Abstract
Physiological cardiac hypertrophy, in response to stimuli such as exercise, is considered adaptive and beneficial. In contrast, pathological cardiac hypertrophy that arises in response to pathological stimuli such as unrestrained high blood pressure and oxidative or metabolic stress is maladaptive and may precede heart failure. We found that the transcript encoding DNA-damage-inducible transcript 4-like (DDiT4L) was expressed in murine models of pathological cardiac hypertrophy, but not in those of physiological cardiac hypertrophy. In cardiomyocytes, DDiT4L localized to early endosomes and promoted stress-induced autophagy through a process involving mechanistic target of rapamycin complex 1 (mTORC1). Exposing cardiomyocytes to various types of pathological stress increased the abundance of DDiT4L, which inhibited mTORC1 but activated mTORC2 signaling. Mice with conditional cardiac-specific overexpression of DDiT4L had mild systolic dysfunction, increased baseline autophagy, reduced mTORC1 activity, and increased mTORC2 activity, all of which were reversed by suppression of transgene expression. Genetic suppression of autophagy also reversed cardiac dysfunction in these mice. Our data showed that DDiT4L may be an important transducer of pathological stress to autophagy through mTOR signaling in the heart and that DDiT4L could be therapeutically targeted in cardiovascular diseases in which autophagy and mTOR signaling play a major role.
Introduction
Autophagy is a critical, highly orchestrated, cellular catabolic process that plays a vital role in cell homeostasis and adaption to stress in the heart. Consequently, dysregulation of autophagy leads to cell damage and death. Autophagy has multiple roles in cardiac diseases and is altered at different points in many cardiac diseases (1). Furthermore, autophagy can be either protective or detrimental in cardiac disease (2). Due to the importance of autophagy in the heart, insight into its regulation would answer an unmet need in the field and potentially identify therapeutic targets for cardiovascular diseases.
An important regulator of autophagy is the mammalian target of rapamycin (mTOR), which responds to various stimuli such as growth factors, cellular energy status, oxygen concentrations, and stress to control cell metabolism and growth (3). mTOR acts in two distinct complexes: the rapamycin sensitive mTOR complex 1 (mTORC1) consisting of mTOR, raptor, and mLST8, which enhances protein synthesis and cell growth but inhibits autophagy, and the rapamycin insensitive complex mTORC2 consisting of mTOR, rictor, sin1, and mLST8, which is implicated in cell proliferation, growth and cytoskeleton rearrangement (4). Like autophagy, mTOR has a complex role in the heart, and its dysregulation has been implicated in many types of heart disease (5–11).
DDiT4L (also known as Redd2 and Rtp801L) and its homolog DDiT4 are upstream inhibitors of mTOR in several tissues and cell models (12–15). In this study, using Deep Serial Analysis of Gene Expression (DSAGE), we identified DDiT4L as a candidate gene with the greatest increase in expression in a murine model of pathological but not in one of physiological hypertrophy. We showed that DDiT4L was expressed in cardiomyocytes, and was increased in murine and cultured neonatal rat ventricular myocyte (NRVM) models of pathological but not physiological stress, and in cardiac tissue from patients with dilated cardiomyopathy. Notably, the abundance of the related DDiT4 was not changed by pathological stress. DDiT4L was transcriptionally controlled by serum response factor (SRF), localized in early endosomes, and was itself a key regulator of stress induced (but not baseline) autophagy through mTORC1 inhibition in NRVMs. Adult mice with conditional cardiac-specific overexpression of DDiT4L had increased autophagy, repressed mTORC1 signaling and decreased left ventricular wall thickness with mild left ventricular dilation. This phenotype was fully reversed with suppression of transgene expression, and relied on autophagy because it was also reversed with genetic suppression of autophagy (which was achieved by breeding DDiT4L transgenic mice to beclin 1 haploinsufficient mice). Finally, DDiT4L also activated mTORC2 signaling. In sum, we showed that DDiT4L promoted autophagy by inhibiting mTOR signaling and that its abundance was increased in response to pathological stressors downstream of SRF in a manner distinct from the related DDiT4. Our results also suggested that an increase in stress-induced autophagy at baseline (in the absence of stress) may lead to cardiac structural changes and mild dysfunction that can be reversed by cessation of autophagy without adverse effects.
Results
DDiT4L gene expression is increased with pathological but not physiological stress
To identify genes that are differentially altered in pathological cardiac hypertrophy and physiological hypertrophy, we used DSAGE, a nanotechnology that facilitates global transcriptional profiling (16). We compared transcriptional profiles from hearts of transgenic mice with cardiac over-expression of constitutively active serum and glucocorticoid regulated kinase-1 (SGK1-CA), a model for pathological hypertrophy (17), constitutively active Akt1 (Akt1-CA), a model of physiological or adaptive hypertrophy (18), and their relative controls (dominant negative (DN) SGK1 or Akt1) (19), and wild-type littermates. Cardiac tissue from 4 week-old mice (before cardiac dysfunction developed in the SGK1-CA mice) was used for the experiments. ~2×106 cDNA clones from each genotype were sequenced, corresponding to >33,000 independent transcripts (data file S1). DDiT4L was the transcript with the greatest increase in expression in SGK1-CA mice compared to wild-type mice (Fig. 1A and Supplementary Table 1), and its abundance did not significantly change in any of the other genotypes (Supplementary Table 1). The expression of DDiT4, the closest homologue to DDiT4L, decreased in the SGK1-CA pathological hypertrophy model and did not change in the Akt1-CA physiological hypertrophy model (Supplementary Table 1). qPCR analysis confirmed that the expression of DDiT4L, but not DDiT4, was increased both in SGK1-CA hearts and in NRVMs with adenoviral over-expression of SGK1-CA (Fig. 1B,C, Fig. S1A, B). DDiT4L expression was not changed in NRVMs with adenoviral overexpression of myr-Akt1 (Fig. 1C).
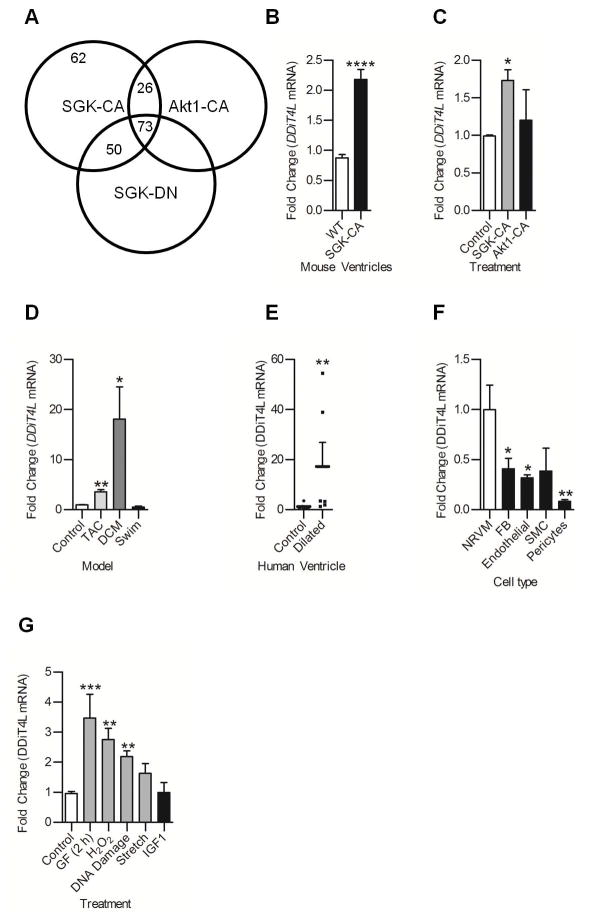
Figure 1. DDiT4L expression is increased in mouse models of pathological hypertrophy and cell models of pathological stress.
(A) DSAGE results showed 62 transcripts increased in mice overexpressing SGK-CA, a model of pathological hypertrophy, which were not changed in mice overexpressing Akt-CA or SGK-DN. For each genotype, DSAGE was run on a pool of 5 mouse heart samples. Parameters were a fold change of at least 1.5, p<0.001, sense, redundancy greater than 2, and for mice overexpressing Akt-CA or SGK-DN these transcripts were not increased more than 1.4 fold.
(B,C) DDiT4L expression was increased in SGK-CA mouse hearts and in neonatal rat ventricular cardiomyocytes (NRVMs) infected with SGK-CA virus but not wild-type mouse hearts or NRVMs infected with myr-Akt virus; n=3–5 mice per genotype, or 4 Ad-infections, 2 dishes per infection. One Way ANOVA followed by Bonferroni posttest, *p<0.05, ****p<0.0001 compared to controls.
(D) DDiT4L expression was increased in transverse aortic constriction (TAC) and genetic dilated cardiomyopathy (DCM) but not in an exercise (swim) model of hypertrophy. n=3–5 mice per model. One Way ANOVA followed by Bonferroni posttest, *p<0.05, **p<0.01 compared to controls.
(E) DDiT4L expression was increased in left ventricular tissue from left ventricle of human dilated cardiomyopathy patients at time of explant compared to controls. n= 6–8 patients. Mann-Whitney nonparametric test, **p<0.01, compared to controls
(F) DDiT4L expression was greatest in cardiomyocytes (NRVM) compared to other cell types in the heart. n=3 preparations, two samples per preparation. One Way ANOVA followed by Bonferroni posttest *p<0.05, **p<0.01 compared to cardiomyocytes. FB – fibroblast, SMC – smooth muscle cell
(G) DDiT4L expression was increased in models of pathological stress in NRVMs and unchanged in a physiological model (IGF-1 treatment). n=3–4 preparations, two samples per preparation. GF: glucose and serum deprivation. One Way ANOVA followed by Bonferroni posttest *p<0.05, **p<0.01, compared to controls
To investigate if DDIT4L is increased in other models of hypertrophy and heart failure, we measured DDiT4L in a genetic model of dilated cardiomyopathy (DCM) caused by a F764L missense mutation in the gene encoding myosin heavy chain (20), the transverse aortic constriction (TAC) model of pressure overload-induced hypertrophy, and a swim-induced physiological model of hypertrophy. DDiT4L expression was increased in both the TAC and DCM models (Fig. 1D) but not in the swim model. In contrast, DDiT4 expression was unchanged (Fig. S1C). Finally, in ventricular tissue from human dilated cardiomyopathy patients obtained at the time of orthotopic transplantation, the expression of DDiT4L expression, but not that of DDiT4, was increased (Fig. 1E and Fig. S1D) compared with age- and sex-matched healthy donors. These data show that DDiT4L expression was increased in different models of cardiac pathology in mice and in human DCM patients, but was unchanged after physiological stress. Moreover, DDiT4L regulation appeared to be distinct from that of its homolog DDiT4.
In the different cell types of the heart, the expression of DDiT4L was greatest in cardiomyocytes compared to fibroblasts, smooth muscle cells, endothelial cells and pericytes (Fig. 1F). In contrast, DDiT4 was expressed at similar amounts between cell types (Fig. S1E). Furthermore, DDiT4L expression in NRVMs was unaltered by IGF1 treatment (a model for physiological stress), but increased in response to various pathological stressors, including glucose deprivation, oxidative stress, DNA damage, and pathological stretch (Fig. 1G). Conversely, DDiT4 was not changed with glucose deprivation or H2O2 treatment, but was decreased in the stretch model (Fig. S1F), further demonstrating the distinct regulation of DDiT4L and DDiT4 expression in NRVMs. These data demonstrated that DDiT4L, but not DDiT4, was selectively expressed in cardiomyocytes and was increased by pathological stress in the heart.
DDiT4L inhibits cellular hypertrophy and mTORC1 signaling in NRVMs
Given its distinct regulation in both in vivo and in vitro models of pathological stress, we next sought to characterize the role of DDiT4L in cardiomyocytes using siRNA-mediated knockdown and adenoviral-mediated overexpression (Ad-DDiT4L) (Fig. 2A). Knockdown of DDiT4L led to a modest, but significant increase in cell size, while Ad-DDiT4L caused a significant decrease in NRVM size at baseline (Fig. 2B), suggesting a possible anti-hypertrophic function for DDiT4L in the absence of external stimuli.
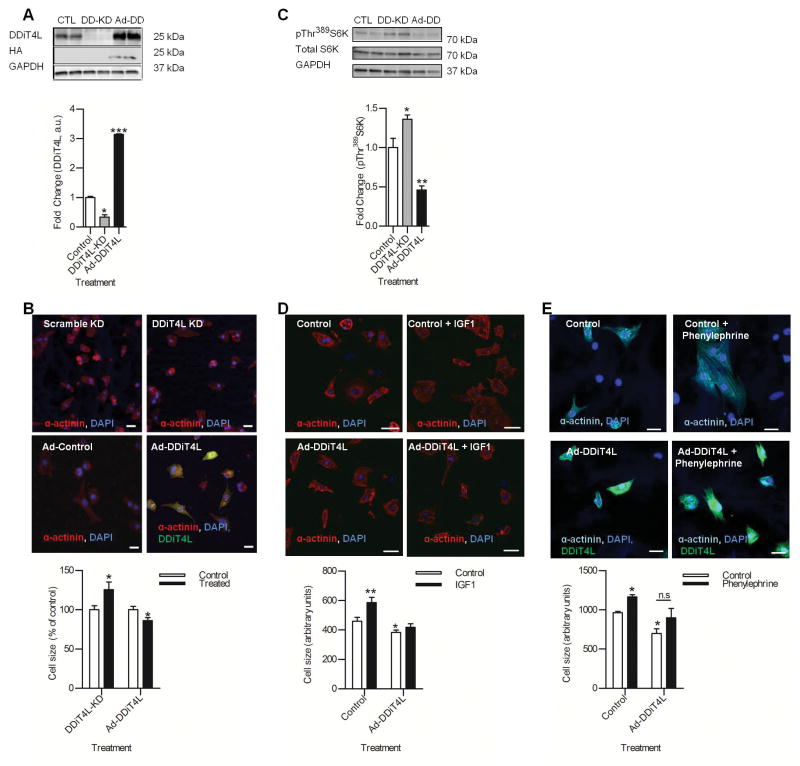
Figure 2. DDiT4L inhibits mTOR signaling in NRVMs in a TSC2 dependent manner.
(A) DDiT4L knockdown (DD-KD) decreased DDiT4L protein, and Ad-DDiT4L (Ad-DD) infection increased DDiT4L protein abundance. n=3 preparations, two samples per preparation. One Way ANOVA followed by Bonferroni posttest *p<0.05, ***p<0.0001.
(B) DDiT4L knockdown increased cardiomyocyte size (25%), and Ad-DDiT4L infection decreased cardiomyocyte size (14%). n=3 preparations, 2 coverslips per experiment,4 images per coverslip, 80–120 cells per treatment. One Way ANOVA followed by Bonferroni posttest *p<0.05. Scale bar is 20 μm.
(C) DDiT4L knockdown (DD-KD) under serum free conditions led to increased mTORC1 signaling as measured by increased phosphorylation of Thr389 in S6K, and Ad-DDiT4L infection (Ad-DD) led to decreased phosphorylation of Thr389 in S6K. n=4–5 preparations, two samples per preparation. One-way ANOVA, Bonferroni posttest *p<0.05, **p<0.01. Phosphorylated (P) proteins were normalized to total protein.
(D) Treatment of control cells with IGF1 led to an increase in NRVM size (27%), which was inhibited in Ad-DDiT4L treated cells. n=3 preparations, 2 samples per preparation, 4 images per coverslip, 40–60 cells per treatment. One Way ANOVA followed by Bonferroni posttest *p<0.05, **p<0.01. Scale bar is 20 μM.
(E) Treatment of control cells with phenylephrine led to an increase in NRVM size (20%) which was inhibited with Ad-DDiT4L. n=4 preparations, 2 samples per preparation, 4 images per coverslip, 60–80 cells per treatment. One Way ANOVA followed by Bonferroni posttest, *p<0.05. Scale bar is 20 μm.
Because DDiT4L inhibits mTOR in other cell types, and because mTOR signaling has been implicated in cardiomyocyte growth (21), we investigated if DDiT4L inhibited mTOR signaling in NRVMs. siRNA-mediated knockdown of DDiT4L in serum-free conditions (low baseline mTORC1 activity) led to increased mTORC1 activity, as measured by increased phosphorylation of the mTORC1 downstream effector P70-S6 kinase (S6K) at Thr389, whereas Ad-DDiT4L led to mTORC1 inhibition as shown by decreased phosphorylation of S6K (Fig. 2C). mTORC1 is an important determinant of protein synthesis and cell size in response to both IGF1 (which induces physiological hypertrophy) and phenylephrine (which induces pathological hypertrophy) (22). Ad-DDiT4L inhibited the increase in NRVM size triggered by treatment with IGF1 (Fig. 2D) and attenuated the phenylephrine effect (Fig. 2E). Together, our results confirmed that DDiT4L inhibited mTORC1, and that increased expression of DDiT4L can partially prevent cardiomyocyte hypertrophy in response to normal and pathological stimuli in NRVMs.
DDiT4L activates autophagy in an mTOR-dependent, TSC2-dependent manner
Because mTOR is a central nodal point in the regulation of autophagy, a cellular process that has been implicated in the response of the heart to pathological stressors (23, 24), we next sought to determine whether DDiT4L activated autophagy in NRVMs. Screening a panel of autophagy-related genes revealed significant increases in Atg7 and MAPLC3 after Ad-DDiT4L overexpression compared to Ad-GFP controls, while the expression of both genes was decreased after DDiT4L knockdown compared to scramble siRNA controls (Fig. 3A).
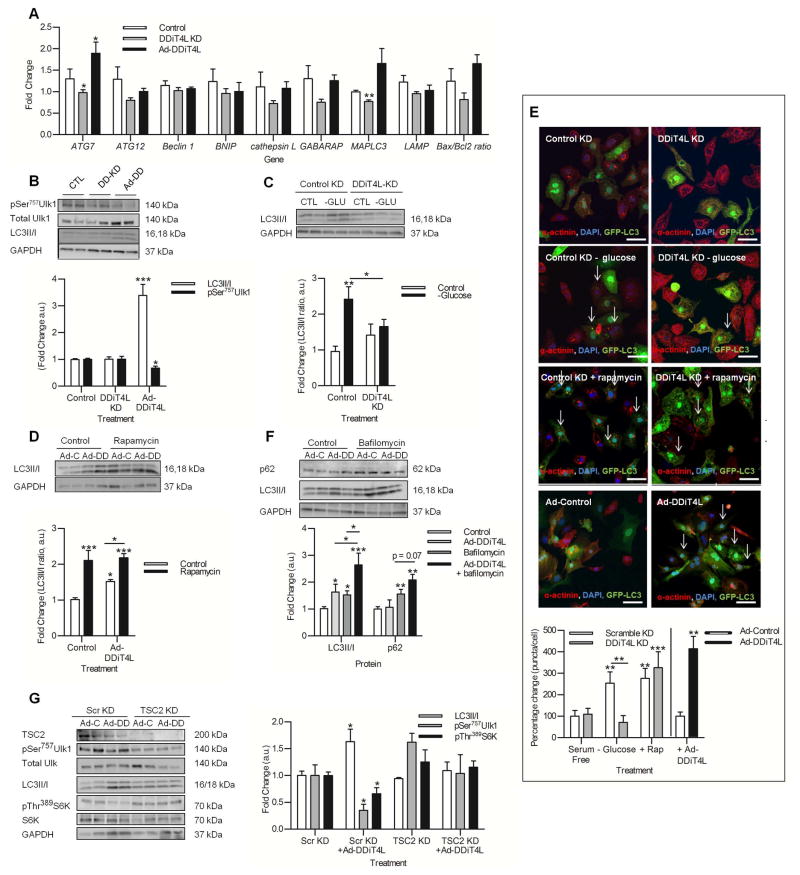
Figure 3. DDiT4L increases autophagic flux in an mTOR-dependent and TSC2-independent manner.
(A) The expression of the autophagy genes ATG7 and MAPLC3 was increased with Ad-DDiT4L infection and decreased with DDiT4L knockdown. n=4 preparations, 2 samples per preparation. Student t-test *p<0.05 compared to control.
(B) Ad-DDiT4L infection (Ad-DD) increased the LC3II/I ratio and decreased phosphorylation of Ser757 in Ulk1 with no changes seen with DDiT4L knockdown (DD-KD). n=4 preparations, 2 samples per preparation. One Way ANOVA followed by Bonferroni posttest *p<0.05, ***p<0.001 compared to control. Phospho-proteins were normalized to total protein.
(C) Glucose deprivation (-GLU) increased autophagy, which was inhibited by DDiT4L knockdown. n=3 preparations, 2 samples per preparation. One-Way ANOVA, Bonferroni post-test, *p<0.05, **p<0.01 compared to control or indicated sample.
(D) Treatment of Ad-DDiT4L-infected (Ad-DD) cells with rapamycin did not lead to a further increase in autophagy. n=4 preparations, 2 samples per preparation. One-Way ANOVA, Bonferroni post-test *p<0.05, ***p<0.001 compared to control or indicated sample.
(E) Glucose deprivation, rapamycin and Ad-DDiT4L infection increased the number of GFP-LC3 autophagosomes in NRVMs, and knockdown of DDiT4L prevented the increase caused by glucose deprivation. n=4 preparations, 2 samples per preparation, 4 images per coverslip, 100–160 cells per treatment. Arrows indicate cells with autophagosomes. One-Way ANOVA, Bonferroni posttest, *p<0.05, **p<0.01, ***p<0.001 compared to control or indicated sample.
(F) Ad-DDiT4L infection and bafilomycin increased the LC3II/I ratio in an additive fashion. n=6–10 preparations, 2 samples per preparation. Bafilomycin treatment, but not Ad-DDiT4L infection alone, significantly increased p62 abundance. n = 4 preparations, 2 samples per preparation. One-Way ANOVA, Bonferroni posttest *p<0.05, **p<0.01, ***p<0.001 compared to control or indicated sample.
(G) TSC2 knockdown attenuated the induction of autophagy by Ad-DDiT4L infection in NRVMs as assessed by LC3II/I or phosphorylation (p) of Ser757 in Ulk1 and inhibited the decrease in pS6K. n=4 preparations, 2 samples per preparation. One-Way ANOVA, Bonferroni post-test *p<0.05 compared to control or indicated sample. Phospho-proteins were normalized to total protein.
To further study the role of DDiT4L and autophagy, we carried out Western blotting for several components in the mTOR-autophagy pathway. Ad-DDiT4L in NRVMs led to an increase in the microtubule-assoicated protein 1A/1B-light chain 3 (LC3II/I) ratio, a marker of functional autophagy (Fig. 3B), and decreased phosphorylation of Ser757 in Unc-51-like kinase 1 (Ulk1), a key downstream effector of mTORC1-dependent inhibition of autophagy. The LC3II/I ratio or phosphorylation of Ulk1 was not changed by DDiT4L siRNA in the absence of stimuli; however, DDiT4L knock-down abolished glucose deprivation-induced mTORC1 inhibition (Fig. S2A) and increased the LC3II/I ratio (Fig. 3C), demonstrating that DDiT4L was necessary for glucose deprivation-induced autophagy, but might not affect baseline autophagy (in the absence of stress).
To explore whether DDiT4L induced autophagy was mTORC1 dependent, NRVMs were treated with the mTORC1 inhibitor rapamycin. Independent addition of Ad-DDiT4L or rapamycin inhibited mTORC1 (Fig. S2B), while together they increased LC3II/I ratio in a non-additive manner (Fig. 3 D), suggesting that DDiT4L promotes autophagy through an mTORC1-dependent pathway. We confirmed these findings by co-transfecting a constitutively-active form of the mTORC1-activating GTPase Rheb (CA-Rheb) in NRVMs treated with Ad-DDiT4L. Activation of mTORC1 by CA-Rheb led to increased phosphorylation of S6K in Ad-DDiT4L-infected cells, and inhibited the increase in the LC3II/I ratio (Fig. S2C,D,E), demonstrating that DDiT4L acts upstream of mTORC1 in activating autophagy. To further confirm the increase in autophagy indicated by the LC3II/I ratio, confocal microscopy was carried out on NRVMs infected with GFP-LC3-encoding virus. Ad-DDiT4L increased the number of autophagosomes, while DDiT4L knockdown decreased the number of autophagosomes in glucose-deprived cells, but not in rapamycin-treated cells (Fig. 3E), confirming that DDiT4L played an important role in stimulating autophagy upstream of mTORC1.
To determine whether DDIT4L increased autophagic flux or inhibited lysosomal fusion to autophagosomes in NRVMs, we used bafilomycin, an inhibitor of autophagosome-lysosome fusion. Addition of bafilomycin or Ad-DDiT4L virus alone led to significant increases in the LC3II/I ratio and the effects of the combination were additive (Fig. 3F), demonstrating that DDiT4L functioned by increasing autophagic flux. As expected, bafilomycin also caused a significant increase in the abundance of the autophagy adaptor protein p62, although this was not further increased in a significant manner by addition of Ad-DDiT4L (Fig. 3F).
Under many conditions, mTORC1 is activated in a Tuberous sclerosis complex 2 (TSC2)-dependent manner; however, mTORC1 activation by amino acids is TSC2-independent (25, 26). To determine whether DDiT4L-mediated autophagy was TSC2 dependent we measured autophagy in NRVMs with TSC2 knockdown. Ad-DDiT4L did not induce autophagy or alter the phosphorylation of Ulk1 or S6K in NRVMs with TSC2 knockdown (Fig. 3G). These findings suggested that in NRVMs, DDiT4L induces autophagic flux in an mTORC1 and TSC2 dependent manner.
DDiT4L is localized to early endosomes and may interact with mTOR
DDiT4L binds to Interferon Regulatory Factor 1 (IRF1) in a punctate pattern in the cytoplasm of bone marrow cells (27). In NRVMs infected with Ad-DDiT4L, immunoreactivity for DDiT4L was found in the cytoplasmic but not nuclear fractions (Fig. 4A). Due to the lack of DDiT4L antibodies that are reliable for immunocytochemistry, we performed HA-immunostaining in Ad-DDiT4L-infected serum-starved NRVMs and found that HA-tagged DDiT4L was present in a punctate pattern in the cytoplasm (Fig. 4B, Fig. S2F). HA-tagged DDiT4L did not localize with the lysosomal protein Lamp1, the autophagosomal protein LC3, the vesicular protein Vamp4, the lipid raft protein Caveolin 3, the endosomal protein Appl, the Golgi associated protein GOPC, or the vesicular protein syntaxin 3 (Fig. S2F) under serum free conditions. However, HA-tagged DDiT4L partially localized with EEA1 (early endosome antigen 1) and Rab5 in these conditions indicating that DDiT4L could be found in early endosomes (Fig. 4B). Under these conditions, DDiT4L also appeared to localize with mTOR, suggesting that trafficking of DDiT4L in early endosomes to compartments where mTOR is present could occur. HA-tagged DDiT4L also co-immunoprecipitated with mTOR under mild detergent conditions, suggesting the presence of DDiT4L in a complex that also contains mTOR (Fig. S2G). This data showed that in NRVMs, DDiT4L was localized on early endosomes and could exert its effect on autophagy by inhibiting mTOR, although the other components of this complex are yet to be elucidated.
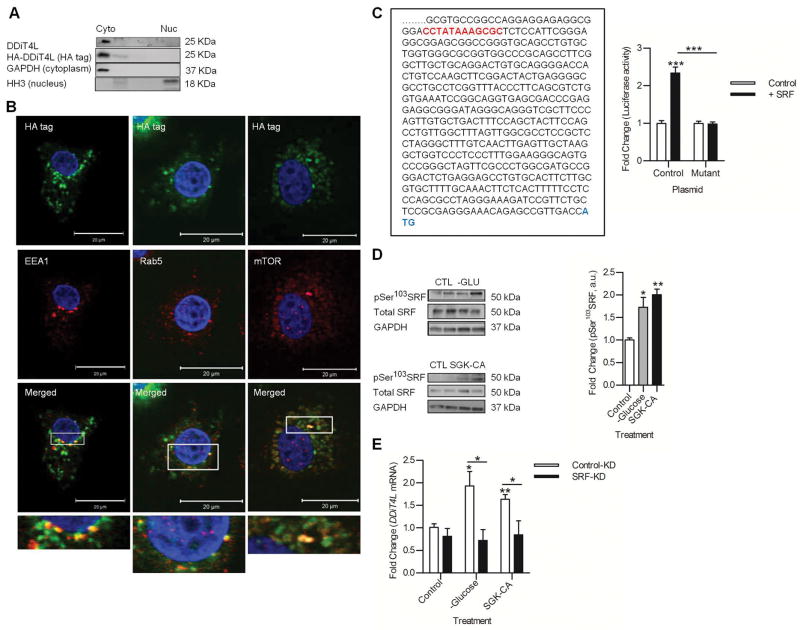
Figure 4. DDiT4L is localized to early endosomes and its gene is transcriptionally activated by SRF.
(A) DDiT4L was in the cytoplasmic fraction of NRVMs, not in the nuclear fraction. n=4 preparations, 2 samples per preparation.
(B) Overexpressed DDiT4L stained in a punctate fashion in NRVMs and colocalized with EEA1 (early endosome antigen 1), Rab5 and mTOR staining in serum starved NRVMs. n=4 preparations, 2 samples per preparation. Scale bar, 20 μm.
(C) Section of the 2000 bp 5′ UTR of DDiT4L cloned into Bgl basic luciferase construct. SRF like cARG box is highlighted in red, and the DDiT4L start codon is highlighted in blue. HEK293 cells transfected with SRF plasmid and the 5′UTR luciferase construct had increased luciferase activity, which was not seen when the cArG box was mutated to CCGGGGAAGCGC. n=5 preparations, 2 samples per preparation. One Way ANOVA followed by Bonferroni posttest, ***p<0.01.
(D) Glucose deprivation or infection with SGK-CA adenovirus increased phosphorylation (p) of SRF in NRVMs. n=4–5 preparations, 2 samples per preparation. Student t-test *p<0.05, **p<0.01 compared to control. Phospho-proteins were normalised to total protein.
(E) Knockdown of SRF in NRVMs prevented SGK-CA adenovirus and glucose deprivation from inducing increased expression of DDiT4L. n = 4 preparations, 2 samples per preparation. One Way ANOVA, Bonferroni posttest, *p<0.05, **p<0.01.
DDiT4L expression is increased by serum response factor (SRF)
DDiT4 is target of the hypoxia-inducible transcription factor HIF1α (28, 29). Analysis of the 5′-UTR of DDiT4L revealed a putative cArG box like serum response element, suggesting possible binding by serum response factor (SRF), a transcription factor that is stimulated by pathological stress in the heart (30, 31). Luciferase assays using a plasmid containing the 2000 bp upstream region of DDiT4L in HEK293 cells demonstrated that SRF cotransfection increased luciferase activity, which was not seen when the cArG box was mutated (Fig 4C). In NRVMs, glucose deprivation or infection with SGK1-CA adenovirus increased the activity of SRF, as indicated by the increased phosphorylation of Ser103 (Fig. 4D), which was associated with increased DDiT4L mRNA expression under these conditions (Fig. 1B,G). Furthermore, silencing of SRF in NRVMs abolished the increase in DDiT4L expression induced by SGK1-CA adenovirus or glucose deprivation (Fig. 4E). These findings showed that SRF was necessary and sufficient to induce DDiT4L in response to certain stressors, and links cellular stress to autophagy and mTORC1 signaling through DDiT4L.
Overexpression of DDiT4L in the heart leads to increased autophagy and mild systolic dysfunction that is reversed by suppressing transgene expression
We next sought to investigate DDiT4L in vivo by conditionally over-expressing DDiT4L in cardiomyocytes using an αMHC tet-off system (32) (Fig. S3A). When mated with the tetracycline-activator driver (Tta), transgene expression is suppressed by doxycycline in the ‘double transgenic’ mice (referred to as DDiT4L/Tta) and activated by removal of doxycycline from the diets of these mice (Fig. S3A). DDIT4L was increased in abundance in the heart (Fig. 5A) to amounts similar to that seen after TAC induced hypertrophy (Fig. S3B), but not at the mRNA level in quadriceps muscle or lung (Fig. S3C). As expected from our NRVM experiments, DDiT4L overexpression triggered an increase in autophagy and a decrease in mTOR signaling (Fig. 5A–F). There were no compensatory changes in DDiT4 in the DDiT4L/Tta mice (Fig. S3D).
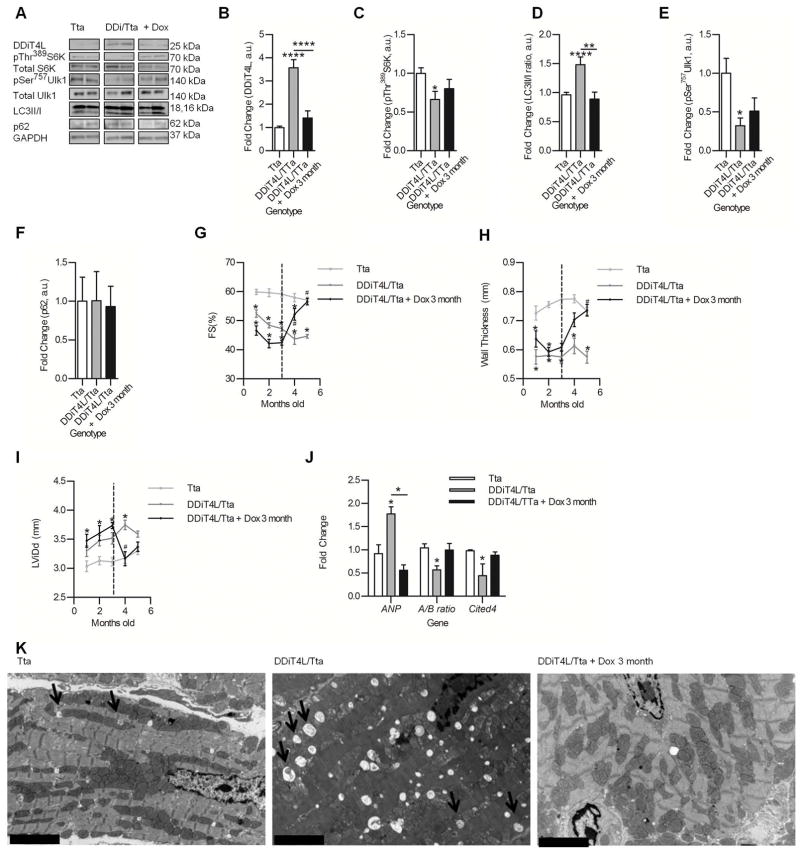
Figure 5. DDiT4L conditional overexpressing mice show a mild, stable systolic phenotype with mTOR inhibition and increased autophagy, which is reversible upon suppression of transgene.
(A to F) Western blots (A) of cardiac lysate from transgenic mice show increased DDiT4L abundance (B), decreased phosphorylation (p) of Thr389 in S6K (C), increased LC3II/I ratio (D), decreased phosphorylation of Ser757 in Ulk1 (E), and no changes in p62 abundance (F). These effects were reversed upon transgene suppression. n=6–9 mice per genotype. One-Way ANOVA, Bonferroni posttest *p<0.05, **p<0.01, ****p<0.0001. Representative Western Blots are from the same blot, but they may not be contiguous Phospho-proteins were normalized to total protein.
(G to I) Mice overexpressing DDiT4L from birth show decreased fractional shortening (FS) (G), thinner ventricular walls (H) and a larger left ventricular diastolic dimension (LVDd) (I) which was reversed upon doxycycline administration. n=6–9 mice per genotype. Two-Way ANOVA, Bonferroni posttest. *p<0.05 compared to Tta control, #p<0.05 compared to DDiT4L/Tta.
(J) ANP expression was increased and the A/B MHC ratio and Cited4 expression was decreased in mice overexpressing DDiT4L which was reversed with transgene suppression. n=6–9 mice per genotype. One-Way ANOVA, Bonferroni posttest, *p<0.05 compared to control or indicated sample.
(K) EM images showed increased numbers of autophagosomes (indicated by the arrows) in cardiomyocytes from DDiT4L overexpressing mice. n=4 mice per genotype. Scale bar represents 5 μM.
Mice with overexpression of DDiT4L from birth (because doxycycline was not added to their diet after birth) showed mild ventricular remodeling, mild left ventricular dilation, and decreased fractional shortening and wall thickness on echocardiography (Fig. 5G,H,I), but no changes in heart weight to body weight ratio (HW/BW) or heart weight to tibia length ratio (HW/TL) at 5 months of age (Supplementary Table 2). The DDiT4L/Tta mice had no changes in myocyte architecture, or changes in cross-sectional cell size as measured by hematoxylin/eosin (H and E) staining and wheat germ agglutinin (WGA) staining of cardiac tissue sections respectively (Fig. S3E), but the length of isolated ventricular cells was decreased compared to cells from Tta controls (Fig. S3G). DDiT4L/Tta mouse sarcomeres were not disorganized, but were shorter when compared with Tta mouse (Fig S3H). The decrease in the wall thickness, cardiomyocyte length, and sarcomere length suggest a mild anti-hypertrophic effect of DDiT4L in the heart at baseline (33, 34). There was no increase in macroscopic fibrosis in the DDiT4L/Tta mice but these mice had an increase in the expression of Col1, Col3 and CTGF, which encode the pro-fibrotic factors collagen 1, collagen 3, and connective tissue growth factor, respectively (Fig. S3F). These mice also had increased ANP (which encodes atrial natriuretic peptide), decreased alpha/beta MHC (which encodes myosin heavy chain) ratio and decreased Cited4 expression (35) (Fig. 5J), suggestive of a fetal gene expression program seen with pathological stress. Consistent with its effects on autophagy and mTORC1 signaling in vitro, the transgenic mice had decreased phosphorylation of S6K (Fig. 5A, C), and increased autophagy as shown by an increase in the LC3II/I ratio (Fig. 5A,D), decreased phosphorylation of Ulk1 (Fig. 5A,E), no change in p62 abundance (Fig. 5A,F), and increased numbers of autophagosomes by electron microscopy imaging (Fig. 5K). Together these data confirmed that DDiT4L expression in cardiomyocytes inhibited mTORC1 and induced autophagy in vivo. Furthermore, it appeared that induction of stress-autophagy pathways at baseline led to a mild ventricular dysfunctional phenotype.
When we fed doxycycline to these mice at 3 months of age to suppress overexpression of the transgene, DDiT4L abundance returned to baseline (Fig, 5A,B), and cardiac structure and function normalized (Fig. 5G,H,I). There was a concomitant normalization of the expression of pathological and fibrosis genes (Fig. 5J, Fig. S3F), de-repression of mTORC1 signaling and normalization of autophagy (Fig. 5A–F, K), suggesting that the phenotype observed was the direct effect of DDiT4L signaling, and not a secondary effect due to developmental changes or adaptive changes in response to the cardiac structural phenotype.
Transgenes under the α-MHC promoter are not substantially expressed prenatally. However, to confirm that the effects were not due to developmental overexpression of DDiT4L, we kept the pups and their mother on a doxycycline diet to suppress transgene expression until one month after birth, then switched the mice to normal chow to allow transgene expression. Echocardiography was performed before cessation of doxycycline, and at one and two months after transgene expression. DDiT4L overexpressing mice showed no changes in their heart function after one month of normal chow compared to Tta controls, but after two months developed a cardiac phenotype similar to the mice with DDiT4L overexpression in the heart for 5 months as described above (Fig. S4A,B,C). Moreover, one month after normal chow there was decreased phosphorylation of S6K (Fig. S4D,E), increased LC3II/I (Fig. S4D,F), and decreased p62 abundance (Fig. S4D,H). These results suggested that inhibited mTORC1 and increased autophagic flux induced by DDiT4L overexpression preceded and was likely responsible for the cardiac structural and functional changes in these mice. Our data suggested that over-expression of DDiT4L at baseline led to mTORC1 inhibition and an increase in autophagic flux, which resulted in mild ventricular dysfunction characterized by cardiac atrophy and a pathological molecular signature, all of which were reversed by suppression of the transgene.
Heterozygous knockout of beclin partially reverses the effect of DDiT4L/Tta overexpression in the heart
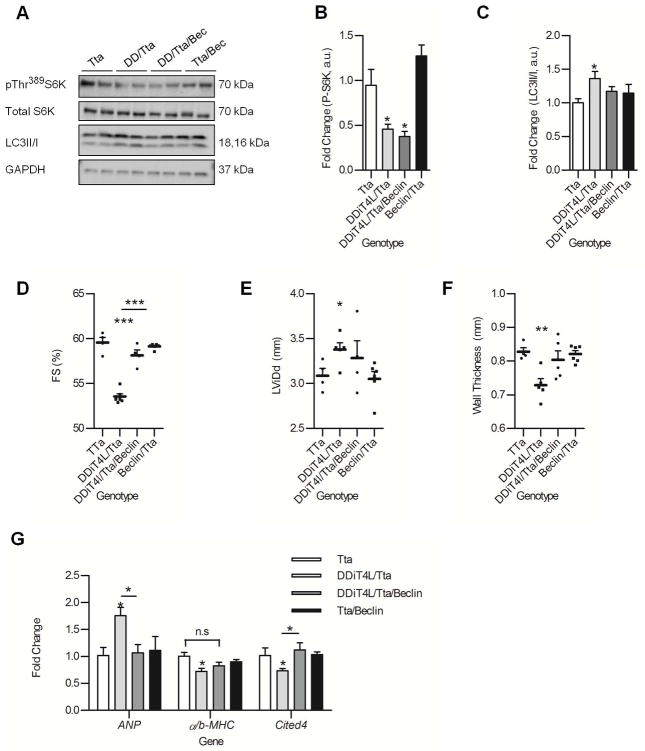
To discriminate the specific role of autophagy from the overall effect of inhibited mTORC1 signaling on the cardiac phenotype, DDiT4L/Tta mice were crossed with beclin 1(+/−) knockout mice, in which autophagy is decreased downstream of mTORC1 signaling (36, 37). As expected, phosphorylation of S6K was decreased in these mice at 2 months of age, consistent with continued mTORC1 inhibition (Fig. 6A,B). However, the increase in autophagy in the DDiT4L/Tta mice was normalized as assessed by the LC3II/I ratio (Fig. 6 C), which was associated with a normalization of the cardiac phenotype, notably a reversal of the effect of DDiT4L/Tta on cardiac fractional shortening (Fig 6D), LViDd (Fig 6E), and wall thickness (Fig 6F). Finally, the suppression of autophagy also correlated with a reversal of the pathological molecular signature induced by DDiT4L over-expression (Fig. 6G). Overall our results suggest that the baseline increase in stress autophagy in the DDiT4L/Tta mice has a detrimental, but reversible effect on cardiac structure and function.
Figure 6. Heterozygous knockout of beclin partially reverses the effect of DDiT4L/Tta overexpression in the heart.
(A to C) Western blots (A) of cardiac lysates showed that DDiT4L/Tta/Beclin mice had no change in phosphorylation (p) of Thr389 in S6K (B) but normalized LC3II/I ratios (C) compared to DDiT4L/Tta mice at 2 months. n = 3–6 mice per genotype. One Way ANOVA followed by Bonferroni posttest. *p<0.05 compared to control. Phospho-proteins were normalised to total protein
(D to F) DDiT4L/Tta/Beclin mice had heart function comparable to Tta controls, with a significant difference in fractional shortening (FS) compared to DDiT4L/Tta mice. n = 3–6 mice per genotype. One Way ANOVA followed by Bonferroni posttest, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 compared to control or indicated sample.
(G) DDiT4L/Tta/Beclin mice had normalized ANP and Cited4 expression compared to DDiT4L/Tta transgenic mice. n = 3–6 mice per genotype. One Way ANOVA followed by Bonferroni posttest *p<0.05 compared to control or indicated sample.
DDiT4L increases phosphorylation of Akt at Ser473
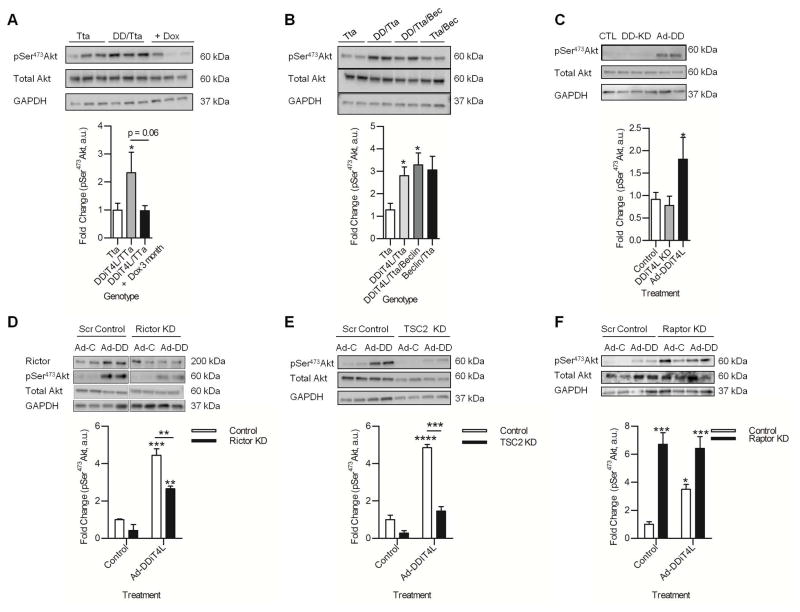
The DDiT4L/Tta mice have a relatively mild phenotype compared to murine models with mTOR ablation, which results in inhibition of both mTORC1 and mTORC2 (38). We therefore investigated the possibility that mTORC2 may be activated as a compensatory mechanism, thereby mitigating some of the deleterious effects of mTORC1 inhibition at baseline (5, 7). Phosphorylation of Akt (also known as protein kinase B) at Ser473 is a downstream marker of mTORC2 activity, and was increased with cardiac overexpression of DDiT4L at 5 months (Fig. 7A), although the increase was not significant at 1 month of transgene expression (Fig. S4G). As with the other cardiac phenotypes described above, this increase was also reversed with transgene suppression (Fig. 7A), but not with suppression of autophagy in the DDiT4L/Tta/Beclin mice (Fig. 7B). These data suggested that Akt phosphorylation was more likely related to DDiT4L-mediated effects on mTOR signaling rather than a secondary response to the cardiac dysfunction. Correspondingly, phosphorylation of Akt in NRVMs was increased by Ad-DDiT4L infection but unaffected by DDiT4L knockdown (Fig. 7C).
Figure 7. DDiT4L activates Akt.
(A) Phosphorylation (p) of Akt was increased in DDiT4L overexpressing mice, which was reversed after transgene suppression. n=6–9 mice per genotype. One-Way ANOVA, Bonferroni post-test, *p<0.05. Phospho-proteins were normalized to total protein.
(B) Phosphorylation of Akt remained increased in DDiT4L/Tta/Beclin mice. n = 3–6 mice per genotype. One Way ANOVA followed by Bonferroni posttest. *p<0.05. Phospho-proteins were normalized to total protein.
(C) Phosphorylation of Akt was increased in NRVMs infected with Ad-DDiT4L (AD-DD), with no change seen with DDiT4L knockdown (DD-KD). n=4–6 preparations, 2 samples per preparation. One Way ANOVA followed by Bonferroni posttest *p<0.05 compared to control. Phospho-proteins were normalized to total protein.
(D) Knockdown of Rictor reduced Ad-DDiT4L infection (Ad-DD) from increasing the phosphorylation of Akt. n = 4 preparations, 2 samples per preparation. One Way ANOVA followed by Bonferroni posttest, *p<0.05, **p<0.01, ***p<0.001 compared to control or indicated sample.
(E) Phosphorylation of Akt was increased in Ad-DDiT4L-infected (Ad-DD) NRVMs, which was blunted after knockdown of TSC2. n=4 preparations, 2 samples per preparation. One Way ANVOA followed by Bonferroni posttest, ***p<0.001, ****p<0.0001 compared to control or indicated sample. Phospho-proteins were normalised to total protein.
(F) Knockdown of Raptor increased phosphorylation of Akt, which was not further increased with Ad-DDiT4L infection (Ad-DD). n=3 preparations, 2 samples per preparation. One Way ANOVA followed by Bonferroni posttest, *p<0.05, ***p<0.001 compared to control or indicated sample. Phospho-proteins were normalised to total protein.
The phosphorylation of Thr1179 in IRS1 was not altered in the hearts of DDiT4L overexpressing mice or in Ad-DDiT4L-infected NRVMs (Fig. S5A,B). Because this phosphorylation event is required for downstream phosphoinositide 3-kinase (PI3K) mediated-phosphorylation of Akt (39), this finding suggested that the increase in the phosphorylation of Akt was not secondary to upstream activation of the IGF/PI3K pathways (Fig. S5A,B). Finally, silencing of Rictor in NRVMs abrogated the increase in the phosphorylation of Akt with Ad-DDiT4L infection (Fig. 7D), providing further evidence that the activation of Akt with DDiT4L overexpression depended on the presence of the mTORC2 complex. Together, these data suggested that DDiT4L overexpression led to mTORC2 activation.
DDiT4L overexpression did not induce an increase in the phosphorylation of Akt in NRVMs with knockdown of TSC2 (Fig. 7E), suggesting that similar to DDiT4L-mediated inhibition of mTORC1, the activation of mTORC2 with DDiT4L overexpression was TSC2-dependent. siRNA mediated ablation of raptor in NRVMs led to increased basal phosphorylation of Akt (Fig. 7F), which was not further augmented by Ad-DDiT4L infection, showing that the DDiT4L-mediated increase in mTORC2 activity proceeded through mTORC1 inhibition. These data are consistent with the inhibition of mTORC2 by mTORC1-mediated phosphorylation of the stress activated protein kinase Sin-1 (40). DDiT4L inhibition of mTORC1 may subsequently derepress this axis, representing another facet of the complex interplay between mTORC1 and mTORC2 signaling.
Discussion
Autophagy in the heart plays a critical role both in normal homeostasis as well as in the response to stressors. Here, we characterized DDiT4L, a regulator of mTOR signaling in the heart that was identified in a discovery screen to delineate signaling pathways that were differentially activated by pathological and physiological stress. We showed that DDiT4L activated stress-induced autophagy and differentially regulated mTORC1 and mTORC2 signaling in cardiomyocytes in a TSC2-dependent mechanism. Expression of DDiT4L in the heart increased basal autophagy, caused mild dysfunction, mild atrophy and a molecular signature of pathological stress. This phenotype was completely reversed by suppression of DDiT4L expression and with genetic suppression of autophagy, raising the possibility of therapeutic targeting of DDiT4L in cardiomyopathies.
This study stemmed from a DSAGE-based discovery, which identified DDiT4L as the top candidate gene that showed increased expression specifically in a transgenic model of pathological hypertrophy, but not in a one of physiological hypertrophy. DDiT4L expression was also increased in other genetic and acquired murine and cultured cardiomyocyte models of pathological stress, but not in physiological stress, and was increased in the left ventricle of human DCM patients. We saw increases in DDiT4L mRNA in TAC after one week during the hypertrophy phase, and in the DCM mouse model, which have dilated ventricles and abnormal wall stress. The temporal expression of DDiT4L in mice subjected to TAC as they progress from hypertrophy to heart failure may be complex and subjected to multiple different interacting signaling pathways. In this regard, regulation of autophagy is complex in the TAC model, with initial increases in autophagy followed by a suppression of autophagy in the chronic heart failure stages (41). In cardiomyocytes DDiT4L expression was increased by the transcription factor SRF, which has previously been implicated in mediating stress responses in the heart (42, 43). Knockdown of DDiT4L at baseline in cardiomyocytes led to activated mTORC1 but not to changes in autophagy signaling, suggesting that SRF increases DDiT4L expression under stress conditions and may specifically regulate stress-induced autophagy rather than basal autophagy.
DDiT4L protein localized with both early endosome markers and mTOR under stress conditions, when DDiT4L is activated and mTOR inhibited, suggesting that DDiT4L may traffic to an mTOR-containing cellular compartment when cells are under stress, leading to inhibition of mTORC1 through a TSC2-dependent manner. However the interaction between DDiT4L and the mTOR complex in this cellular compartment and the exact mechanism of interaction is yet to be elucidated. In the absence of stressors, a basal increase in DDiT4L in the heart increased autophagy, led to a mild reversible cardiomyopathy with mild cardiac atrophy and inhibited cardiomyocyte growth. These effects were not observed when autophagy was genetically inhibited in these mice, demonstrating that it was due to increased autophagy, rather than overall inhibition of mTORC1. Whether these ‘anti-hypertrophic’ effects of DDiT4L are adaptive or additionally maladaptive in the presence of pathological stressors will be the subject of future investigations in murine models of heart disease.
In both cultured cardiomyocytes and the DDiT4L transgenic mouse we saw increased autophagy, measured through Western blotting for LC3II/I, phosphorylated Ulk1, confocal and EM imaging. Through treatment of cardiomyocytes with bafilomycin and measuring LC3II/I and p62 we concluded this was due to increased autophagic flux rather than inhibited lysosomal fusion. Although the abundance of p62 was unchanged (at 5 months) or slightly decreased (1 month) in the transgenic mouse model, this was not surprising as p62 undergoes complex transcriptional regulation over time, altering protein abundance independently of autophagy (44–46). There is a distinction between classic (mediated through atg5/atg7) and alternative (mediated through Ulk/beclin/rab9) pathways (47, 48). Given the implication of Ulk-1 and beclin in DDiT4L-induced autophagy, it is possible that DdiT4L may couple cellular stress to the alternative autophagy pathway. However, further complementation experiments with critical components of these pathways would be necessary to fully delineate these mechanisms.
Fully reversible cardiac phenotypes are rare and can shed light on pathways that may be targeted for therapeutic development for heart disease. Our DDiT4L transgenic mouse had complete reversal of both the cardiac phenotype and the molecular signature of DDiT4L overexpression with inhibition of transgene expression, and when autophagy was inhibited through beclin 1 haploinsufficiency, confirming that the reversible nature of this phenotype was due to autophagy. It is possible that the reversal of this phenotype has to occur within a certain time window, and prolonged periods of increased baseline autophagy may ultimately lead to irreversible changes. Another mouse model of increased autophagy and cardiomyocyte atrophy shares features with our model, demonstrating similar reversibility upon cessation of transgene expression (34). These findings highlight the importance of autophagy in basal and disease conditions in the heart, and puts forward DDiT4L and autophagy as therapeutic targets for heart disease.
We postulate that the mild phenotype observed in our mouse model (compared to ablation of models of mTOR ablation) was due to the counteracting protective role for mTORC2 signaling. We found a significant increase in the phosphorylation of the specific mTORC2 target Ser473 in Akt in our transgenic mouse model, as well as in NRVMs overexpressing DDiT4L. Phosphorylation of Akt at Ser473 is typically seen in physiological models of cardiac hypertrophy (49, 50) and acute overexpression of active Akt prevents progression to heart failure in mice that have undergone pressure overload-induced hypertrophy (51), suggesting one possible mechanism for cardioprotection by DDiT4L overexpression. This activation of Akt was not due to increased autophagy because the DDiT4L/Tta/Beclin mice still showed increased phosphorylation of Akt despite decreased autophagy. DDiT4L modulation of both mTORC1 and mTORC2 pathways adds another facet to a complex feed-back regulatory pathway where the exact balance between mTORC1 and mTORC2 activity may ultimately determine the cardiac phenotype.
Finally, our studies showed that unlike DDiT4, DDiT4L may be implicated more in stress responses than in basal ones. Unlike DDiT4, the DDiT4L protein structure has not been solved, and apart from its binding to 14-3-3, little is known about its functional domains. The crystal structure of DDiT4 has revealed a conserved functionally important hot spot, which when mutated prevents DDiT4 from inhibiting mTOR (52). This conserved site is not present on DDiT4L, suggesting that DDiT4L inhibits mTOR through other binding partners. Furthermore, mTORC1 activation depends on DDIT4 translocation to the plasma membrane by G-protein coupled receptors, and this translocation was not observed with DDiT4L (53).
In summary, our current model demonstrated that DDiT4L was activated by pathological stress in the heart by SRF, leading to induction of autophagy through inhibition of mTORC1 signaling. Additionally DDiT4L led to activation of mTORC2. Overexpression of DDiT4L in the heart in vivo resulted in increased autophagy, inhibited mTORC1, activated mTORC2, increased markers of pathological hypertrophy and mild functional and structural changes. This phenotype was completely reversed by suppression of transgene expression or genetic inhibition of autophagy. Due to the importance of autophagy and mTOR signaling in the heart and disease, further investigation into DDiT4L and its regulation of autophagy and mTOR may lead to novel therapeutic targets for treatment of heart pathologies.
Methods
Generation of adenovirus
HA-DDiT4L adenovirus was generated with the AdEasy system following published protocols (54), infected into HEK293T cells and isolated using Vivapure Adenopack (Sartorius stedim biotech, Germany).
Cardiac-specific HA-DDiT4L overexpression in mice
All animals were housed in the BIDMC animal facility and experiments were approved and carried out under IACUC guidelines. Mice expressing HA-DDiT4L were generated by injecting linear DNA into FVB oocytes. Mice with tetracycline-controlled cardiac overexpression of HA-DDiT4L were generated in a FVB background by crossing mice with HA-DDiT4L downstream of an Inducible a-MHC vector with mice expressing a myocardial Tta (gift from Jeff Robbins, Cincinnati Children’s Hospital Medical Center (32)). Transgene was either constitutively expressed, or inhibited with 200 mg/kg of doxycycline in food from mating. Tta littermates were used as experimental controls. To generate DDiT4L/Tta/Beclin mice, heterozygote beclin knockout mice (B6.129X1-Becn1tm1Blev/J mice purchased from Jackson lab) were crossed with DDiT4L/Tta expressing mice. (32)). Transgenes were either constitutively expressed, or inhibited with 200 mg/kg of doxycycline in food from mating. Tta littermates were used as experimental controls. To generate DDiT4L/Tta/Beclin mice, heterozygote beclin knockout mice (B6.129X1-Becn1tm1Blev/J mice purchased from Jackson lab) were crossed with DDiT4L/Tta expressing mice. (32)). (32)). Transgenes were either constitutively expressed, or inhibited with 200 mg/kg of doxycycline in food from mating. Tta littermates were used as experimental controls. To generate DDiT4L/Tta/Beclin mice, heterozygote beclin knockout mice (B6.129X1-Becn1tm1Blev/J mice purchased from Jackson lab) were crossed with DDiT4L/Tta expressing mice. (32)). Transgenes were either constitutively expressed, or inhibited with 200 mg/kg of doxycycline in food from mating. Tta littermates were used as experimental controls. To generate DDiT4L/Tta/Beclin mice, heterozygote beclin knockout mice (B6.129X1-Becn1tm1Blev/J mice purchased from Jackson lab) were crossed with DDiT4L/Tta expressing mice. (32)).
Exercise model of physiological hypertrophy
Mice aged 12–16 weeks were forced to swim in a tank (50 cm diameter, ~2000 cm2 surface area) containing water maintained at 30–32° C to avoid thermal stress. Mice initially swam for 10 minutes twice per day, with the sessions separated by a 4 hour break. The swim sessions were increased by 10 minutes each day until the sessions reached a maximum of 90 minutes each, which were maintained for the remainder of the program. The mice swam 7 days/week for 4 weeks to produce a robust cardiac hypertrophic response (10% increase in HW/BW).
Transverse Aortic Constriction Model
Mice aged 8–12 weeks were anesthetized either with ketamine (80–100 mg/kg) and xylazine (12 mg/kg), or with inhaled isofluorane (1–2% isofluorane in oxygen, dosed to effect). Mice were intubated using a 22GA Angiocath and put on a small animal ventilator CWE, and anesthesia with 1–2% isofluorane in oxygen was delivered through the ventilator. Ventilation parameters were: Tidal volume 0.25 ml, Respiratory rate 110 br/min, Insp time 0.3 sec. The operative field was shaved, cleaned with 10% Betadine and 70% EtOH. A lateral thoracotomy was performed with hemostasis achieved using the Gemini microcautery system. The ascending aorta was dissected from the surrounding connective tissue and a 6.0 silk Prolene suture without needle was placed around the transverse aorta. The suture was then tied down against a blunted 25G) needle and the needle was removed to create a chronic model of pressure overload hypertrophy. The chest cavity was closed using continuous silk 6.0 Prolene suture and 9 mm wound clips. The animal was maintained on a warm pad throughout the procedure and kept on the pad until recovery from anesthesia. Once regular spontaneous respirations resumed, the mouse was disconnected from the ventilator and extubated. The animal was observed until it could ambulate and then transferred back to a new cage. Wound clips were removed 10 days post-operatively, and animals received buprenorphine for 3 days, after getting the first dose preemptively.
DSAGE screen
RNA was extracted from transgenic mice with cardiac specific overexpression of SGK1-CA and SGK1-DN mice (17) as well as transgenic mice with cardiac specific overexpression of Akt1-CA (18) and Akt1-DN (19) mice. Total RNA was prepared from 5 hearts from male mice of each genotype at 4 weeks using Trizol (Invitrogen). RNA from each genotype was pooled in equal proportion to provide 10μg of total RNA for the generation of cDNA libraries (16, 55) and subjected to DSAGE as previously described (16, 55). For DSAGE results, statistical comparisons of gene expression were performed as previously described (56) using the Audic and Claverie alternative to Fisher’s exact test (57).(16, 49).
Human studies
Discarded, de-identified (anonymized) pathological tissues were obtained and studied under an institutional review board–approved protocol.
Cardiac function
Echocardiograms were performed on anaesthetized mice using a GE Vivid 7 ultrasound machine (GE Healthcare) with a frequency of 14.0 mHz, 131.5 frames per second and a depth of 1 cm for 2-dimensional images. M-mode images were taken at the midpapillary muscle level and were used for measurements. Echocardiograms were analyzed using Vivid 7 software.
Tissue Harvesting
Mice were sacrificed by overdose of ketamine/xylazine (80/12 mg/kg). Body weight, dry lung, heart, and left ventricular weight was measured along with tibia length. The left ventricle was cut into three pieces for RNA extraction, protein extraction and immunohistochemistry.
Immunohistochemistry
Cardiac slices stored in 4% PFA were embedded in paraffin and sectioned and stained for Mason Trichrome stain (for fibrosis), Hematoxylin/Eosin (for histology) and Wheat Germ Agglutinin (for cell size) (BIDMC histology core). Slides were imaged on either an upright fluorescent microscope (Leica CTR5000) or Zeiss confocal (LSM 510 meta) and cell size was measured using ImageJ software.
Electron Microscopy
1 mm slices of tissue were fixed in EM fixation buffer (2.5% Glutaraldehyde, 2.0% Paraformaldehyde, in 0.1M Cacodylate Buffer). Samples were prepared and imaged for autophagasomes (at the Beth Israel Deaconess Medical Center HDDC imaging core B, courtesy of Andrea Calhoun and Susan Hagen, Beth Israel Deaconess Medical Center, Boston, MA).
Isolation and culture of neonatal rat ventricular cardiomyocytes and other primary cells
NRVMs and fibroblasts were isolated from post-natal day 1 rat pups using enzymatic digestion. 24 h after plating, media was changed to serum and antibiotic free, and 24 h following cells were transfected with DDiT4L siRNA (Qiagen), TSC2 GapmeR antisense oligonucleotide (Exiqon), CA-Rheb plasmid (Addgene # 32520, (58)) or infected with HA-DDiT4L adenovirus, or GFP-LC3 adenovirus (gift from Dr. J. Sadoshima,, Rutgers, New Jersey). 48 h after transfection or infection cells were used for experiments. The smooth muscle cell line pac1 was purchased from ATCC. Endothelial cells and pericytes were isolated from mouse hearts and skeletal muscle using Dynabeads (59, 60). The following treatments were carried out: glucose deprivation for 2 h, oxidative stress (100 μM H2O2 for 18 h), pathological stretch (20% stretch over 6 h), DNA damage (doxorubicin 2.5 μM 6 h), IGF1 (20 ng/mL.. h), phenylephrine (10 μM 24 h), rapamycin (10 nM, 2h, high dose 50 nM 2 h), bafilomycin (20 nM, 6 h).
Real time PCR
RNA was isolated from both tissue and cell samples using Trizol (Qiagen), and phase separation followed by column purification (Qiagen). 1 μg of RNA was added to RT-PCR reactions (Applied Bioscience) and qPCR was carried out using cyber green (Kapa). Primers used are listed in the supplemental information tables S3 and S4.
Western blotting
Protein lysates from both tissue and cells were made using lysis buffer (Cell signaling) containing PMSF, protease, and phosphatase inhibitors for 1 h at 4 degrees with rocking. Protein (50 μg) was run on a 4–11% SDS page gel (Criterion, Bio-Rad), transferred onto nitrocellulose, blocked in 5% BSA or milk and probed with primary antibodies, (DDiT4L (Fitzgerald), DDiT4L (Abcam) P-S6K, S6K, LC3II/I, HA, P-Ulk1,Ulk1, Histone H3, P-Akt, Akt (Cell Signaling), GAPDH, TSC2, SRF, IRS1 (Santa Cruz), P-SRF, P-IRS1 (Abcam))Proteins were detected using an anti-goat secondary antibody conjugated to HRP (Dako) and a Bio-Rad ChemiDoc MP. Blots were analysed using ImageJ software, with proteins normalised to the housekeeper GAPDH, and phospho-proteins normalised to their respective total protein.
Confocal microscopy
Cells were plated on 15 mm laminin coated coverslips and treated as above, fixed, stained and imaged on a Ziess 710 confocal microscope. For autophagasome experiments cells were infected with GFP-LC3 virus 48 h before fixation. Cells were fixed with 4% PFA for 20 min, permeabilized for 20 min with 0.5% Triton X-100, stained with α-actinin (Abcam), mounted using DAPI hardset mounting media (Vector Laboratories) and imaged using a Zeiss 710 confocal microscope. For localization experiments the following antibodies were used at a concentration fo 1:100: HA, LC3II/I, mTOR, caveolin3, Appl, GOPC, Syntaxin6 (Cell Signaling), LAMP1 (Abgent), and VAMP4 (Novus biologicals). Image analysis was carried out using ImageJ software.
Luciferase assay
A luciferase reporter plasmid was made by inserting about 2100 bp of the 5′ UTR of mouse DDiT4L into a pGL3 basic luciferase reporter vector. Luciferase assays were carried out using Promega Dual Luciferase assay kits. SRF plasmid was purchased from Addgene.
mTOR immunoprecipitation
mTOR immunoprecipitations were carried out using 0.3% CHAPs as previously described (61).
Statistics
Unless otherwise specified, data are expressed as mean±SEM. All statistics were carried out using GraphPad Prism 5. Mean comparisons between two groups were compared using the unpaired Student’s t-test. For multiple comparisons, One- or Two-way ANOVA were performed, followed by Bonferroni posttest as specified. For data that were not normally distributed, non-parametric testing was done as indicated in the figure legends.
Supplementary Material
Fig. S1. DDiT4 is differentially regulated compared to DDiT4L
Fig. S2. Regulation of mTOR signaling by DDiT4L under different conditions in NRVMs.
Fig. S3. Conditional heart-specific overexpression of DDiT4L
Fig. S4. Acute increases in DDiT4L lead to alterations in mTORC1 signaling but not in systolic function.
Fig. S5. Overexpression of DDiT4L does not alter the phosphorylation of IRS1
Table S1. Top 20 candidates from DSAGE screen of genes with increased expression in SGK-CA mice compared to other transgenic mice
Table S2. DDiT4L/Tta mice have mild diastolic dysfunction which is reversed upon transgene suppression
Table S3. Mouse primers
Table S4. Rat primers
Data File S1. Raw DSAGE data file.
Acknowledgments
We thank Christine Seidman and Jonathan Seidman for their help with DSAGE, Karyn Esser (University of Florida) for the DDiT4L plasmid, Dr. Jeffrey Robbins (University of Cincinnati) for the tTA driver transgenic mice, and Dr. J. Sadoshima for the GFP-LC3 adenovirus (Rutgers New Jersey).
Funding: SD was funded by the Klarman Scholars Foundation and a grant from NHLBI (RO1 HL122547), MK was funded by a grant from NHLBI (RO1-HL102368, RO1-HL114775) and TR was funded by grants from NHLBI (RO1 HL094677 and RO1 HL135886). Electron microscopy was done at the BIDMC HDDC Core by Andrea Calhoun and Susan Hagen (NIH P30 DK034854,
Footnotes
Author Contributions: SD designed, contributed to data analysis and wrote the paper. BS designed, carried out and analyzed experiments and wrote the paper. VS, HF, MCC, MM, DH conducted experiments and analyzed data. MK, AR, TSK contributed reagents and critical review of the manuscript, AR, PO, conducted the DSAGE screen and provided critical review of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The DDiT4L transgenic mouse has been deposited in Jackson Laboratories.
References and Notes
- 1.Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death Differ. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- 2.Gottlieb RA, Mentzer RM. Autophagy: an affair of the heart. Heart Fail Rev. 2013;18:575–584. doi: 10.1007/s10741-012-9367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114:549–564. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 2013;38:233–242. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shende P, et al. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation. 2011;123:1073–1082. doi: 10.1161/CIRCULATIONAHA.110.977066. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Y, et al. Mechanistic target of rapamycin (Mtor) is essential for murine embryonic heart development and growth. PLoS One. 2013;8:e54221. doi: 10.1371/journal.pone.0054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamai T, et al. Rheb (Ras homologue enriched in brain)-dependent mammalian target of rapamycin complex 1 (mTORC1) activation becomes indispensable for cardiac hypertrophic growth after early postnatal period. J Biol Chem. 2013;288:10176–10187. doi: 10.1074/jbc.M112.423640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song X, et al. mTOR attenuates the inflammatory response in cardiomyocytes and prevents cardiac dysfunction in pathological hypertrophy. Am J Physiol Cell Physiol. 2010;299:C1256–1266. doi: 10.1152/ajpcell.00338.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Völkers M, et al. Mechanistic target of rapamycin complex 2 protects the heart from ischemic damage. Circulation. 2013;128:2132–2144. doi: 10.1161/CIRCULATIONAHA.113.003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sciarretta S, et al. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–1146. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marin TM, et al. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest. 2011;121:1026–1043. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 13.Miyazaki M, Esser KA. REDD2 is enriched in skeletal muscle and inhibits mTOR signaling in response to leucine and stretch. Am J Physiol Cell Physiol. 2009;296:C583–592. doi: 10.1152/ajpcell.00464.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004;18:2879–2892. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai J, et al. DSAGE Identifies Osteopontin as a Downstream Effector of Integrin-Linked Kinase (ILK) in Cardiac-Specific ILK Knockout Mice. Circ Heart Fail. 2013 doi: 10.1161/CIRCHEARTFAILURE.113.000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das S, et al. Pathological role of serum- and glucocorticoid-regulated kinase 1 in adverse ventricular remodeling. Circulation. 2012;126:2208–2219. doi: 10.1161/CIRCULATIONAHA.112.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsui T, et al. Phenotypic spectrum caused by transgenic overexpression of activated Akt in the heart. J Biol Chem. 2002;277:22896–22901. doi: 10.1074/jbc.M200347200. [DOI] [PubMed] [Google Scholar]
- 19.Nagoshi T, et al. PI3K rescues the detrimental effects of chronic Akt activation in the heart during ischemia/reperfusion injury. J Clin Invest. 2005;115:2128–2138. doi: 10.1172/JCI23073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt JP, et al. Cardiac myosin missense mutations cause dilated cardiomyopathy in mouse models and depress molecular motor function. Proc Natl Acad Sci U S A. 2006;103:14525–14530. doi: 10.1073/pnas.0606383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 22.Rolfe M, McLeod LE, Pratt PF, Proud CG. Activation of protein synthesis in cardiomyocytes by the hypertrophic agent phenylephrine requires the activation of ERK and involves phosphorylation of tuberous sclerosis complex 2 (TSC2) Biochem J. 2005;388:973–984. doi: 10.1042/BJ20041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu H, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maejima Y, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013;19:1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta M, Rath PC. Interferon regulatory factor-1 (IRF-1) interacts with regulated in development and DNA damage response 2 (REDD2) in the cytoplasm of mouse bone marrow cells. Int J Biol Macromol. 2014;65:41–50. doi: 10.1016/j.ijbiomac.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Jin HO, et al. Hypoxic condition- and high cell density-induced expression of Redd1 is regulated by activation of hypoxia-inducible factor-1alpha and Sp1 through the phosphatidylinositol 3-kinase/Akt signaling pathway. Cell Signal. 2007;19:1393–1403. doi: 10.1016/j.cellsig.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Regazzetti C, Bost F, Le Marchand-Brustel Y, Tanti JF, Giorgetti-Peraldi S. Insulin induces REDD1 expression through hypoxia-inducible factor 1 activation in adipocytes. J Biol Chem. 2010;285:5157–5164. doi: 10.1074/jbc.M109.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am J Physiol Heart Circ Physiol. 2001;280:H1782–1792. doi: 10.1152/ajpheart.2001.280.4.H1782. [DOI] [PubMed] [Google Scholar]
- 31.Lauriol J, et al. RhoA signaling in cardiomyocytes protects against stress-induced heart failure but facilitates cardiac fibrosis. Sci Signal. 2014;7:ra100. doi: 10.1126/scisignal.2005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanbe A, et al. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92:609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 33.Cao DJ, et al. Mechanical unloading activates FoxO3 to trigger Bnip3-dependent cardiomyocyte atrophy. J Am Heart Assoc. 2013;2:e000016. doi: 10.1161/JAHA.113.000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schips TG, et al. FoxO3 induces reversible cardiac atrophy and autophagy in a transgenic mouse model. Cardiovasc Res. 2011;91:587–597. doi: 10.1093/cvr/cvr144. [DOI] [PubMed] [Google Scholar]
- 35.Bostrom P, et al. C/EBPbeta Controls Exercise-Induced Cardiac Growth and Protects against Pathological Cardiac Remodeling. Cell. 2010;143:1072–1083. doi: 10.1016/j.cell.2010.11.036. (*co-senior, co-corresponding, equal contributing authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsui Y, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 37.Riehle C, et al. Insulin receptor substrate signaling suppresses neonatal autophagy in the heart. J Clin Invest. 2013;123:5319–5333. doi: 10.1172/JCI71171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D, et al. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J Clin Invest. 2010;120:2805–2816. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassan B, Akcakanat A, Holder AM, Meric-Bernstam F. Targeting the PI3-kinase/Akt/mTOR signaling pathway. Surg Oncol Clin N Am. 2013;22:641–664. doi: 10.1016/j.soc.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu P, Guo J, Gan W, Wei W. Dual phosphorylation of Sin1 at T86 and T398 negatively regulates mTORC2 complex integrity and activity. Protein Cell. 2014;5:171–177. doi: 10.1007/s13238-014-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirakabe A, et al. Drp1-Dependent Mitochondrial Autophagy Plays a Protective Role Against Pressure Overload-Induced Mitochondrial Dysfunction and Heart Failure. Circulation. 2016;133:1249–1263. doi: 10.1161/CIRCULATIONAHA.115.020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank D, et al. Mice with cardiac-restricted overexpression of Myozap are sensitized to biomechanical stress and develop a protein-aggregate-associated cardiomyopathy. J Mol Cell Cardiol. 2014;72:196–207. doi: 10.1016/j.yjmcc.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Shyu KG, Cheng WP, Wang BW, Chang H. Hypoxia activates muscle-restricted coiled-coil protein (MURC) expression via transforming growth factor-β in cardiac myocytes. Clin Sci (Lond) 2014;126:367–375. doi: 10.1042/CS20130260. [DOI] [PubMed] [Google Scholar]
- 44.Baudot AD, Haller M, Mrschtik M, Tait SW, Ryan KM. Using enhanced-mitophagy to measure autophagic flux. Methods. 2015;75:105–111. doi: 10.1016/j.ymeth.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu YT, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10:431–441. doi: 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honda S, et al. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat Commun. 2014;5:4004. doi: 10.1038/ncomms5004. [DOI] [PubMed] [Google Scholar]
- 48.Maejima Y, et al. Recent progress in research on molecular mechanisms of autophagy in the heart. Am J Physiol Heart Circ Physiol. 2015;308:H259–268. doi: 10.1152/ajpheart.00711.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kemi OJ, et al. Activation or inactivation of cardiac Akt/mTOR signaling diverges physiological from pathological hypertrophy. J Cell Physiol. 2008;214:316–321. doi: 10.1002/jcp.21197. [DOI] [PubMed] [Google Scholar]
- 50.McMullen JR, et al. Phosphoinositide 3-kinase(p110alpha) plays a critical role for the induction of physiological, but not pathological, cardiac hypertrophy. Proc Natl Acad Sci U S A. 2003;100:12355–12360. doi: 10.1073/pnas.1934654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ceci M, et al. Cardiac-specific overexpression of E40K active Akt prevents pressure overload-induced heart failure in mice by increasing angiogenesis and reducing apoptosis. Cell Death Differ. 2007;14:1060–1062. doi: 10.1038/sj.cdd.4402095. [DOI] [PubMed] [Google Scholar]
- 52.Vega-Rubin-de-Celis S, et al. Structural analysis and functional implications of the negative mTORC1 regulator REDD1. Biochemistry. 2010;49:2491–2501. doi: 10.1021/bi902135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Michel G, et al. Plasma membrane translocation of REDD1 governed by GPCRs contributes to mTORC1 activation. J Cell Sci. 2014;127:773–787. doi: 10.1242/jcs.136432. [DOI] [PubMed] [Google Scholar]
- 54.Luo J, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 55.Kim JB, et al. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 56.Christodoulou DC, Gorham JM, Herman DS, Seidman JG. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Current protocols in molecular biology/edited by Frederick M. Ausubel … [et al.] 2011;Chapter 4(Unit4):12. doi: 10.1002/0471142727.mb0412s94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome research. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 58.Nie D, et al. Tsc2-Rheb signaling regulates EphA-mediated axon guidance. Nat Neurosci. 2010;13:163–172. doi: 10.1038/nn.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sawada N, et al. Endothelial PGC-1α mediates vascular dysfunction in diabetes. Cell Metab. 2014;19:246–258. doi: 10.1016/j.cmet.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowe GC, et al. Disconnecting mitochondrial content from respiratory chain capacity in PGC-1-deficient skeletal muscle. Cell Rep. 2013;3:1449–1456. doi: 10.1016/j.celrep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarbassov DD, Sabatini DM. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J Biol Chem. 2005;280:39505–39509. doi: 10.1074/jbc.M506096200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. DDiT4 is differentially regulated compared to DDiT4L
Fig. S2. Regulation of mTOR signaling by DDiT4L under different conditions in NRVMs.
Fig. S3. Conditional heart-specific overexpression of DDiT4L
Fig. S4. Acute increases in DDiT4L lead to alterations in mTORC1 signaling but not in systolic function.
Fig. S5. Overexpression of DDiT4L does not alter the phosphorylation of IRS1
Table S1. Top 20 candidates from DSAGE screen of genes with increased expression in SGK-CA mice compared to other transgenic mice
Table S2. DDiT4L/Tta mice have mild diastolic dysfunction which is reversed upon transgene suppression
Table S3. Mouse primers
Table S4. Rat primers
Data File S1. Raw DSAGE data file.