Abstract
Objective
We performed single-variant and gene-based association analyses of plasma amyloid-β (aβ) concentrations using whole exome sequence from 1,414 African and European Americans. Our goal was to identify genes that influence plasma aβ42 concentrations and aβ42:aβ40 ratios in late middle age (mean = 59 years), old age (mean = 77 years), or change over time (mean = 18 years).
Methods
Plasma aβ measures were linearly regressed onto age, gender, APOE ε4 carrier status, and time elapsed between visits (fold-changes only) separately by race. Following inverse normal transformation of the residuals, seqMeta was used to conduct race-specific single-variant and gene-based association tests while adjusting for population structure. Linear regression models were fit on autosomal variants with minor allele frequencies (MAF)≥1%. T5 burden and Sequence Kernel Association (SKAT) gene-based tests assessed functional variants with MAF≤5%. Cross-race fixed effects meta-analyses were Bonferroni-corrected for the number of variants or genes tested.
Results
Seven genes were associated with aβ in late middle age or change over time; no associations were identified in old age. Single variants in KLKB1 (rs3733402; p = 4.33x10-10) and F12 (rs1801020; p = 3.89x10-8) were significantly associated with midlife aβ42 levels through cross-race meta-analysis; the KLKB1 variant replicated internally using 1,014 additional participants with exome chip. ITPRIP, PLIN2, and TSPAN18 were associated with the midlife aβ42:aβ40 ratio via the T5 test; TSPAN18 was significant via the cross-race meta-analysis, whereas ITPRIP and PLIN2 were European American-specific. NCOA1 and NT5C3B were associated with the midlife aβ42:aβ40 ratio and the fold-change in aβ42, respectively, via SKAT in African Americans. No associations replicated externally (N = 725).
Conclusion
We discovered age-dependent genetic effects, established associations between vascular-related genes (KLKB1, F12, PLIN2) and midlife plasma aβ levels, and identified a plausible Alzheimer’s Disease candidate gene (ITPRIP) influencing cell death. Plasma aβ concentrations may have dynamic biological determinants across the lifespan; plasma aβ study designs or analyses must consider age.
Introduction
Alzheimer’s disease (AD) is a major public health burden, afflicting 5.1 million Americans aged 65 or older; the number of cases is expected to triple by 2050, costing the nation $1.1 trillion [1]. Although there are no effective treatments to prevent, slow, or cure AD, researchers have made considerable progress in dissecting its genetic etiology and revealing biological pathways that may contain druggable targets. The International Genomics of Alzheimer’s Project leveraged results from genome-wide association studies (GWAS) of late-onset AD (LOAD) to identify candidate therapeutic targets in the immune response, endocytosis, cholesterol transport, and protein ubiquitination pathways [2]. GWAS and sequencing studies have nominated variants (representing ≈30 genes) from the whole allele frequency spectrum [3–7], yet there is a substantial proportion of LOAD heritability unexplained [6, 7]. The identification of additional therapeutic candidates is hindered by the reduced power of case-control outcomes [8].
More recent studies have employed endophenotypes of particular facets of AD pathophysiology to identify known and novel candidate genes using moderate sample sizes [9–12]. Capitalizing on the critical role that amyloid-β (aβ) plays in AD pathophysiology [12], along with functional information linking most AD-associated genes to aβ production and clearance [4], genome-wide association studies have employed cerebrospinal fluid (CSF) and plasma aβ concentrations as endophenotypes [9–12]. Plasma levels are less expensive and invasive to measure than CSF levels, are associated with brain aβ [13–16], and may capture any shared mechanisms regulating aβ processes (production, secretion, degradation, and clearance) throughout the body [12, 17]. However, their utility as AD endophenotypes is questionable. Only a handful of prospective studies and systematic meta-analyses have associated plasma aβ42 levels or the aβ42:aβ40 ratio with LOAD [18–23], while a paucity of associations have been documented between plasma aβ traits and AD linkage and GWAS findings [17, 24, 25]. Furthermore, plasma aβ concentrations reflect a wide array of tissue sources [26] and age-related health conditions, such as subcortical white matter lesions, cerebral microbleeds, hypertension, diabetes, infarcts, ischemic heart disease, and chronic kidney disease [14, 27, 28], a few of which are themselves associated with AD. This fosters ambiguity in the pathophysiological interpretation (AD or non-AD) of any findings.
Therefore, before exploiting plasma aβ concentrations in genetic or epidemiological studies of AD, a more comprehensive assessment of their age-dependent biological determinants is needed [27]. Deciphering the dynamic genetic architecture of plasma aβ concentrations could simultaneously implicate pathophysiological processes, pathways, and mechanisms contributing to aβ plaque accumulation in the asymptomatic preclinical and progressive phases of AD [29, 30] while suggesting novel health conditions that alter the association between plasma aβ and AD in epidemiological studies. This vital information may clarify the inconsistencies in reported associations between plasma aβ and AD across studies, as well as provide insight into the role (if any) of plasma aβ in AD pathogenesis and the utility of plasma aβ concentrations as AD biomarkers.
To systematically assess the dynamic biological determinants of plasma aβ, we performed an exome-wide association study of plasma aβ concentrations measured during two different life stages. The goal was to identify genes influencing plasma aβ42 concentrations and aβ42:aβ40 ratios in late middle age, old age, or change over time. We performed single-variant and gene-based association analyses using whole exome sequence from 1,414 Atherosclerosis Risk in Communities-Neurocognitive Study (ARIC-NCS) participants. This study was well-suited for this investigation because: 1) participants had two amyloid measurements spaced an average of 18 years apart, with mean ages of 59 and 77 years for the two blood draws; 2) the sample included both European (EAs) and African Americans (AAs), allowing exploration of rare population-specific variants that may contribute to health disparities; 3) the sample was enriched for dementia and mild cognitive impairment, and hence should be enriched for variants contributing to aβ accumulation; and 4) the availability of whole exome sequence permitted the analysis of both common and rare variants, enabling the first systematic interrogation of rare variants for plasma aβ levels. This investigation expanded upon the lone published GWAS meta-analysis of plasma aβ concentrations [12] which focused on cross-sectional measurements in non-demented elderly participants of European ancestry.
Subjects and methods
Subjects
ARIC was initiated in 1987 as a population-based cohort study of 15,792 middle-aged (45–64 years) participants drawn from four US communities (Washington County, MD; Forsyth County, NC; Jackson, MS; and suburban Minneapolis, MN) [31]. Four study visits were completed by 1999, with a fifth visit (ARIC-NCS; N = 6,538) conducted in 2011–2013 [32]. Plasma aβ, the phenotype for this investigation, was quantified on a subset (N = 2,588) of ARIC-NCS enriched for cognitive impairment. The plasma aβ sample included all individuals exhibiting impaired cognitive status (defined as low mini-mental status exam score or low standardized score on any of five cognitive domains accompanied by cognitive decline on longitudinally administered tests) during the fifth exam, all participants with a brain MRI from a prior ARIC exam, and an age-stratified (<80 years, ≥80 years) random sample of the remaining cognitively normal participants from each field center. This investigation focused on 1,414 AA and EA participants with whole exome sequence, covariates, and amyloids measured at the third (1993–1995) and fifth visits. Of these participants, 152 (72 AAs and 80 EAs) had dementia and 560 (152 AAs and 408 EAs) had mild cognitive impairment by the fifth visit; 79% of these dementia and cognitive impairment cases were ascribed to AD as the primary etiology. Cognitive diagnoses were adjudicated at the fifth visit using cognitive, neurologic, and brain imaging assessments (comprehensive diagnostic details are given in [32]). Cognitive status was not available on the whole sample at the third visit. The ARIC study has been approved by the Institutional Review Board at each field center, namely Wake Forest Baptist Medical Center (Forsyth County, NC), University of Mississippi Medical Center (Jackson, MS), University of Minnesota (suburban Minneapolis, MN), and Johns Hopkins University (Washington County, MD). Participants provided written informed consent prior to each examination.
Plasma amyloid-β ascertainment
Amyloid quantification was performed by the Department of Molecular Pharmacology and Experimental Therapeutics at Mayo Clinic, Jacksonville, FL, from August to December 2014. The INNO-BIA assay (INNOGENETICS N.V, Ghent, Belgium) required 69 plates to measure aβ42 and aβ40 levels in both races; for each participant, the same plate was used to simultaneously measure the amyloid levels at the third and fifth visit. Beads (xMAP microspheres; conjugate 1A) bound to aβ40 and aβ42 emitted fluorescence detected by the Luminex 200 IS Total system. A five-parameter logistic regression model related the fluorescence intensities of six standards to their known amyloid concentrations. The resultant model predicted the concentrations of aβ40 and aβ42 from the measured fluorescence intensities in the samples. Intensities outside the range of the standards could not be inferred. For the single-visit analyses of aβ42, samples with intensities below the minimal detectable level were assigned the threshold concentrations (12 pg/ml). These individuals were omitted from the analysis of fold-changes in aβ42 and all analyses of the aβ42:aβ40 ratio since their ranks relative to those with measured values were inconclusive; 98 (36 AAs, 62 EAs), 17 (6 AAs, 11 EAs), 107 (41 AAs, 66 EAs), and 105 (41 AAs, 64 EAs) participants were omitted from the ratio at visit 3, the ratio at visit 5, the fold-change in the ratio, and the fold-change in aβ42 analyses, respectively, due to subthreshold intensities.
Whole exome sequencing
DNA samples were assembled into Illumina paired-end pre-capture libraries; the oligonucleotide sequences and protocol are available on the Baylor College of Medicine Human Genome Sequencing Center (HGSC) website (http://www.hgsc.bcm.edu/content/protocols-sequencing-library-construction). Two, four, or six pre-capture libraries were pooled together, hybridized to the HGSC VCRome 2.1 design [33] (42Mb, NimbleGen), and sequenced in a single lane on the Illumina HiSeq 2000 or the HiSeq 2500 platform. The HGSC Mercury pipeline (https://www.hgsc.bcm.edu/content/mercury) conducted the Illumina sequence analysis while the Consensus Assessment of Sequence and Variation program de-multiplexed the pooled samples. The Burrows-Wheeler Alignment [34] algorithm mapped reads to the Genome Reference Consortium Human Build 37 (GRCh37) sequence, producing Binary Alignment/Map (BAM) files. Aligned reads were then recalibrated using the Genome Analysis ToolKit [35], BAM sorting, duplicate read marking, and realignment near insertions or deletions (indels). The Atlas2 [36] suite called both single nucleotide variants (SNVs) and indels, generating high-quality variant call files.
The ARIC exome sequence was quality controlled as part of the Cohorts for Hearts and Aging Research in Genomic Epidemiology (CHARGE) consortium. SNVs and indels were centrally filtered for posterior probabilities<0.95, variant read counts<3, variant read ratios <0.25 or >0.75, total read depths of the references<10-fold, mappability scores<0.8, missing rates>20%, mean coverage depths>500-fold, race-specific Hardy-Weinberg Equilibrium p-values<5x10-6, and strand bias (>99% variant reads in a single strand direction). In addition, SNVs with total coverage<10-fold and indels with total coverage<30-fold were excluded. Individuals were excluded for sex-mismatch, missingness>20%, singleton counts of 0, or values>6 standard deviations from the mean for singleton counts, race-specific mean depths, TiTv ratios, or heterozygote to homozygote ratios. The mean depth of coverage was 109.6 and 82.7 for AAs and EAs, respectively.
The annotation file included quality-controlled variants observed in at least one available exome sequencing project (e.g. CHARGE, the NHLBI Exome Sequencing Project). Variants were annotated using ANNOVAR [37] and dbNSFP v2.0 according to GRCh37 and RefSeq. This multiple study SNPinfo file was used as a component of the R package seqMeta (http://cran.rproject.org/web/packages/seqMeta/index.html).
Covariates
We included age, sex, and apolipoprotein-E (APOE) as covariates in this investigation. APOE genotypes were determined using TaqMan assays and the ABI 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). APOE ε4 carrier status (0 = No, 1 = Yes) indicated whether an individual carried at least one copy of the ε4 allele; this carrier status was strongly associated (minimum p-value of 1x10-8 for aβ42 at visit 5) with age- and gender-adjusted plasma aβ levels in EAs and was moderately (p-values in the range of 0.1 to 0.2) associated in AAs. Although the sample included APOE ε4 homozygotes (23 (6%) and 20 (2%) in the AAs and EAs, respectively), APOE ε4 carrier status tended to fit better (by Akaike Information Criterion in models of age- and gender-adjusted plasma aβ) than separate coefficients for ε4 heterozygotes and homozygotes or the number of ε4 alleles.
Population structure
Race-specific principal components (PCs), the first ten of which were utilized to control for population stratification in the statistical analysis, were calculated from the genotype data using Eigenstrat [38]. Variants with minor allele frequencies (MAFs)<0.05, missing rates>0.05, or Hardy-Weinberg Equilibrium p-values<1x10-5 were excluded before pruning the variants for linkage disequilibrium (r2) >0.3. In total, 29,551 and 16,323 SNVs were used to construct the PCs in AAs and EAs, respectively.
Statistical methods
Single-visit amyloid measures were linearly regressed onto age, gender, and APOE ε4 carrier status separately by race (field center was not statistically significant). The fold-change amyloid traits were regressed onto the age at the third visit, the time elapsed between visits, gender, and APOE ε4 carrier status. Following rank-based inverse normal transformation of the residuals, we used seqMeta (version 1.5; http://cran.r-project.org/web/packages/seqMeta/) to conduct race-specific single-variant and gene-based association tests while adjusting for population structure (the first ten PCs). Linear regression models, assuming additive genetic effects, were fit on all autosomal SNVs with MAF≥0.01. T5 burden and Sequence Kernel Association (SKAT; using default “Wu” weights) gene-based tests were conducted on functional (nonsynonymous, splicing, stop gain, stop loss, or frameshift) autosomal variants with MAF ≤ 0.05. For both the single-variant and gene-based tests, we meta-analyzed the African and European American results (score statistics and genotype covariance matrices) using fixed effects models in seqMeta. We applied a Bonferroni correction for the number of unique variants (up to 113,423) or genes (up to 16,733) tested per trait (S1 Table contains the number of variants/genes tested per trait), yielding exome-wide significance thresholds of 4.41x10-7 (0.05/113,423) and 2.99 x10-6 (0.05/16,733) for the single-variant and gene-based tests, respectively. The quantile-quantile plots from the race-specific analyses and the cross-race meta-analyses exhibited minimal inflation and were well-behaved for both the single-variant (genomic inflation factors ranged from 0.99 to 1.03; S1 Fig) and gene-based tests (both overall and by minor allele count (MAC) thresholds; S2–S7 Figs).
Replication
We internally replicated significant single-variant tests using 1,014 ARIC participants with exome chip (Illumina Human Exome BeadChip v1.0) but not exome sequence data; these individuals had plasma aβ measured at both the third and fifth visits and were used to replicate both cross-sectional and change over time traits. The genotype calling and quality-control procedures have been described elsewhere [39], while the statistical methods matched those of the sequence analyses. We did not internally replicate significant gene-based tests because few functional SNVs (≤ 2) were available in the genes of interest using exome chip participants from the appropriate racial group. We externally replicated both the significant single-variant and gene-based tests using 725 participants from the Framingham Heart Study (FHS) Offspring and Third Generation (Gen 2 and Gen 3) cohorts [40, 41]; these participants had amyloids measured once and were used to replicate cross-sectional findings only. All significant cross-sectional associations from the discovery phase were with midlife plasma aβ levels (i.e. from the third visit in ARIC), thus we restricted the FHS replication sample to participants aged 50–70 years (mean age of 60±6 years, 48% female). The amyloid assessment was consistent between FHS and ARIC (the INNO-BIA assay conducted at the same lab), as was the exome sequence calling, quality control, and annotation (both members of CHARGE). The single-variant and gene-based analyses of the inverse-transformed amyloids incorporated a kinship matrix to account for FHS family structure and included significant PCs.
Results
The discovery analysis included 406 AAs and 1,008 EAs with whole exome sequence (Table 1; for summary statistics by cognitive status, see S2 and S3 Tables). Only one-third of AAs were male, whereas about half (47%) of EAs were male. Both races had similar age distributions, with the third visit corresponding to late middle age and the fifth visit corresponding to older age, and high APOE ε4 carriage rates compared to the general population (20–25%) [42]. In both AAs and EAs, mean aβ42 levels increased between visits and mean aβ42:aβ40 ratios decreased between visits. Although the third visit aβ42 levels differed between the races, the mean fold-changes were similar. The internal (ARIC exome chip) and external (FHS exome sequence) replication samples had lower APOE ε4 carriage rates (34% for ARIC AAs, 18% for ARIC EAs, and 22% for FHS EAs) than the discovery samples (S4 Table). The FHS replication sample included only EAs and had more males (52%) and higher midlife plasma aβ levels (both aβ42 and aβ42:aβ40 ratio) than all ARIC samples.
Table 1. Descriptive statistics of sequenced ARIC participants.
| Characteristic | African Americans (N = 406) | European Americans (N = 1,008) |
|---|---|---|
| % Female | 67.7% | 53.4% |
| % APOE ε4 carriers | 44.1% | 33.0% |
| Time between amyloid measurements (in years) | ||
| Mean (SD) | 18 (1) | 18 (1) |
| Range | 16–20 | 16–20 |
| Age (in years) | ||
| Visit 3 Mean (SD) | 59 (5) | 60 (5) |
| Range | 50–71 | 50–70 |
| Visit 5 Mean (SD) | 77 (5) | 77 (5) |
| Range | 67–89 | 67–90 |
| Plasma aβ42 (in pg/ml) | ||
| Visit 3 Mean (SD) | 26.69 (9.30) | 30.06 (9.64) |
| Interquartile Interval | 20.10–31.80 | 23.20–36.34 |
| Visit 5 Mean (SD) | 33.57 (12.06) | 38.45 (11.23) |
| Interquartile Interval | 25.50–39.70 | 30.70–45.20 |
| Fold-change Mean (SD) | 1.30 (0.49) | 1.31 (0.41) |
| Interquartile Interval | 1.04–1.44 | 1.06–1.47 |
| aβ42:aβ40 ratio | ||
| Visit 3 Mean (SD) | 0.20 (0.11) | 0.19 (0.07) |
| Interquartile Interval | 0.15–0.22 | 0.15–0.23 |
| Visit 5 Mean (SD) | 0.17 (0.07) | 0.17 (0.06) |
| Interquartile Interval | 0.13–0.20 | 0.13–0.19 |
| Fold-change Mean (SD) | 0.95 (0.33) | 0.93 (0.31) |
| Interquartile Interval | 0.75–1.11 | 0.73–1.08 |
NOTE: SD = Standard deviation. The amyloid measures were non-normal, thus the interquartile interval conveyed the amyloid values in the 25th to 75th percentile.
Single-variant analysis
ARIC exome sequence analyses
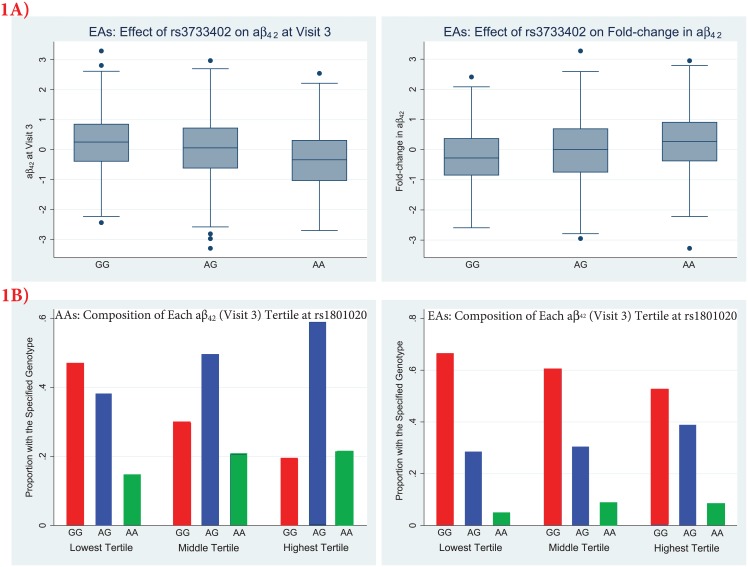
Kallikrein B, plasma (Fletcher factor) 1 [KLKB1] and coagulation factor XII (Hageman factor) [F12] contained common SNVs significantly (p≤4.41x10-7) associated with aβ42 traits (Table 2). Nonsynonymous SNV rs3733402 in KLKB1 was associated with aβ42 levels at the third visit and the fold-change in aβ42 across visits; these associations were significant using the EAs alone but had the same directions of effects and moderate statistical support (p<0.09) in AAs. EAs with the A allele had lower aβ42 levels at the third visit but an increased fold-change in aβ42 over visits (Fig 1A); individuals with this allele may start late-midlife with lower levels but have larger increases over the next 16–20 years. A second SNV in KLKB1 (rs925453) was associated with the third visit aβ42 levels but became insignificant after conditioning on rs3733402 (p-values of 0.27 and 0.43 in AAs and EAs, respectively), undermining its claim as an independent finding.
Table 2. Significant single-variant results.
| Trait | Gene | SNV Rs Number (Functional Region) | Coded Allele | Chr | Position in base pairs (Build 37) | Analysis | Race | N | CAF | β | se(β) | p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aβ42 at visit 3 | KLKB1 | rs3733402 (Nonsynonymous) | A | 4 | 187,158,034 | ARIC Exome Sequence | AA | 406 | 0.73 | -0.18 | 0.08 | 0.03 |
| EA | 1,007 | 0.51 | -0.26 | 0.04 | 3.55E-09 | |||||||
| Meta | 1,413 | 0.57 | -0.24 | 0.04 | 4.33E-10 | |||||||
| ARIC Exome Chip | AA | 149 | 0.69 | -0.12 | 0.14 | 0.40 | ||||||
| EA | 865 | 0.51 | -0.19 | 0.05 | 4.02E-05 | |||||||
| Meta | 1,014 | 0.54 | -0.19 | 0.04 | 3.07E-05 | |||||||
| Meta-ARIC | ALL | 2,427 | 0.56 | -0.22 | 0.03 | 9.90E-14 | ||||||
| FHS | EA | 725 | 0.53 | 0.04 | 0.05 | 0.41 | ||||||
| F12 | rs1801020 (UTR5) | G | 5 | 176,836,532 | ARIC Exome Sequence | AA | 406 | 0.57 | -0.28 | 0.07 | 7.71E-05 | |
| EA | 1,007 | 0.76 | -0.20 | 0.05 | 8.37E-05 | |||||||
| Meta | 1,413 | 0.71 | -0.23 | 0.04 | 3.89E-08 | |||||||
| FHS | EA | 725 | 0.75 | -0.05 | 0.06 | 0.39 | ||||||
| Fold-change in aβ42 | KLKB1 | rs3733402 (Nonsynonymous) | A | 4 | 187,158,034 | ARIC Exome Sequence | AA | 364 | 0.71 | 0.14 | 0.08 | 0.09 |
| EA | 941 | 0.50 | 0.27 | 0.05 | 9.28E-09 | |||||||
| Meta | 1,305 | 0.56 | 0.24 | 0.04 | 5.03E-09 | |||||||
| ARIC Exome Chip | AA | 132 | 0.69 | 0.19 | 0.15 | 0.20 | ||||||
| EA | 818 | 0.50 | 0.16 | 0.05 | 8.88E-04 | |||||||
| Meta | 950 | 0.53 | 0.16 | 0.05 | 3.80E-04 | |||||||
| Meta- ARIC | ALL | 2,255 | 0.55 | 0.20 | 0.03 | 1.58E-11 |
NOTE: Chr = Chromosome; N = Sample size; CAF = Coded allele frequency; β = Effect of each copy of the coded allele on the trait; se(β) = Standard error of the effect of each copy of the coded allele on the trait. SNV rs1801020 was not available on the exome chip in ARIC and was poorly imputed in the 1000 Genomes GWAS data in ARIC (IMPUTE2 imputation qualities of 0.707 and 0.606 in AAs and EAs, respectively).
Fig 1. KLKB1 and F12 single-variant associations in ARIC participants with exome sequence.
Panel A shows boxplots of the third visit aβ42 levels and the fold-changes in aβ42 stratified by the rs3733402 genotype in EAs. Panel B depicts the relative frequencies of the rs1801020 genotypes within each aβ42 (visit 3) tertile in AAs and EAs. The plotted values are inverse normal transformed amyloid values adjusted for age, gender, time between visits (fold-change aβ42 only), and APOE ε4 carriage status.
Rs1801020 in the 5’ untranslated region of F12 was significantly associated with the third visit aβ42 levels through cross-race meta-analysis (Table 2). Both races provided evidence of association (p<0.0001), with lower aβ42 levels for the G allele. The lowest tertile of aβ42 had the largest proportion of participants with the GG genotype in both races (Fig 1B). Eight suggestive associations (4.41x10-7<p ≤8.82x10-6) with aβ were identified (S5 Table), including SNVs in PPP5C (rs917948), FUT9 (rs9499636), ECHS1 (rs2230260), SMAP1 (rs576516), PDZD8 (rs35664484), RRP12 (rs6584122), CACNA2D4 (rs202022529) and ADGRF5 (rs678312).
Replication analyses
We internally validated the significant KLKB1 (rs3733402) associations using 1,014 ARIC participants who had exome chip but not exome sequence data. The directions of effects on the third visit aβ42 levels and the fold-change in aβ42 levels were consistent between participants with exome chip and sequence (Table 2). The rs3733402 associations replicated in EAs alone, with strengthened evidence in the cross-race meta-analysis. We could not internally replicate rs1801020 in F12; it was not available on the exome chip and was poorly imputed in the 1000 Genomes GWAS data (AAs and EAs had imputation qualities of 0.707 and 0.606, respectively). Neither the KLKB1 or F12 variants externally replicated in FHS.
Gene-based tests
ARIC exome sequence analyses
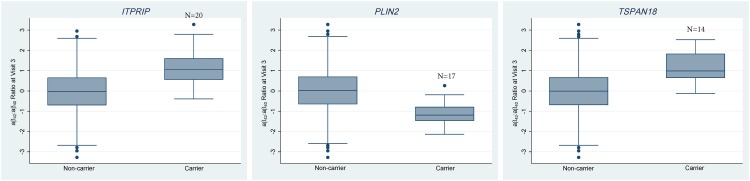
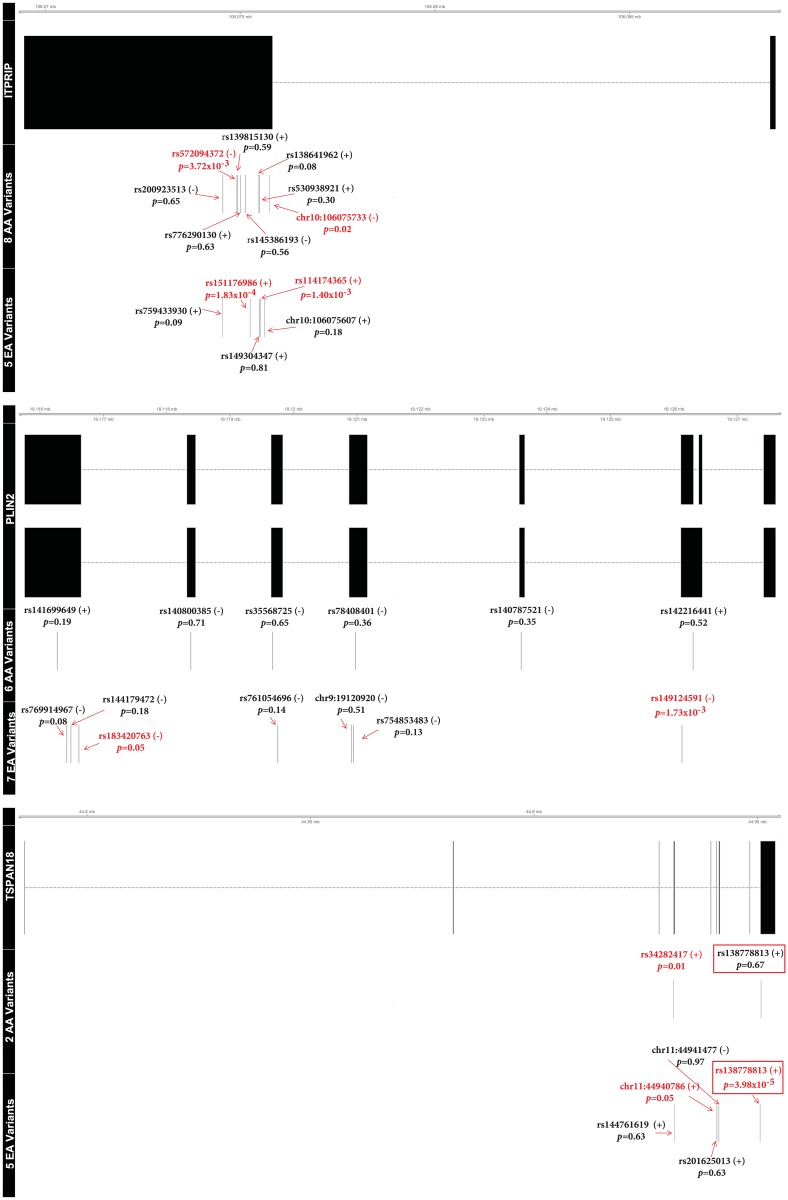
Inositol 1,4,5-triphosphate receptor interacting protein [ITPRIP], perilipin-2 [PLIN2], tetraspanin 18 [TSPAN18], nuclear receptor coactivator 1 [NCOA1], and 5’-nucleotidase, cytosolic IIIB [NT5C3B] were associated (p≤2.99x10-6) with plasma aβ via gene-based tests (Table 3) and had cumulative MAFs ranging from 0.76% to 1.49% in the significant race-specific- or meta-analysis. ITPRIP, PLIN2, and TSPAN18 were significantly associated with the third visit aβ42:aβ40 ratio via the T5 burden test; the ITPRIP and PLIN2 associations were EA-specific with no shared variants across races, whereas TSPAN18 showed nominal association (p<0.05) in both races, had one variant (rs138778813) shared across races, and was significant in the cross-race meta-analysis. Carrying a minor allele in ITPRIP or TSPAN18 increased the third visit aβ42:aβ40 ratio in EAs whereas carrying a minor allele in PLIN2 decreased the ratio (Fig 2). These patterns were reflected in the single-variant model coefficients of the contributing variants (Fig 3 and S6–S8 Tables); all PLIN2 variants had negative coefficients in EAs and all but one of the TSPAN18 and ITPRIP variants had positive coefficients. The lone overlapping TSPAN18 variant (rs138778813) increased the ratio in both races but showed greater evidence of association in EAs than AAs (single-variant p-values of 3.98x10-5 and 0.67, respectively), possibly due to differences in the number of copies of the minor allele in the two groups (10 and 1 minor allele copies, respectively).
Table 3. Significant gene-based (T5 and SKAT) results.
| Significant Test in ARIC | Trait | Gene | Chr | Analysis | Race | N | MAC | # SNVs | T5 Test | SKAT | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | se(β) | p-value | p-value | |||||||||
| T5 | aβ42:aβ40 ratio at visit 3 | ITPRIP | 10 | ARIC | AA | 370 | 47 | 8 | -0.11 | 0.15 | 0.48 | 0.15 |
| EA | 945 | 20 | 5 | 1.13 | 0.23 | 5.40E-07 | 9.68E-05 | |||||
| Meta | 1,315 | 67 | 13 | 0.28 | 0.13 | 0.03 | 3.57E-03 | |||||
| FHS | EA | 725 | 10 | 7 | 0.10 | 0.32 | 0.75 | 0.03 | ||||
| PLIN2 | 9 | ARIC | AA | 370 | 18 | 6 | -0.14 | 0.25 | 0.57 | 0.65 | ||
| EA | 945 | 17 | 7 | -1.15 | 0.25 | 2.94E-06 | 1.02E-03 | |||||
| Meta | 1,315 | 146a | 13 | -0.33 | 0.08 | 8.59E-05 | 1.05E-03 | |||||
| FHS | EA | 725 | 18 | 6 | -0.28 | 0.24 | 0.24 | 0.44 | ||||
| TSPAN18 | 11 | ARIC | AA | 370 | 6 | 2 | 1.00 | 0.42 | 0.02 | 0.02 | ||
| EA | 945 | 14 | 5 | 1.15 | 0.27 | 2.04E-05 | 4.29E-05 | |||||
| Meta | 1,315 | 20 | 6 | 1.11 | 0.23 | 1.08E-06 | 1.74E-05 | |||||
| FHS | EA | 725 | 25 | 5 | -0.14 | 0.21 | 0.51 | 0.27 | ||||
| SKAT | aβ42:aβ40 ratio at visit 3 | NCOA1 | 2 | ARIC | AA | 370 | 11 | 5 | 0.90 | 0.31 | 4.02E-03 | 1.34E-06 |
| EA | 945 | 85 | 15 | -0.11 | 0.11 | 0.31 | 0.08 | |||||
| Meta | 1,315 | 96 | 19 | 0.00 | 0.10 | 0.99 | 0.65 | |||||
| FHS | EA | 725 | 58 | 10 | -0.06 | 0.13 | 0.64 | 0.46 | ||||
| Fold-change in aβ42b | NT5C3B | 17 | ARIC | AA | 364 | 7 | 4 | 1.35 | 0.38 | 4.40E-04 | 5.67E-07 | |
| EA | 941 | 9 | 5 | 0.02 | 0.31 | 0.94 | 0.88 | |||||
| Meta | 1,305 | 16 | 9 | 0.54 | 0.24 | 0.02 | 3.18E-06 | |||||
NOTE: Chr = Chromosome; N = Sample size; MAC = Number of copies of the minor alleles; # SNVs = Number of SNVs contributing to the gene-based test; β = Effect of the minor alleles on the trait; se(β) = Standard error of the effect of the minor alleles.
aOne of the PLIN2 SNVs (rs35568725) had a minor allele frequency of 0.059 in EAs but 0.005 in AAs, thus was included in the meta-analysis (combined minor allele frequency 0.044) for both cohorts by default; we performed the meta-analysis both including and omitting this variant in the EA group. Including this variant in both racial groups yielded a MAC of 146, a T5 meta-analysis p-value of 8.59E-05 (β = -0.33, se(β) = 0.08), and a SKAT meta-analysis p-value of 1.05E-03. Excluding this variant in the EAs yielded a minor allele count of 35, a T5 meta-analysis p-value of 2.12E-04 (β = -0.64, se(β) = 0.17), and a SKAT meta-analysis p-value of 3.36E-03.
b Fold-change in aβ42 was not available in FHS.
Fig 2. Effects of carrying a minor allele in TSPAN18, ITPRIP, and PLIN2 on the third visit aβ42: aβ40 ratio in EAs.
The plotted values are the inverse normal transformed ratios adjusted for age, gender, and APOE ε4 carriage status.
Fig 3. Variants contributing to the significant T5 gene-based tests.
This figure depicts the single-variant model results for variants contributing to the T5 tests of ITPRIP, PLIN2, and TSPAN18 on the third visit aβ42: aβ40 ratio. One isoform was drawn unless the regions harboring variants differed substantially. Each contributing variant is labeled by name, single-variant association p-value, and direction of the minor allele effect (in parentheses). Variants with nominal (p<0.05) levels of association are in red. Overlapping variants in the two racial groups are encapsulated by a red box.
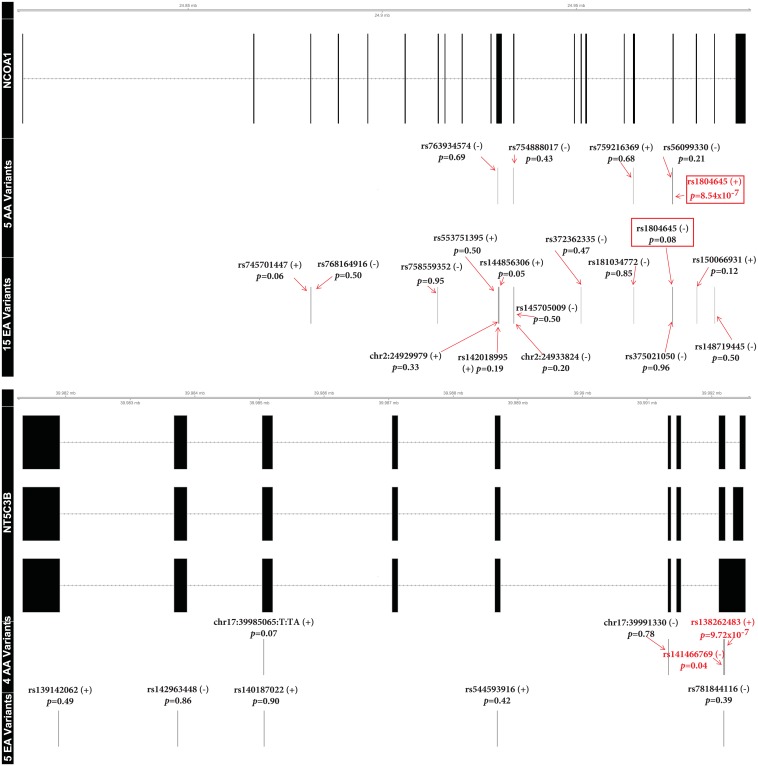
NCOA1 and NT5C3B were associated with the third visit aβ42:aβ40 ratio and the fold-change in aβ42 levels, respectively, by SKAT (Table 3). These associations were AA-specific and had one major contributing SNV each. Rs1804645 (NCOA1; p = 8.54x10-7; 6 minor allele copies) and rs138262483 (NT5C3B; p = 9.72x10-7; 4 minor allele copies) were the primary contributors (Fig 4 and S9 and S10 Tables). No NT5C3B variants overlapped races but NCOA1 had one variant (rs1804645) with opposite directions of effects and different allele frequencies in AAs and EAs (MAF of 0.008 and 0.037, respectively). Sixteen additional genes (ABL2, ATP2A1, CCDC102B, CSHL1, DSG4, FGF23, FLNB, GALNT13, LMF2, MED25, NIPAL2, NUDT7, SNAPC1, TARSL2, TMEM202, TRUB2) were suggestively (2.99 x10-6 < p ≤ 5.98x10-5) associated with amyloids through the T5 or SKAT tests (S11 and S12 Tables).
Fig 4. Variants contributing to the significant SKAT gene-based tests.
This figure displays the single-variant model results for variants contributing to the SKAT tests of NCOA1 and NT5C3B on the third visit aβ42: aβ40 ratio and fold-change in aβ42, respectively. One isoform was drawn unless the regions harboring variants differed substantially. Each contributing variant is labeled by name, single-variant association p-value, and direction of the minor allele effect. Variants with nominal (p<0.05) levels of association are in red. Overlapping variants in the two racial groups are encapsulated by a red box.
Replication analyses
Of the four genes associated with the third visit aβ42:aβ40 ratio, none strictly replicated in FHS cohorts (Table 3). However, ITPRIP was nominally (p-value≤0.05) associated using SKAT instead of the T5 burden test. Rs114174365, the only ITPRIP variant with more than one minor allele copy in both ARIC and FHS EAs, had similar effect sizes (0.89 and 1.01) in both, with p-values 0.001 and 0.045, respectively (S13 Table). PLIN2, NCOA1, and TSPAN18 did not even nominally replicate. The largest contributing TSPAN18 variant (rs138778813; 10 and 21 minor allele copies in ARIC and FHS EAs) had opposite directions of effects and different allele frequencies (0.005 and 0.015, respectively) in the two cohorts, possibly due to study design issues (relateds in FHS, required survival to older age and oversampling for cognitive impairment in ARIC). We could not replicate the association between NT5C3B and the fold-change in aβ42 because FHS had only one amyloid assessment available.
Temporal sensitivity of associations
We compared the exome-wide p-values from the third and fifth visits using scatterplots but found no systemic patterns (such as the third visit yielding lower p-values than the fifth visit, etc.) for either aβ42 or the ratio using single-variant or gene-based tests. The ranks of the p-values at the two time points were only weakly correlated (Spearman correlation coefficients ranging from 0.14 to 0.34). Our significant and suggestive findings demonstrated temporal sensitivity; no findings were significant or suggestive at both visits (S14–S18 Tables). Only half (F12, PLIN2, NCOA1) of the significant third visit associations showed nominal evidence at the fifth visit. The effect of rs3733402 (KLKB1) on aβ42 levels was near zero (p-values>0.620) for all fifth visit analyses. Rs1801020 (F12) was nominally associated with aβ42 in the fifth visit EA- and meta-analyses (p-values 0.014 and 0.036, respectively) but the effect estimates were attenuated (-0.126 and -0.088, respectively) compared to the third visit (-0.201 and -0.229, respectively). Similar results were observed for the significant gene-based tests.
Phenotype-specificity of associations
All significant associations were specific to one type of amyloid measure (aβ42 or the ratio; see S19 and S20 Tables) and failed to produce even suggestive evidence for the other. For the two common variants associated with aβ42 traits (rs3733402 in KLKB1 and rs1801020 in F12), the fold-change was the ratio measure with the strongest cross-race evidence (meta-analysis p-values of 0.005 and 1.46x10-4 and consistent direction of effects across AAs and EAs). We also note that two of the ratio-associated genes (ITPRIP and NCOA1) failed to produce nominal associations (p-value<0.05) for any aβ42 trait in any analysis (AA-specific, EA-specific, or meta-analysis).
Discussion
Leveraging late-midlife plasma aβ concentrations and fold-changes from a moderate-sized biracial sample enabled the identification of seven genes that were obscured in cross-sectional analyses of the elderly. These findings echoed those of the published GWAS meta-analysis of plasma aβ concentrations which failed to identify significant genetic variants in non-demented elderly participants [12]. Plasma aβ concentrations change with age, at least until dementia or brain plaques appear [22, 43], and could have a dynamic genetic architecture across the lifespan. Such age-dependent genetic associations have been reported for other complex traits, including blood pressure [44, 45] and lipids [46], and can represent genes that are active at specific ages or genes that are active across the age spectrum with varying effect magnitudes [45]. Accounting for age-dependent genetic effects, whether through model-based approaches or age stratification, can enhance gene discovery efforts and our understanding of intraindividual variation in plasma aβ levels [44–46]. Such analyses may suggest the optimal age range to capture particular facets of disease (e.g. AD, vascular) pathophysiology while negating the need for massive sample sizes.
Numerous phenomena could reshape the genetic landscape with aging. Gene expression and posttranslational protein modification are dynamic processes that change with age [45]. An accumulation of behavioral and environmental exposures can cause epigenetic changes, such as DNA methylation, histone modification, and microRNA expression, thereby altering gene expression (and hence the detectable genetic effects present) across the lifespan [47]. Age-related oxidative damage may mediate an accumulation of posttranslational modifications to proteins [48], altering their function and the effects of genes encoding them. More specific to our phenotypes, age-related health conditions may increase the heterogeneity of genetic effects contributing to plasma aβ levels, thereby changing the genetic underpinnings of these traits with age; subcortical white matter lesions, cerebral microbleeds, hypertension, diabetes, infarcts, ischemic heart disease, and chronic kidney disease have been associated with plasma aβ42 levels or the ratio [14, 27, 28]. Gene-age interactions could arise from differences in the aβ processes perturbed (production, degradation, transportation across the blood brain barrier, reabsorption through the CSF, or clearance from the body [49–53]) or tissue sources (platelets, skeletal muscle, pancreas, kidney, liver, vascular wall, lung, intestine, skin, glands, and brain [26]) across the lifespan. For example, the genetic underpinnings of plasma aβ could mimic the pattern hypothesized for early- versus late-onset AD [26]; genes involved in aβ production may have a greater impact on plasma concentrations in the young, while genes involved in aβ clearance may have a greater impact in the elderly.
Three gene-based associations (ITPRIP, TSPAN18, and PLIN2) had decent minor allele counts in the discovery sample and plausible connections to plasma aβ. The protein encoded by ITPRIP binds to the inositol-1,4,5-triphosphate receptor (IP3R) and allosterically downregulates IP3-induced calcium release from the endoplasmic reticulum [54]. Oligomeric aβ42 alters IP3-triggered calcium release [55] and, in return, IP3-induced calcium release may influence amyloid precursor protein (APP) cleavage to aβ40 and aβ42 [56]. Thus, ITPRIP may interact with IP3R to modulate both the aβ concentration and synaptic plasticity through calcium signaling [54]. A pathway analysis of plasma aβ GWAS results lent credibility to our ITPRIP finding [12]; eighteen of twenty-seven Ingenuity canonical pathways associated with plasma aβ contained the receptor (IP3R) modulated by ITPRIP. Located near a previously reported AD linkage peak [57], ITPRIP is an attractive AD candidate therapeutic target because of its role in cell death; it binds and inhibits the activity of death-associated protein kinase, a key component in cell death signaling pathways [58]. Itprip knock-out mice show increased cell death [58]; the effect on neuronal death is quite pronounced, with Itprip knock-out hippocampal neurons exhibiting 28% survival of wild-types after exposure to stimuli [58]. Reduction of IP3R–mediated calcium signaling rescued presenilin-associated AD pathogenesis in mouse models [59], thus drugs that enhance ITPRIP activity may prevent cell death in neurodegenerative diseases and stroke [58].
TSPAN18, supported by both races in ARIC, encodes a four-transmembrane protein from a highly conserved family known to influence the spatial organization of membrane proteins through interactions with each other, signaling proteins, enzymes, transmembrane receptors, and adhesion molecules [60]. Tetraspanins also participate in cellular trafficking through endocytic and recycling organelles, lysosomes, or secreted vesicles [61, 62]. Up- or down-regulation of tetraspanin proteins and microdomains influences aβ production, specifically through α- and γ-secretase processing of APP [63]. The tetraspanin associated with plasma aβ in this investigation, TSPAN18, has been inconsistently associated with schizophrenia in Han Chinese [64, 65], while other tetraspanins have been associated with schizophrenia, bipolar disorder, and X-linked mental retardation [66, 67].
PLIN2 encodes perilipin-2, the most prevalent lipid-droplet-associated protein in nonadipose tissue [68]. Perilipin-2 is associated with lipid droplet biogenesis, cholesterol efflux, plasma very low-density lipoprotein cholesterol levels, and intracellular and plasma triglyceride levels [68–70]. The association between aβ and PLIN2 is plausible since cholesterol concentrations and trafficking impact APP processing and aβ degradation [71, 72] and lipid droplet presence is positively correlated with aβ levels in brain neurons from AD patients [73]. The association between PLIN2 and plasma aβ may be due to atherosclerosis [70, 74, 75]; perilipin-2 aids foam cell formation [74] and is overexpressed in atherosclerotic plaques [70] which produce aβ [76]. The overexpression of PLIN2 in macrophages increases the expression of monocyte chemoattractant protein-1 [77], which itself elevates aβ accumulation in AD mice [78]. The association between PLIN2 and plasma aβ may also be due to a host of metabolic disorders, such as fatty liver disease and obesity, which are associated with perilipin-2 and aβ levels [79, 80]. We note that PLIN2 is near a well-known AD linkage peak on chromosome 9p22.1 [81].
Single-variant tests linked two key members (F12 and KLKB1) of the kallikrein-kinin system to plasma aβ. F12 and KLKB1 encode proteins that participate in complement activation, blood clotting, neutrophil aggregation, fibrinolysis (through plasminogen activation), and the bioprocessing of vasoactive peptides [82]. Most notably, contact activation of coagulation factor XII (FXII; encoded by F12) causes cleavage of plasma prekallikrein (encoded by KLKB1) to kallikrein, liberating bradykinin from high molecular weight kininogen [83]. The significant variants in F12 (rs1801020) and KLKB1 (rs3733402) have documented biological consequences. The A allele at Rs1801020, which is four bases upstream of the translation initiation codon, decreases translation efficiency and plasma FXII activity [84]. The G allele at rs3733402 substitutes serine for asparagine in an apple domain that mediates the binding of plasma prekallikrein to high molecular weight kininogen [83]. Thus, rs3733402 and rs1801020 are expected to alter bradykinin levels; they have also been associated with levels of plasma renin [85], biological surrogates of endothelin-1 and adrenomedullin [86], and B-type natriuretic peptide (rs3733402 only)[87]. The renin-angiotensin and endothelin-1 signaling pathways have been associated with plasma aβ via a pathway analysis of GWAS results [12], somewhat corroborating our common variant associations.
There are several potential mechanisms connecting the kallikrein-kinin system to aβ. Bradykinin, endothelin-1, and angiotensin-1 (a product of the renin cascade) are substrates for the aβ-degrading proteases neprilysin, endothelin-converting enzyme, and angiotensin converting enzyme, respectively [53], possibly influencing aβ degradation rates. Or, perhaps, bradykinin could participate in a feedback loop to regulate aβ42 levels; aβ42 may interact with FXII to increase bradykinin levels while bradykinin may decrease the formation of aβ42 through α-secretase processing of APP [88–91]. Whether the association between plasma aβ and the kallikrein-kinin system reflects any AD pathophysiology is unknown [92]. Variants in KLKB1 or F12 were not identified in the largest GWAS of AD to date [3], although the kallikrein-kinin system is overactivated in the plasma, CSF, and frontal and temporal cortices of AD cases [90, 92, 93] and is most prominently expressed in brain regions with the earliest signs of AD [94]. FXII, its binding sites, and components of its proteolytic cascades are present in aβ plaques of autopsied brains [95]. In addition, studies have shown that bradykinin can incite τau protein phosphorylation and subsequent learning and memory impairments [96], induce IP3 accumulation and mobilization of intracellular calcium [97], and impact inducible nitric oxide synthase, resulting in cognitive impairment [94].
We surmise that KLKB1 and F12 could be associated with both vascular disease and aβ deposition, with subsequent effects on AD. Pairwise associations between kallikrein-kinin system genes (KLKB1 and F12), complex vascular traits (hypertension, myocardial infarction, and stroke), and plasma aβ (aβ42 or the ratio) concentrations have been reported [14, 27, 98–101]. In turn, midlife vascular risk factors have been linked to both cognitive decline and brain amyloid deposition in late-life [102, 103], while the latter has been associated with plasma aβ concentrations [13–16]. The association of vascular-related genes with midlife, but not late-life, plasma aβ and the association of midlife, but not late-life, vascular risk factors with late-life brain amyloid deposition [103], bolsters the assertion that genetic studies of midlife plasma aβ levels can capture contributors to late-life amyloid deposition.
Although several of the observed plasma aβ genetic associations were supported by in vitro studies and animal models, external replication is required. This is particularly true for the tenuous AA-specific findings in NT5C3B and NCOA1 which had smaller minor allele counts (S21 Table details the functions of these genes and their possible connections to aβ). Unfortunately, our replication efforts were hindered by the paucity of studies with plasma aβ and exome sequence in a large number of middle-aged (especially underrepresented minority) participants. Relying on a single sample of 725 EAs exacerbated the already difficult task of replicating single-variant and gene-based tests [6]. Few functional variants overlapped across ARIC and FHS (≤3 per gene within race, ≤1 per gene across races) and the study designs differed. ARIC included AAs and EAs who survived until the fifth examination and were oversampled for cognitive impairment. In contrast, FHS included EAs who attended the visit of interest, without enriching for future cognitive status. Thus, non-replication could indicate false positives due to the complicated ARIC sample selection strategy and attrition. As is common practice in agnostic gene identification studies, we ignored these issues in the analyses which biased the estimated effects but allowed us to find and prioritize candidate genes for further study.
Our investigation had a few limitations. The blood samples from the third visit were stored for two decades before amyloid assessment whereas the fifth visit samples were only stored short term. Therefore the genetic associations observed in the third visit and the change over time may not be due to the age effects but rather genes influencing plasma aβ levels (via degradation) over long term storage. This concern is somewhat minimized by the fact that aβ levels are stable for at least one year and three freeze-thaw cycles [104] and INNO-BIA assays have been used on samples stored for one to two decades [19, 105]. Some participants, potentially containing rare aβ-associated variants with large effects, had amyloid concentrations below the minimal detection threshold. The exclusion of these participants from the ratio and fold-change analyses may have precluded the identification of additional aβ-associated genes. Our primary analysis ignored the amyloid assay plate effects, included low-frequency (0.01<MAF≤0.05) variants in the burden test, and ignored interactions between variants/genes and APOE ε4 carriage or future cognitive status. Sensitivity analyses showed that these minimally impacted the significant findings (see S22–S26 Tables for analyses incorporating batch effects, restricting to rare variants (MAF≤0.01), and stratifying by APOE ε4 and cognitive impairment), although the magnitude of the single-variant effects tended to be larger among those who became cognitively impaired. Lastly, we used residuals of fold changes as a simplistic outcome in this investigation but will conduct future studies using more sophisticated longitudinal analyses.
Overall, this investigation highlighted the potential age-dependency of plasma aβ genetic associations, established connections between midlife plasma aβ levels and vascular-associated genes, and suggested novel candidate AD genes for further study. Our findings implicate complex traits (e.g. hypertension) that are associated with both plasma aβ and AD. Therefore, we must be cognizant that differences in non-AD complex trait distributions may confound the association between plasma aβ and AD across studies. A more comprehensive understanding of the biological contributors to plasma aβ across the lifespan is critical to understand their role (if any) in AD pathogenesis and their utility as AD biomarkers.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to extend our deepest appreciation to the participants and staffs of the Atherosclerosis Risk in Communities (ARIC) Study and the Framingham Heart Study (FHS) for their important contributions.
Data Availability
ARIC study participants did not consent to have their data publicly available. Therefore data underlying the results of this investigation are subject to ethical and legal restrictions by the Institutional Review Boards at each field center, namely Wake Forest Baptist Medical Center (Forsyth County, NC), University of Mississippi Medical Center (Jackson, MS), University of Minnesota (suburban Minneapolis, MN), and Johns Hopkins University (Washington County, MD). However, there are well-established procedures to request this data from the ARIC Coordinating Center at the University of North Carolina—Chapel Hill. Instructions for submitting a data request can be found at: http://www2.cscc.unc.edu/aric/distribution-agreements.
Funding Statement
The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). ARIC Neurocognitive data was collected through the joint support of the National Heart, Lung, and Blood Institute; National Institute of Neurological Disorders and Stroke; National Institute on Aging; and the National Institute on Deafness and Other Communication Disorders (U01 HL096812, HL096814, HL096899, HL096902, HL096917), with previous brain MRI examinations funded by R01 HL70825 from the National Heart, Lung, and Blood Institute. This work was supported by the Framingham Heart Study (NHLBI/NIH contract HHSN268201500001I to the Boston University School of Medicine) and by grants from the National Institute on Aging (R01 AG008122, R01 AG054076, R01 AG049607, R01 AG033193, U01 AG049505, U01 AG052409) and the National Institute of Neurological Disorders and Stroke (NS017950 and UH2 NS100605). Funding support for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium” was provided by the National Institutes of Health through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Sequencing for both studies was carried out at the Baylor College of Medicine Human Genome Sequencing Center (U54 HG003273). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alzheimer's Association. 2015 Alzheimer's disease facts and figures. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2015;11(3):332–84. Epub 2015/05/20. [DOI] [PubMed] [Google Scholar]
- 2.International Genomics of Alzheimer's Disease Consortium. Convergent genetic and expression data implicate immunity in Alzheimer's disease. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2015;11(6):658–71. Epub 2014/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature genetics. 2013;45(12):1452–8. Epub 2013/10/29. doi: 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karch CM, Goate AM. Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biological psychiatry. 2015;77(1):43–51. Epub 2014/06/22. doi: 10.1016/j.biopsych.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakobsdottir J, van der Lee SJ, Bis JC, Chouraki V, Li-Kroeger D, Yamamoto S, et al. Rare Functional Variant in TM2D3 is Associated with Late-Onset Alzheimer's Disease. PLoS genetics. 2016;12(10):e1006327 Epub 2016/10/21. doi: 10.1371/journal.pgen.1006327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lord J, Lu AJ, Cruchaga C. Identification of rare variants in Alzheimer's disease. Frontiers in genetics. 2014;5:369 Epub 2014/11/13. doi: 10.3389/fgene.2014.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calero M, Gomez-Ramos A, Calero O, Soriano E, Avila J, Medina M. Additional mechanisms conferring genetic susceptibility to Alzheimer's disease. Frontiers in cellular neuroscience. 2015;9:138 Epub 2015/04/29. doi: 10.3389/fncel.2015.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glahn DC, Knowles EE, McKay DR, Sprooten E, Raventos H, Blangero J, et al. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2014;165B(2):122–30. Epub 2014/01/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han MR, Schellenberg GD, Wang LS, Alzheimer's Disease Neuroimaging I. Genome-wide association reveals genetic effects on human Abeta42 and tau protein levels in cerebrospinal fluids: a case control study. BMC neurology. 2010;10:90 Epub 2010/10/12. doi: 10.1186/1471-2377-10-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Swaminathan S, Shen L, Risacher SL, Nho K, Foroud T, et al. Genome-wide association study of CSF biomarkers Abeta1-42, t-tau, and p-tau181p in the ADNI cohort. Neurology. 2011;76(1):69–79. Epub 2010/12/03. doi: 10.1212/WNL.0b013e318204a397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramirez A, van der Flier WM, Herold C, Ramonet D, Heilmann S, Lewczuk P, et al. SUCLG2 identified as both a determinator of CSF Abeta1-42 levels and an attenuator of cognitive decline in Alzheimer's disease. Human molecular genetics. 2014;23(24):6644–58. Epub 2014/07/17. doi: 10.1093/hmg/ddu372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chouraki V, De Bruijn RF, Chapuis J, Bis JC, Reitz C, Schraen S, et al. A genome-wide association meta-analysis of plasma Abeta peptides concentrations in the elderly. Molecular psychiatry. 2014;19(12):1326–35. Epub 2014/02/19. doi: 10.1038/mp.2013.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzen KY, Yang SY, Chen TF, Cheng TW, Horng HE, Wen HP, et al. Plasma Abeta but not tau is related to brain PiB retention in early Alzheimer's disease. ACS chemical neuroscience. 2014;5(9):830–6. Epub 2014/07/24. doi: 10.1021/cn500101j [DOI] [PubMed] [Google Scholar]
- 14.Janelidze S, Stomrud E, Palmqvist S, Zetterberg H, van Westen D, Jeromin A, et al. Plasma beta-amyloid in Alzheimer's disease and vascular disease. Scientific reports. 2016;6:26801 Epub 2016/06/01. doi: 10.1038/srep26801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaminathan S, Risacher SL, Yoder KK, West JD, Shen L, Kim S, et al. Association of plasma and cortical amyloid beta is modulated by APOE epsilon4 status. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2014;10(1):e9–e18. Epub 2013/04/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rembach A, Faux NG, Watt AD, Pertile KK, Rumble RL, Trounson BO, et al. Changes in plasma amyloid beta in a longitudinal study of aging and Alzheimer's disease. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2014;10(1):53–61. Epub 2013/03/16. [DOI] [PubMed] [Google Scholar]
- 17.Schupf N, Lee A, Park N, Dang LH, Pang D, Yale A, et al. Candidate genes for Alzheimer's disease are associated with individual differences in plasma levels of beta amyloid peptides in adults with Down syndrome. Neurobiology of aging. 2015;36(10):2907 e1–10. Epub 2015/07/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koyama A, Okereke OI, Yang T, Blacker D, Selkoe DJ, Grodstein F. Plasma amyloid-beta as a predictor of dementia and cognitive decline: a systematic review and meta-analysis. Archives of neurology. 2012;69(7):824–31. Epub 2012/03/28. doi: 10.1001/archneurol.2011.1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chouraki V, Beiser A, Younkin L, Preis SR, Weinstein G, Hansson O, et al. Plasma amyloid-beta and risk of Alzheimer's disease in the Framingham Heart Study. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2015;11(3):249–57 e1. Epub 2014/09/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert JC, Schraen-Maschke S, Richard F, Fievet N, Rouaud O, Berr C, et al. Association of plasma amyloid beta with risk of dementia: the prospective Three-City Study. Neurology. 2009;73(11):847–53. Epub 2009/09/16. doi: 10.1212/WNL.0b013e3181b78448 [DOI] [PubMed] [Google Scholar]
- 21.Graff-Radford NR, Crook JE, Lucas J, Boeve BF, Knopman DS, Ivnik RJ, et al. Association of low plasma Abeta42/Abeta40 ratios with increased imminent risk for mild cognitive impairment and Alzheimer disease. Archives of neurology. 2007;64(3):354–62. Epub 2007/03/14. doi: 10.1001/archneur.64.3.354 [DOI] [PubMed] [Google Scholar]
- 22.Mayeux R, Honig LS, Tang MX, Manly J, Stern Y, Schupf N, et al. Plasma A[beta]40 and A[beta]42 and Alzheimer's disease: relation to age, mortality, and risk. Neurology. 2003;61(9):1185–90. Epub 2003/11/12. [DOI] [PubMed] [Google Scholar]
- 23.van Oijen M, Hofman A, Soares HD, Koudstaal PJ, Breteler MM. Plasma Abeta(1–40) and Abeta(1–42) and the risk of dementia: a prospective case-cohort study. The Lancet Neurology. 2006;5(8):655–60. Epub 2006/07/22. doi: 10.1016/S1474-4422(06)70501-4 [DOI] [PubMed] [Google Scholar]
- 24.Chou CT, Liao YC, Lee WJ, Wang SJ, Fuh JL. SORL1 gene, plasma biomarkers, and the risk of Alzheimer's disease for the Han Chinese population in Taiwan. Alzheimer's research & therapy. 2016;8(1):53. Epub 2016/12/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reitz C, Cheng R, Schupf N, Lee JH, Mehta PD, Rogaeva E, et al. Association between variants in IDE-KIF11-HHEX and plasma amyloid beta levels. Neurobiology of aging. 2012;33(1):199 e13–7. Epub 2010/08/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toledo JB, Shaw LM, Trojanowski JQ. Plasma amyloid beta measurements—a desired but elusive Alzheimer's disease biomarker. Alzheimer's research & therapy. 2013;5(2):8. Epub 2013/03/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toledo JB, Vanderstichele H, Figurski M, Aisen PS, Petersen RC, Weiner MW, et al. Factors affecting Abeta plasma levels and their utility as biomarkers in ADNI. Acta neuropathologica. 2011;122(4):401–13. Epub 2011/08/02. doi: 10.1007/s00401-011-0861-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gronewold J, Klafki HW, Baldelli E, Kaltwasser B, Seidel UK, Todica O, et al. Factors Responsible for Plasma beta-Amyloid Accumulation in Chronic Kidney Disease. Molecular neurobiology. 2016;53(5):3136–45. Epub 2015/05/29. doi: 10.1007/s12035-015-9218-y [DOI] [PubMed] [Google Scholar]
- 29.Poljak A, Sachdev PS. Plasma amyloid beta peptides: an Alzheimer's conundrum or a more accessible Alzheimer's biomarker? Expert review of neurotherapeutics. 2017;17(1):3–5. Epub 2016/07/28. doi: 10.1080/14737175.2016.1217156 [DOI] [PubMed] [Google Scholar]
- 30.Jack CR Jr., Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. The Lancet Neurology. 2010;9(1):119–28. Epub 2010/01/20. doi: 10.1016/S1474-4422(09)70299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. American journal of epidemiology. 1989;129(4):687–702. Epub 1989/04/01. [PubMed] [Google Scholar]
- 32.Knopman DS, Gottesman RF, Sharrett AR, Wruck LM, Windham BG, Coker L, et al. Mild Cognitive Impairment and Dementia Prevalence: The Atherosclerosis Risk in Communities Neurocognitive Study (ARIC-NCS). Alzheimer's & dementia. 2016;2:1–11. Epub 2016/03/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bainbridge MN, Wang M, Wu Y, Newsham I, Muzny DM, Jefferies JL, et al. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome biology. 2011;12(7):R68 Epub 2011/07/27. doi: 10.1186/gb-2011-12-7-r68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. Epub 2009/05/20. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43(5):491–8. Epub 2011/04/12. doi: 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC bioinformatics. 2012;13:8 Epub 2012/01/14. doi: 10.1186/1471-2105-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38(16):e164 Epub 2010/07/06. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–9. Epub 2006/07/25. doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 39.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PloS one. 2013;8(7):e68095 Epub 2013/07/23. doi: 10.1371/journal.pone.0068095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Preventive medicine. 1975;4(4):518–25. Epub 1975/12/01. [DOI] [PubMed] [Google Scholar]
- 41.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, et al. The Third Generation Cohort of the National Heart, Lung, and Blood Institute's Framingham Heart Study: design, recruitment, and initial examination. American journal of epidemiology. 2007;165(11):1328–35. Epub 2007/03/21. doi: 10.1093/aje/kwm021 [DOI] [PubMed] [Google Scholar]
- 42.Musiek ES, Holtzman DM. Three dimensions of the amyloid hypothesis: time, space and 'wingmen'. Nature neuroscience. 2015;18(6):800–6. Epub 2015/05/27. doi: 10.1038/nn.4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho SM, Lee S, Yang SH, Kim HY, Lee MJ, Kim HV, et al. Age-dependent inverse correlations in CSF and plasma amyloid-beta(1–42) concentrations prior to amyloid plaque deposition in the brain of 3xTg-AD mice. Scientific reports. 2016;6:20185 Epub 2016/02/03. doi: 10.1038/srep20185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi G, Gu CC, Kraja AT, Arnett DK, Myers RH, Pankow JS, et al. Genetic effect on blood pressure is modulated by age: the Hypertension Genetic Epidemiology Network Study. Hypertension. 2009;53(1):35–41. Epub 2008/11/26. doi: 10.1161/HYPERTENSIONAHA.108.120071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simino J, Shi G, Bis JC, Chasman DI, Ehret GB, Gu X, et al. Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. American journal of human genetics. 2014;95(1):24–38. Epub 2014/06/24. doi: 10.1016/j.ajhg.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simino J, Kume R, Kraja AT, Turner ST, Hanis CL, Sheu WH, et al. Linkage analysis incorporating gene-age interactions identifies seven novel lipid loci: the Family Blood Pressure Program. Atherosclerosis. 2014;235(1):84–93. Epub 2014/05/14. doi: 10.1016/j.atherosclerosis.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berdasco M, Esteller M. Hot topics in epigenetic mechanisms of aging: 2011. Aging cell. 2012;11(2):181–6. Epub 2012/02/11. doi: 10.1111/j.1474-9726.2012.00806.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oien DB, Osterhaus GL, Latif SA, Pinkston JW, Fulks J, Johnson M, et al. MsrA knockout mouse exhibits abnormal behavior and brain dopamine levels. Free radical biology & medicine. 2008;45(2):193–200. Epub 2008/05/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patterson BW, Elbert DL, Mawuenyega KG, Kasten T, Ovod V, Ma S, et al. Age and amyloid effects on human central nervous system amyloid-beta kinetics. Annals of neurology. 2015;78(3):439–53. Epub 2015/06/05. doi: 10.1002/ana.24454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jawaid A, Kremer EA, Piatek A, Schulz PE. Improvement of age-related memory impairment with infusion of young plasma: a role for the peripheral amyloid sink? Journal of the American Geriatrics Society. 2015;63(2):419–20. Epub 2015/02/18. doi: 10.1111/jgs.13262 [DOI] [PubMed] [Google Scholar]
- 51.Zlokovic BV, Deane R, Sagare AP, Bell RD, Winkler EA. Low-density lipoprotein receptor-related protein-1: a serial clearance homeostatic mechanism controlling Alzheimer's amyloid beta-peptide elimination from the brain. Journal of neurochemistry. 2010;115(5):1077–89. Epub 2010/09/22. doi: 10.1111/j.1471-4159.2010.07002.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Song M, Liu X, Su Kang S, Duong DM, Seyfried NT, et al. Delta-secretase cleaves amyloid precursor protein and regulates the pathogenesis in Alzheimer's disease. Nature communications. 2015;6:8762 Epub 2015/11/10. doi: 10.1038/ncomms9762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baranello RJ, Bharani KL, Padmaraju V, Chopra N, Lahiri DK, Greig NH, et al. Amyloid-beta protein clearance and degradation (ABCD) pathways and their role in Alzheimer's disease. Current Alzheimer research. 2015;12(1):32–46. Epub 2014/12/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen C, Huo LR, Zhao XL, Wang PR, Zhong N. Novel interactive partners of neuroligin 3: new aspects for pathogenesis of autism. Journal of molecular neuroscience: MN. 2015;56(1):89–101. Epub 2014/12/04. doi: 10.1007/s12031-014-0470-9 [DOI] [PubMed] [Google Scholar]
- 55.Jensen LE, Bultynck G, Luyten T, Amijee H, Bootman MD, Roderick HL. Alzheimer's disease-associated peptide Abeta42 mobilizes ER Ca(2+) via InsP3R-dependent and -independent mechanisms. Frontiers in molecular neuroscience. 2013;6:36 Epub 2013/11/10. doi: 10.3389/fnmol.2013.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kyratzi E, Efthimiopoulos S. Calcium regulates the interaction of amyloid precursor protein with Homer3 protein. Neurobiology of aging. 2014;35(9):2053–63. Epub 2014/05/06. doi: 10.1016/j.neurobiolaging.2014.03.019 [DOI] [PubMed] [Google Scholar]
- 57.Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, et al. Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science. 2000;290(5500):2302–3. Epub 2000/12/23. doi: 10.1126/science.290.5500.2302 [DOI] [PubMed] [Google Scholar]
- 58.Kang BN, Ahmad AS, Saleem S, Patterson RL, Hester L, Dore S, et al. Death-associated protein kinase-mediated cell death modulated by interaction with DANGER. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30(1):93–8. Epub 2010/01/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shilling D, Muller M, Takano H, Mak DO, Abel T, Coulter DA, et al. Suppression of InsP3 receptor-mediated Ca2+ signaling alleviates mutant presenilin-linked familial Alzheimer's disease pathogenesis. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34(20):6910–23. Epub 2014/05/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuidscherwoude M, Gottfert F, Dunlock VM, Figdor CG, van den Bogaart G, van Spriel AB. The tetraspanin web revisited by super-resolution microscopy. Scientific reports. 2015;5:12201 Epub 2015/07/18. doi: 10.1038/srep12201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andreu Z, Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Frontiers in immunology. 2014;5:442 Epub 2014/10/04. doi: 10.3389/fimmu.2014.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8(2):89–96. Epub 2006/12/22. doi: 10.1111/j.1600-0854.2006.00515.x [DOI] [PubMed] [Google Scholar]
- 63.Seipold L, Saftig P. The Emerging Role of Tetraspanins in the Proteolytic Processing of the Amyloid Precursor Protein. Frontiers in molecular neuroscience. 2016;9:149 Epub 2017/01/10. doi: 10.3389/fnmol.2016.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yue WH, Wang HF, Sun LD, Tang FL, Liu ZH, Zhang HX, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nature genetics. 2011;43(12):1228–31. Epub 2011/11/01. doi: 10.1038/ng.979 [DOI] [PubMed] [Google Scholar]
- 65.Yuan J, Jin C, Qin HD, Wang J, Sha W, Wang M, et al. Replication study confirms link between TSPAN18 mutation and schizophrenia in Han Chinese. PloS one. 2013;8(3):e58785 Epub 2013/03/19. doi: 10.1371/journal.pone.0058785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scholz CJ, Jacob CP, Buttenschon HN, Kittel-Schneider S, Boreatti-Hummer A, Zimmer M, et al. Functional variants of TSPAN8 are associated with bipolar disorder and schizophrenia. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2010;153B(4):967–72. Epub 2010/01/07. [DOI] [PubMed] [Google Scholar]
- 67.Zemni R, Bienvenu T, Vinet MC, Sefiani A, Carrie A, Billuart P, et al. A new gene involved in X-linked mental retardation identified by analysis of an X;2 balanced translocation. Nature genetics. 2000;24(2):167–70. Epub 2000/02/02. doi: 10.1038/72829 [DOI] [PubMed] [Google Scholar]
- 68.Magne J, Aminoff A, Perman Sundelin J, Mannila MN, Gustafsson P, Hultenby K, et al. The minor allele of the missense polymorphism Ser251Pro in perilipin 2 (PLIN2) disrupts an alpha-helix, affects lipolysis, and is associated with reduced plasma triglyceride concentration in humans. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2013;27(8):3090–9. Epub 2013/04/23. [DOI] [PubMed] [Google Scholar]
- 69.Najt CP, Lwande JS, McIntosh AL, Senthivinayagam S, Gupta S, Kuhn LA, et al. Structural and functional assessment of perilipin 2 lipid binding domain(s). Biochemistry. 2014;53(45):7051–66. Epub 2014/10/23. doi: 10.1021/bi500918m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larigauderie G, Furman C, Jaye M, Lasselin C, Copin C, Fruchart JC, et al. Adipophilin enhances lipid accumulation and prevents lipid efflux from THP-1 macrophages: potential role in atherogenesis. Arteriosclerosis, thrombosis, and vascular biology. 2004;24(3):504–10. Epub 2004/01/07. doi: 10.1161/01.ATV.0000115638.27381.97 [DOI] [PubMed] [Google Scholar]
- 71.Zinser EG, Hartmann T, Grimm MO. Amyloid beta-protein and lipid metabolism. Biochimica et biophysica acta. 2007;1768(8):1991–2001. Epub 2007/04/10. doi: 10.1016/j.bbamem.2007.02.014 [DOI] [PubMed] [Google Scholar]
- 72.Lee CY, Tse W, Smith JD, Landreth GE. Apolipoprotein E promotes beta-amyloid trafficking and degradation by modulating microglial cholesterol levels. The Journal of biological chemistry. 2012;287(3):2032–44. Epub 2011/12/02. doi: 10.1074/jbc.M111.295451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomez-Ramos P, Asuncion Moran M. Ultrastructural localization of intraneuronal Abeta-peptide in Alzheimer disease brains. Journal of Alzheimer's disease: JAD. 2007;11(1):53–9. Epub 2007/03/16. [DOI] [PubMed] [Google Scholar]
- 74.Paul A, Chang BH, Li L, Yechoor VK, Chan L. Deficiency of adipose differentiation-related protein impairs foam cell formation and protects against atherosclerosis. Circulation research. 2008;102(12):1492–501. Epub 2008/05/17. doi: 10.1161/CIRCRESAHA.107.168070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gupta A, Iadecola C. Impaired Abeta clearance: a potential link between atherosclerosis and Alzheimer's disease. Frontiers in aging neuroscience. 2015;7:115 Epub 2015/07/03. doi: 10.3389/fnagi.2015.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Meyer GR, De Cleen DM, Cooper S, Knaapen MW, Jans DM, Martinet W, et al. Platelet phagocytosis and processing of beta-amyloid precursor protein as a mechanism of macrophage activation in atherosclerosis. Circulation research. 2002;90(11):1197–204. Epub 2002/06/18. [DOI] [PubMed] [Google Scholar]
- 77.Chen FL, Yang ZH, Wang XC, Liu Y, Yang YH, Li LX, et al. Adipophilin affects the expression of TNF-alpha, MCP-1, and IL-6 in THP-1 macrophages. Molecular and cellular biochemistry. 2010;337(1–2):193–9. Epub 2009/10/24. doi: 10.1007/s11010-009-0299-7 [DOI] [PubMed] [Google Scholar]
- 78.Lai AY, McLaurin J. Clearance of amyloid-beta peptides by microglia and macrophages: the issue of what, when and where. Future neurology. 2012;7(2):165–76. Epub 2012/06/28. doi: 10.2217/fnl.12.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shie FS, Shiao YJ, Yeh CW, Lin CH, Tzeng TT, Hsu HC, et al. Obesity and Hepatic Steatosis Are Associated with Elevated Serum Amyloid Beta in Metabolically Stressed APPswe/PS1dE9 Mice. PloS one. 2015;10(8):e0134531 Epub 2015/08/06. doi: 10.1371/journal.pone.0134531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conte M, Franceschi C, Sandri M, Salvioli S. Perilipin 2 and Age-Related Metabolic Diseases: A New Perspective. Trends in endocrinology and metabolism: TEM. 2016;27(12):893–903. Epub 2016/09/24. doi: 10.1016/j.tem.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 81.Kunkle BW, Jaworski J, Barral S, Vardarajan B, Beecham GW, Martin ER, et al. Genome-wide linkage analyses of non-Hispanic white families identify novel loci for familial late-onset Alzheimer's disease. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2015. Epub 2015/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siegerink B, Rosendaal FR, Algra A. Antigen levels of coagulation factor XII, coagulation factor XI and prekallikrein, and the risk of myocardial infarction and ischemic stroke in young women. Journal of thrombosis and haemostasis: JTH. 2014;12(5):606–13. Epub 2014/07/01. [DOI] [PubMed] [Google Scholar]
- 83.Katsuda I, Maruyama F, Ezaki K, Sawamura T, Ichihara Y. A new type of plasma prekallikrein deficiency associated with homozygosity for Gly104Arg and Asn124Ser in apple domain 2 of the heavy-chain region. European journal of haematology. 2007;79(1):59–68. Epub 2007/06/30. doi: 10.1111/j.1600-0609.2007.00871.x [DOI] [PubMed] [Google Scholar]
- 84.Kanaji T, Okamura T, Osaki K, Kuroiwa M, Shimoda K, Hamasaki N, et al. A common genetic polymorphism (46 C to T substitution) in the 5'-untranslated region of the coagulation factor XII gene is associated with low translation efficiency and decrease in plasma factor XII level. Blood. 1998;91(6):2010–4. Epub 1998/04/16. [PubMed] [Google Scholar]
- 85.Biswas N, Maihofer AX, Mir SA, Rao F, Zhang K, Khandrika S, et al. Polymorphisms at the F12 and KLKB1 loci have significant trait association with activation of the renin-angiotensin system. BMC medical genetics. 2016;17:21 Epub 2016/03/13. doi: 10.1186/s12881-016-0283-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verweij N, Mahmud H, Mateo Leach I, de Boer RA, Brouwers FP, Yu H, et al. Genome-wide association study on plasma levels of midregional-proadrenomedullin and C-terminal-pro-endothelin-1. Hypertension. 2013;61(3):602–8. Epub 2013/02/06. doi: 10.1161/HYPERTENSIONAHA.111.203117 [DOI] [PubMed] [Google Scholar]
- 87.Musani SK, Fox ER, Kraja A, Bidulescu A, Lieb W, Lin H, et al. Genome-wide association analysis of plasma B-type natriuretic peptide in blacks: the Jackson Heart Study. Circulation Cardiovascular genetics. 2015;8(1):122–30. Epub 2015/01/07. doi: 10.1161/CIRCGENETICS.114.000900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shibayama Y, Joseph K, Nakazawa Y, Ghebreihiwet B, Peerschke EI, Kaplan AP. Zinc-dependent activation of the plasma kinin-forming cascade by aggregated beta amyloid protein. Clinical immunology. 1999;90(1):89–99. Epub 1999/01/13. doi: 10.1006/clim.1998.4621 [DOI] [PubMed] [Google Scholar]
- 89.Bergamaschini L, Donarini C, Foddi C, Gobbo G, Parnetti L, Agostoni A. The region 1–11 of Alzheimer amyloid-beta is critical for activation of contact-kinin system. Neurobiology of aging. 2001;22(1):63–9. Epub 2001/02/13. [DOI] [PubMed] [Google Scholar]
- 90.Zamolodchikov D, Chen ZL, Conti BA, Renne T, Strickland S. Activation of the factor XII-driven contact system in Alzheimer's disease patient and mouse model plasma. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(13):4068–73. Epub 2015/03/17. doi: 10.1073/pnas.1423764112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nitsch RM, Kim C, Growdon JH. Vasopressin and bradykinin regulate secretory processing of the amyloid protein precursor of Alzheimer's disease. Neurochemical research. 1998;23(5):807–14. Epub 1998/05/05. [DOI] [PubMed] [Google Scholar]
- 92.Ashby EL, Love S, Kehoe PG. Assessment of activation of the plasma kallikrein-kinin system in frontal and temporal cortex in Alzheimer's disease and vascular dementia. Neurobiology of aging. 2012;33(7):1345–55. Epub 2010/11/16. doi: 10.1016/j.neurobiolaging.2010.09.024 [DOI] [PubMed] [Google Scholar]
- 93.Bergamaschini L, Parnetti L, Pareyson D, Canziani S, Cugno M, Agostoni A. Activation of the contact system in cerebrospinal fluid of patients with Alzheimer disease. Alzheimer disease and associated disorders. 1998;12(2):102–8. Epub 1998/07/03. [DOI] [PubMed] [Google Scholar]
- 94.Prediger RD, Medeiros R, Pandolfo P, Duarte FS, Passos GF, Pesquero JB, et al. Genetic deletion or antagonism of kinin B(1) and B(2) receptors improves cognitive deficits in a mouse model of Alzheimer's disease. Neuroscience. 2008;151(3):631–43. Epub 2008/01/15. doi: 10.1016/j.neuroscience.2007.11.009 [DOI] [PubMed] [Google Scholar]
- 95.Yasuhara O, Walker DG, McGeer PL. Hageman factor and its binding sites are present in senile plaques of Alzheimer's disease. Brain research. 1994;654(2):234–40. Epub 1994/08/22. [DOI] [PubMed] [Google Scholar]
- 96.Wang Q, Wang J. Injection of bradykinin or cyclosporine A to hippocampus induces Alzheimer-like phosphorylation of Tau and abnormal behavior in rats. Chinese medical journal. 2002;115(6):884–7. Epub 2002/07/19. [PubMed] [Google Scholar]
- 97.Huang HM, Ou HC, Hsueh SJ. Amyloid beta peptide enhanced bradykinin-mediated inositol (1,4,5)trisphosphate formation and cytosolic free calcium. Life sciences. 1998;63(3):195–203. Epub 1998/08/11. [DOI] [PubMed] [Google Scholar]
- 98.Lu X, Zhao W, Huang J, Li H, Yang W, Wang L, et al. Common variation in KLKB1 and essential hypertension risk: tagging-SNP haplotype analysis in a case-control study. Human genetics. 2007;121(3–4):327–35. Epub 2007/02/24. doi: 10.1007/s00439-007-0340-4 [DOI] [PubMed] [Google Scholar]
- 99.Guella I, Duga S, Ardissino D, Merlini PA, Peyvandi F, Mannucci PM, et al. Common variants in the haemostatic gene pathway contribute to risk of early-onset myocardial infarction in the Italian population. Thrombosis and haemostasis. 2011;106(4):655–64. Epub 2011/09/09. doi: 10.1160/TH11-04-0247 [DOI] [PubMed] [Google Scholar]
- 100.Santamaria A, Mateo J, Tirado I, Oliver A, Belvis R, Marti-Fabregas J, et al. Homozygosity of the T allele of the 46 C->T polymorphism in the F12 gene is a risk factor for ischemic stroke in the Spanish population. Stroke; a journal of cerebral circulation. 2004;35(8):1795–9. Epub 2004/07/03. [DOI] [PubMed] [Google Scholar]
- 101.Lambert JC, Dallongeville J, Ellis KA, Schraen-Maschke S, Lui J, Laws S, et al. Association of plasma Aβ peptides with blood pressure in the elderly. PloS one. 2011;6(4):e18536 Epub 2011/04/29. doi: 10.1371/journal.pone.0018536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gottesman RF, Schneider AL, Albert M, Alonso A, Bandeen-Roche K, Coker L, et al. Midlife hypertension and 20-year cognitive change: the atherosclerosis risk in communities neurocognitive study. JAMA neurology. 2014;71(10):1218–27. Epub 2014/08/05. doi: 10.1001/jamaneurol.2014.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gottesman RF, Schneider AL, Zhou Y, Coresh J, Green E, Gupta N, et al. Association Between Midlife Vascular Risk Factors and Estimated Brain Amyloid Deposition. Jama. 2017;317(14):1443–50. Epub 2017/04/12. doi: 10.1001/jama.2017.3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lachno DR, Emerson JK, Vanderstichele H, Gonzales C, Martenyi F, Konrad RJ, et al. Validation of a multiplex assay for simultaneous quantification of amyloid-beta peptide species in human plasma with utility for measurements in studies of Alzheimer's disease therapeutics. Journal of Alzheimer's disease: JAD. 2012;32(4):905–18. Epub 2012/08/14. doi: 10.3233/JAD-2012-121075 [DOI] [PubMed] [Google Scholar]
- 105.Yaffe K, Weston A, Graff-Radford NR, Satterfield S, Simonsick EM, Younkin SG, et al. Association of plasma beta-amyloid level and cognitive reserve with subsequent cognitive decline. Jama. 2011;305(3):261–6. Epub 2011/01/20. doi: 10.1001/jama.2010.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
ARIC study participants did not consent to have their data publicly available. Therefore data underlying the results of this investigation are subject to ethical and legal restrictions by the Institutional Review Boards at each field center, namely Wake Forest Baptist Medical Center (Forsyth County, NC), University of Mississippi Medical Center (Jackson, MS), University of Minnesota (suburban Minneapolis, MN), and Johns Hopkins University (Washington County, MD). However, there are well-established procedures to request this data from the ARIC Coordinating Center at the University of North Carolina—Chapel Hill. Instructions for submitting a data request can be found at: http://www2.cscc.unc.edu/aric/distribution-agreements.