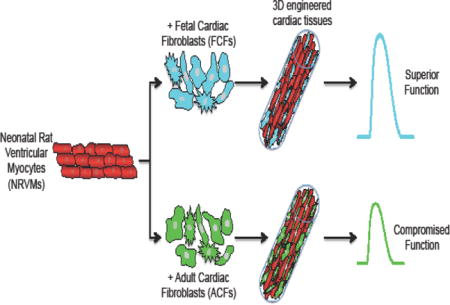
Abstract
Complex heterocellular interactions between cardiomyocytes and fibroblasts in the heart involve their bidirectional signaling via cell-cell contacts, paracrine factors, and extracellular matrix (ECM). These interactions vary with heart development and pathology leading to changes in cardiac structure and function. Whether cardiac fibroblasts of different ages interact differentially with cardiomyocytes to distinctly impact their function remains unknown. Here, we explored the direct structural and functional effects of fetal and adult cardiac fibroblasts on cardiomyocytes using a tissue-engineered 3D co-culture system. We show that the age of cardiac fibroblasts is a strong determinant of the structure, function, and molecular properties of co-cultured tissues. In particular, in vitro expanded adult, but not fetal, cardiac fibroblasts significantly deteriorated electrical and mechanical function of the co-cultured cardiomyocytes, as evidenced by slower action potential conduction, prolonged action potential duration, weaker contractions, higher tissue stiffness, and reduced calcium transient amplitude. This functional deficit was associated with structural and molecular signatures of pathological remodeling including fibroblast proliferation, interstitial collagen deposition, and upregulation of pro-fibrotic markers. Our studies imply critical roles of the age of supporting cells in engineering functional cardiac tissues and provide a new physiologically relevant in vitro platform to investigate influence of heterocellular interactions on cardiomyocyte function, development, and disease.
Keywords: Cardiac fibroblasts, Tissue engineering, Interstitial fibrosis, Extracellular matrix, Co-culture model
Graphical abstract
1. Introduction
Cardiac function is orchestrated by complex and dynamic crosstalk among different types of cells and their interactions with extracellular matrix (ECM) [1]. These cell-cell and cell-ECM relationships have been shown to vary with the cardiac development, physiological changes, and different types of pathology [1–4]. Although cardiomyocytes are the functional cells of the heart that conduct electrical impulses and mechanically contract, cardiac fibroblasts (CFs) are traditionally recognized as the most abundant cell population in the heart [5–11]. Until recently, cardiac fibroblasts have been considered as a passive cell type merely providing a structural support to the contracting cardiomyocytes. However, a growing body of evidence suggests that fibroblasts in the heart are active and critical modulators of cardiac development, homeostasis, and function, and could therefore represent an exciting target for future therapeutic interventions [3, 12].
Cardiomyocytes and CFs in the heart can communicate through direct cell-cell contacts, soluble paracrine factors, and ECM-mediated interactions [12, 13]. We, and others, have demonstrated that neonatal cardiac fibroblasts can alter electrophysiological properties of neonatal cardiomyocytes in standard 2D (monolayer) culture [10, 14–20]. However, 2D cultures for the most part fail to faithfully recapitulate structural, biochemical, and mechanical milieu of the contracting heart, including the complex cell-cell and cell-matrix interactions [21, 22]. Use of 3D co-culture systems, on the other hand, may allow systematic studies of the cardiomyocyte-fibroblast interactions within a physiologically relevant environment akin to native contracting myocardium [23–26]. Furthermore, such systems can provide a versatile in vitro platform for optimizing engineered cardiac tissue function, studying cardiac disease, and testing candidate therapeutics.
Specifically, how the age of supporting non-cardiomyocytes within engineered cardiac tissues affect cardiomyocyte function and maturation remains to be explored. Previously, distinct roles of fetal and adult fibroblasts in hyperplastic and hypertrophic growth of embryonic cardiomyocytes have been suggested from experiments in 2D co-cultures [27]. In the current study, we hypothesized that cardiac fibroblasts from fetal and adult heart would distinctly influence the structure and function of engineered cardiac tissues. To test this hypothesis, we employed a 3D culture system consisting of defined fractions of neonatal rat cardiomyocytes and fetal or adult cardiac fibroblasts and applied immunofluorescence, electrophysiological, biomechanical, and gene expression analyses to investigate the fibroblast-induced changes in structural, functional, and molecular properties of cardiomyocytes. We report that unlike fetal cardiac fibroblasts, adult cardiac fibroblasts induced adverse changes in engineered cardiac tissues resembling those found in cardiac fibrotic disease.
2. Materials and Methods
2.1. Cell isolation and culture
All animal procedures were performed in compliance with the Institutional Animal Care and Use Committee at Duke University and the NIH Guide for the Care and Use of Laboratory Animals. Neonatal rat ventricular cells were dissociated from 2-day-old Sprague Dawley rats and preplated in a T175 flask in two 50-minute steps to remove non-myocytes and enrich for neonatal rat ventricular myocyte (NRVM) population as previously described [17, 28, 29]. The cells collected after the second 50-minute preplate were named PP2 (preplate 2), and reproducibly contained a high number (89.6 ± 1.8%) of cardiac Troponin-T (cTnT)+ cardiomyocytes (Suppl. Fig. 1). Fetal cardiac fibroblasts (FCFs) were isolated from embryonic day 13.5 CD-1 murine hearts. Briefly, fetal ventricles were removed under a dissection microscope and cut into 3–4 pieces. The minced tissues were digested with 2 mg/ml collagenase type II (Worthington Biochemicals) in HBSS, incubated at 37 °C for 3 min with gentle vortexing and pipetting. The supernatant was collected and diluted in FBS on ice. The 3-min digestion step was repeated a total of 5 times to collect all the cells. Adult cardiac fibroblasts (ACFs) were isolated from 3–5-month-old CD-1 murine hearts. The ventricles were trimmed and minced into smaller pieces less than 1 mm3. The minced tissues were digested with 1 mg/ml collagenase/dispase (Roche Life Science). The tissues were incubated at 37 °C for 20 min on a rocking platform followed by pipetting and collecting the supernatant into another tube with equal volume of FBS on ice. The procedure was repeated for 4 additional times followed by homogenization in gentleMACS Dissociator. For both fetal and adult fibroblasts, after collecting supernatants from 5 serial digestions, the cells were filtered through 100 μm nylon cell strainerand centrifuged at 500×g for 5 min, and then resuspended in fibroblast medium (DMEM with 10% FBS and Penicillin/streptomycin). Fibroblasts were then seeded into T75 tissue culture flasks coated with 0.1% gelatin (EMD Millipore) and cultured until confluence. The rat neonatal cardiac fibroblasts (rNCFs) were collected as the fast-adhering cells during preplating steps in NRVM isolation, and rat adult cardiac fibroblasts (rACFs) were isolated from ventricles of 3–5-month-old Sprague Dawley rats using the same procedure described for mouse ACFs. All fibroblasts were subcultured at 1:3 ratio and used at passage 2–4.
2.2. Fabrication of co-cultured cardiac tissue bundles
NRVMs and cardiac fibroblasts (fetal or adult) were combined at a ratio of 10:3 and encapsulated in cylindrical hydrogel constructs (bundles) by modifying our previously published methods [28, 30–34]. Briefly, each bundle was made using a solution of 2.25 × 105 NRVMs with or without 0.675 × 105 cardiac fibroblasts (fetal or adult), 20 μl of culture media (DMEM, 10% (v/v) horse serum, 1% (v/v) chick embryo extract, 100 U/ml penicillin G, 1 mg/ml Aminocaproic Acid and 50 μg/ml Ascorbic Acid), 8 μl of 10 mg/ml Fibrinogen (Akron), 4 μl of Matrigel, 8 μl of 2× media, and 0.4 μl of 50 unit/ml thrombin in 0.1% BSA in PBS) [17]. The cell-hydrogel mixture was injected into polydimethylsiloxane (PDMS) molds cast from Teflon masters that were placed in 12-well plates. The molds were pre-treated with 0.2% (w/v) pluronic F-127 (Invitrogen) and fitted with laser-cut Cerex frames (9.2 × 9.5 mm outer dimensions, 6.8 × 8.3 mm inner dimensions). The cell-hydrogel mixture was polymerized within PDMS molds for 45 min at 37°C followed by addition of 2 ml culture media per well. Frames with polymerized cardiac bundles were removed from the molds the next day and cultured dynamically at a rocking platform in suspension for 14 days. Culture media were changed every other day.
2.3. Measurement of electrical propagation
Action potential propagation in cardiac bundles was optically mapped using our previously established methods [28, 31, 34]. Briefly, bundles were stained with a voltage-sensitive dye, Di-4 ANEPPS (10 μM), for 6 min and then incubated in Tyrode’s solution supplemented with 10 μM blebbistatin to prevent motion artifacts. Electrical activity was stimulated with a point electrode at a bundle end and was recorded in microscopic mode at 4× magnification using a 504-channel photodiode array (RedShirt Imaging). Data analysis for conduction velocity (CV) and action potential duration (APD) were performed by customized MATLAB software [17, 28, 31, 34–36].
2.4. Measurement of engineered tissue force
Passive tension and contractile force generation in response to electrical stimulation were recorded by mounting cardiac bundles onto a custom-made setup with force transducer and a computer-controlled linear actuator, as previously described [31, 32, 34, 37]. Briefly, frames cut to contain a single cardiac bundle were transferred to a chamber with Tyrode’s solution maintained at 37°C. The side of the frame was cut to allow cardiac bundle stretch by linear actuator to 24% above the resting culture length in 4% increments. Two Hz electrical stimulation was applied by a pair of platinum electrodes and generated isometric contractile force was measured after tissue was equilibrated for 5 min at each stretch increment. Contractile force traces were analyzed for maximum peak contractile force, twitch rise time and decay time, and passive force using a custom MATLAB program [28, 31, 34].
2.5. Imaging of calcium transients
For measurement of calcium transients, cardiac bundles were stained with intracellular calcium indicator Rhod-2 AM as previously described[38–41]. Briefly, cardiac bundles were washed in Tyrode’s solution, incubated with 6.25μg/ml Rhod-2 AM and 0.02% pluronic F-127 in media 199 for 45 minutes at 37°C, and then incubated with 10 μM blebbistatin in Tyrode’s solution to prevent motion artifacts. Cardiac bundles were placed in a heated live imaging chamber, stimulated at 2Hz using a point electrode and videos were acquired using a fast EMCCD camera (iXonEM., Andor) affixed to a Nikon microscope under 4× objective. Videos were analyzed using Andor Solis software and relative changes in fluorescence signal were calculated by a formula ΔF/F = (Fmax-Fbase)/(Fbase-Fbackground) [32].
2.6. Immunostaining and image analysis
Cardiac bundles were fixed with 2%v/v paraformaldehyde (Electron Microscopy Sciences) on a rocking platform overnight at 4°C. Fixed cardiac bundles were washed in PBS, blocked in antibody buffer (5%w/v chick serum, 0.5%v/v Triton X-100, in PBS) for 1 hr at room temperature, and then incubated with primary antibodies overnight at 4°C in antibody buffer. The following primary antibodies were used at indicated dilutions: α-sarcomeric actinin (Sigma A7811, 1:200), Vimentin (Abcam ab92547, 1:400), Collagen I (Abcam ab34710, 1:200), Connexin 43 (Abcam Ab 11370, 1:200), N-cadherin (Abcam Ab12221, 1:200), DsRed (Clontech, 1:200). Alexa-Fluor conjugated secondary antibodies (Invitrogen) and DAPI were applied at a 1:400 dilution in antibody buffer for 3hrs at room temperature. Cardiac bundles were washed in PBS, mounted on slides, and imaged using a Leica inverted SP5 confocal microscope. Image analysis was performed using ImageJ software.
2.7. Lentivirus production and transduction
High-titer lentivirus was produced using a second-generation lentiviral packaging system, as previously described [33, 35]. Briefly, 293FT cells (Life Technologies, R700-07) were co-transfected with lentiviral plasmid pRRL-CAG-mCherry-Puro (constructed from pRRL-CMV vector, a gift from Dr. Inder Verma, Salk Institute), packaging plasmid psPAX2 (Addgene, #12260), and envelope plasmid pMD2.G (Addgene, #12259) at 2:1:1 mass ratios using Lipofectamine 2000 (Life Technologies). Lentiviral particles containing supernatants were collected 3 days after transfection, centrifuged at 500 g for 10 minutes and filtered through 0.45 μm cellulose acetate filter (Corning), followed by incubation with Lenti-X Concentrator (Clontech) at 3:1 volume ratio overnight at 4°C. Concentrated lentiviral particles were harvested after centrifugation at 1500g for 45 minutes at 4°C, resuspended in DMEM, and stored in −80°C. Fetal or adult cardiac fibroblasts were transduced with the lentivirus at the density of 104 cells/cm2 in culture media (DMEM, 10% FBS, penicillin/streptomycin) with 8 ug/ml polybrene. Five days after transduction, 5ug/ml puromycin was added to select for successfully transduced cells for 5 days.
2.8. Real-time qPCR
Total RNA was extracted using RNeasy Fibrous Tissue Mini Kit according to the manufacturer’s instructions (Qiagen). Total RNA was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad). Standard qPCR reactions were performed with iTaq Universal SYBR Green Supermix (Bio-Rad) in the Applied Biosystems 7900HT Fast Real-time PCR System at the core facility of Duke Center for Genomic and Computational Biology. The primers used are listed in Supplementary Tables 1 and 2.
2.9. Statistics
Statistical significances were evaluated on the normalized data to the controls from each independent experiment by one-way ANOVA with post hoc Tukey’s test or unpaired t-test using GraphPad Prism. P<0.05 was considered statistically significant. Results are presented as mean ± SEM.
3. Results
3.1. Effect of fetal cardiac fibroblasts (FCFs) and adult cardiac fibroblasts (ACFs) on structure of cardiac bundles
In order to engineer a physiologically relevant 3D cardiac culture system, we encapsulated NRVMs in fibrin-based 3D tissue bundles (Suppl. Fig. 2A) that contracted vigorously (Suppl. Video 1) and exhibited advanced functional properties, as we recently reported [28]. The NRVMs in the interior of the bundles were uniformly aligned, cross-striated, and electrically and mechanically coupled via Connexin 43 and N-cadherin junctions (Suppl. Fig. 2B, C). Both control NRVM bundles and those with supplemented FCFs or ACFs exhibited a layer of vimentin+ cardiac fibroblasts at the outer bundle surface (Suppl. Fig. 3A–C), however, the number of fibroblasts residing interstitially between cardiomyocytes in the bundle interior was increased in the FCF and ACF compared to NRVM group. (Suppl. Fig. 3A′–C′). To investigate whether the exogenously added CFs were those that preferably located in the bundle interior, we lentivirally labeled FCFs and ACFs with a fluorescent protein mCherry and puromycin resistance gene. After puromycin selection, more than 90% of FCFs and ACFs were mCherry+ (Suppl. Fig. 4). When used to construct co-cultured bundles, almost all mCherry+ fibroblasts were found to reside throughout the bundle interior (Suppl. Fig. 5A–C), which was further confirmed in cross-sectional immunostainings (Suppl. Fig. 5D–F). Thus, upon initial bundle assembly with a uniform cell mixture, the exogenous FCFs and ACFs remained in interstitial spaces in the bundle interior, whereas the endogenous fibroblasts from NRVM isolation mainly occupied the outer surface of the bundle.
From quantitative analysis of bundle transverse sections (Fig. 1A–C), we found that the diameters of the +FCF and +ACF bundles were significantly larger compared to that of NRVM control bundles, with the +ACF bundles being the largest in size (Fig. 1D). Simultaneously, the +FCF bundles exhibited 37.3% more nuclei, whereas the +ACF bundles had 91.9% more nuclei than NRVM control bundles (Fig. 1E–H). Analysis of F-actin+ staining in the interior of the bundles demonstrated similar cardiac muscle areas in all bundle groups (Fig. 1I–L), with +ACF bundles exhibited significantly higher Vimentin+ area than +FCF or NRVM control bundles (Fig. 1M–P). To further assess FCF and ACF induced changes in cellular composition, we counted nuclei in: 1) the F-actin+ area (corresponding to cardiomyocytes [28]), 2) the vimentin+ area at the periphery (dominated by endogenous CFs) and 3) the mCherry+ area (corresponding to exogenous CFs, Suppl. Fig. 5D–F) of bundle cross-sections. We found that adding FCFs or ACFs did not alter CM nuclei counts in bundles (Suppl. Fig. 5G). While adding FCFs slightly but insignificantly increased numbers of endogenous CFs, this increase was significant in the presence of ACFs (Suppl. Fig. 5H). Furthermore, the number of ACFs in bundles was significantly higher than the number of FCFs (Supl. Fig. 5I). From the results in Suppl. Figs. 1 and 5G–I, we could also estimate relative changes in cell numbers during the 2-week bundle culture. Specifically, endogenous CF:CM ratio was only marginally increased in control bundles (from ~1:9 to ~1:8) and significantly increased in +ACF bundles (from ~1:9 to ~2:7). While the FCF:CM ratio remained unchanged (~1:3) during bundle culture, the ACF:CM ratio doubled (from ~1:3 to ~2:3). Overall, these analyses suggested that the presence of FCFs in cardiac bundles had minor to no effects on the number of CMs or endogenous CFs, while relative fractions of both ACFs and endogenous CFs were increased in the +ACF bundles.
Figure 1. Effect of FCFs and ACFs on cellular composition of cardiac bundles.
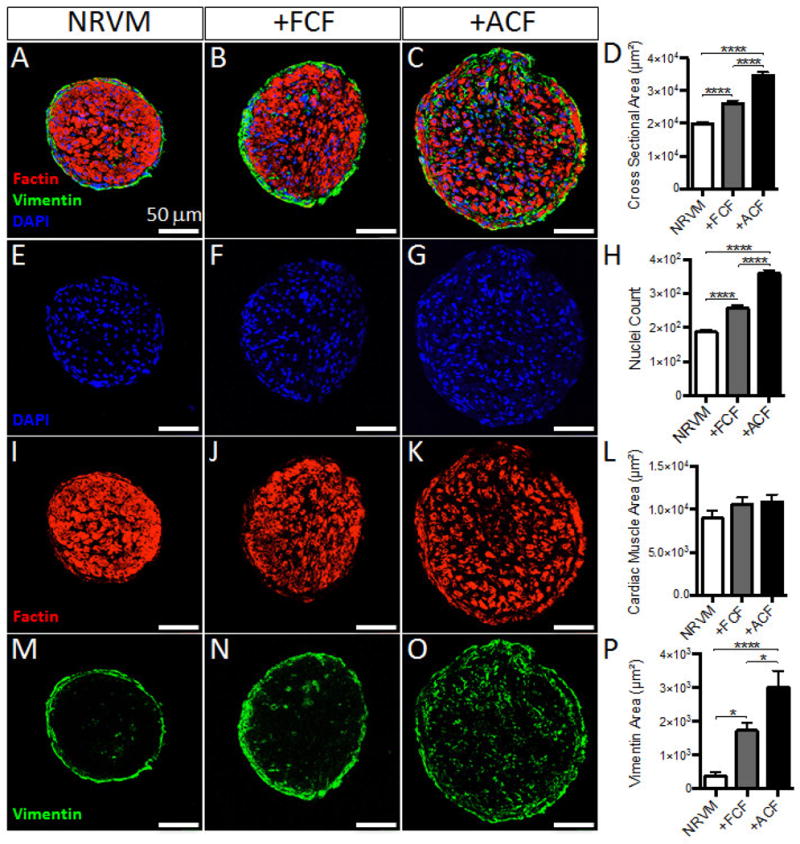
(A–C) Representative cross sections of NRVM, +FCF and +ACF bundles, stained for filamentous actin (Factin, red), Vimentin (green) and DNA (DAPI, blue). (D) Total cross sectional areas of cardiac bundles. (E–H) DAPI stainings from A–C, shown as example images used for quantification of nuclei counts per bundle cross-section (H). (I–L) Factin stainings from A–C, shown as example images used for quantification of cardiac muscle area (excluding the positive staining within outer vimentin+ area) per bundle cross-section (L). (M–P) Vimentin stainings from A–C, shown as example images used for quantification of Vimentin+ area per bundle cross-section (n=9 bundles per group). *P < 0.05, ****P < 0.0001.
3.2. Effect of FCFs and ACFs on electrical and mechanical function of cardiac bundles
We optically mapped action potential propagation in cardiac bundles using a voltage-sensitive membrane dye (di-4 ANEPPS) and applying point stimulus at one end of the bundle (Fig. 2A–B). The +ACF bundles exhibited significantly slower conduction velocity (CV) and longer action potential duration (APD) than both +FCF and NRVM bundles (32.95 ± 2.33 cm/s vs. 40.34 ± 2.70 cm/s and 39.43 ± 1.20 cm/s and 228.90 ± 19.81 ms vs. 164.90 ± 12.58 ms and 176.00 ± 7.42 ms, respectively, Fig. 2C–D). On the other hand, all 3 groups had similar maximum capture rates of ~6 Hz (Fig. 2E). To assess the mechanical function of the engineered bundles, we measured their contractile forces (Fig. 3A) and the force-length relationships (Fig. 3B). The +ACF bundles generated significantly lower contractile forces than +FCF and NRVM bundles (0.82 ± 0.20 mN vs. 1.54 ± 0.18 mN and 1.31 ± 0.13 mN, respectively, Fig. 3C) and significantly higher passive forces at 20% stretch (6.05 ± 1.63 mN vs. 2.29 ± 0.64 mN and 3.21 ± 0.86 mN, respectively, Fig. 3D). Consistent with increased stiffness, +ACF bundles exhibited significantly higher collagen I deposition (Fig. 3E–H). Notably, the collagen deposition in +ACF bundles was present in between aligned NRVMs, closely resembling the histology of interstitial fibrosis. Furthermore, the +ACF bundles exhibited significantly prolonged twitch kinetics than +ACF and NRVM bundles (rise time: 62.19 ± 0.85 ms vs. 44.13 ± 0.73 ms and 46.53 ± 0.40 ms, respectively, Fig. 3I and decay time: 147.56 ± 2.44 ms vs. 120.47 ± 2.77 ms and 117.12 ± 1.71 ms, respectively, Fig. 3J). We further examined whether altered calcium handling could be one of underlying causes of the lower force generation capacity of +ACF bundles, and found that they exhibited a significantly reduced Ca2+ transient amplitude (Fig. 3K, L). Overall, ACFs, unlike FCFs, exerted detrimental effects on the electrical and mechanical function of 3D engineered cardiac tissue bundles.
Figure 2. Effect of FCFs and ACFs on electrical function of cardiac bundles.
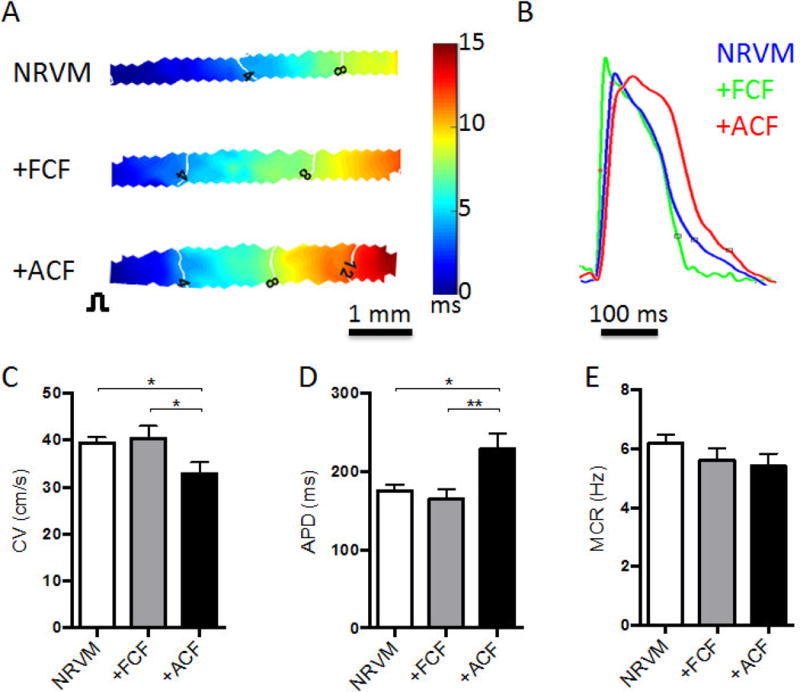
(A) Representative isochrone maps of action potential propagation in response to 2 Hz point stimulation (pulse sign, left) in NRVM, +FCF, and +ACF bundles. (B) Representative optical action potential traces from cardiac bundles stained with di-4 ANEPPS and electrically stimulated at 2 Hz. (C–E) Conduction velocity (CV, C), action potential duration (APD, D), and maximum capture rate (MCR, E) in bundles (N=4 independent experiments, n=10–12 bundles per group). *P < 0.05, **P < 0.01.
Figure 3. Effects of FCFs and ACFs on mechanical function of cardiac bundles.
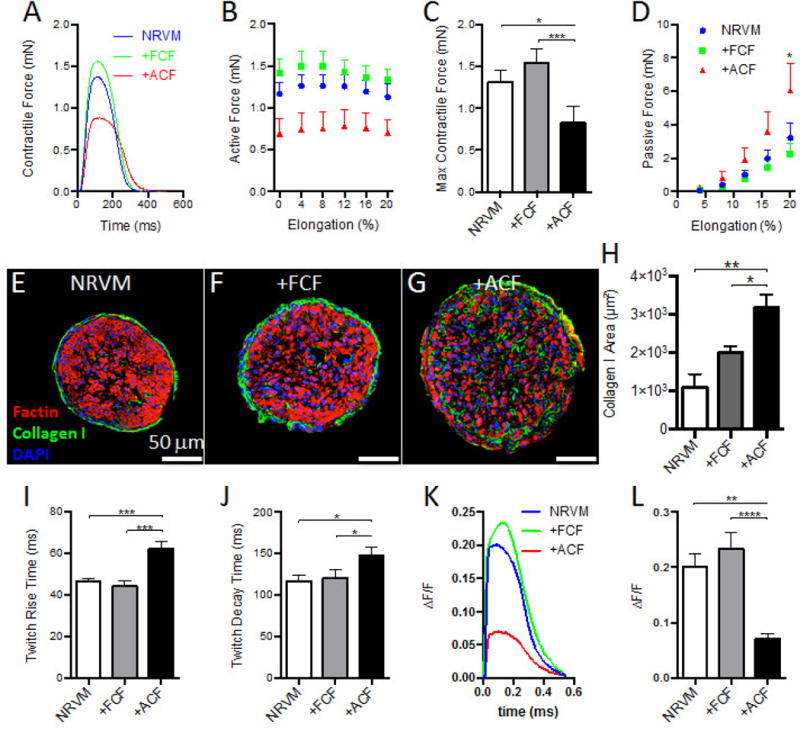
(A) Representative traces of contractile forces recorded from NRVM (blue trace), +FCF (green trace) and +ACF (red trace) bundles paced at 1 Hz by field electrodes. (B) Active force curves as a function of tissue elongation. (C) Maximum contractile forces recorded during 1 Hz pacing at optimal tissue length. (D) Passive force curves as a function of tissue elongation (N=4 independent experiments, n=15–18 bundles per group for B–D). (E–G) Representative cross sections of NRVM, +FCF and +ACF bundles stained for filamentous actin (Factin, red), Collagen I (green), and DNA (DAPI, blue). (H) Quantified Collagen I+ area per bundle cross-section (n=4–6 bundles per group). (I) Twitch rise time measured between 10% and 90% of peak amplitude. (J) Twitch decay time measured between 90% and 10% of peak amplitude (N=4 independent experiments, n=15–18 bundles per group for I–J). (K) Representative traces of calcium transients measured by ΔF/F of Rhod2-AM florescent signals recorded from NRVM, +FCF, and +ACF bundles at 2 Hz by field electrodes. (L) Calcium transient amplitude (N=3 independent experiments, n=10–12 bundles per group). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
3.3. Gene expression changes in cardiomyocytes co-cultured with FCFs and ACFs
We assessed expression of cardiomyocyte genes associated with sarcomeric structure, electromechanical function, and calcium handling (Fig. 4A). Consistent with decreased force generating capacity and Ca2+ transient amplitude in +ACF bundles, the expression levels of genes associated with mechanical function, calcium handing, and E–C coupling, including SERCA2 (Atp2a2; Fig. 4B), Ryanodine receptor 2 (Ryr2; Fig. 4C), Phospholamban (Pln; Fig. 4D), NCX (Slc8a1; Fig. 4E), Cav1.2 (Cacna1c; Fig. 4F) and Cav3.2 (Cacna1h; Fig. 4G), were all significantly decreased in +ACF compared to +FCF and NRVM bundles. In addition, the sarcomeric gene alpha myosin heavy chain (Myh6; Fig. 4H) was significantly downregulated in +ACF compared to +FCF bundles. Furthermore, consistent with significantly slower CV in +ACF bundles, the gap junctional Connexin 43 (Gja1; Fig. 4K) and cardiac sodium channel Nav1.5 (Scn5a; Fig. 4I) were significantly downregulated in +ACF vs. +FCF bundles. No statistical difference was found for potassium channel Kv4.3 (Kcnd3; Fig. 4J) expression. Notably, compared to NRVM control bundles, most genes associated with cardiomyocyte mechanical function and structure were significantly upregulated in +FCF bundles (Fig. 4), indicating a likely beneficial effect of FCFs on cardiomyocyte maturation.
Figure 4. Effect of FCFs and ACFs on gene expression profile of NRVMs in cardiac bundles.
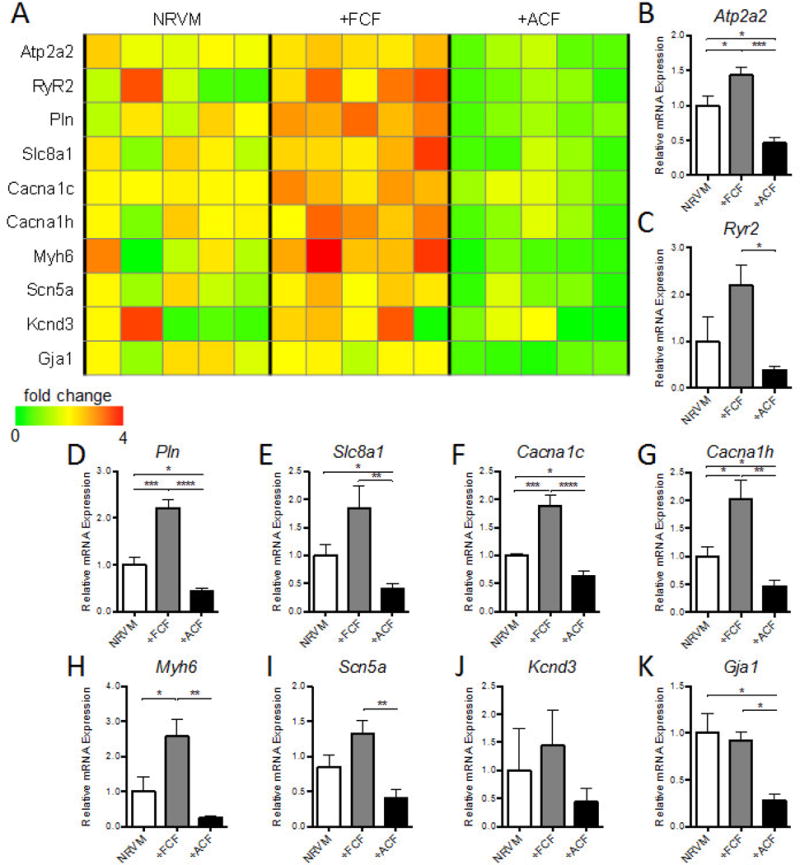
(A) Heat map of cardiac structural and functional gene expression in NRVM, +FCF, and +ACF bundles, with bar graphs showing summarized data for Atp2a2 (B), Ryr2 (C), Pln (D), Slc8a1 (E), Cacna1c (F), Cacna1h (G), Myh6 (H), Scn5a (I), Kcnd3 (J) and Gja1 (K). The relative mRNA expression was normalized to house keeping gene B2M and shown relative to NRVM controls as fold change (N=5 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
3.4. Molecular differences between cultured FCFs and ACFs
To reveal potential molecular differences between ACFs and FCFs that may have contributed to their distinct effects on cardiomyocytes, we examined expression of key genes coding ECM proteins and select secreted factors after fibroblasts were expanded in monolayers for 2–4 passages and prior to bundle culture (Fig. 5A). We found that ECM genes associated with fibrosis including Collagen I (Col1a1; Fig. 5B) and Collagen III (Col3a1; Fig. 5C), but not Fibronectin (Fn1; Fig. 5D) were strongly upregulated in ACFs vs. FCFs, which was consistent with the elevated collagen I deposition observed in the +ACF bundles (Fig. 3E–H). Notably, Periostin (Postn; Fig. 5E) and Smooth muscle actin (Acta2; Fig. 5F), key markers of activated fibroblasts [14, 42–45], were dramatically upregulated in ACFs. Connective tissue growth factor (Ctgf; Fig. 5G), a secreted factor associated with fibrosis, also showed strong upregulation in ACFs, whereas heparin-binding EGF like growth factor (Hbegf; Fig. 5H) and insulin-like growth factor 1 (Igf1; Fig. 5I) showed decreased trends without reaching significant difference. We further examined if the expanded FCFs and ACFs differed in their expression of senescence-related genes including Glb1 encoding beta-galactosidase [46, 47], Cdkn1a encoding p21 [48], proliferation marker Mki67, and several senescence associated secretory phenotype (SASP) genes [49] (Suppl. Fig. 6). The Glb1 expression was very low (Suppl. Fig. 6A) and comparable between FCFs and ACFs, which together with their similar expressions of Mki67 (Suppl. Fig. 6C), suggested that both cell types remained non-senescent. Furthermore, expressions of p21 and SASP related genes (Suppl. Figs. 6B and D–L) did not show a consistent trend in FCFs vs. ACFs, suggesting no major differences in their senescent state. Of note is also that some of the observed gene expression differences could stem from the inherently distinct expression profiles of FCFs and ACFs [50]. Overall, while monolayer expansion for 2–4 passages did not appear to induce cell senescence, ACFs, relative to FCFs, exhibited molecular signatures of an activated fibroblast phenotype found in cardiac fibrotic diseases [4].
Figure 5. Differential gene expression in cultured FCFs and ACFs.
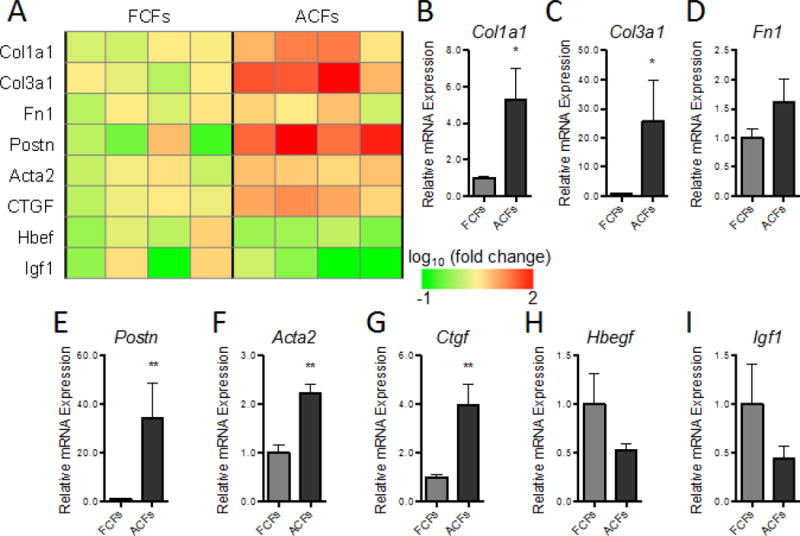
(A) Heat map of gene expression for a panel of ECM and growth factor genes in FCF and ACF monolayers expanded for 2–4 passages, with bar graphs showing summarized data for Col1a1 (B), Col3a1 (C), Fn1 (D), Postn (E), Acta2 (F), Ctgf (G), Hbegf (H), and Igf1 (I). The relative mRNA expression was normalized to house keeping gene B2M and shown relative to FCFs as fold change (N=4 independent experiments). *P < 0.05, **P < 0.01.
4. Discussion
We engineered cardiac tissue bundles to investigate whether the age of cardiac fibroblasts is a factor that can significantly influence the functional and molecular properties of co-cultured cardiomyocytes. The 3D bundles represent the native myocardium more faithfully than traditional 2D culture platforms [21, 22], while allowing the quantification of both electrophysiological and mechanical function at the tissue level. Using this system, we showed for the first time that adult cardiac fibroblasts deteriorate electrophysiological and mechanical function of NRVM tissues via downregulated expression of important ion channels, electrical coupling, calcium handling, and contraction related genes. In contrast, fetal cardiac fibroblasts either improved or did not change various functional and molecular parameters in co-cultured cardiomyocytes. Functional deterioration in cardiac bundles containing adult fibroblasts was associated with pronounced interstitial fibrosis and could be at least in part attributed to a pathological activation of fibroblasts that showed increased expression of extracellular pro-fibrotic factors.
Others and we have previously shown the beneficial effects of fetal or neonatal fibroblasts on the formation and function of 3D engineered tissues made of cardiomyocytes derived from mouse or human pluripotent stem cells [31, 34, 51–56]. Specifically, within a hydrogel environment, highly purified stem cell-derived cardiomyocytes are generally unable to spread and form functional 3D syncytium without addition of stromal cells, such as cardiac fibroblasts. In contrast, primary neonatal rat ventricular tissue isolates used in this study already contain a low percentage of endogenous fibroblasts sufficient to support 3D cardiac tissue formation and advanced electrical and mechanical function akin to those of native adult myocardium [28]. Thus, the observed functional effects from adding exogenous fetal or adult cardiac fibroblasts to cardiac bundles were primarily a consequence of their distinct action on cardiomyocytes rather than on bundle formation.
It is well documented that the heart undergoes complex structural remodeling to develop from an immature fetal to a mature adult myocardium [57] and cardiac fibroblasts are known to play an important role in that process [4, 58]. It was tempting to speculate that adult cardiac fibroblasts would promote cardiomyocyte function and maturation by providing an adult-like microenvironment, a notion also of direct relevance for implantation of immature stem cell-derived cardiomyocytes into adult myocardium. However, our results demonstrated that while fetal cardiac fibroblasts subcultured in vitro enhanced expression of functional cardiomyocyte genes, similarly cultured adult cardiac fibroblasts exerted a pro-fibrotic action on cardiac bundles as evidenced by the slower conduction velocity, prolonged action potential duration, attenuated contractile force generation, higher stiffness, and decreased calcium transient amplitude. The deteriorated cardiac function was associated with structural and molecular remodeling, including significant interstitial presence of adult cardiac fibroblasts and deposited collagen, as well as downregulated expression of cardiac functional and structural genes. Overall, our findings suggest that fetal, rather than adult, cardiac fibroblasts are superior supporting cell source for cardiac tissue engineering, based on their favorable expression of growth factors and ECM proteins and the lack of pathological activation during 2D expansion or 3D culture. Of note, the miniaturized cardiac tissue bundles used in this study require relatively low cell numbers (e.g. 2.25 × 105 NRVMs and 0.675 × 105 fibroblasts) and can be readily applied to investigate how other non-myocyte sources (e.g. vascular cells, other stromal cells, mesenchymal stem cells) directly interact with cardiomyocytes in a 3D biomimetic environment to impact cardiac structure, function, or maturation.
Different behaviors of fetal and adult fibroblasts within cardiac bundles may be attributed to their distinct phenotypes attained during 2D expansion (Fig. 5) and/or specific reactions to the 3D microenvironment (ECM composition, oxygen tension). Furthermore, their resulting effects on cardiomyocytes are likely multifarious, potentially involving paracrine, cell contact, and ECM mediated mechanisms [59–61]. To date, only 2 studies suggested the age-dependent effects of cardiac fibroblast secreted factors or ECM proteins on cardiomyocytes.
Using a 2D co-culture system, Ieda et al. reported that fetal cardiac fibroblasts induced embryonic cardiomyocyte proliferation through secretion of fibronectin, collagen III, and HBEGF, whereas adult cardiac fibroblasts stimulated embryonic cardiomyocyte hypertrophy [27]. Williams et al. showed that compared to adult cardiac ECM coated substrates, fetal cardiac ECM coated substrates enhanced adhesion and proliferation of NRVMs in vitro, and that fetal cardiac ECM is dominated by fibronectin, whereas adult cardiac ECM is dominated by collagen I [62]. Similar to these studies, our gene expression analysis showed increased expression of collagen I and decreased expression of HBEGF in adult compared to fetal cardiac fibroblasts; however, we observed increased expression of collagen III and no significant differences in their fibronectin expression or effect on cardiomyocyte proliferation. Furthermore, adult (but not fetal) cardiac fibroblasts significantly proliferated in bundles and appeared to stimulate the proliferation of endogenous fibroblasts.
Fibroblast number and percentage in the heart increase with development [50, 63, 64]. While in the fetal and neonatal hearts fibroblasts are proliferative, in the healthy adult heart, they are mostly quiescent [63, 65]; however, in response to pathological stimuli, adult fibroblasts activate and differentiate into proliferative, α-smooth muscle actin (αSMA) expressing myofibroblasts, leading to dysregulated ECM deposition and fibrosis, and eventually, deterioration of cardiac function[1, 45, 66, 67]. Previous studies have shown that even during in vitro monolayer culture, adult cardiac fibroblasts from normal hearts can attain activated phenotype [14, 68, 69]. Cartledge et al. specifically reported that in monolayer cultures, adult fibroblasts isolated from normal rat hearts affected the morphology and calcium transient of cardiomyocytes in a similar manner as the freshly isolated myofibroblasts from pressure-overload induced hypertrophic hearts [14]. Most recently, periostin has emerged to be an important marker for activated cardiac fibroblasts (myofibroblasts) in different cardiac pathologies [42, 70]. In our study, the most dramatically elevated gene in adult vs. fetal cardiac fibroblast monolayers was periostin (more than 30-fold), which along with upregulated expression of Acta2 (αSMA), CTGF, Collagen I and Collagen III, suggested that the cultured adult cardiac fibroblasts attained an activated, pathological-like phenotype [42, 70–73]. This activated phenotype was further evidenced by increased fibroblast proliferation, interstitial collagen deposition, and functional deficit in 3D bundles. Detailed mechanistic studies to uncover key factors for increased propensity of adult vs. fetal cardiac fibroblasts to attain a pathological phenotype in vitro and negatively affect cardiomyocyte function warrant further investigations and may lead to fibroblast-specific interventions to reduce or halt progression of cardiac fibrosis.
The neonatal rat ventricular myocytes have been widely used for in vitro studies of cardiac development, signaling, and pathophysiology in both 2D and 3D culture settings [28, 74–76]. While our 3D co-cultures of neonatal rat ventricular myocytes and mouse cardiac fibroblasts represent a cross-species platform, they are expected to enable future studies of how cardiac non-myocytes from various mouse models of congenital or acquired heart diseases affect cardiomyocyte function in vitro. Cross-species 2D co-cultures of cardiac myocytes and fibroblasts have been utilized previously in numerous studies [77–81]. Although there may exist some incompatibilities between mouse and rat cells, our same-species bundle co-cultures of NRVMs with rat neonatal or adult cardiac fibroblasts replicated the key functional findings we observed in the mouse-rat co-culture setting (Suppl. Fig. 7). Furthermore, comparing the results for adult, neonatal, and fetal fibroblasts in Suppl. Figs. 7A–C, and Figs. 3C and 2C&D suggests a general negative trend in the effect of fibroblast age on in vitro NRVM function.
In summary, we engineered a physiologically relevant, 3D co-culture system, which allowed us to investigate direct cardiomyocyte-fibroblast functional crosstalk in a cardiomimetic tissue-like microenvironment [23–26]. Through these studies, we provided the first evidence that the age of cardiac fibroblasts is a strong determinant of the structure, function, and molecular properties of engineered cardiac tissues and that key features of fibrotic myocardium can be replicated in vitro by supplementing NRVMs with cultured adult rather than fetal cardiac fibroblasts. These results open doors to future mechanistic studies of how age and source of non-myocytes impact cardiac maturation, function, and pathology.
Supplementary Material
Statement of Significance.
Previous studies have shown that cardiomyocytes and fibroblasts in the heart interact through direct contacts, paracrine factors, and matrix-mediated crosstalk. However, whether cardiac fibroblasts of different ages distinctly impact cardiomyocyte function remains elusive. We employed a tissue-engineered hydrogel-based co-culture system to study interactions of cardiomyocytes with fetal or adult cardiac fibroblasts. We show that the age of cardiac fibroblasts is a strong determinant of the structure, function, and molecular properties of engineered cardiac tissues and that key features of fibrotic myocardium are replicated by supplementing cardiomyocytes with adult but not fetal fibroblasts. These findings relate to implantation of stem cell-derived cardiomyocytes in adult myocardium and warrant further studies of how age and source of non-myocytes impact cardiac function and maturation.
Acknowledgments
We acknowledge the Light Microscopy Core Facility (LMCF), the Flow Cytometry Shared Resource (FCSR), and Genomic and Computational Biology (GCB) at Duke University for the use of their equipment as well as C. Jackman, M. Juhas, S. S. Bjergaard, A. Krol, L. Li, and S. Okuwa for their technical assistance. This study was supported by the National Institutes of Health grants [HL104326, HL126524, and HL132389, and HL134764] to N.B. and Morton Friedman Fellowship from Duke Biomedical Engineering and American Heart Association Pre-doctoral Fellowship to Y. L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None.
Uncategorized References
- 1.Baudino TA, Carver W, Giles W, Borg TK. Cardiac fibroblasts: friend or foe? Am J Physiol Heart Circ Physiol. 2006;291:H1015–1026. doi: 10.1152/ajpheart.00023.2006. [DOI] [PubMed] [Google Scholar]
- 2.Bursac N. Cardiac fibroblasts in pressure overload hypertrophy: the enemy within? J Clin Invest. 2014;124:2850–2853. doi: 10.1172/JCI76628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furtado MB, Nim HT, Boyd SE, Rosenthal NA. View from the heart: cardiac fibroblasts in development, scarring and regeneration. Development. 2016;143:387–397. doi: 10.1242/dev.120576. [DOI] [PubMed] [Google Scholar]
- 4.Gourdie RG, Dimmeler S, Kohl P. Novel therapeutic strategies targeting fibroblasts and fibrosis in heart disease. Nat Rev Drug Discov. 2016 doi: 10.1038/nrd.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nag AC. Study of non-muscle cells of the adult mammalian heart: a fine structural analysis and distribution. Cytobios. 1980;28:41–61. [PubMed] [Google Scholar]
- 6.Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, Malm T, Andra M, Jashari R, Nyengaard JR, Possnert G, Jovinge S, Druid H, Frisen J. Dynamics of Cell Generation and Turnover in the Human Heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol. 2007;293:H1883–1891. doi: 10.1152/ajpheart.00514.2007. [DOI] [PubMed] [Google Scholar]
- 8.Souders CA, Borg TK, Banerjee I, Baudino TA. Pressure overload induces early morphological changes in the heart. Am J Pathol. 2012;181:1226–1235. doi: 10.1016/j.ajpath.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camelliti P, Borg TK, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res. 2005;65:40–51. doi: 10.1016/j.cardiores.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 10.Kohl P, Gourdie RG. Fibroblast-myocyte electrotonic coupling: does it occur in native cardiac tissue? J Mol Cell Cardiol. 2014;70:37–46. doi: 10.1016/j.yjmcc.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter KE, Turner NA. Cardiac fibroblasts: at the heart of myocardial remodeling. Pharmacol Ther. 2009;123:255–278. doi: 10.1016/j.pharmthera.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Zhang P, Su J, Mende U. Cross talk between cardiac myocytes and fibroblasts: from multiscale investigative approaches to mechanisms and functional consequences. Am J Physiol Heart Circ Physiol. 2012;303:H1385–1396. doi: 10.1152/ajpheart.01167.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogle BM, Bursac N, Domian I, Huang NF, Menasche P, Murry CE, Pruitt B, Radisic M, Wu JC, Wu SM, Zhang J, Zimmermann WH, Vunjak-Novakovic G. Distilling complexity to advance cardiac tissue engineering. Sci Transl Med. 2016;8:342ps313. doi: 10.1126/scitranslmed.aad2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cartledge JE, Kane C, Dias P, Tesfom M, Clarke L, McKee B, Al Ayoubi S, Chester A, Yacoub MH, Camelliti P, Terracciano CM. Functional crosstalk between cardiac fibroblasts and adult cardiomyocytes by soluble mediators. Cardiovasc Res. 2015;105:260–270. doi: 10.1093/cvr/cvu264. [DOI] [PubMed] [Google Scholar]
- 15.Yue L, Xie J, Nattel S. Molecular determinants of cardiac fibroblast electrical function and therapeutic implications for atrial fibrillation. Cardiovasc Res. 2011;89:744–753. doi: 10.1093/cvr/cvq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasquez C, Mohandas P, Louie KL, Benamer N, Bapat AC, Morley GE. Enhanced fibroblast-myocyte interactions in response to cardiac injury. Circ Res. 2010;107:1011–1020. doi: 10.1161/CIRCRESAHA.110.227421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedrotty DM, Klinger RY, Kirkton RD, Bursac N. Cardiac fibroblast paracrine factors alter impulse conduction and ion channel expression of neonatal rat cardiomyocytes. Cardiovasc Res. 2009;83:688–697. doi: 10.1093/cvr/cvp164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zlochiver S, Munoz V, Vikstrom KL, Taffet SM, Berenfeld O, Jalife J. Electrotonic myofibroblast-to-myocyte coupling increases propensity to reentrant arrhythmias in two-dimensional cardiac monolayers. Biophys J. 2008;95:4469–4480. doi: 10.1529/biophysj.108.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miragoli M, Salvarani N, Rohr S. Myofibroblasts induce ectopic activity in cardiac tissue. Circ Res. 2007;101:755–758. doi: 10.1161/CIRCRESAHA.107.160549. [DOI] [PubMed] [Google Scholar]
- 20.Gaudesius G, Miragoli M, Thomas SP, Rohr S. Coupling of cardiac electrical activity over extended distances by fibroblasts of cardiac origin. Circ Res. 2003;93:421–428. doi: 10.1161/01.RES.0000089258.40661.0C. [DOI] [PubMed] [Google Scholar]
- 21.Abbott A. Cell culture: biology’s new dimension. Nature. 2003;424:870–872. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 22.Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerecht-Nir S, Radisic M, Park H, Cannizzaro C, Boublik J, Langer R, Vunjak-Novakovic G. Biophysical regulation during cardiac development and application to tissue engineering. Int J Dev Biol. 2006;50:233–243. doi: 10.1387/ijdb.052041sg. [DOI] [PubMed] [Google Scholar]
- 24.Hirt MN, Hansen A, Eschenhagen T. Cardiac tissue engineering: state of the art. Circ Res. 2014;114:354–367. doi: 10.1161/CIRCRESAHA.114.300522. [DOI] [PubMed] [Google Scholar]
- 25.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 26.Kurokawa YK, George SC. Tissue engineering the cardiac microenvironment: Multicellular microphysiological systems for drug screening. Adv Drug Deliv Rev. 2016;96:225–233. doi: 10.1016/j.addr.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackman CP, Carlson AL, Bursac N. Dynamic culture yields engineered myocardium with near-adult functional output. Biomaterials. 2016;111:66–79. doi: 10.1016/j.biomaterials.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bursac N, Papadaki M, Cohen RJ, Schoen FJ, Eisenberg SR, Carrier R, Vunjak-Novakovic G, Freed LE. Cardiac muscle tissue engineering: toward an in vitro model for electrophysiological studies. Am J Physiol. 1999;277:H433–444. doi: 10.1152/ajpheart.1999.277.2.H433. [DOI] [PubMed] [Google Scholar]
- 30.Bian W, Liau B, Badie N, Bursac N. Mesoscopic hydrogel molding to control the 3D geometry of bioartificial muscle tissues. Nat Protoc. 2009;4:1522–1534. doi: 10.1038/nprot.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D, Shadrin IY, Lam J, Xian HQ, Snodgrass HR, Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials. 2013;34:5813–5820. doi: 10.1016/j.biomaterials.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juhas M, Engelmayr GC, Jr, Fontanella AN, Palmer GM, Bursac N. Biomimetic engineered muscle with capacity for vascular integration and functional maturation in vivo. Proc Natl Acad Sci U S A. 2014;111:5508–5513. doi: 10.1073/pnas.1402723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madden L, Juhas M, Kraus WE, Truskey GA, Bursac N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. eLife. 2015;4:e04885. doi: 10.7554/eLife.04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liau B, Christoforou N, Leong KW, Bursac N. Pluripotent stem cell-derived cardiac tissue patch with advanced structure and function. Biomaterials. 2011;32:9180–9187. doi: 10.1016/j.biomaterials.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen HX, Kirkton RD, Bursac N. Engineering prokaryotic channels for control of mammalian tissue excitability. Nat Commun. 2016;7:13132. doi: 10.1038/ncomms13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkton RD, Bursac N. Engineering biosynthetic excitable tissues from unexcitable cells for electrophysiological and cell therapy studies. Nat Commun. 2011;2:300. doi: 10.1038/ncomms1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juhas M, Bursac N. Roles of adherent myogenic cells and dynamic culture in engineered muscle function and maintenance of satellite cells. Biomaterials. 2014;35:9438–9446. doi: 10.1016/j.biomaterials.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christoforou N, Liau B, Chakraborty S, Chellapan M, Bursac N, Leong KW. Induced pluripotent stem cell-derived cardiac progenitors differentiate to cardiomyocytes and form biosynthetic tissues. PLoS One. 2013;8:e65963. doi: 10.1371/journal.pone.0065963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin N, Badie N, Yu L, Abraham D, Cheng H, Bursac N, Rockman HA, Wolf MJ. A method to measure myocardial calcium handling in adult Drosophila. Circ Res. 2011;108:1306–1315. doi: 10.1161/CIRCRESAHA.110.238105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Sun AY, Kim JJ, Graham V, Finch EA, Nepliouev I, Zhao G, Li T, Lederer WJ, Stiber JA, Pitt GS, Bursac N, Rosenberg PB. STIM1-Ca2+ signaling modulates automaticity of the mouse sinoatrial node. Proc Natl Acad Sci U S A. 2015;112:E5618–5627. doi: 10.1073/pnas.1503847112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liau B, Zhang D, Bursac N. Functional cardiac tissue engineering. Regen Med. 2012;7:187–206. doi: 10.2217/rme.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, SC JL, Aronow BJ, Tallquist MD, Molkentin JD. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun. 2016;7:12260. doi: 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaur H, Takefuji M, Ngai CY, Carvalho J, Bayer J, Wietelmann A, Poetsch A, Hoelper S, Conway SJ, Mollmann H, Looso M, Troidl C, Offermanns S, Wettschureck N. Targeted Ablation of Periostin-Expressing Activated Fibroblasts Prevents Adverse Cardiac Remodeling in Mice. Circ Res. 2016;118:1906–1917. doi: 10.1161/CIRCRESAHA.116.308643. [DOI] [PubMed] [Google Scholar]
- 44.Moore-Morris T, Cattaneo P, Puceat M, Evans SM. Origins of cardiac fibroblasts. J Mol Cell Cardiol. 2016;91:1–5. doi: 10.1016/j.yjmcc.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeisberg EM, Kalluri R. Origins of cardiac fibroblasts. Circ Res. 2010;107:1304–1312. doi: 10.1161/CIRCRESAHA.110.231910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, Kleijer WJ, DiMaio D, Hwang ES. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 47.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 48.Jurk D, Wang C, Miwa S, Maddick M, Korolchuk V, Tsolou A, Gonos ES, Thrasivoulou C, Saffrey MJ, Cameron K, von Zglinicki T. Postmitotic neurons develop a p21-dependent senescence-like phenotype driven by a DNA damage response. Aging Cell. 2012;11:996–1004. doi: 10.1111/j.1474-9726.2012.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kensah G, Roa Lara A, Dahlmann J, Zweigerdt R, Schwanke K, Hegermann J, Skvorc D, Gawol A, Azizian A, Wagner S, Maier LS, Krause A, Drager G, Ochs M, Haverich A, Gruh I, Martin U. Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro. European heart journal. 2013;34:1134–1146. doi: 10.1093/eurheartj/ehs349. [DOI] [PubMed] [Google Scholar]
- 52.Xi J, Khalil M, Spitkovsky D, Hannes T, Pfannkuche K, Bloch W, Saric T, Brockmeier K, Hescheler J, Pillekamp F. Fibroblasts support functional integration of purified embryonic stem cell-derived cardiomyocytes into avital myocardial tissue. Stem Cells Dev. 2011;20:821–830. doi: 10.1089/scd.2010.0398. [DOI] [PubMed] [Google Scholar]
- 53.Matsuura K, Masuda S, Haraguchi Y, Yasuda N, Shimizu T, Hagiwara N, Zandstra PW, Okano T. Creation of mouse embryonic stem cell-derived cardiac cell sheets. Biomaterials. 2011;32:7355–7362. doi: 10.1016/j.biomaterials.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 54.Pfannkuche K, Neuss S, Pillekamp F, Frenzel LP, Attia W, Hannes T, Salber J, Hoss M, Zenke M, Fleischmann BK, Hescheler J, Saric T. Fibroblasts facilitate the engraftment of embryonic stem cell-derived cardiomyocytes on three-dimensional collagen matrices and aggregation in hanging drops. Stem Cells Dev. 2010;19:1589–1599. doi: 10.1089/scd.2009.0255. [DOI] [PubMed] [Google Scholar]
- 55.Thavandiran N, Dubois N, Mikryukov A, Masse S, Beca B, Simmons CA, Deshpande VS, McGarry JP, Chen CS, Nanthakumar K, Keller GM, Radisic M, Zandstra PW. Design and formulation of functional pluripotent stem cell-derived cardiac microtissues. Proc Natl Acad Sci U S A. 2013;110:E4698–4707. doi: 10.1073/pnas.1311120110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaaf S, Shibamiya A, Mewe M, Eder A, Stohr A, Hirt MN, Rau T, Zimmermann WH, Conradi L, Eschenhagen T, Hansen A. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PloS one. 2011;6:e26397. doi: 10.1371/journal.pone.0026397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2014;114:511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furtado MB, Costa MW, Pranoto EA, Salimova E, Pinto AR, Lam NT, Park A, Snider P, Chandran A, Harvey RP, Boyd R, Conway SJ, Pearson J, Kaye DM, Rosenthal NA. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res. 2014;114:1422–1434. doi: 10.1161/CIRCRESAHA.114.302530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tirziu D, Giordano FJ, Simons M. Cell communications in the heart. Circulation. 2010;122:928–937. doi: 10.1161/CIRCULATIONAHA.108.847731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takeda N, Manabe I. Cellular Interplay between Cardiomyocytes and Nonmyocytes in Cardiac Remodeling. Int J Inflam. 2011;2011:535241. doi: 10.4061/2011/535241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan D, Takawale A, Lee J, Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair. 2012;5:15. doi: 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams C, Quinn KP, Georgakoudi I, Black LD., 3rd Young developmental age cardiac extracellular matrix promotes the expansion of neonatal cardiomyocytes in vitro. Acta biomaterialia. 2014;10:194–204. doi: 10.1016/j.actbio.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen W, Frangogiannis NG. Fibroblasts in post-infarction inflammation and cardiac repair. Biochim Biophys Acta. 2013;1833:945–953. doi: 10.1016/j.bbamcr.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baum J, Duffy HS. Fibroblasts and myofibroblasts: what are we talking about? J Cardiovasc Pharmacol. 2011;57:376–379. doi: 10.1097/FJC.0b013e3182116e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circ Res. 2009;105:1164–1176. doi: 10.1161/CIRCRESAHA.109.209809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circ Res. 2009;105:1164–1176. doi: 10.1161/CIRCRESAHA.109.209809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leslie KO, Taatjes DJ, Schwarz J, vonTurkovich M, Low RB. Cardiac myofibroblasts express alpha smooth muscle actin during right ventricular pressure overload in the rabbit. Am J Pathol. 1991;139:207–216. [PMC free article] [PubMed] [Google Scholar]
- 68.Rohr S. Cardiac fibroblasts in cell culture systems: myofibroblasts all along? J Cardiovasc Pharmacol. 2011;57:389–399. doi: 10.1097/FJC.0b013e3182137e17. [DOI] [PubMed] [Google Scholar]
- 69.Dawson K, Wu CT, Qi XY, Nattel S. Congestive Heart Failure Effects on Atrial Fibroblast Phenotype: Differences between Freshly-Isolated and Cultured Cells. PLoS One. 2012;7:e52032. doi: 10.1371/journal.pone.0052032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaur H, Takefuji M, Ngai C, Carvalho J, Bayer J, Wietelmann A, Poetsch A, Holper S, Conway SJ, Mollmann H, Looso M, Troidl C, Offermanns S, Wettschureck N. Targeted Ablation of Periostin-Expressing Activated Fibroblasts Prevents Adverse Cardiac Remodeling in Mice. Circ Res. 2016 doi: 10.1161/CIRCRESAHA.116.308643. [DOI] [PubMed] [Google Scholar]
- 71.Biernacka A, Frangogiannis NG. Aging and Cardiac Fibrosis. Aging Dis. 2011;2:158–173. [PMC free article] [PubMed] [Google Scholar]
- 72.Kong P, Christia P, Frangogiannis NG. The pathogenesis of cardiac fibrosis. Cell Mol Life Sci. 2014;71:549–574. doi: 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horn MA, Trafford AW. Aging and the cardiac collagen matrix: Novel mediators of fibrotic remodelling. J Mol Cell Cardiol. 2016;93:175–185. doi: 10.1016/j.yjmcc.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McSpadden LC, Nguyen H, Bursac N. Size and ionic currents of unexcitable cells coupled to cardiomyocytes distinctly modulate cardiac action potential shape and pacemaking activity in micropatterned cell pairs, Circulation. Arrhythmia and electrophysiology. 2012;5:821–830. doi: 10.1161/CIRCEP.111.969329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedrotty DM, Klinger RY, Kirkton RD, Bursac N. Cardiac fibroblast paracrine factors alter impulse conduction and ion channel expression of neonatal rat cardiomyocytes. Cardiovasc Res. 2009;83:688–697. doi: 10.1093/cvr/cvp164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shadrin IY, Khodabukus A, Bursac N. Striated muscle function, regeneration, and repair. Cell Mol Life Sci. 2016;73:4175–4202. doi: 10.1007/s00018-016-2285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kizana E, Ginn SL, Smyth CM, Boyd A, Thomas SP, Allen DG, Ross DL, Alexander IE. Fibroblasts modulate cardiomyocyte excitability: implications for cardiac gene therapy. Gene Ther. 2006;13:1611–1615. doi: 10.1038/sj.gt.3302813. [DOI] [PubMed] [Google Scholar]
- 78.Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, Lorts A, Brunskill EW, Dorn GW, 2nd, Conway SJ, Aronow BJ, Robbins J, Molkentin JD. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res. 2007;101:313–321. doi: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mohamed TM, Abou-Leisa R, Stafford N, Maqsood A, Zi M, Prehar S, Baudoin-Stanley F, Wang X, Neyses L, Cartwright EJ, Oceandy D. The plasma membrane calcium ATPase 4 signalling in cardiac fibroblasts mediates cardiomyocyte hypertrophy. Nat Commun. 2016;7:11074. doi: 10.1038/ncomms11074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pijnappels DA, van Tuyn J, de Vries AA, Grauss RW, van der Laarse A, Ypey DL, Atsma DE, Schalij MJ. Resynchronization of separated rat cardiomyocyte fields with genetically modified human ventricular scar fibroblasts. Circulation. 2007;116:2018–2028. doi: 10.1161/CIRCULATIONAHA.107.712935. [DOI] [PubMed] [Google Scholar]
- 81.Engels MC, Askar SF, Jangsangthong W, Bingen BO, Feola I, Liu J, Majumder R, Versteegh MI, Braun J, Klautz RJ, Ypey DL, De Vries AA, Pijnappels DA. Forced fusion of human ventricular scar cells with cardiomyocytes suppresses arrhythmogenicity in a co-culture model. Cardiovasc Res. 2015;107:601–612. doi: 10.1093/cvr/cvv191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


