Abstract
Leptospirosis is the most widespread zoonosis and is considered a major public health problem worldwide. Currently, there is no widely available vaccine against leptospirosis for use in humans. A purified, recombinant subunit vaccine that includes the last six immunoglobulin-like (Ig-like) domains of the leptospiral protein LigA (LigA7’-13) protects against lethal infection but not renal colonization after challenge by Leptospira interrogans. In this study, we examined whether the addition of the first seven Ig-like domains of LigB (LigB0-7) to LigA7’-13, can enhance immune protection and confer sterilizing immunity in the Golden Syrian hamster model of acute leptospirosis. Hamsters were subcutaneously immunized with soluble, recombinant LigA7’-13, LigB0-7, or a combination of LigA7’-13 and LigB0-7 in Freund’s adjuvant. Immunization with Lig proteins generated a strong humoral immune response with high titers of IgG that recognized homologous protein, and cross-reacted with the heterologous protein as assessed by ELISA. LigA7’-13 alone, or in combination with LigB0-7, protected all hamsters from intraperitoneal challenge with a lethal dose of L. interrogans serovar Copenhageni strain Fiocruz L1-130. However, bacteria were recovered from the kidneys of all animals. Of eight animals immunized with LigB0-7, only three survived Leptospira challenge, one of which lacked renal colonization and had antibodies to native LigB by immunoblot. In addition, sera from two of the three LigB0-7 immunized survivors cross-reacted with LigA11-13, a region of LigA that is sufficient for protection. In summary, we confirmed that LigA7’-13 protects hamsters from death but not infection, and immunization with LigB0-7, either alone or in combination with LigA7’-13, did not confer sterilizing immunity.
Introduction
Leptospirosis is the most widespread zoonotic disease affecting both humans and animals. Infection by pathogenic strains of Leptospira spp. commonly occurs through direct contact with an infected animal’s urine or indirectly through contaminated water. Almost all mammals can harbor Leptospira in the proximal renal tubules, from which spirochetes are excreted in urine. Rats serve as a major carrier in human leptospirosis particularly in urban settings. Infection in rats is mostly asymptomatic, with bacteria cleared from the bloodstream and organs except the renal tubules leading to urinary shedding of up to 107 leptospires per milliliter months after initial infection [1, 2]. Humans, on the other hand, serve as accidental hosts with whom infection is acute and occasionally fatal. Leptospirosis in humans can present as an acute, self-limited disease characterized by an abrupt onset of fever, headache, chills, nausea and vomiting, myalgia, and less commonly skin rashes. A severe disease form is Weil’s syndrome, characterized by multiorgan system complications including jaundice, acute hepatic and renal dysfunction, and hemorrhage [1–5]. An infected individual may also be asymptomatic and shed leptospires, similar to maintenance animal hosts. Asymptomatic renal colonization and urinary shedding were observed among a group of individuals in the Peruvian Amazon [6]. However, the prevalence of persistently infected individuals and their relevance to disease transmission are unknown.
Although killed, whole-cell leptospiral vaccines are available in some countries including France, Cuba, and Japan [7–9], there is currently no widely available vaccine against leptospirosis for use in humans. Most vaccines available in the market are for veterinary use and are either inactivated leptospires (heat-killed or formalin-killed) consisting of a limited panel of serovars or outer membrane fractions. Immunity is directed mainly against leptospiral lipopolysaccharide (LPS), whose structure differs among the several hundred serovars of Leptospira. These vaccines produce short-term immunity and do not confer cross-protection against all pathogenic serovars [10, 11]. Therefore, there is a great need for development of vaccines that elicit a robust humoral and cellular immunity against pathogenic Leptospira spp., providing immunoprotection across serovars and sterilizing immunity against bacterial infection.
Surface-exposed outer membrane proteins (OMPs) that play a role in bacterial virulence and can be recognized by the host immune response early during the infection are attractive vaccine candidates. They have the potential to elicit cross-protection against different serovars of pathogenic Leptospira [12]. A wide range of OMPs have been tested for their protective ability including LipL32 [13, 14], OmpL1 and LipL41 [15, 16], and leptospiral immunoglobulin (Ig)-like proteins A (LigA) and B (LigB) [8, 17–27].
LigA and LigB consist of a lipoprotein signal peptide, followed by a series of 12–13 Ig-like domains [28, 29] similar to those found in bacterial virulence determinants invasin in Yersinia pseudotuberculosis [30], and intimin in Escherichia coli [31]. The first six domains and a portion of the seventh domain (conserved region) are nearly identical between the two Lig proteins, with an amino acid sequence identity of 99%. LigB, unlike LigA, has a unique carboxy-terminal non-repeat domain, and is found in all pathogenic strains of Leptospira [28, 29]. While LigA is not found in all pathogenic species, it is expressed by most characterized strains of L. interrogans, a major human pathogen [10]. Lig proteins are leptospiral surface components, and have been shown to bind extracellular matrix (ECM) components fibronectin, elastin, tropoelastin, laminin, and collagens I, III, and IV [32–37]. Therefore, these proteins may be involved in bacterial attachment to host tissues and colonization. Additionally, heterologous expression of ligA and ligB partially protects the non-pathogen L. biflexa from killing by human serum. Cleavage of the complement components C3b and C4b by complement regulators bound by LigA and LigB is believed to be responsible for this observation [38–40]. The Lig proteins also acquire key host proteins that participate in hemostasis. They bind to plasminogen, which is converted to plasmin in the presence of urokinase-type plasminogen activator, leading to cleavage of C3b, C5, and fibrinogen [39]. LigB also suppresses coagulation by capturing fibrinogen and factor XIII [33, 41, 42]. In virulent strains, ligA and ligB are upregulated at physiological osmolarity [32, 43, 44] and temperature [45] while expression is lost when strains are culture attenuated [28]. These observations suggest that the expression of ligA and ligB during infection may facilitate bacterial survival and dissemination. While the genetic knockout of ligB in L. interrogans serovar Copenhageni does not affect the virulence in hamsters or kidney colonization in rats [46], the targeted repression of both ligA and ligB in L. interrogans serovar Manilae results in attenuation in hamsters and failure to recover bacteria from tissues [47]. These findings suggest the overlapping or redundant roles of LigA and LigB in pathogenesis.
A number of studies have shown that LigA subunit or DNA vaccines generated from different serovars protected rodent models against homologous challenge infection [18–21, 23–26]. For the Copenhageni serovar, a carboxy-terminal LigA fragment starting within the seventh Ig domain is protective in the hamster model [26]. Ig domains 11–12, along with 10 or 13, are sufficient for protection [18]. Our group has also demonstrated that LigA domains 7–13 expressed as a lipoprotein in live E. coli and administered orally improved the hamster’s survival following L. interrogans lethal challenge [8]. However, LigA did not confer sterilizing immunity in any of these studies; renal colonization was observed in all immunized animals.
LigB also has potential as subunit vaccine [17, 23, 27]. The ligB gene, unlike ligA, is present in all pathogenic serovars of Leptospira that have been examined [29, 48]. Yan et al. [27] showed that subcutaneous immunization with the region of LigB that is nearly identical to LigA (first seven domains) improved the survival of hamsters challenged with L. interrogans serovar Pomona with corresponding reduction in kidney, liver, and spleen lesions. Immunization with LigB0-7 also increased splenic lymphocyte proliferation and upregulated transcripts of the Th1 cytokines IL-12 and IFN-γ, suggesting activation of cell-mediated immunity. This response is thought to be necessary to prevent leptospiral renal colonization and urinary shedding in cattle vaccinated with L. borgpetersenii serovar Hardjo [49].
In the current study, we determined if addition of LigB to a LigA vaccine can confer sterilizing immunity. We cloned and expressed soluble Ig-like domains 7–13 of LigA (LigA7’-13) and the N-terminal region including Ig domains 1–7 of LigB (LigB0-7) in E. coli. Using a hamster model of acute leptospirosis, we immunized animals with the recombinant proteins alone or in combination, to assess their ability to protect the animals from fatal infection with Leptospira, and provide sterilizing immunity of the kidney.
Materials and methods
Bacterial cultures
Leptospira interrogans serovar Copenhageni (pathogenic, strain Fiocruz L1-130) were grown in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium [10, 50, 51] at 30°C. This strain has a median lethal dose (LD50) range of 37–104 [52, 53], and a reported median endpoint dose (ED50) of ~20 [8] in hamsters challenged intraperitoneally. The genome sequence was previously reported by Nascimento et al. [54].
Chemically competent E. coli strain DH5α (New England Biolabs, Ipswich, MA) was used for gene cloning, while strain BLR (DE3) pLysS (EMD Millipore, Charles, MO) was used for protein expression and purification. E. coli were grown in Luria-Bertani (LB) or 2x YT medium (BD Biosciences, Sparks, MD) supplemented with 0.2% v/v dextrose and 100 μg/ml ampicillin (Sigma-Aldrich, St. Louis, MO). LB agar, containing 15 g/L agar formulation, was used for plating (Mo Bio Laboratories, Carlsbad, CA).
Animals
The work involving animals in this study was carried out according to the regulations of, and approved by, the Institutional Animal Care and Use Committee (IACUC, Protocol 09018–14) at the Veterans Affairs Greater Los Angeles Healthcare System (VAGLAHS), Los Angeles, CA. All animal experiments were performed under Animal Biosafety Level (ABSL-2) conditions. Female Golden Syrian hamsters (strain HsdHan:AURA) were purchased at the age of 3–4 weeks from Envigo RMS (Indianapolis, IN). The hamsters were housed in individually ventilated microisolator cages (4 per cage) with sterile, absorbent beddings changed twice weekly. The animals were fed and watered ad libitum throughout the course of the experiment. Following Leptospira challenge, hamsters were weighed and monitored daily for endpoint criteria as previously described [8, 55] including loss of appetite, gait or difficulty in breathing, prostration, ruffled fur, and weight loss of ≥ 10% of maximum weight. Hamsters that exhibited these manifestations were euthanized by isoflurane narcosis followed by thoracotomy.
Plasmid construction
The plasmid construct described in Coutinho et al. [18] was used to express LigA Ig-like domains 7’-13 (LIC10465; amino acids 631–1224). The ligB (lic10464) gene region encoding the amino acids after signal peptide followed by the first seven Ig-like domains (amino acids 19–672) was amplified from L. interrogans sv. Copenhageni str. Fiocruz L1-130 using primer pair LigB0-7For and LigB0-7Rev (Table 1). The DNA amplicons, which included NdeI and XhoI restriction sites were ligated into pET-20b(+) (Novagen), provided a carboxy-terminal 6xHis tag. Plasmid constructs were verified by restriction digest and nucleotide sequencing (Table 1). Both LigA7’-13 and LigB0-7 plasmids were transformed into E. coli expression strain BLR (DE3) pLysS (Novagen). A plasmid containing a shorter gene fragment, ligA11-13 (encoding amino acids 943–1224, [18]) was also prepared as described earlier using primers LigA11-13For and LigA11-13Rev (Table 1).
Table 1. List of oligonucleotides used in this study.
| Oligonucleotides (5’ → 3’) | Reference | |
|---|---|---|
| Primers used for cloning Lig proteins | ||
| LigA7’-13 For | AAC ATA TCT CAT ATG CTT ACC GTT TCC AAC ACA AAC GCC AA | [18] |
| LigA7’-13 Rev | TTC CTC GAG TGG CTC CGT TTT AAT AGA GGC TAA T | [18] |
| LigB0-7 For | AAA GTC ATA TGT CTT GGC CAC TTT TAA | This study |
| LigB0-7 Rev | GAA ACT CGA GTG CCG GAG TTA CCT TTA AAA TT | This study |
| LigA11-13 For | CAT CAA TGA CAT ATG AGA ATA GCT TCA ATC GAA GTA ACA CC | [18] |
| LigA11-13 Rev | TTC CTC GAG TGG CTC CGT TTT AAT AGA GGC TAA T | [18] |
| Primers used for sequencing | This study | |
| LigA oF2 | CCG GTA TCT TCA CCG ACC AC | |
| LigA oR3 | GGA TAG AGG TCA GAA CCG CC | |
| LigA oF4 | CCC GAC CTC TTC TCA CAA AGC | |
| LigA oR5 | CGT TCG GGT CAG AAG AAG ACC | |
| LigA oF6 | TCA AAT CTA ACC GTG CGT GG | |
| LigB oR1 | CGA TGG CTG AGA ATT GAC GA | |
| LigB oF2 | CAC TGT TTC TGC TTC TAG CGA G | |
| LigB oR3 | GAA GTG CGA GTT GGA GAG AC | |
| LigB oR4 | GCT TAC GGA GCA GCT ACA GG | |
| qPCR amplification | [56] | |
| LipL32 45F | AAG CAT TAC CGC TTG TGG TG | |
| LipL32 268F | GAA CTC CCA TTT CAG CGA TT | |
| LipL32 189P | FAM AA AGC CAG GAC AAG CGC CG BHQ1 | |
Recombinant protein expression and purification
Two-hundred fifty milliliters of 2x YT supplemented with 0.2% dextrose and 100 μg/ml ampicillin was inoculated with a 1:50 dilution of E. coli containing pET-20b(+) + LigA7’-13 or LigB0-7 overnight culture. The E. coli cultures were grown at 37°C until the optical density at 600 nm (OD600) reached 0.5–0.6, and isopropyl-β-D-thiogalactopyranoside (IPTG) (Apex Biotechnology Products) was added at a final concentration of 0.5–1 mM to induce protein expression. The temperature was shifted to 30°C, and cultures were incubated for 2 h with agitation at 225 rpm. Cells were harvested by centrifugation at 5,000 x g at 4°C for 10 min. The bacterial pellet was washed with phosphate buffered saline (PBS) pH 7.2 (Gibco, Grand Island, NY) followed by centrifugation at 5,000 x g at 4°C for 10 min.
The harvested cells were lysed using BugBuster Protein Extraction Reagent (EMD Millipore, Billerica, MA) in the presence of 0.25 mM phenylmethanesulfonyl fluoride (PMSF) and 10 U/ml DNase I (Thermo Scientific, Rockford, IL), and incubated at room temperature (RT) for 20 min while shaking. The lysate was subjected to centrifugation at 15,000 x g for 20 min at 4°C. The supernatant was collected, and soluble histidine-tagged proteins were purified by affinity chromatography using Ni-NTA agarose per the manufacturer’s instructions (Qiagen, Valencia, CA) at 4°C. After collecting the flow-through, the column was washed 10 times with 20 mM imidazole in 50 mM NaH2PO4 and 500 mM NaCl (pH 8.0) to remove unbound proteins. His-tagged proteins were eluted from the column with 500 mM imidazole in 50 mM NaH2PO4 and 500 mM NaCl (pH 8.0). The eluted fractions were analyzed by SDS-PAGE, pooled, and dialyzed extensively in buffer containing 50 mM NaH2PO4 and 50 mM NaCl (pH 8.0) to remove imidazole. The protein concentration was determined by bicinchoninic acid (BCA) analysis (Pierce, Rockford, IL). When necessary, proteins were concentrated in Amicon centrifugal filter units (EMD Millipore) with a molecular weight cut-off of 10 kDa. The purified recombinant proteins were used immediately for immunization or stored at -80°C for subsequent ELISA and western blot analysis.
Hamster immunization
Recombinant LigA7’-13 and LigB0-7 proteins were expressed and purified prior to every immunization and were delivered as soluble proteins. Animals were immunized as previously described [18]. Groups of five to eight female hamsters, 3–4 weeks of age, were immunized subcutaneously with 100 μg recombinant LigA7’-13, LigB0-7, or 50:50 mixture of LigA7’-13 and LigB0-7, in a total volume of 200 μl with Freund’s complete adjuvant (Sigma-Aldrich) in a 1:1 ratio. The hamsters were boosted at days 14 and 28 with the same amount of protein with Freund’s incomplete adjuvant (Sigma-Aldrich) in a 1:1 ratio. Control groups included animals that were immunized with PBS alone or PBS with adjuvant. Blood was collected via retro-orbital route 3 days before the first immunization (pre-bleed) and weekly thereafter (days 4, 11, 18, 25, and 32) until animals were challenged with L. interrogans at day 35 post-immunization. Serum was prepared from collected blood by centrifugation at 21,100 x g for 15 min, and was stored at -80°C until use.
Leptospira challenge and sample collection
One week after the last immunization, hamsters were challenged intraperitoneally with 1 ml of 1 x 104 L. interrogans sv. Copenhageni st. Fiocruz L1-130 (≥ 500-fold over ED50) [8]. The animals were weighed daily and monitored for endpoint criteria [8, 55] as described earlier. Hamsters that survived infection were euthanized 28 days post-challenge. At the time of sacrifice, one kidney was collected for culture in EMJH, while the other kidney and liver were used for quantification of L. interrogans by quantitative PCR (qPCR). Serum prepared from blood collected by post-mortem cardiac puncture were used for serologic analyses and microscopic agglutination test (MAT).
Collected kidney tissues were pulverized by passing them through a 3 ml syringe and inoculated into a tube containing semisolid EMJH supplemented with 100 μg/ml 5-fluorouracil. A 1:100 dilution was passaged to a new semisolid medium and the tubes were incubated at 30°C. The cultures were checked weekly for the presence of L. interrogans by dark field microscopy for up to 28 days, before being designated negative. Positive cultures generally show up between 7–14 days.
Quantitative PCR
Hamster kidney and liver tissues were prepared for DNA extraction by dicing 50–80 mg of tissue and suspending in 500 μl PBS. Following tissue homogenization for 1 min at 5 movements per second (OMNI Bead Ruptor 24, Omni International, Kennesaw, GA), total genomic DNA was extracted from equivalents of 25 mg tissue using the DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s instructions but eluted with 100 μl of elution buffer. DNA was prepared for the standard curve by growing serovar Copenhageni in EMJH culture medium to a density of 4 x 105 spirochetes/ml. DNA was extracted from 5 ml culture using the same kit. The concentration of eluted DNA was determined using a spectrophotometer at OD260 (GE NanoVue Spectrophotometer). All DNA samples were kept at -80°C until use.
Leptospira DNA (1 x 100 to 1 x 107 genomic equivalents (GEq) /5 μl) was prepared as previously described [57]. L. interrogans genome was quantified by qPCR using Bio-Rad iTaq Universal Probes supermix (Bio-Rad, Hercules, CA) supplemented with 250 nM lipL32 forward (45F) and reverse (268R) primers, and 150 nM lipL32 probe (189P) [56] (Table 1). Five microliters of standard or sample DNA was added to 15 μl PCR mix. The amplification protocol is as follows: 3 min at 95°C, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. All standards and samples were run in duplicate. A standard curve was generated using Bio-Rad iQCycler5 software, and the number of GEq was extrapolated from the threshold cycle (CT) values. A negative result was assigned where no amplification occurred or if the CT value was greater than 36 [56]. Data are presented as number of L. interrogans GEq per gram of tissue. Uninfected hamster liver and kidney tissues were used as negative controls to determine the limit of detection (LOD) of the assay.
Microscopic agglutination test (MAT)
Kidney culture-negative samples were subjected to MAT as described by Goris and Hartskeerl [58]. Sera were heat-inactivated at 56°C for 30 min. Serial two-fold dilutions of serum (1:10 to 1:10,240) were prepared in PBS pH 7.2, and incubated at 30°C for 30 min with equal volumes of L. interrogans sv. Copenhageni st. Fiocruz L1-130 (optical density at 420 nm (OD420) = 0.2) in a 96-well plate. Cultures were examined under dark-field microscopy for bacterial agglutination. The titer was determined as the highest dilution > 50% of the bacteria are freely moving compared to control suspension without sera. Sera from uninfected hamsters were used as controls.
Enzyme-linked immunosorbent assay (ELISA)
Serum antibody responses to recombinant protein immunization were quantified by ELISA as previously described [8, 18] with few modifications. ELISA was performed by coating microtiter plates (Immulon 4HBX, Thermo Scientific) with 1 μg of recombinant protein per well (100 μl diluted in PBS, pH 7.2) overnight at 4°C. The wells were washed three times with 200 μl PBS, blocked with 200 μl protein-free blocking buffer in PBS (PFB) (Pierce) for 1 h at RT, then incubated with 100 μl 1:6,400 dilutions of hamster sera in PFB for 1 h at 37°C. The plates were washed three times with PBS and incubated with 100 μl horse radish peroxidase (HRP)-conjugated goat anti-Syrian hamster IgG (1:5,000 in PFB) (Jackson ImmunoResearch, West Grove, PA) for 30 min at RT. After 3 washes with PBS, plates were developed with 100 μl 3,3′,5,5′-tetramethylbenzidine (1-step Turbo TMB HRP substrate, Thermo Scientific) for 15 min at RT in the dark. The absorbance at 655 nm (OD655) was determined using an automated plate reader (Bio-Rad 550 Microplate Reader, Bio-Rad). The IgG response was expressed as relative α-Lig antibody level, determined by subtracting the absorbance of the pre-immune bleed (baseline or background read) from the immune sera collected at different time points. All experiments were done in triplicate.
Immunoblot analysis
Immunoblot analysis were performed to determine if sera from immunized animals recognize purified His-tagged or native Lig proteins, and to determine the antigenicity of the subunit vaccines. L. interrogans lysate was prepared by growing cultures to mid-log phase, followed by induction with 120 mM NaCl for 4 h at 30°C, 150 rpm. Bacteria were harvested by centrifugation at 15,000 x g for 15 min, washed with PBS, and centrifuged again to collect the pellet. The bacterial pellet was resuspended in SDS protein sample buffer supplemented with dithiothreitol (DTT) (Thermo Scientific), boiled for 5 min, and kept at -20°C until use.
Fifteen microliters of L. interrogans whole cell lysate or 100–200 ng/well purified recombinant Lig proteins or control leptospiral lipoprotein LipL32 were separated by 10% or 4–12% SDS-PAGE (NuPage Bis-Tris protein gels, Life Technologies, Carlsbad, CA) and transferred to polyvinylidene difluoride (PVDF) membrane (Immobilon, EMD Millipore) for western blotting. The membranes were blocked with 5% skim milk in PBS-Tween 20 (Thermo Scientific) overnight at 4°C. The membranes were cut into strips and incubated in pooled or individual hamster sera (pre-bleed or last bleed) from immunized animals or control rabbit α-Lig antiserum (at dilutions 1:2,000–1:10,000 in 5% milk in PBS-Tween 20) for 1 h at RT over a rocker. The Lig antiserum, which reacts with LigA and LigB, was raised against amino acids 342–1224 of L. kirschneri LigA and has been described previously [28]. As loading control for Leptospira lysate, membranes were probed with rabbit α-LipL41 (1:10,000 dilution). As loading controls for recombinant LipL32, membranes were probed with rabbit α-Lip32 (1:5,000 dilution) and mouse α-6xHis epitope (1:1,000 dilution, Thermo Scientific). To test the antigenicity of subunit vaccines, membrane strips were incubated with 1:250 dilution of the following: pooled sera from hamsters infected intraperitoneally with Copenhageni serovar, and corresponding pre-bleeds as control; pooled sera obtained from convalescent patients diagnosed with leptospirosis (provided by Dr. Albert Ko, Yale University, New Haven, CT), and normal human serum (Millipore) as control.
After incubation with primary antisera, the membranes were washed three times with PBS-Tween 20 and were probed with horseradish peroxidase (HRP)-conjugated α-hamster (1:5,000 dilution, Jackson ImmunoResearch), α-rabbit (1:5,000–1:10,000 dilution, GE Healthcare), α-mouse (1:5,000 dilution, GE Healthcare), or α-human (1:20,000 dilution, Sigma-Aldrich) antibodies for 40 min at RT. Membranes were washed and the antigen-antibody binding was detected by using enhanced chemiluminescence (ECL) system. Blots were incubated in ECL reagent (Pierce) for 1 min, exposed to a film (Amersham HyperFilm ECL), and developed (Konica SRX-101A).
Statistical tests
The differences in survival and mortality rates among experimental groups were determined using the Log-rank (Mantel-Cox) test and Fisher’s exact test, respectively. Kruskal-Wallis test was used to determine whether there is a significant difference in the number of bacteria in kidney or liver among the survivors from different immunization groups while one-way analysis of variance (ANOVA) was used to compare immune responses among animals in the same treatment group. For all tests, statistical significance was set to P ≤ 0.05. All analyses were performed using Graph Prism 5 (GraphPad Software, Inc., La Jolla, CA).
Results
Preparation of purified recombinant Lig proteins
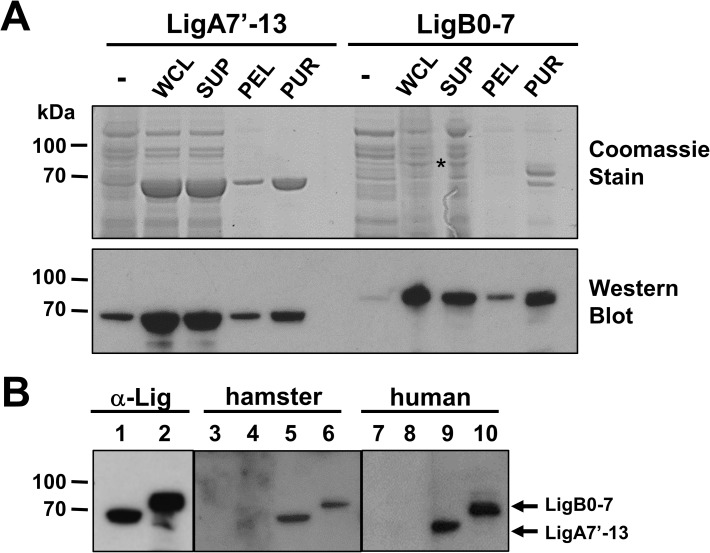
To test the protective properties of Lig proteins, recombinant LigA7’-13 and LigB0-7 proteins were prepared. The segment of ligA encoding LigA7’-13 was cloned and expressed as previously described [18]. The LigB0-7 construct starts after the signal sequence and spans the first through seventh Ig-like repeat domains of the LigB protein. Comparison of the amino acid sequences of the LigA7’-13 and LigB0-7 using Basic Local Alignment Search Tool (BLAST) shows that the sequence identity between the two proteins is 66%. Both LigA7’-13 and LigB0-7 were expressed as soluble proteins, although LigA7’-13 expression in E. coli was more robust compared to LigB0-7 (Fig 1). The expressed proteins contain 6xHis tag at the C-terminal region and were purified under non-denaturing conditions by affinity chromatography using a nickel-charged resin. Both proteins were dialyzed extensively to remove imidazole in the solution. Purified, recombinant LigA7’-13 was stable in solution, however, precipitation was observed in purified LigB0-7 when kept for prolonged periods at 4°C. We evaluated the antigenicity of the purified Lig proteins by immunoblot analysis and demonstrated that both LigA7’-13 and LigB0-7 were recognized by sera from hamsters infected intraperitoneally with L. interrogans, and by sera of leptospirosis patients from Brazil in convalescent phase (Fig 1).
Fig 1. Expression, purification, and antigenicity of recombinant His-tagged Lig proteins.
(A) E. coli BLR (DE3) pLysS transformed with pET-20b(+) containing ligA7’-13 or ligB0-7 were grown to an OD600 = 0.5–0.6 prior induction with 0.5–1 mM IPTG for 2 h at 30°C. Bacterial cells were harvested by centrifugation, followed by lysis in the presence of protease inhibitors. Supernatant was separated from pellet fraction by centrifugation at 16,000 x g at 4°C. His-tagged Lig proteins were purified from the supernatant fraction by affinity chromatography using Ni-NTA resin. Fifteen microliters of whole cell lysates from uninduced (-) and induced cultures (WCL), supernatant (SUP) and pellet (PEL) fractions, and 2 μg purified protein (PUR) were separated by 4–12% SDS-PAGE followed by Coomassie staining. A replicate gel was also run loaded with 5 μl cell fractions and 200 ng purified proteins for immunoblot analysis. The proteins were transferred to a PVDF membrane and the blot was probed with rabbit α-Lig antiserum (dilution 1:10,000). The expressed His-tagged LigA7’-13 and LigB0-7 were found predominantly in the supernatant. Asterisk shows LigB0-7, produced at lower levels compared to LigA7’-13. (B) Two hundred nanograms of LigA7’-13 (lanes 1, 3, 5, 7, 9) or LigB0-7 (lanes 2, 4, 6, 8, 10) were run in 4–12% SDS-PAGE gel and transferred to PVDF. Membranes were probed with 1:10,000 control α-Lig antiserum (lanes 1, 2), or 1:250 of the following: pooled sera from hamsters before (lanes 3, 4) and after (lanes 5, 6) intraperitoneal challenge with L. interrogans, normal human serum (lanes 7, 8), and pooled sera from leptospirosis patients in convalescent stage (lanes 9, 10). Both Lig proteins were recognized by sera from Leptospira-infected hamsters and humans.
Humoral response of Lig immunized hamsters
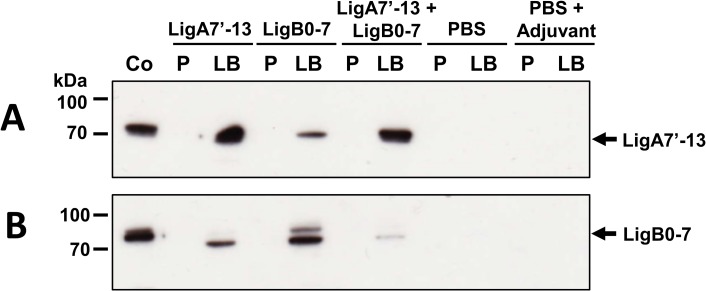
To assess whether animals immunized with Lig proteins developed an IgG antibody response, sera were collected weekly during the immunization protocol and examined by ELISA using LigA7’-13 and LigB0-7 antigens. As presented in Fig 2, there was no detectable IgG against either purified protein in serum from hamsters in both control groups, PBS and PBS with adjuvant. Immunization with LigA7’-13 or LigB0-7 produced IgG that recognizes the homologous recombinant protein fragment. Sera from LigA7’-13 + LigB0-7 immunized hamsters react with both Lig protein fragments. The antibody responses observed were directed towards the immunized Lig protein, and not to the epitope tag, as sera from both control and immunized groups did not recognize a non-relevant 6xHis-tagged leptospiral lipoprotein LipL32 (S1 Fig). However, cross-reactivity was observed as sera from LigA7’-13 vaccinated animals reacted against recombinant LigB0-7 immobilized on ELISA plates and vice versa. Cross-reactions were confirmed by immunoblot analysis, probing recombinant Lig proteins immobilized on PVDF membrane with pooled sera from immunized animals (Fig 3).
Fig 2. IgG response to immunization with purified recombinant Lig proteins.
Serum samples were collected from hamsters weekly during the immunization protocol. Anti-LigA7’-13 (A) or anti-LigB0-7 (B) antibody levels were measured in triplicate by ELISA. Each data line represents the IgG response of an individual animal over time; in black are animals that survived the challenge while in red are animals that met the endpoint criteria. Vertical dotted lines indicate immunization days (blue) or challenge with L. interrogans (red). Each data point represents the mean IgG level read at OD655 minus pre-bleed read. Error bars indicate standard deviation. There was no significant difference in the IgG response among hamsters that received the same vaccine treatment at all time points (One-way ANOVA, P = 0.9).
Fig 3. Recognition of purified, recombinant LigA7’-13 and LigB0-7 proteins by pooled sera from immunized hamsters.
One to two hundred nanograms per well of purified His-tagged LigA7’-13 (A) or LigB0-7 (B) were run in 4–12% SDS-PAGE then transferred to PVDF membrane for western blot analysis. The membrane was cut to strips and probed with 1:5,000 pooled sera collected before immunization (pre-bleed, P) and at day 32 (last bleed, LB) from immunized and control groups. Another membrane strip was incubated with 1:5,000 rabbit α-Lig as positive control (Co). Sera collected from LigA7’-13-vaccinated animals cross reacted with recombinant LigB0-7 and vice versa.
The IgG response of individual animals that received the same vaccine treatment were comparable at all time points (one-way ANOVA, P = 0.9) (Fig 2). A substantial increase in α-LigA7’-13 and α-LigB0-7 antibody titers was observed at day 18, after the first immunization boost and IgG antibody levels were maintained until the leptospiral challenge at day 35 (Fig 2 and S2 Fig). There was no significant difference in the α-LigA7’-13 IgG levels among treatment groups LigA7’-13, LigB0-7, or combination, at all time points (S2 Fig). The α-LigB0-7 IgG level in LigB0-7 immunized hamsters was significantly higher one week after the first immunization (day 11 bleed) compared to animals of the other vaccine groups. However, antibody titers were comparable to other vaccine groups at later time points (S2 Fig). These results suggest that both LigA7’-13 and LigB0-7 immunizations elicit humoral immune response in hamsters.
Protective properties of Lig vaccines against L. interrogans challenge
Recombinant Lig antigens were evaluated for their protective properties using a hamster model of acute leptospirosis. Animals were challenged intraperitoneally one week after the last immunization with 1x104 L. interrogans sv. Copenhageni str. Fiocruz L1-130 (~500-fold the ED50 [8]). The challenge strain used was at a low passage (2) and expresses LigA and LigB when incubated at physiological osmolarity (S3 Fig).
Hamsters were observed and their weights monitored daily after challenge, as weight loss has been found to be an early objective sign of clinical leptospirosis [18]. PBS or PBS with adjuvant sham immunized animals started to lose weight as early as day 7 post-challenge and by day 15, all hamsters in both groups met the endpoint criteria (Fig 4 and Table 2). Two hamsters from the PBS with adjuvant group succumbed to Leptospira challenge before showing signs of the disease or reaching the 10% weight loss designated as our endpoint. In contrast, all LigA7’-13 and LigA7’-13 + LigB0-7 immunized hamsters survived the intraperitoneal bacterial challenge. LigB0-7 vaccinated animals exhibited 37.5% survival; five of the eight LigB-immunized animals showed signs of leptospirosis 8–12 days post-challenge. The difference between the survival rates of LigB0-7 vaccinated animals and control animals (PBS and PBS with adjuvant groups) was not statistically different (Fisher’s exact test, P = 0.2088 and P = 0.2308, respectively) (Fig 4). In addition, no difference was observed in IgG response among all LigB0-7 immunized animals whether they survived infection or met endpoint (Fig 2). Two of the LigB0-7 immunized survivors exhibited 4.1% and 4.2% weight loss starting at day 7 or 8, but did not meet the endpoint criteria set for weight loss at ≥ 10%, and were able to recover their weights by day 11. We did not observe any significant weight loss among LigA7’-13 or LigA7’-13 + LigB0-7 immunized hamsters following bacterial challenge.
Fig 4. Survival of hamsters immunized with purified recombinant Lig proteins after lethal challenge.
Groups of 5–8 female hamsters were immunized subcutaneously three times with 100 μg LigA7’-13 (●), LigB0-7 (■), LigA7’-13 + LigB0-7 (◆), or controls PBS (○) and PBS with adjuvant (□). Animals were challenged intraperitoneally with 1 x 104 L. interrogans at day 0 and post-challenge survival was followed until day 28. Asterisks indicate significant difference between mortality rates compared (Fisher’s exact test, *P < 0.05, ***P < 0.001, ns not significant).
Table 2. Protection conferred by immunization with recombinant Lig proteins against intraperitoneal L. interrogans challenge.
| Vaccine groups | No. animals that survived challenge/total animals (%) | No. animals that met endpoint criteria/total animals (endpoint days) | No. animals that are culture positive/total animals |
|---|---|---|---|
| LigA7’-13 | 8/8 (100%) | 0/8 | 8/8 |
| LigB0-7 | 3/8 (37.5%) | 5/8 (10,10,12,12,12) | 7/8* |
| LigA7’-13 + LigB0-7 | 8/8 (100%) | 0/8 | 8/8 |
| PBS | 0/6 (0%) | 6/6 (10,10,10, 13,15,15) | 6/6 |
| PBS + Adjuvant | 0/5 (0%) | 5/5 (8,10,10, 10,10) | 5/5 |
*One of the hamsters that survived the intraperitoneal challenge; qPCR was below LOD; negative MAT result.
Sterilizing immunity conferred by Lig protein immunization
Pathogenic Leptospira spp. disseminate in the blood and reach target organs including the kidney [1, 2]. To determine if Lig protein immunization provides protection against renal colonization, kidneys collected from hamsters were cultured in semisolid EMJH supplemented with 5-fluorouracil, and checked weekly under dark field microscope for the presence of motile Leptospira. As presented in Table 2, all kidney tissue cultures collected from sham-immunized animals were positive. Similar to previous studies [8, 18, 22, 26], kidneys of LigA7’-13 immunized survivors were colonized with leptospires. The addition of LigB0-7 to LigA7’-13 vaccine did not assist in eradication of the bacteria from the kidneys; all LigA7’-13 + LigB0-7 vaccinated animals had positive kidney cultures. One of the hamsters in the LigB0-7-immunized group was culture-negative, and found to be MAT-negative when tested at serum dilutions 1:10 to 1:10,240.
Leptospira in both liver and kidney were quantified by qPCR to accurately assess the difference in bacterial load among survivors of the three vaccine groups. As shown in Fig 5, we did not detect L. interrogans in the liver of hamsters 28 days post-challenge. On the other hand, we observed high bacterial burdens in kidneys with up to 108 GEq per gram of tissue among survivors. However, there was no significant difference in the kidney burdens among hamsters from the three different treatment groups (Kruskal-Wallis test, P = 0.4050). The bacterial kidney load in one of the LigB0-7 immunized hamster was below the set limit of detection; this hamster was also kidney culture-negative and MAT-negative. These results suggest that LigB0-7 does not protect hamsters from L. interrogans challenge, nor does the addition of LigB0-7 to LigA7’-13 lower bacterial burden in kidneys.
Fig 5. Bacterial load in kidney and liver of hamsters that survived the L. interrogans challenge.
Total genomic DNA was extracted from kidney (A) and liver (B), and analyzed by qPCR performed in duplicates with lipL32 primers and probes (Table 1) to quantify leptospiral tissue load. Bacterial burden was expressed as genomic equivalents (GEq) per gram of tissue. Black lines show mean bacterial load with error bars representing standard deviation, while dotted lines indicate limit of detection. There was no difference in the bacterial burden in kidneys among the different vaccine groups (Kruskal-Wallis test, P = 0.4050).
Recognition of native Lig proteins by sera from immunized animals
To determine if IgG produced from Lig protein immunization of hamsters recognize native proteins, we incubated L. interrogans in the presence of 120 mM NaCl for 4 h at 30°C. After osmotic induction, the bacteria were harvested by centrifugation, washed with PBS, and resuspended in sample buffer. Leptospira lysates were immobilized on PVDF membranes and probed with pooled or individual sera from control and treatment groups collected prior to leptospiral challenge.
Fig 6A shows that pooled sera from LigA7’-13 immunized animals bind to native LigA. On the other hand, pooled sera from LigB0-7 vaccinated hamsters recognize both native LigA and LigB proteins, although LigA signal was weak. The cross-reactivity with native LigA is possibly due to high sequence similarity (94%) between LigB0-7 and the corresponding LigA0-7 fragment (amino acids 19–672). However, immunization with both proteins generated antibodies that recognize LigA only. Since these hamsters were immunized with half the amount of each protein, we speculate the LigB0-7 dosage used was too low to elicit a robust production of antibodies that bind native LigB protein. Leptospira proteins were not detected by pre-bleed sera nor by sera collected from mock-immunization groups.
Fig 6. Recognition of native LigA and LigB in Leptospira lysate by sera from immunized hamsters.
A L. interrogans culture at an OD420 = 0.29 was incubated in EMJH supplemented with 120 mM NaCl for 4 h at 30°C. Bacteria was harvested by centrifugation, washed with PBS, resuspended in sample buffer, and boiled for 5 min. Fifteen microliters of bacterial lysate was separated in 4–12% SDS-PAGE, then transferred to a PVDF membrane for western blot analysis. (A) The membrane was cut to strips and probed with pooled sera (1:2,000 dilution) collected before immunization (pre-bleed, PB), and at day 32 (last bleed, LB) from immunized and control hamsters. (B) The membrane strips were probed with sera (1:2,000 dilution) from individual LigB0-7 vaccinated hamsters (1–8). The prominent band recognized by serum from hamster 7 migrates slower than LigB. The membranes were also probed with rabbit α-Lig (Co, 1:2,000 dilution) and rabbit α-LipL41 (1:10,000 dilution) as controls. Asterisks (*) represent hamsters that survived the challenge, while pound sign (#) designates animal that survived and was culture- and MAT-negative.
We examined individual sera from LigB0-7 immunized hamsters to determine if there is a difference in the recognition of native Lig proteins among animals that survived or met the endpoint criteria. None of the sera from hamsters that succumbed to the bacterial challenge recognized native LigA or LigB (Fig 6B). Serum from only one survivor (hamster 1) recognized both LigA and LigB; serum is from the animal from which we were not able to recover Leptospira from the kidney. Sera from the two other survivors (hamsters 6 and 7) did not recognize LigB. The prominent band observed when probed with serum from hamster 7 had a higher molecular weight compared to LigB. However, the serum from this animal recognized the native LigA. These results indicate that when leptospiral lysate was probed by pooled sera from LigB0-7 immunized animals (Fig 6A), the higher molecular weight band observed is predominantly from a non-relevant protein recognized by hamster 7 sera and in part contributed by hamster 1 sera binding to native LigB. We repeated the assay using a higher concentration of sera (1:500 dilution) in an effort to enhance some possibly weak signals of the other sera samples but observed the same results as shown in Fig 6B.
Recognition of domains 11–13 of LigA is by sera from immunized hamsters
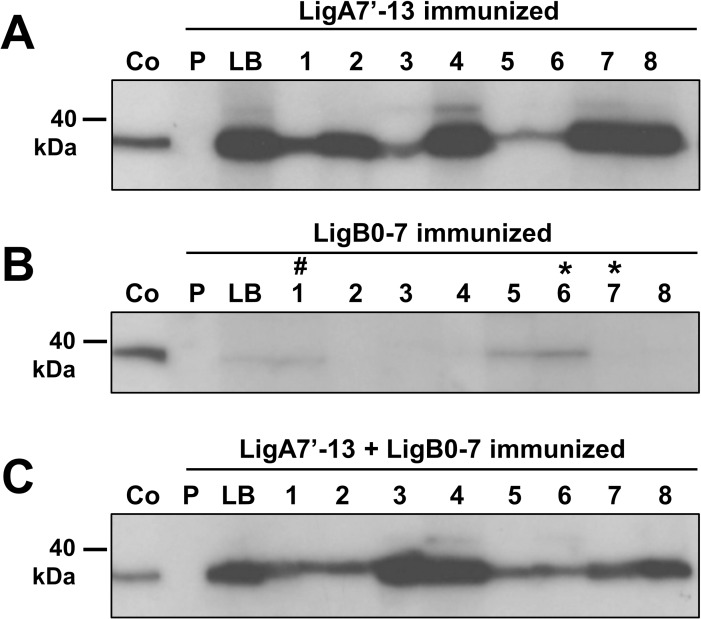
Both ELISA and immunoblot analysis showed that IgG produced by vaccinating with purified His-tagged LigA7’-13 or LigB0-7 strongly recognizes homologous protein, and cross-reacts with heterologous protein. We prepared a shorter construct of ligA, consisting of domains 11–13, which has been described as sufficient for immunoprotection [18]. By immunoblot analysis, we observed that sera from LigA7’-13 or LigA7’-13 + LigB0-7 immunized survivors recognized recombinant LigA11-13 protein (Fig 7). This construct has a 41% sequence identity with LigB0-7 protein (BLAST). Only three sera from eight LigB0-7 immunized animals recognized LigA11-13 (Fig 7). Interestingly, two (hamster 1 and 6) out of the three sera are from hamsters that survived the L. interrogans challenge.
Fig 7. Recognition of purified, recombinant LigA11-13 proteins by sera from immunized hamsters.
One hundred nanograms per well of purified His-tagged LigA11-13 were run in 4–12% SDS-PAGE then transferred to a PVDF membrane for western blot analysis. The membrane was cut to strips and probed with 1:3,000 pooled sera collected before immunization (pre-bleed, P) and at day 32 (last bleed, LB), and sera from individual hamsters immunized with LigA7’-13 (A), LigB0-7 (B), and LigA7’-13 + LigB0-7 (C). Another membrane strip was incubated with 1:3,000 rabbit α-Lig as positive control (Co). In Panel B, asterisks (*) represent hamsters that survived the challenge, while pound sign (#) designates animal that survived and was culture- and MAT-negative. All animals immunized with LigA7’-13 alone, or in combination with LigB0-7, survived the challenge.
Discussion
In an effort to develop a vaccine that confers protection across all pathogenic serovars and protects the kidneys from bacterial colonization, we tested the immunoprotective properties of LigA7’-13 in combination with LigB0-7. We confirmed results of previous studies that the immunization of hamsters with LigA7’-13 protected hamsters from death but not from infection, while animals that received LigB0-7 protein were mostly susceptible to leptospiral infection [18, 26]. We found that addition of LigB0-7 to LigA7’-13 neither conferred sterilizing immunity nor reduced the bacterial burden in kidneys.
Earlier studies suggested that the mechanism of protection against leptospirosis is mediated largely by the humoral immune response [3, 59]. We observed high titers of antibodies recognizing the homologous protein, and a strong cross-reactivity of IgG antibodies against the heterologous recombinant Lig protein due to the high sequence conservation between the two constructs. Although hamsters immunized with LigB0-7 generated antibodies against the protective fragment LigA7’-13, immunization with LigB0-7 did not protect animals from death. We therefore tested sera from immunized animals for the recognition of domains 11–13 of LigA as this was described to be sufficient in protecting hamsters from infection [18]. These domains are found in the unique region of LigA described to attach to the gelatin binding domain of fibronectin [37]. In addition, LigA 11–13 overlaps with the domains that facilitate binding to the complement regulator C4b-binding protein (C4BP) [60]. Sera from two out of the three LigB0-7 immunized animals that survived the challenge cross-reacted with LigA11-13 by immunoblot assay. These findings raise the possibility that LigA domains that mediate attachment to the extracellular matrix and involved in the capture of a complement regulator are sufficient to protect animals from infection and warrant further studies to determine the mechanisms involved in LigA immunoprotection.
Despite a strong humoral response of all LigB0-7 immunized animals, only three of the eight survived Leptospira challenge, with sterilizing immunity achieved in one case. A possible explanation for this observation is that the other seven animals failed to generate antibody that reacts against native LigB produced by L. interrogans. Previous reports have showed LigBrep (corresponding to our LigB0-7 construct) protected kidneys from bacterial colonization when administered as a DNA vaccine [21, 61]. In this context, it would be of interest to determine whether sera from animals immunized with LigBrep DNA recognize the native LigB protein. However in these studies, bacterial burden in kidneys was determined primarily by culture or by imprinting tissue samples on glass plate and probing with anti-Leptospira antibodies–methods that may be less sensitive and can generate false negative results. Using qPCR, we concluded that the addition of LigB0-7 to LigA7’-13 failed to lower bacteria burdens in kidneys.
Although LigB0-7 was found predominantly in the soluble fraction and purified under non-denaturing conditions, we noticed protein precipitation during long term storage. The instability of LigB0-7 in solution may have resulted in the loss of native structure and subsequent loss of integrity of epitopes necessary to elicit protective antibodies. In the study conducted by Murray and colleagues [62], out of the 223 L. borgpetersenii proteins predicted to be surface-exposed, only 12% were expressed and purified in E. coli as soluble proteins. The remaining proteins were purified under denaturing conditions and none of these antigens were found to be protective in a hamster model of infection. In fact, the disparity in the reported claims of protection conferred by LigB as subunit vaccine may in part be due to the differences in the solubility and/or epitopes of proteins used for immunization [17, 26, 27, 61]. It should be noted that the protection conferred by soluble rLigBcon (corresponding to LigB0-7 construct) immunization described by Yan et al. [27] has some caveats, as discussed elsewhere [10]. Their group used a less virulent Leptospira strain at a very high challenge dose and were unable to induce lethal infection in all control animals in three independent experimental trials.
A recent study by Conrad et al. [63] demonstrated that a subunit vaccine preparation of the conserved region of LigB (amino acids 131–645) conferred sterilizing immunity. Similar to other Lig vaccine studies, the expressed protein was found mostly in the insoluble fraction and purified under denaturing conditions followed by a dialysis step to refold the recombinant protein. The LigB (131–645) construct described lacks the region between the signal peptide and Ig-like domain 1 and the C-terminal segment of the Ig-like domain 7 present in our LigB0-7 (19–762) that potentially rendered our protein predominantly soluble. In our study, the hamsters were immunized subcutaneously with a mixture of LigB0-7 and Freund’s adjuvant. Conrad et al. described that intramuscular immunization of LigB (131–645) adsorbed on aluminum hydroxide protected 80–100% hamsters against intraperitoneal challenge of 200 (10x ED50) L. interrogans sv. Copenhageni str. Fiocruz L1-130. Additionally, the kidneys of hamsters that survived bacterial challenge were Leptospira-negative as assessed by culture method and/or by qPCR. This is the first evidence of protection both from lethal infection and renal colonization conferred by a Lig subunit vaccine. The findings of their group, along with our results, highlight the importance of immunogen selection, recombinant protein preparation method, route of administration, adjuvant selection, immuogen dosage, and frequency of immunization as determinants in design of potential subunit vaccines.
Studies in cattle generated a mounting interest in exploring the role of cell-mediated responses in immunity against Leptospira infection. In the work of Bolin et al. [64], vaccination of whole cell-pentavalent vaccine failed to protect cattle from infection with L. borgpetersenii serovar Hardjo despite high titers of circulating antibodies against LPS. Immunization with whole cell monovalent serovar Hardjo vaccine however, protected cattle from infection, renal colonization, and urinary shedding following Leptospira challenge. The protection conferred by this vaccine correlates with its ability to stimulate Th1 response and IFN-γ production [49, 65, 66]. Findings from these studies indicate that anti-LPS humoral immune response is likely to be insufficient for protective immunity in cattle [64, 65, 67], and perhaps in other species. Due to the limited commercial availability of immunological reagents for use in hamsters, we were not able to determine the cell-mediated immune response of hamsters to Lig protein immunizations.
Our results confirm previous studies showing the immunoprotective property of LigA [18–21, 23–26]. Although different serovars and challenge doses were used in these studies, leptospiral challenge was performed through intraperitoneal inoculation. This method allows reproducible amounts of bacteria to be introduced into the animal. However, it does not mimic the natural entry of leptospires into hosts. Future experiments should employ challenge methods that reflect the natural transmission of the pathogen including infecting animals through skin [68, 69] or mucous membranes [57, 65]. Despite the limited number of tools available to measure both humoral and cellular immunity, hamsters remain to be one of the best animal models for use in Leptospira vaccine studies owing to their high susceptibility to infection and ability to exhibit clinical features that mimic severe human leptospirosis symptoms [55].
A major challenge in the leptospirosis field is the development of vaccines that confer cross-protection across all pathogenic serovars, protect kidneys from bacterial colonization, and induce long lasting immune protection. An ideal subunit vaccine must have the ability to elicit both humoral and cell-mediated immune responses. Although we have demonstrated the ability of subcutaneous immunization with LigA7’-13 to protect hamsters from lethal infection, renal colonization still persists. In our study, the addition of LigB0-7 did not protect the kidneys from bacterial colonization nor decrease the bacterial load in the renal tubules. However, it is interesting to note that one of the LigB0-7-immunized animals that survived bacterial challenge with sterilizing immunity also generated antibodies recognizing native LigB and cross-reacting with the protective domains of LigA. It may be possible to achieve sterilizing immunity through immunization with a subunit vaccine that generates an immune response that recognizes native epitopes.
Supporting information
One hundred nanograms per well of purified His-tagged LipL32 were separated in 10% SDS-PAGE then transferred to PVDF membrane for western blot analysis. The membrane was cut to strips and probed with 1:5,000 pooled sera collected before immunization (pre-bleed, P) and at day 32 (last bleed, LB) from immunized and control groups (A). Another membrane strip was incubated with 1:5,000 rabbit α-LipL32 as positive control (Co). As loading controls, the membrane strips were reprobed with 1:5,000 α-LipL32 (B) and 1:1,000 α-6xHis epitope (C). Sera collected from LigA7’-13 and/or LigB0-7 immunized animals do not recognize the unrelated protein nor the epitope tag.
(TIF)
Serum samples were collected from hamsters weekly during the immunization protocol. Anti-LigA7’-13 (A) or anti-LigB0-7 (B) antibody levels were measured in triplicate by ELISA. Each data line represents the average IgG response (minus pre-bleed read) of 5–8 animals over time while error bars indicate standard deviation. Dotted lines indicate vaccine immunization days (blue) or challenge with Leptospira (red). There was no difference in the immune response among treatment groups (LigA7’-13, LigB0-7, and LigA7’-13 + LigB0-7) or between the control groups (PBS and PBS + Adjuvant). Anti-LigA and anti-LigB IgG levels of treatment groups were statistically higher compared to control groups starting at day 11 post-immunization (Bonferroni multiple comparison test, *P<0.05, ***P<0.001, ****P<0.0001). Anti-LigB response of LigB-immunized hamsters was statistically higher than LigA7’-13 and LigA7’-13 + LigB0-7 immunized animals at day 11 (#P<0.05).
(TIF)
A 1:50 dilution of L. interrogans from hamster kidney culture was prepared in liquid EMJH (passage 2), and incubated at 30°C at 150 rpm. To determine Lig protein expression, bacteria at an OD420 = 0.1 was induced with 120 mM NaCl for 4 h at 30°C. Western blot analysis of salt-induced or uninduced L. interrogans using α-Lig antibody (dilution 1:2,000) show expression of both LigA and LigB by the challenge strain. Membrane was also probed with α-LipL41 (1:10,000 dilution) as loading control.
(TIF)
Acknowledgments
We are grateful to Jane Babbitt for the helpful discussions and critical review of this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a Veterans Affairs Merit Award to DAH, and National Institutes of Health grant R01 AI 034431 to DAH.
References
- 1.Adler B, de la Peña Moctezuma A. Leptospira and leptospirosis. Vet Microbiol. 2010; 140(3–4):287–96. doi: 10.1016/j.vetmic.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 2.Ko AI, Goarant C, Picardeau M. Leptospira: The dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009; 7(10):736–47. doi: 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evangelista KV, Coburn J. Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiol. 2010; 5(9):1413–25. doi: 10.2217/fmb.10.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14(2):296–326. doi: 10.1128/CMR.14.2.296-326.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Athanazio DA, Silva EF, Santos CS, Rocha GM, Vannier-Santos MA, McBride AJA, et al. Rattus norvegicus as a model for persistent renal colonization by pathogenic Leptospira interrogans. Acta Tropica. 2008; 105(2):176–80. doi: 10.1016/j.actatropica.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 6.Ganoza CA, Matthias MA, Saito M, Cespedes M, Gotuzzo E, Vinetz JM. Asymptomatic renal colonization of humans in the Peruvian Amazon by Leptospira. PLoS Negl Trop Dis. 2010; 4(2):e612 doi: 10.1371/journal.pntd.0000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koizumi N, Watanabe H. Leptospirosis vaccines: past, present, and future. J Postgrad Med. 2005; 51(3):210–4. [PubMed] [Google Scholar]
- 8.Lourdault K, Wang LC, Vieira A, Matsunaga J, Melo R, Lewis MS, et al. Oral immunization with Escherichia coli expressing a lipidated form of LigA protects hamsters against challenge with Leptospira interrogans serovar Copenhageni. Infect Immun. 2014; 82(2):893–902. doi: 10.1128/IAI.01533-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez Sánchez R OFA, Pérez Sierra A, Baly Gil A, Díaz González M, Baró Suárez M, Menéndez Capote R, et al. The reactogenicity and immunogenicity of the first Cuban vaccine against human leptospirosis. Rev Cub Med Trop. 1998; 50(2):159–66. [PubMed] [Google Scholar]
- 10.Adler B (Ed). Leptospira and leptospirosis Heidelberg: Springer-Verlag; 2014. [Google Scholar]
- 11.Faine S, Adler B, Bolin C, Perolat P. Leptospira and leptospirosis Melbourne: Medicsci; 1999. [Google Scholar]
- 12.Srikram A, Zhang K, Bartpho T, Lo M, Hoke DE, Sermswan RW, et al. Cross-protective immunity against leptospirosis elicited by a live, attenuated lipopolysaccharide mutant. J Infect Dis. 2011; 203(6):870–9. doi: 10.1093/infdis/jiq127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphryes PC, Weeks ME, AbuOun M, Thomson G, Nunez A, Coldham NG. Vaccination with leptospiral outer membrane lipoprotein LipL32 reduces kidney invasion of Leptospira interrogans serovar Canicola in hamsters. Clin Vaccine Immunol. 2014; 21(4):546–51. doi: 10.1128/CVI.00719-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branger C, Chatrenet B, Gauvrit A, Aviat F, Aubert A, Bach JM, et al. Protection against Leptospira interrogans sensu lato challenge by DNA immunization with the gene encoding Hemolysin-Associated Protein 1. Infect Immun. 2005; 73(7):4062–9. doi: 10.1128/IAI.73.7.4062-4069.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X, Sun A, Ruan P, Zhang Z, Yan J. Characterization of conserved combined T and B cell epitopes in Leptospira interrogans major outer membrane proteins OmpL1 and LipL41. BMC Microbiol. 2011; 11(1):21 doi: 10.1186/1471-2180-11-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haake DA, Mazel MK, McCoy AM, Milward F, Chao G, Matsunaga J, et al. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect Immun. 1999; 67(12):6572–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Y, Faisal SM, Yan W, Chang YC, McDonough SP, Zhang N, et al. Evaluation of novel fusion proteins derived from extracellular matrix binding domains of LigB as vaccine candidates against leptospirosis in a hamster model. Vaccine. 2011; 29(43):7379–86. doi: 10.1016/j.vaccine.2011.07.070 [DOI] [PubMed] [Google Scholar]
- 18.Coutinho ML, Choy HA, Kelley MM, Matsunaga J, Babbitt JT, Lewis MS, et al. A LigA three-domain region protects hamsters from lethal infection by Leptospira interrogans. PLoS Negl Trop Dis. 2011; 5(12):e1422 doi: 10.1371/journal.pntd.0001422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faisal S, Yan W, Chen C, Palaniappan R, McDonough SP, Chang Y-F. Evaluation of protective immunity of Leptospira immunoglobulin like protein A (LigA) DNA vaccine against challenge in hamsters. Vaccine. 2008; 26(2):277–87. doi: 10.1016/j.vaccine.2007.10.029 [DOI] [PubMed] [Google Scholar]
- 20.Faisal SM, Yan W, McDonough SP, Chang YF. Leptospira immunoglobulin-like protein A variable region (LigAvar) incorporated in liposomes and PLGA microspheres produces a robust immune response correlating to protective immunity. Vaccine. 2009; 27(3):378–87. doi: 10.1016/j.vaccine.2008.10.089 [DOI] [PubMed] [Google Scholar]
- 21.Forster KM, Hartwig DD, Seixas FK, Bacelo KL, Amaral M, Hartleben CP, et al. A conserved region of leptospiral immunoglobulin-like A and B proteins as a DNA vaccine elicits a prophylactic immune response against leptospirosis. Clin Vaccine Immunol. 2013; 20(5):725–31. doi: 10.1128/CVI.00601-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartwig DD, Bacelo KL, de Oliveira PD, Oliveira TL, Seixas FK, Amaral MG, et al. Mannosylated LigANI produced in Pichia pastoris protects hamsters against leptospirosis. Curr Microbiol. 2014; 68(4):524–30. doi: 10.1007/s00284-013-0505-4 [DOI] [PubMed] [Google Scholar]
- 23.Koizumi N, Watanabe H. Leptospiral immunoglobulin-like proteins elicit protective immunity. Vaccine. 2004; 22(11–12):1545–52. doi: 10.1016/j.vaccine.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 24.Monaris D, Sbrogio-Almeida ME, Dib CC, Canhamero TA, Souza GO, Vasconcellos SA, et al. Protective immunity and reduced renal colonization induced by vaccines containing recombinant Leptospira interrogans outer membrane proteins and flagellin adjuvant. Clin Vaccine Immunol. 2015; 22(8):965–73. doi: 10.1128/CVI.00285-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palaniappan RUM, McDonough SP, Divers TJ, Chen C, Pan M, Matsumoto M, et al. Immunoprotection of recombinant leptospiral immunoglobulin-like protein A against Leptospira interrogans serovar Pomona infection. Infect Immun. 2006; 74(3):1745–50. doi: 10.1128/IAI.74.3.1745-1750.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva EF, Medeiros MA, McBride AJ, Matsunaga J, Esteves GS, Ramos JG, et al. The terminal portion of leptospiral immunoglobulin-like protein LigA confers protective immunity against lethal infection in the hamster model of leptospirosis. Vaccine. 2007; 25(33):6277–86. doi: 10.1016/j.vaccine.2007.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan W, Faisal SM, McDonough SP, Divers TJ, Barr SC, Chang CF, et al. Immunogenicity and protective efficacy of recombinant Leptospira immunoglobulin-like protein B (rLigB) in a hamster challenge model. Microbes Infect. 2009; 11(2):230–7. doi: 10.1016/j.micinf.2008.11.008 [DOI] [PubMed] [Google Scholar]
- 28.Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, et al. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol Microbiol. 2003; 49(4):929–45. doi: 10.1046/j.1365-2958.2003.03619.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride AJ, Cerqueira GM, Suchard MA, Moreira AN, Zuerner RL, Reis MG, et al. Genetic diversity of the leptospiral immunoglobulin-like (Lig) genes in pathogenic Leptospira spp. Infect Genet Evol. 2009; 9(2):196–205. doi: 10.1016/j.meegid.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamburger ZA, Brown MS, Isberg RR, Bjorkman PJ. Crystal structure of invasin: a bacterial integrin-binding protein. Science. 1999; 286(5438):291–5. [DOI] [PubMed] [Google Scholar]
- 31.Luo Y, Frey EA, Pfuetzner RA, Creagh AL, Knoechel DG, Haynes CA, et al. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature. 2000; 405(6790):1073–7. doi: 10.1038/35016618 [DOI] [PubMed] [Google Scholar]
- 32.Choy HA, Kelley MM, Chen TL, Moller AK, Matsunaga J, Haake DA. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect Immun. 2007; 75(5):2441–50. doi: 10.1128/IAI.01635-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choy HA, Kelley MM, Croda J, Matsunaga J, Babbitt JT, Ko AI, et al. The multifunctional LigB adhesin binds homeostatic proteins with potential roles in cutaneous infection by pathogenic Leptospira interrogans. PloS One. 2011; 6(2):e16879 doi: 10.1371/journal.pone.0016879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin YP, Chang YF. A domain of the Leptospira LigB contributes to high affinity binding of fibronectin. Biochem Biophys Res Commun. 2007; 362(2):443–8. doi: 10.1016/j.bbrc.2007.07.196 [DOI] [PubMed] [Google Scholar]
- 35.Lin YP, Chang YF. The C-terminal variable domain of LigB from Leptospira mediates binding to fibronectin. J Vet Sci. 2008; 9(2):133–44. doi: 10.4142/jvs.2008.9.2.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin YP, Lee DW, McDonough SP, Nicholson LK, Sharma Y, Chang YF. Repeated domains of Leptospira immunoglobulin-like proteins interact with elastin and tropoelastin. J Biol Chem. 2009; 284(29):19380–91. doi: 10.1074/jbc.M109.004531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y-P, McDonough SP, Sharma Y, Chang Y-F. The terminal immunoglobulin-like repeats of LigA and LigB of Leptospira enhance their binding to gelatin binding domain of fibronectin and host cells. PloS One. 2010; 5(6):e11301 doi: 10.1371/journal.pone.0011301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castiblanco-Valencia MM, Fraga TR, Breda LCD, Vasconcellos SA, Figueira CP, Picardeau M, et al. Acquisition of negative complement regulators by the saprophyte Leptospira biflexa expressing LigA or LigB confers enhanced survival in human serum. Immunol Lett. 2016; 173:61–8. doi: 10.1016/j.imlet.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castiblanco-Valencia MM, Fraga TR, Pagotto AH, de Toledo Serrano SM, Abreu PAE, Barbosa AS, et al. Plasmin cleaves fibrinogen and the human complement proteins C3b and C5 in the presence of Leptospira interrogans proteins: A new role of LigA and LigB in invasion and complement immune evasion. Immunobiology. 2016; 221(5):679–89. doi: 10.1016/j.imbio.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 40.Choy HA. Multiple activities of LigB potentiate virulence of Leptospira interrogans: Inhibition of alternative and classical pathways of complement. PloS One. 2012; 7(7):e41566 doi: 10.1371/journal.pone.0041566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh CL, Chang E, Tseng A, Ptak C, Wu LC, Su CL, et al. Leptospira immunoglobulin-like protein B (LigB) binds to both the C-terminal 23 amino acids of fibrinogen αC domain and factor XIII: Insight into the mechanism of LigB-mediated blockage of fibrinogen α chain cross-linking. PLoS Negl Trop Dis. 2016; 10(9):e0004974 doi: 10.1371/journal.pntd.0004974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin YP, McDonough SP, Sharma Y, Chang YF. Leptospira immunoglobulin-like protein B (LigB) binding to the C-terminal fibrinogen αC domain inhibits fibrin clot formation, platelet adhesion and aggregation. Mol Microbiol. 2011; 79(4):1063–76. doi: 10.1111/j.1365-2958.2010.07510.x [DOI] [PubMed] [Google Scholar]
- 43.Lo M, Cordwell SJ, Bulach DM, Adler B. Comparative transcriptional and translational analysis of leptospiral outer membrane protein expression in response to temperature. PLoS Negl Trop Dis. 2009; 3(12):e560 doi: 10.1371/journal.pntd.0000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsunaga J, Sanchez Y, Xu X, Haake DA. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect Immun. 2005; 73(1):70–8. doi: 10.1128/IAI.73.1.70-78.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsunaga J, Coutinho ML. Positive regulation of Leptospira interrogans kdp expression by KdpE as demonstrated with a novel β-galactosidase reporter in Leptospira biflexa. Appl Environ Microbiol. 2012; 78(16):5699–707. doi: 10.1128/AEM.00713-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Croda J, Figueira CP, Wunder EA, Santos CS, Reis MG, Ko AI, et al. Targeted mutagenesis in pathogenic Leptospira species: disruption of the ligB gene does not affect virulence in animal models of leptospirosis. Infect Immun. 2008; 76(12):5826–33. doi: 10.1128/IAI.00989-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pappas CJ, Picardeau M. Control of gene expression in Leptospira spp. by Transcription Activator-Like Effectors demonstrates a potential role for LigA and LigB in Leptospira interrogans virulence. Appl Environ Microbiol. 2015; 81(22):7888–92. doi: 10.1128/AEM.02202-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fouts DE, Matthias MA, Adhikarla H, Adler B, Amorim-Santos L, Berg DE, et al. What makes a bacterial species pathogenic?: Comparative genomic analysis of the genus Leptospira. PLoS Negl Trop Dis. 2016; 10(2):e0004403 doi: 10.1371/journal.pntd.0004403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown RA, Blumerman S, Gay C, Bolin C, Duby R, Baldwin CL. Comparison of three different leptospiral vaccines for induction of a type 1 immune response to Leptospira borgpetersenii serovar Hardjo. Vaccine. 2003; 21(27–30):4448–58. [DOI] [PubMed] [Google Scholar]
- 50.Ellinghausen HC Jr., McCullough WG. Nutrition of Leptospira pomona and growth of 13 other serotypes: Fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am J Vet Res. 1965; 26:45–51. [PubMed] [Google Scholar]
- 51.Johnson RC, Harris VG. Differentiation of pathogenic and saprophytic leptospires I. Growth at low temperatures. J Bacteriol. 1967; 94(1):27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barocchi MA, Ko AI, Reis MG, McDonald KL, Riley LW. Rapid translocation of polarized MDCK cell monolayers by Leptospira interrogans, an invasive but nonintracellular pathogen. Infect Immun. 2002; 70(12):6926–32. doi: 10.1128/IAI.70.12.6926-6932.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva ÉF, Santos CS, Athanazio DA, Seyffert N, Seixas FK, Cerqueira GM, et al. Characterization of virulence of Leptospira isolates in a hamster model. Vaccine. 2008; 26(31):3892–6. doi: 10.1016/j.vaccine.2008.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, et al. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004; 186(7):2164–72. doi: 10.1128/JB.186.7.2164-2172.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haake DA. Hamster model of leptospirosis. Curr Protoc Microbiol. 2006; Chapter 12:Unit 12E 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoddard RA. Detection of pathogenic Leptospira spp. through real-time PCR (qPCR) targeting the LipL32 gene. Methods Mol Biol. 2013; 943:257–66. doi: 10.1007/978-1-60327-353-4_17 [DOI] [PubMed] [Google Scholar]
- 57.Wunder EA, Figueira CP, Santos GR, Lourdault K, Matthias MA, Vinetz JM, et al. Real-time PCR reveals rapid dissemination of Leptospira interrogans after intraperitoneal and conjunctival inoculation of hamsters. Infect Immun. 2016; 84(7):2015–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goris MGA, Hartskeerl RA. Leptospirosis serodiagnosis by the microscopic agglutination test Curr Protoc Microbiol: John Wiley & Sons, Inc.; 2005. [DOI] [PubMed] [Google Scholar]
- 59.Adler B, Faine S. Host immunological mechanisms in the resistance of mice to leptospiral infections. Infect Immun. 1977; 17(1):67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breda LC, Hsieh CL, Castiblanco Valencia MM, da Silva LB, Barbosa AS, Blom AM, et al. Fine mapping of the interaction between C4b-binding protein and outer membrane proteins LigA and LigB of pathogenic Leptospira interrogans. PLoS Negl Trop Dis. 2015; 9(10):e0004192 doi: 10.1371/journal.pntd.0004192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forster KM, Hartwig DD, Oliveira TL, Bacelo KL, Schuch R, Amaral MG, et al. DNA prime-protein boost based vaccination with a conserved region of leptospiral immunoglobulin-like A and B proteins enhances protection against leptospirosis. Mem Inst Oswaldo Cruz. 2015; 110(8):989–95. doi: 10.1590/0074-02760150222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murray GL, Lo M, Bulach DM, Srikram A, Seemann T, Quinsey NS, et al. Evaluation of 238 antigens of Leptospira borgpetersenii serovar Hardjo for protection against kidney colonisation. Vaccine. 2013; 31(3):495–9. doi: 10.1016/j.vaccine.2012.11.028 [DOI] [PubMed] [Google Scholar]
- 63.Conrad NL, Cruz McBride FW, Souza JD, Silveira MM, Felix S, Mendonca KS, et al. LigB subunit vaccine confers sterile immunity against challenge in the hamster model of leptospirosis. PLoS Negl Trop Dis. 2017; 11(3):e0005441 doi: 10.1371/journal.pntd.0005441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolin CA, Zuerner RL, Trueba G. Effect of vaccination with a pentavalent leptospiral vaccine containing Leptospira interrogans serovar Hardjo type Hardjo-Bovis on type Hardjo-Bovis infection of cattle. Am J Vet Res. 1989; 50(12):2004–8. [PubMed] [Google Scholar]
- 65.Bolin CA, Alt DP. Use of a monovalent leptospiral vaccine to prevent renal colonization and urinary shedding in cattle exposed to Leptospira borgpetersenii serovar Hardjo. Am J Vet Res. 2001; 62(7):995–1000. [DOI] [PubMed] [Google Scholar]
- 66.Naiman BM, Alt D, Bolin CA, Zuerner R, Baldwin CL. Protective killed Leptospira borgpetersenii vaccine induces potent Th1 immunity comprising responses by CD4 and gamma delta T lymphocytes. Infect Immun. 2001; 69(12):7550–8. doi: 10.1128/IAI.69.12.7550-7558.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolin CA, Cassells JA, Zuerner RL, Trueba G. Effect of vaccination with a monovalent Leptospira interrogans serovar hardjo type hardjo-bovis vaccine on type hardjo-bovis infection of cattle. Am J Vet Res. 1991; 52(10):1639–43. [PubMed] [Google Scholar]
- 68.Coutinho ML, Matsunaga J, Wang LC, de la Pena Moctezuma A, Lewis MS, Babbitt JT, et al. Kinetics of Leptospira interrogans infection in hamsters after intradermal and subcutaneous challenge. PLoS Negl Trop Dis. 2014; 8(11):e3307 doi: 10.1371/journal.pntd.0003307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Truccolo J, Charavay F, Merien F, Perolat P. Quantitative PCR assay to evaluate ampicillin, ofloxacin, and doxycycline for treatment of experimental leptospirosis. Antimicrob Agents Chemother. 2002; 46(3):848–53. doi: 10.1128/AAC.46.3.848-853.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
One hundred nanograms per well of purified His-tagged LipL32 were separated in 10% SDS-PAGE then transferred to PVDF membrane for western blot analysis. The membrane was cut to strips and probed with 1:5,000 pooled sera collected before immunization (pre-bleed, P) and at day 32 (last bleed, LB) from immunized and control groups (A). Another membrane strip was incubated with 1:5,000 rabbit α-LipL32 as positive control (Co). As loading controls, the membrane strips were reprobed with 1:5,000 α-LipL32 (B) and 1:1,000 α-6xHis epitope (C). Sera collected from LigA7’-13 and/or LigB0-7 immunized animals do not recognize the unrelated protein nor the epitope tag.
(TIF)
Serum samples were collected from hamsters weekly during the immunization protocol. Anti-LigA7’-13 (A) or anti-LigB0-7 (B) antibody levels were measured in triplicate by ELISA. Each data line represents the average IgG response (minus pre-bleed read) of 5–8 animals over time while error bars indicate standard deviation. Dotted lines indicate vaccine immunization days (blue) or challenge with Leptospira (red). There was no difference in the immune response among treatment groups (LigA7’-13, LigB0-7, and LigA7’-13 + LigB0-7) or between the control groups (PBS and PBS + Adjuvant). Anti-LigA and anti-LigB IgG levels of treatment groups were statistically higher compared to control groups starting at day 11 post-immunization (Bonferroni multiple comparison test, *P<0.05, ***P<0.001, ****P<0.0001). Anti-LigB response of LigB-immunized hamsters was statistically higher than LigA7’-13 and LigA7’-13 + LigB0-7 immunized animals at day 11 (#P<0.05).
(TIF)
A 1:50 dilution of L. interrogans from hamster kidney culture was prepared in liquid EMJH (passage 2), and incubated at 30°C at 150 rpm. To determine Lig protein expression, bacteria at an OD420 = 0.1 was induced with 120 mM NaCl for 4 h at 30°C. Western blot analysis of salt-induced or uninduced L. interrogans using α-Lig antibody (dilution 1:2,000) show expression of both LigA and LigB by the challenge strain. Membrane was also probed with α-LipL41 (1:10,000 dilution) as loading control.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.