide MHC class I monomers and multimers through ligand exchange. Curr Protoc Immunol. 2009 doi: 10.1002/0471142735.im1816s87. [DOI] [PubMed] [Google Scholar]
- 26.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Roder G, Peters B, Sette A, Lund O, Buus S. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2(8):e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boegel S, Lower M, Bukur T, Sahin U, Castle JC. A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines. Oncoimmunology. 2014;3(8):e954893. doi: 10.4161/21624011.2014.954893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomsen MC, Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 2012;40(Web Server issue):W281–W287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14(1):9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 32.Rapin N, Hoof I, Lund O, Nielsen M. MHC motif viewer. Immunogenetics. 2008;60(12):759–765. doi: 10.1007/s00251-008-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen M, Harndahl M, Stryhn A, Boucherma R, Nielsen LL, Lemonnier FA, Nielsen M, Buus S. Uncovering the peptide-binding specificities of HLA-C: a general strategy to determine the specificity of any MHC class I molecule. J Immunol. 2014;193(10):4790–4802. doi: 10.4049/jimmunol.1401689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apps R, Meng Z, Del Prete GQ, Lifson JD, Zhou M, Carrington M. Relative expression levels of the HLA class-I proteins in normal and HIV-infected cells. J Immunol. 2015;194(8):3594–3600. doi: 10.4049/jimmunol.1403234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 37.Weide B, Zelba H, Derhovanessian E, Pflugfelder A, Eigentler TK, Di Giacomo AM, Maio M, Aarntzen EH, de Vries IJ, Sucker A, Schadendorf D, Buttner P, Garbe C, Pawelec G. Functional T cells targeting NY-ESO-1 or Melan-A are predictive for survival of patients with distant melanoma metastasis. J Clin Oncol. 2012;30(15):1835–1841. doi: 10.1200/JCO.2011.40.2271. [DOI] [PubMed] [Google Scholar]
- 38.Haen SP, Rammensee HG. The repertoire of human tumor-associated epitopes–identification and selection of antigens and their application in clinical trials. Curr Opin Immunol. 2013;25(2):277–283. doi: 10.1016/j.coi.2013.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.