Abstract
Signal transduction of the conserved transforming growth factor-β (TGFβ) family signaling pathway functions through two distinct serine/threonine transmembrane receptors, the type I and type II receptors. Endocytosis orchestrates the assembly of signaling complexes by coordinating the entry of receptors with their downstream signaling mediators. Recently, we showed that the C. elegans type I bone morphogenetic protein (BMP) receptor SMA-6, part of the TGFβ family, is recycled through the retromer complex while the type II receptor, DAF-4 is recycled in a retromer-independent, ARF-6 dependent manner. From genetic screens in C. elegans aimed at identifying new modifiers of BMP signaling, we reported on SMA-10, a conserved LRIG (leucine-rich and immunoglobulin-like domains) transmembrane protein. It is a positive regulator of BMP signaling that binds to the SMA-6 receptor. Here we show that the loss of sma-10 leads to aberrant endocytic trafficking of SMA-6, resulting in its accumulation in distinct intracellular endosomes including the early endosome, multivesicular bodies (MVB), and the late endosome with a reduction in signaling strength. Our studies show that trafficking defects caused by the loss of sma-10 are not universal, but affect only a limited set of receptors. Likewise, in Drosophila, we find that the fly homolog of sma-10, lambik (lbk), reduces signaling strength of the BMP pathway, consistent with its function in C. elegans and suggesting evolutionary conservation of function. Loss of sma-10 results in reduced ubiquitination of the type I receptor SMA-6, suggesting a possible mechanism for its regulation of BMP signaling.
Introduction
The TGFβ family comprises a large family of protein ligands that is involved in many developmental decisions and is often associated with some diseases and cancers [1–4]. It is an ancient signaling family not present in yeast or plants, but is found in the most simpler animals such as sponges and trichoplax [5–7]. The ligands form a continuum of homology but can be roughly grouped into the TGFβ, the BMP, and the activin subfamilies [8, 9]. C. elegans has two complete signaling pathways, the Small and Male tail abnormal (Sma/Mab) pathway and the dauer pathway [10–12]. The Sma/Mab pathway, which is BMP-like, controls body size by regulating cell size rather than cell number, as well as male tail development, mesoderm development, and some aspects of innate immunity [10–13].
Unlike most tyrosine kinase signaling pathways, BMP signals through two related serine-threonine kinase transmembrane receptors [1, 9]. For signaling to occur, both receptors are required at the cell surface to physically interact with each other and their ligands. This interaction results in the phosphorylation of a downstream receptor activated-Smad (R-Smad) protein. This phosphorylation event triggers the binding of a common mediator Smad (Co-Smad) to form a heteromeric complex between R-Smad proteins and the Co-Smad. These Smad complexes accumulate in the nucleus where they can bind additional co-factors to activate or repress transcription.
After signaling, most transmembrane receptors are either recycled for new rounds of signaling or are degraded by the lysosome [14–19]. Once receptors enter early endosomes, they are either sorted into recycling pathways that will return the molecule to the plasma membrane for another round of signaling or are sorted into a degradative pathway [14, 16, 17, 20, 21]. This bifurcation represents an important point of regulation for signaling pathways, although little is known about the regulation of this post-endocytic process.
In the case of the BMP receptors, we have shown that the type I BMP receptor in C. elegans, SMA-6, is recycled through the retromer, whereas the type II receptor DAF-4 is recycled via a separate and distinct recycling pathway regulated by ARF-6 [22]. Two other studies indicate that the mammalian type I BMP receptor may also recycle through the retromer, suggesting this is a conserved mechanism for BMP type I receptors [23, 24].
Distinct sorting pathways may be required for BMP receptors, given the unique property that signaling requires two related receptors. Physically separating the two BMP receptors into molecularly distinct sorting complexes may be necessary to separate the type II receptor from the type I receptor in order to reduce signaling. Whether signaling has ceased by the time the receptors have separated is not known, but at minimum, separation of the receptors into distinct recycling pathways provides another avenue of BMP regulation.
Understanding how BMP signaling components are regulated is of considerable interest. Since BMP is a potent growth factor, cells have devised many different mechanisms to control its activity, such as N-glycosylation, ubiquitylation, sumolation, neddylation, and methylation [25, 26]. Previously, we reported on sma-10, which encodes a member of the LRIG family [27]. LRIG members have a characteristic set of 15 leucine-rich repeats and three immunoglobulin repeats in the ectodomain, juxtaposed to a transmembrane domain and a cytoplasmic domain [27–29]. Leucine-rich and immunoglobulin domains generally participate in protein-protein interactions [30]. Biochemical analysis of SMA-10 shows it binds to both the type I and type II receptors, SMA-6 and DAF-4, respectively [27]. Likewise, the human LRIG1 binds to the human type I and type II receptors, but not to the BMP ligands [27]. The three members of the vertebrate LRIG family (Lrig1/2/3) and the single Drosophila member, lbk, show only modest homology to each other in their ~300 amino acid cytoplasmic domains [27–29] in contrast to their more highly conserved extracellular domains. The C. elegans ortholog has a very short cytoplasmic domain, further highlighting the high divergence in this domain [27]. Expression of the human LRIG1, LRIG2, and LRIG3 paralogs are widespread in vertebrates and show some differential regulation in human tissues. All three have been characterized as tumor suppressors and/or proto-oncogenes [28, 31], consistent with their regulation of signaling pathways.
In this report, we address three questions: 1) where does SMA-10 act to regulate receptor trafficking, 2) is the effect of SMA-10 on BMP function conserved in other organisms, and 3) is SMA-10 specific for BMP signaling. In this work, we show that mutations in sma-10 alter the intracellular trafficking of the type I (SMA-6) receptor in C. elegans. In sma-10 mutants, the receptors enter the cell, but accumulate intracellularly after entry in distinct endocytic compartments. This receptor accumulation results in minimal BMP signaling. Ubiquitination of the BMP receptors is well documented for proper trafficking and signaling [32–34], but how that process is regulated is not well understood. We show that mutations in sma-10 reduce the ubiquitination of the type I receptor, leading to mis-trafficking of the receptor and a decrease in signaling.
Methods
General methods and strains
All C. elegans strains are derived from the Bristol strain N2 [35] and were grown at 20°C on standard nematode growth media plates seeded with OP50 E. coli. Worm cultures, genetic crosses, and other C. elegans husbandry were performed according to standard protocols [35]. A complete list of strains used in this study can be found in S1 Table. RNAi was performed using the feeding method [36]. Feeding constructs were obtained from an RNAi library [37] or from cloned genomic DNA fragments. Larval stage L4 animals were treated for 24h and imaged as young adults.
Plasmids and transgenic lines
Expression of TagRFP- and GFP- tagged versions of SMA-10 in the worm intestine was driven with the intestinal-specific promoter pVha-6 [22]. C. elegans sma-10 genomic DNA, lacking the terminal stop codon, was cloned into the Gateway™ entry vector pDONR 221 (Invitrogen), and subsequently transferred into expression vectors by recombination cloning to generate C-terminal fusions. Microparticle bombardment of unc-119(ed3) mutant animals [38–40] was done to obtain low-copy integrated transgenic lines.
Generation of truncation mutants for the sma-10(wk88) body size rescue was achieved using mutagenesis using the Q5 mutagenesis kit (Life technologies) as per manufacturer’s instructions. A GFP-tagged full length sma-10 expressed from a hypodermal promoter (elt-3) was mutagenized to remove the cytoplasmic tail (ΔCyto) using the forward primer 5’- TAGCATTCGTAGAATTCC -3’ and reverse primer 5’- AATGCAAATTGAAGTGATAACTC -3’. The linearized PCR products were then subjected to the KLD reaction to circularize products and plasmids were confirmed by sequence.
Microscopy and image analysis
Live worms were mounted on 2% (wt/vol) agarose pads with tetramisole. Using argon 488-nm excitation, we used the spectral profile function of the Leica SP5 confocal microscope to establish a spectral profile of the intestinal autofluorescence to separate it from the experimentally determined GFP spectrum. The worm intestine consists of 20 individual epithelial cells with distinct apical, lateral, and basal regions, positioned as bilaterally symmetric pairs to form a long tube around the lumen. The focal planes captured in this study are designated as the Top plane, which captures the top of the intestinal tube (basolateral surface), and the Middle plane, which captures the midsagittal cross section of the apical and basolateral surfaces.
Quantification of images was performed using the open-source Fiji software [41]. Within any set of comparable images, the image capture, scaling conditions and threshold values were identical for all images within a given experiment. For each experiment, at least six animals were analyzed with three randomly selected regions-of-interest (ROI) per animal. All the images were taken from the same cells of the intestine (either the second or third pair of cells relative to the anterior end of the intestine). Quantification of the fluorescence in each ROI was performed. Colocalization images were performed on L4 staged animals, using a confocal microscope equipped with the confocal imager (CARV II; BD Biosciences). Colocalization analysis was conducted using the Costes method to establish a threshold in Fiji software [41] which also outputs the Mander’s and Pearson’s correlation values.
Plasmid constructs expressing full length sma-10 [sma-10(+),red] and sma-10 lacking the cytoplasmic tail [sma-10(Δcyto)] from a hypodermal-specific promoter elt-3 were injected into the sma-10(wk88) mutant background. Transformed animals were assayed for body size at the L4 stage+ 24 hr. Transformants were selected based on presence of GFP in the pharynx and imaged at 10X magnification using standard epifluorescent microscopes. Fiji software [41] was then used to measure body size from tip of the mouth (anterior) to the tip of the tail (posterior) using the Segmented Line tool.
Protein expression and blots
Worms were synchronized by alkaline bleaching and grown on standard NGM plates until young adult stage (L4 + 24 hours). Animals were collected and washed in 5 ml M9 (three times at 1500 rpm). After the final wash, animals were resuspended in 750 μl lysis buffer (50 mm HEPES, 150 mm NaCl, 0.5 mM EDTA supplemented with protease inhibitor tablets (Roche Scientific Ltd). Approximately 500 μl of glass beads (#G8772-500G, Sigma Aldrich) were added and bead beating was carried out for 30 seconds at 4°C with a one minute incubation on ice between each of two bead beatings. Crude lysate was centrifuged at 18,000 rpm for 10 minutes at 4°C and the supernatant was transferred to a fresh pre-cooled microcentrifuge tube and centrifuged again. Supernatant was transferred to a pre-cooled microcentrifuge tube. Protein concentration was determined using the Bradford assay (500–0006, Bio-rad), and 50 μg of protein was mixed with 4x protein sample buffer (Amresco Inc.) and boiled for 10 minutes. These input samples were then stored at –80°C until used for immunoblotting. 20 μl of resuspended Agarose A/G beads (Santa Cruz Biotech) were added to the remaining samples and incubated on a rocker at 4°C for 1 hour followed by a centrifugation at 1500 rpm for 5 minutes at 4°C. Supernatant was collected and incubated with mouse anti-GFP (Roche) for 1–4 hours at 4°C on a rocker platform. 20 μl of agarose A/G beads (Santa Cruz Biotechnology) were added to the samples and incubated at 4°C for 1 hour on a rocker platform. Immunoprecipitates were collected by centrifugation at 1500 rpm for 5 minutes at 4°C while supernatant was discarded. Precipitates were washed three times with 1 ml lysis buffer and centrifuged at 4°C. 75 μl of 2x protein sample buffer was added and the mixture boiled for 10 minutes. Samples were immunoblotted and membranes were probed with mouse anti-GFP (Roche), mouse anti-Ub (Enzo), mouse anti-actin (Santa Cruz Biotechnology), and visualized on a Li-Cor Infrared Imaging System.
Statistical analysis
Statistical significance was determined by a two-tailed t test with significant being P<0.05. Unless otherwise specified, bar graphs represent Mean ± S.E.M. Colocalization analysis was conducted using the Costes method to establish a threshold in Fiji software [41] which also outputs the Mander’s and Pearson’s correlation values. A One-way ANOVA was performed for the series. *** indicates p < 0.0001.
Drosophila strains and genetics
Drosophila melanogaster stocks were grown on standard media at 25°C. The Gal4/UAS system was used to over-express transgenes [42, 43]. Flip-out clones were made by crossing hs-Flp[122]; act>y+>Gal-4 UAS-GFP virgin females with UAS-hairpin RNAi transgenic males from the VDRC (#106679). Larvae were heat shocked at 38°C for 10 minutes. UAS-Dicer2 VDRC (#60007) was combined with UAS-RNAi genes to obtain more efficient target gene knock down for all RNAi experiments [44]. The engrailed-Gal4 (en-Gal4) flies were obtained from Bloomington (#30564). The MARCM system (mosaic analysis with a repressible cell marker) was used to positively identify clones with RNAi lbk expression [45]. Animals were heat shocked at 37°C for one hour after 72 hours of development. Third-instar wing discs were dissected for staining with antibodies to pMad [46].
Generation of Drosophila transgenes
The cDNA of the Drosophila type II receptor punt was inserted into the fly Gateway vector pTWF (Drosophila Genomics Resource Center) and transgenic flies were generated, P[w+ UAS-punt-3xFLAG]. This pTWF vector contains the UASt promoter and P elements end for integration, along with a C-terminal 3xFLAG sequence that was fused in frame to the punt cDNA. The UAS-RNAi-lbk RNAi strain was obtained from Vienna (#106679).
Drosophila antibody staining
Third instar larvae were dissected in chilled 1x Ringers solution. Disc tissue was fixed in formalin (Sigma) for 10 minutes at room temperature. PBST (0.1% Triton X-100 in 1x PBS) was used for the following washing and antibody incubation. Primary antibodies used for staining were rabbit anti-P-MAD (diluted as 1:4000) [46], and mouse monoclonal ANTI-FLAG® M2 antibody (diluted as 1:10000, Sigma). Secondary antibodies, conjugated to Cy3 (diluted 1:200, Jackson ImmunoResearch Lab), and Alexa fluor 633 (diluted 1:100, Invitrogen), were used for detection. All primary antibodies were diluted in PBST and incubated with tissue samples at 4°C overnight. Secondary antibodies were typically incubated with tissue samples for 2 hours at room temperature. Wing imaginal discs were mounted in Vectashield mounting medium (Vector Laboratories) and analyzed using confocal microscopy.
Results
Our previous work has shown that SMA-10 binds directly to both C. elegans receptors, SMA-6 and DAF-4 [22]. Importantly, this interaction is conserved in mammals as shown by binding of LRIG1 to both type I and II mammalian BMP receptors [27], but not the ligand. Receptor binding by SMA-10 suggests several possible modes of action of SMA-10, one of which involves trafficking of BMP receptors. In order to investigate the role of SMA-10, we chose to study it in the well characterized, polarized epithelial cells of the C. elegans intestine, where BMP signaling is required and where there are many cell biological tools available [22, 27, 47].
SMA-10 colocalizes preferentially with the type I receptor
Although SMA-10 and its human ortholog LRIG1 physically interact with both receptors, it appears that there is a greater affinity of the two proteins for the type I receptors as compared to the type II receptors [27]. Given this difference, we wanted to determine if SMA-10 preferentially colocalizes with SMA-6 in vivo. In order to identify colocalization patterns of SMA-10 with the receptors in vivo, we performed confocal microscopy of tagged proteins within the intestine of C. elegans. Consistent with our biochemical findings [27], we observe colocalization of SMA-10 with both SMA-6 and DAF-4 in sub-cellular compartments (Fig 1A and S1C Fig). Importantly, we show that SMA-10 colocalizes disproportionately with SMA-6 as compared to DAF-4 (42.1% with SMA-6 in Fig 1A and 7.0% with DAF-4 in S1 Fig). These data and our previous biochemical studies of physical interactions suggest [27] that sma-10 may exert its effects on BMP signaling primarily through regulation of the type I receptor. Given that we have uncovered two separate trafficking pathways for the receptors, these data suggest that SMA-10 may interact or traffic with SMA-6 in its distinct pathway.
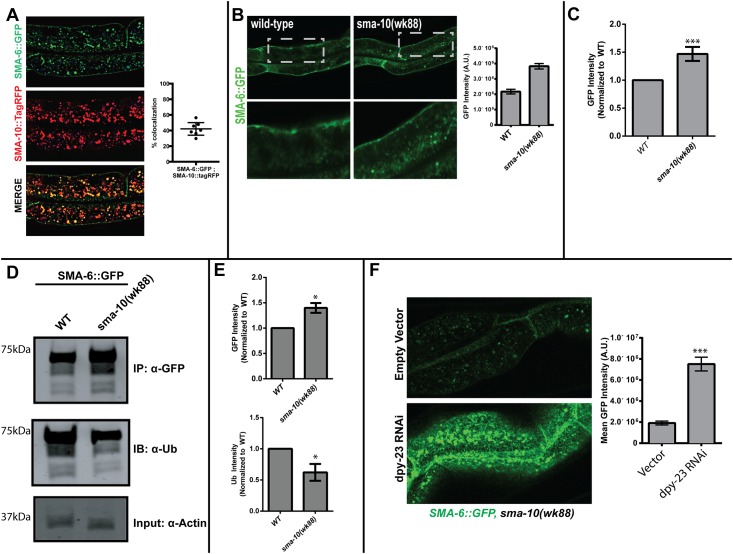
Fig 1. SMA-10 is required for the intracellular trafficking of the type I receptor, SMA-6.
(A) SMA-10 colocalizes preferentially with SMA-6 (type I) intracellularly: SMA-10 shows overlap with both receptors, although the fraction of colocalization with SMA-6 is higher. SMA-6::GFP and SMA-10::tagRFP were expressed in the C. elegans intestine using the vha-6 promoter. Young adult animals (L4+24 hrs) were imaged using the confocal microscope and colocalization was performed. Mander’s overlap coefficient for SMA-10::tagRFP and SMA-6::GFP is 0.512. (B) Loss of sma-10 leads to intracellular accumulation of SMA-6. SMA-6 accumulates within intracellular compartments in the sma-10(wk88) mutant from three regions from six animals, using a total of 18 data points. (C) Total GFP intensity measured from worm lysates from either SMA-6::GFP or SMA-6::GFP,sma-10(wk88) mutant background. (D) Western blot of ubiquitination state of SMA-6 in wildtype and sma-10 mutant background. (E) Quantification of ubiquitination data, (F) Loss of SMA-10 does not affect the biosynthesis or transport of SMA-6 to the plasma membrane. Loss of dpy-23 prevents internalization of clathrin-dependent cargos. SMA-6 biosynthesis and subsequent trafficking to the plasma membrane is unaffected in sma-10(wk88) animals.
SMA-10 is necessary for intracellular trafficking of SMA-6
To study the effect of loss of sma-10 on the receptors, we visualized GFP-tagged receptors in wild-type and in sma-10(wk88) (Fig 1B), an allele that results in a premature stop codon in the ninth leucine-rich domain [27]. We observe that SMA-6 accumulated intracellularly in sma-10(wk88) compared to wild type. A second allele, sma-10(wk66) shows the same defects found in sma-10(wk88)(data not shown) [27]. We confirmed these accumulations by measuring total GFP fluorescence from worm lysates in the wild-type and sma-10(wk88) animals (Fig 1C).
Mammalian LRIG1 is known to regulate EGF by binding to the EGF receptor (EGFR), recruiting the E3 ubiquitin ligase c-Cbl and ubiquitination of EGFR and LRIG1. This leads to trafficking of the receptor (and bound LRIG1) into the lysosome for degradation abolishing further signaling [48]. To determine whether C. elegans SMA-10 affects ubiquitination of the SMA-6 receptor, we performed an immunoprecipitation of GFP-tagged receptor in the wild-type and sma-10(wk88) mutant background (Fig 1D). We observed increased levels of SMA-6 in the sma-10(wk88) mutant (Fig 1D, Upper panel) similar to what we observe in our confocal imaging (Fig 1B) and fluorescence measurements (Fig 1C). Further, we show that loss of sma-10 leads to a significantly decreased ubiquitination signal on SMA-6 (Fig 1D, middle panel). Thus, sma-10 is required for the maximal ubiquitination of the C. elegans type I BMP receptor and this ubiquitination may be required for the normal trafficking of SMA-6.
In order to rule out the possibility that SMA-6 accumulations were due to any synthesis and transport to the plasma membrane of the receptors, we examined the accumulation of SMA-6 at the plasma membrane by knocking down dpy-23. dpy-23 is the μ-2 adaptin member of the AP2 complex and is required for the clathrin-dependent endocytosis of various transmembrane receptors [22, 49–51]. Loss of dpy-23 by RNAi leads to the suppression of clathrin-mediated endocytosis, resulting in accumulation of SMA-6 at the plasma membrane and inhibition of BMP signaling [22]. If sma-10(wk88) were to affect the biosynthesis and trafficking of SMA-6 to the plasma membrane, we would expect that SMA-6 would be unable to accumulate at the surface with the depletion of dpy-23. We observe that SMA-6 is able to efficiently accumulate at the plasma membrane even in the absence of sma-10, suggesting that sma-10 is required for endocytosis/trafficking once SMA-6 has reached the plasma membrane (Fig 1F).
Taken together, these data suggest that SMA-10 is required for proper endocytosis/trafficking of the receptors via a mechanism that requires ubiquitination of the type I receptor, and loss of sma-10 leads to receptor accumulation in compartments that are incompatible with signaling.
Having established that sma-10 is involved with trafficking SMA-6, we asked whether sma-10 also has a broader role in regulating movement of other cellular receptors. We chose two well-studied cargoes that utilize distinct trafficking pathways to study the effect of sma-10 on the trafficking of these cargoes—human transferrin receptor (hTfR) and human IL-2 receptor α-chain (hTAC). hTfR enters cells via clathrin-dependent endocytosis while hTAC is internalized in a clathrin-independent manner [16]. We report that the loss of sma-10 did not alter the localization pattern of these cargoes, although total levels were slightly lower, but not significantly (S2 Fig). EGFR signaling in C. elegans, mediated by a single EGFR (LET-23), directs several embryonic and larval cell fates and also ovulatory contractions in adult hermaphrodites [52]. We do not observe any of the phenotypes associated with loss of EGF signaling in the sma-10 animals (data not shown). These data suggest sma-10 is not a general regulator of receptors, but rather affects a limited number of receptors.
Drosophila LRIG also affects BMP pathways
Having established the specificity of sma-10 regulation of BMP signaling, we wanted to identify if this interaction was conserved in other organisms. We turned to the Drosophila lbk gene, an ortholog of sma-10. In Drosophila, the decapentaplegic BMP pathway (dpp) is well-studied [53, 54], but the role of lbk is not well-defined. Using the MARCM method [45], we made clones expressing lbk RNAi in the wing imaginal disc and assayed dpp output using the phosophorylated Smad (pMad), a reporter for activation of BMP signaling [46]. In clones expressing lbk RNAi, we observed significantly less pMad (Fig 2A and 2B), suggesting that lbk is needed for proper signaling of the BMP pathway in Drosophila. To determine if a reduction of lbk alters signaling strength of the BMP pathway, we over-expressed punt, a type II receptor of the Drosophila BMP pathway, in the posterior half of the wing imaginal disc using the en-Gal4 driver. Over-expression of many BMP components in the wing disc, including punt, cause overgrowth in the wing discs. We would expect depletion of lbk to reduce the overgrowth of the wing disk if it were necessary for BMP signaling. When lbk RNAi was simultaneously expressed along with punt in the posterior compartment of the wing disc, significantly smaller wing discs were observed (p<0.05) (Fig 2D and 2E) (S2 Table). These experiments show that lbk is required for optimal signaling of the BMP pathway in Drosophila.
Fig 2. LBK is required for dpp signaling in the Drosophila wing disc.
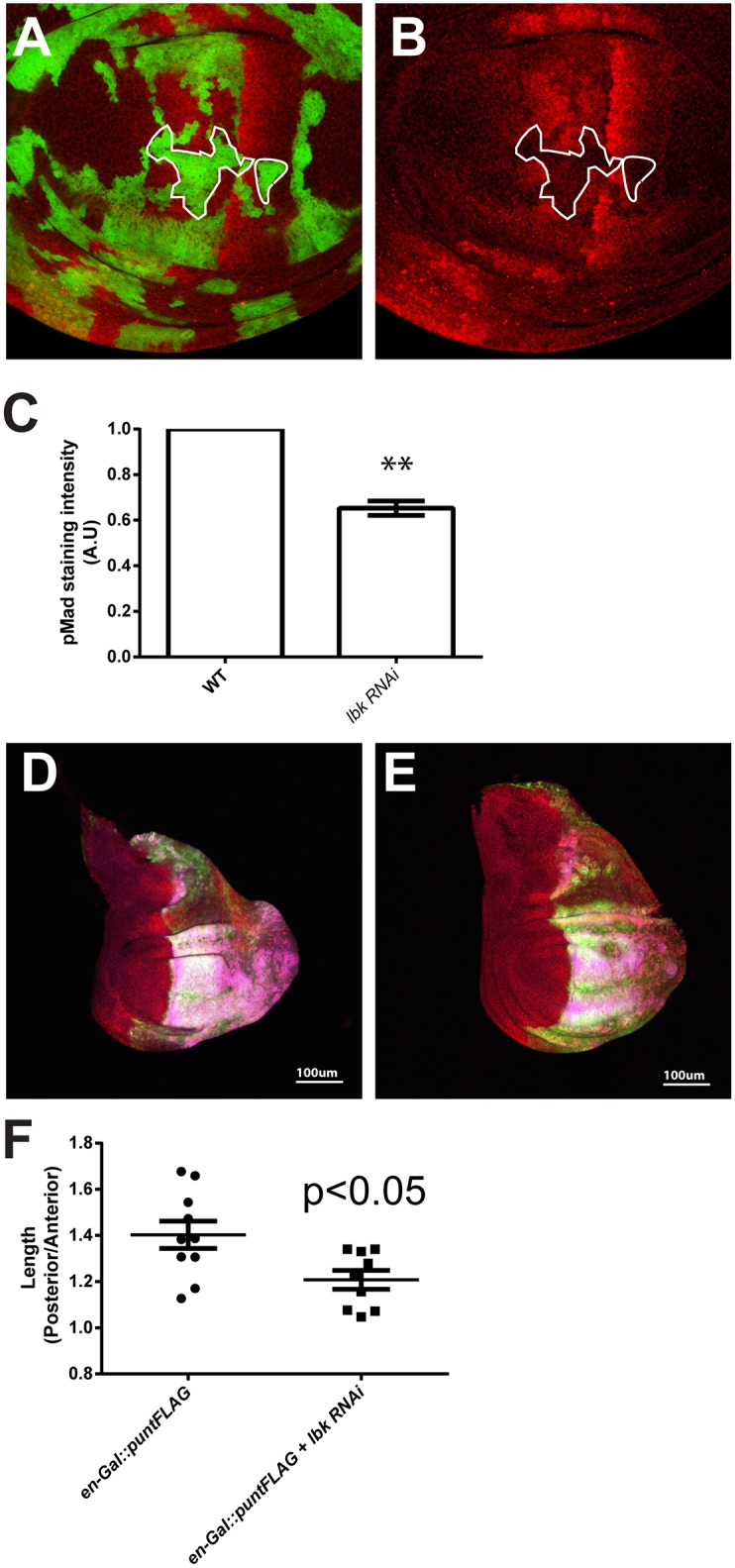
(A, B) Loss of lbk results in reduced phosphorylated MAD (pMAD) levels. RNAi-lbk clones were made with flip-out technology [45]. (A) Green labels clones where lbk is reduced, while red stains for pMad. (B) In RNAi-lbk clones (green), pMAD (red) was significantly decreased (C) compared to the neighboring cells (non-green cells). The genotype of these flies is hs-Flp[122]/+; act>y+>Gal-4 UAS-GFP/UAS-RNAi-lbk. (D) punt (type II receptor) was over expressed in the posterior compartment of the third instar larval wing disc using an en-Gal4 driver. The posterior compartment is labeled by GFP expression (green). punt expression was labeled by its FLAG tag staining (magenta). The genotype of these flies is UAS-Dcr2/+; en-Gal4 UAS-GFP/+; UAS-punt-3xFLAG/+. (E) punt was over expressed while the lbk gene was knocked down in the posterior compartment of the third instar larval wing disc. The posterior compartment was labeled by GFP expression (green). punt expression was labeled by its FLAG tag staining (magenta), and pMad is labeled red. The genotype of these flies is UAS-Dcr2/+; en-Gal4 UAS-GFP/ UAS-RNAi-lbk; UAS-punt-3xFLAG/+. Overexpression of punt caused the enlargement of posterior compartment, but the effect of punt was alleviated by RNAi-lbk. (F) The size of posterior compartment was significantly diminished. The average ratios of diameters of the anterior to posterior compartments are 1.40 and 1.15 (p<0.05) (S2 Table).
SMA-6 accumulates in the MVB in sma-10 mutants
Changes in posttranslational modifications of transmembrane receptors have been shown to direct internalization, regulate the sorting of receptors into the MVB, and promote receptor recycling [55]. For example, a ubiquitinated EGFR can be actively internalized into MVBs and degraded, while removing ubiquitin from the receptor at an earlier stage in sorting promotes its recycling [55]. Blocking either the sorting of receptors into MVBs or the recycling of receptors could result in the accumulations we identified of SMA-6 and DAF-4 in sma-10 mutants.
Our previous work has shown that SMA-6 is internalized through clathrin-dependent endocytosis, and is recycled back to the surface via the retromer complex [22]. Loss of retromer-mediated trafficking leads to degradation of cargo via the lysosome [56–59]. Since we observe intracellular accumulation of SMA-6 in the sma-10 mutant animal (Fig 1), it is unlikely that sma-10 affects retromer-mediated trafficking of the type I receptor. Identifying the endosomal compartments where the type I receptor normally traffics and pinpointing compartments where it accumulates in the sma-10(wk88) animal, will allow us to define the role of sma-10 in regulation of receptor trafficking. In order to determine which intracellular compartments SMA-6 traffics to and how this changes with the loss of sma-10, we performed colocalization experiments of SMA-6 with various endosomal markers [20, 60] in wild-type and in sma-10 mutants (Fig 3 and S3 Fig).
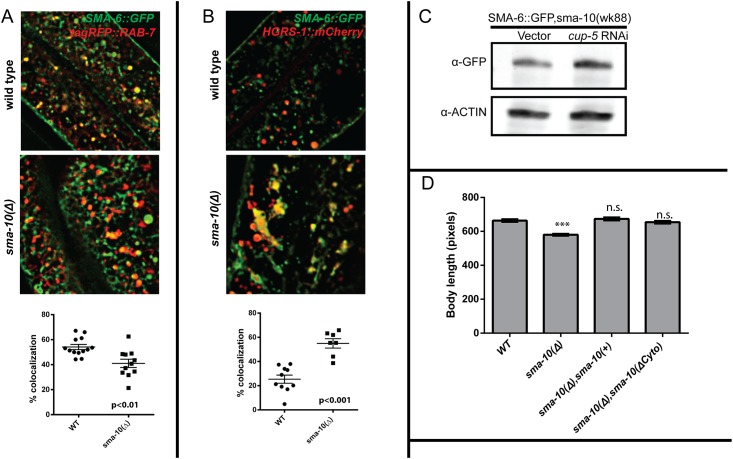
Fig 3. sma-10 is a critical regulator of SMA-6 trafficking.
(A) Loss of sma-10 leads to decreased colocalization of SMA-6 within late endosomes. (B) Loss of sma-10 leads to increased colocalization of SMA-6 within MVBs. (C) Mutants of cup-5, a necessary component of the lysosome, result in accumulation of cargos destined for the lysosome. cup-5 RNAi shows accumulation of SMA-6::GFP in a sma-10 mutant. (D) Body size rescue experiments in C. elegans. The C-terminal cytoplasmic tail of SMA-10 is not required for body size determination in C. elegans. Plasmid constructs expressing full length sma-10 [sma-10(+),red] and sma-10 lacking the cytoplasmic tail [sma-10(Δcyto),blue] from a hypodermal-specific promoter were injected into the sma-10(wk88) mutant background. At least 30 transformed animals were assayed for body size at L4 + 24 hr for each genotype. As observed, full length sma-10 (red) as well as sma-10 lacking the cytoplasmic tail (blue) fully rescue the small body size of sma-10(wk88) indicating that the cytoplasmic tail is not required for the rescue of the body size defects. Bar graphs represent Mean ± S.E.M. A One-way ANOVA was performed for the series. *** indicates p < 0.0001 as compared to the wild-type. A minimum of 15 animals per line were assayed for body size.
We first demonstrated that in wild-type animals SMA-6 colocalizes to several compartments in the endocytic trafficking pathway, including early (tagRFP::RAB-5) and late endosomes (tagRFP::RAB-7), recycling endosomes (RME-1::tagRFP), MVBs (HGRS-1::mCherry) and lysosomes (LMP-1::tagRFP) (Fig 3A, 3B and S3 Fig). We then compared the change, if any, in the colocalization pattern of SMA-6 in the sma-10(wk88) animal to identify the sites at which SMA-6 is mis-localized. We observe that loss of sma-10 leads to a significantly reduced colocalization of SMA-6 within late endosomes (Fig 3A) and an increase in the colocalization with MVBs (Fig 3B). MVBs contain cargo that can be recycled, or once recruited into intraluminal vesicles, are destined for degradation through subsequent fusion to the lysosome [14, 16]. Accumulation within the MVB suggests that at least some of the mis-trafficked SMA-6 may be degraded within the lysosome. Indeed, when lysosome function is blocked using RNAi against cup-5, a gene necessary for lysosome function, we see an increased accumulation of SMA-6 in the sma-10 background (Fig 3C) [47].
The mammalian homolog, LRIG1, recruits the E3-ubiquitin ligase, c-Cbl through interaction with regions in LRIG1’s cytoplasmic tail. Compared to the mammalian LRIG1 protein, C. elegans SMA-10 has a very short cytoplasmic tail (20 amino acids compared to approximately 280 amino acids in mammals), and we were interested to identify whether it had a role in signaling. Intriguingly, the 20 amino acids present in the cytoplasmic tail domain of SMA-10 show a limited homology with a region in the LRIG1 cytoplasmic tail necessary for the interaction with an E3 ubiquitin-protein ligase, c-Cbl (S4 Fig) (These sequences are missing in LRIG2 and LRIG3). We used the rescue of the small body size of the sma-10(wk88) mutant animal as a functional readout for activity. As expected, we observe a complete rescue when the full length genomic sma-10 was injected into the sma-10(wk88) mutant animal (Fig 3D). However, we also observe a complete rescue of the body size for three different transgenic lines expressing a truncated version of sma-10 that lacks the cytoplasmic tail (Fig 3D). These data suggest that, unlike the mammalian LRIG1 cytoplasmic tail’s requirement for EGF regulation, nematode sma-10 does not require the cytoplasmic tail for regulating BMP signaling.
Subcellular localization of SMA-10 suggests early stages of trafficking
Given that loss of sma-10 leads to the aberrant accumulation of SMA-6 within the MVB, it suggests that SMA-10 acts either upstream of or at the MVB. In order to determine the cellular localization patterns of SMA-10, we performed a series of colocalization studies using TagRFP-tagged SMA-10 and a set of GFP-tagged endosomal markers (Fig 4).
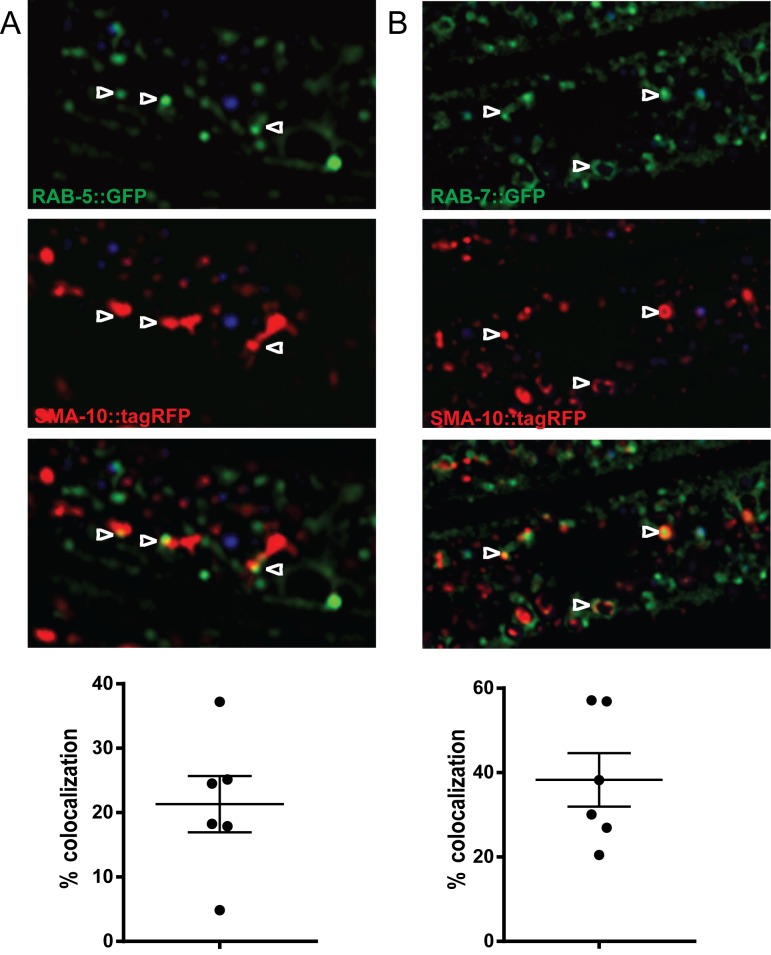
Fig 4. SMA-10 colocalizes at the surface, early and late endosomes.
SMA-10::tagRFP was co-expressed with early endosomal marker, RAB-5::GFP (A), late endosomal marker RAB-7 (B). Images were taken at the L4 + 24 hr stage using a confocal microscope and colocalization was performed.
The presence of a signal peptide and transmembrane domain in SMA-10 suggests that it is present at the plasma membrane, at least transiently. SMA-6 requires an RME-1 to recycle back to the plasma membrane, and we first tested whether SMA-10 localized with the recycling endosome marker, RME-1 [22]. We did not find significant overlap of the two markers, suggesting that SMA-10 does not mediate recycling of SMA-6 through recycling endosomes (data not shown). To test if SMA-10 localizes to the early and late endosomes, we tested colocalization of SMA-10::TagRFP to early endosome marker GFP::RAB-5 and to late endosome marker GFP::RAB-7 (Fig 4A and 4B). Significant colocalization of SMA-10 was identified with both the early and late endosomes. Its cell surface localization and presence at early endosomes suggests an early function for SMA-10. The significant late accumulation in late endosomes could indicate a late function or simply a consequence of its early function.
Discussion
Our previous work has identified C. elegans sma-10 as a conserved positive regulator of BMP signaling and shown that both worm,SMA-10, and human, LRIG1, bind to their receptors respectively [27], with more binding to the type I receptor compared to the type II receptor. In our current study, we show that nematode SMA-10 preferentially colocalizes with the type I receptor SMA-6 (Fig 1A). These results suggest that the accumulations of SMA-6 we observe in the sma-10(wk88) mutant may be a direct consequence of loss of sma-10, whereas the trafficking defect in the type II receptor DAF-4 may be indirect. The binding of SMA-10 to SMA-6 suggests a limited number of mechanisms of action. Another type I receptor, DAF-1, functions with DAF-4 in the dauer pathway [61]. We have not assessed whether SMA-10 affects DAF-1. In vertebrates, LRIGs have been shown to affect receptors through different mechanisms. LRIGs enhance EGF receptor degradation through interactions with c-Cbl [48], alter recruitment of GDNF receptors to lipid rafts [62], inhibit ligand-receptor interactions [63, 64], or regulate receptor shedding [65].
Clues to the molecular function of sma-10 come from observations of altered ubiquitination levels in SMA-6 when SMA-10 is absent. We observe reduced ubiquitination in SMA-6 when SMA-10 is absent (Fig 1D and 1E), increased colocalization of SMA-6 within the MVB (Fig 3B), and the overall accumulation of SMA-6 (Fig 1). Ubiquitin networks regulate endocytic trafficking and/or degradation of a growing number of receptors [66]. In addition, many E3 ligases contain transmembrane (TM) domains to physically tether the ubiquitin complex to specific organelles to compartmentalize the ligase activity of these enzymes. For some E3 ligases that do not contain this TM domain, scaffold proteins exist to restrict their activity to specific organelles [67]. SMA-10 may function as such a transmembrane scaffolding protein. SMA-10 regulation of SMA-6 localization may be through ubiquitination-mediated sorting into the MVB. Alternatively, the loss of ubiquitination of SMA-6 could be a downstream effect of the primary function of SMA-10. LRIG1, the mammalian ortholog of SMA-10, regulates EGF signaling by first binding to the EGFR and then recruiting the E3-Ubiquitin ligase, c-Cbl through interactions with its C-terminus. c-Cbl then ubiquitinates both LRIG1 and EGFR, leading to their degradation within the lysosome. The short cytoplasmic tail is dispensable for body size determination in C. elegans (Fig 3E) as well as for stimulation of BMP signaling in mammalian cells [27]. However, since there is a short stretch of homology of the C. elegans cytoplasmic tail sequences with human Lrig1 and Drosophila lbk (S4 Fig), this does not preclude the importance of the tail for other functions.
Since SMA-10 is present at the surface with SMA-6, it is possible that SMA-10 may be required to bring the receptors in proximity or to protect surface proteins from modification. In C. elegans, we know that the clathrin-dependent pathway is the major entry for the type I receptor. Blocking this pathway with dpy-23 (a key component of the AP2 adapter complex), we do not find surface accumulation of the type II receptor [22], suggesting its entry through non-clathrin pathways. Since signaling occurs when the receptors come together, it is necessary for the different rafts carrying the type I and II receptors to merge during the early phase of signal transduction. Multiple examples exist in which molecules are known to bring together microdomains. A third mammalian TGFβ receptor, TβRIII, has been shown to direct the type I and type II receptors into clathrin-dependent endocytosis and to increase trafficking of the type II receptor into early endosomal compartments, thereby increasing TGFβ signaling [68]. Likewise, betulinic acid induces translocation of receptors from lipid-raft/caveolae-mediated domains to non-caveolae domains, resulting in a rapid increase in reporter gene activation [69]. The opposite effect is seen with Euphol, which induces degradation of the receptors by causing the receptors to move into lipid-raft microdomains [69]. Tetraspanin-enriched microdomains are important for BMP signaling and may also participate in bringing signaling molecules together [70].
These data support the idea that the placement of receptors in plasma surface microdomains strongly influences signaling strength. We have previously shown that SMA-10 binds to both receptors [27] and that each receptor recycles back to the surface through distinct pathways [22]. Inappropriate fusion of their raft microdomains at the surface may lead to intracellular trafficking problems and aberrant accumulations in their distinct routes of trafficking. In sma-10 animals, we observe that the two receptors accumulate in distinct intracellular compartments (Figs 1 and 3, data not shown for DAF-4). Given that SMA-6 is unaffected by arf-6 (a key regulator of the endocytic recycling compartment to plasma membrane) mutants, while DAF-4 accumulates [22], it suggests that the two receptors have not separated at the stage where SMA-10 acts, possibly an earlier step of trafficking. This is consistent with a possible function at the surface for bringing the microdomains together.
Regulation of BMP signaling can also occur via a process called ectodomain shedding [34, 71, 72]. Recently, LRIG2 was shown to inhibit ectodomain shedding of neogenin [65], a gene necessary for BMP signaling. Given that several LRIG binding partners are also ADAM substrates, such as ErbB4, this has led to the suggestion that an additional function of Lrig is to prevent ectodomain shedding of bound proteins [65]. It is interesting to note that some members of the TGFβ family undergo ectodomain shedding and known membrane-associated proteases regulate signaling [34, 71, 72]. Consistent with this, in C. elegans, neogenin (UNC-40), a positive regulator of BMP signaling, undergoes ectodomain shedding by an ADAM protease (SUP-17) [73]. It was hypothesized that tetraspanins in the worm protect UNC-40 cleavage by SUP-17 [73] and thereby promote BMP signaling. It is possible that SMA-10 performs a role similar to that of LRIG2 by protecting one or more of the surface proteins necessary for efficient BMP signaling from ectodomain shedding. DRAG-1 is also part of the BMP cell surface complex in C. elegans [74]. However, it is unlikely to be involved in the core SMA-10 function, since it is missing from Drosophila (but present in mammals), where we show that the SMA-10 ortholog (lbk) affects BMP signaling.
The human homolog of SMA-10, LRIG1, negatively regulates the epidermal growth factor (EGF) signaling (Gur et al 2005). The C. elegans EGFR, let-23, is responsible for vulval development, and perturbations in the pathway lead to distinct vulval developmental phenotypes [75]. A loss-of-function phenotype leads to an animal that is vulvaless (Vuv) while a gain-of-function mutation leads to an animal that is multivulva (Muv) [75]. If SMA-10 is involved in the regulation of EGF signaling in the worm, we would expect that a loss-of-function allele of sma-10 would lead to an animal that is Vuv. We do not observe any obvious vulval defects in the sma-10 mutant strain (data not shown). Based on these observations, sma-10, at least in the nematode, does not affect the EGF signaling pathway, or if so, the effects are minimal or too subtle to be observed without a sensitized background [76–78]. Further, in order to rule out that sma-10 is a general regulator of trafficking, we asked whether SMA-10 also regulated other receptors in C. elegans. The human transferrin receptor (hTfr) is a cargo that has been shown to traffic via a clathrin-dependent mechanism, while the α-chain of the human IL-2 receptor TAC (hTAC) traffics via a clathrin-independent mechanism [16]. We find that SMA-10 does not significantly alter the trafficking of these receptors (S3 Fig) and thus likely functions to regulate BMP, and possibly a small group of receptors.
Our work shows that the function of sma-10 as a positive regulator of BMP signaling is conserved in other phyla. We show that pMad levels are altered in Drosophila lbk mutants (Fig 2) and have previously shown that the Drosophila ortholog can rescue C. elegans sma-10 mutants [27]. Additionally, we have shown that expression of SMA-10 in mammalian cells stimulates BMP signaling [27]. Our current work expands on the function of this conserved protein and identifies its role as a critical regulator of trafficking and ubiquitination of the type I receptor in C. elegans. Whether these phenotypes are a direct or indirect consequence of SMA-10 function is currently being investigated.
Supporting information
(DOCX)
(DOCX)
(A) Loss of sma-10 leads to intracellular accumulation of DAF-4. (B) Total GFP intensity measured from worm lysates from either DAF-4::GFP or DAF-4::GFP,sma-10(wk88) mutant background. (C) The colocalization between DAF-4 and SMA-10 is minimal.
(TIF)
(A). hTAC::GFP and hTfr::GFP [16] were expressed in the intestinal cells of wild-type and sma-10 mutant animals. Fluorescence confocal microscopy was performed and intensity measured using Fiji software in the same manner as in Fig 1. As observed, there is no significant decrease in the total fluorescence of GFP in the sma-10 animals. Further, there is not gross change in the localization patterns of the GFP-tagged cargos. (B) Western blots of animals from (A).
(TIF)
Colocalization data in early endosomes (A), recycling endosomes (B), or lysosomes (C) as detected by the colocalization of the endocytic markers tagRFP::RAB-5, RME-1::tagRFP and LMP-1::tagRFP with SMA-6::GFP respectively.
(TIF)
(A) The cytoplasmic tails vary greatly in length among the three organisms. Green boxes indicate the regions which show most conservation with the small 20 aa tail of C. elegans. (B) Alignments of the amino acids from the green region from panel A.
(TIF)
Acknowledgments
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by GM103995 (https://www.nih.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Massagué J. TGFβ Signal Transduction. Annu Rev Biochem. 1998;67:753–91. doi: 10.1146/annurev.biochem.67.1.753 [DOI] [PubMed] [Google Scholar]
- 2.Massagué J. TGFβ in Cancer. Cell. 2008;134(2):215–30. Epub 2008/07/30. doi: 10.1016/j.cell.2008.07.001 .18662538 [Google Scholar]
- 3.Miyazono K, Ten Dijke P, Ichijo H, Heldin CH. Receptors for transforming growth factor-β. Adv Immunol. 1994;55:181–220. [PubMed] [Google Scholar]
- 4.Padgett RW. TGFβ Signaling Pathways and Disease. Cancer and Metastasis Reviews. 1999;18:247–59. [DOI] [PubMed] [Google Scholar]
- 5.Huminiecki L, Goldovsky L, Freilich S, Moustakas A, Ouzounis C, Heldin CH. Emergence, development and diversification of the TGF-β signalling pathway within the animal kingdom. BMC Evol Biol. 2009;9:28 doi: 10.1186/1471-2148-9-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454(7207):955–60. doi: 10.1038/nature07191 [DOI] [PubMed] [Google Scholar]
- 7.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466(7307):720–6. doi: 10.1038/nature09201 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massague J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–30. doi: 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moustakas A, Heldin CH. The regulation of TGFβ signal transduction. Development. 2009;136(22):3699–714. doi: 10.1242/dev.030338 . [DOI] [PubMed] [Google Scholar]
- 10.Padgett RW, Patterson GI. TGFβ signaling in the nematode In: Derynck R, K M, editors. The TGFβ Family: Cold Spring Harbor Press; 2008. p. 527–45. [Google Scholar]
- 11.Patterson GI, Padgett RW. TGFβ-related pathways. Roles in Caenorhabditis elegans development. Trends in Genetics. 2000;16(1):27–33. [DOI] [PubMed] [Google Scholar]
- 12.Savage-Dunn C. TGF-β signaling (September 9, 2005) http://wormbook.org/; 2005. http://www.wormbook.org/.
- 13.Savage-Dunn C, Padgett RW. The TGF-β Family in Caenorhabditis elegans. 2017. January 17 In: Cold Spring Harb Perspect Biol [Internet]. https://www.ncbi.nlm.nih.gov/pubmed/28096268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burd C, Cullen PJ. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6(2). doi: 10.1101/cshperspect.a016774 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cendrowski J, Maminska A, Miaczynska M. Endocytic regulation of cytokine receptor signaling. Cytokine Growth Factor Rev. 2016;32:63–73. doi: 10.1016/j.cytogfr.2016.07.002 . [DOI] [PubMed] [Google Scholar]
- 16.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10(9):597–608. doi: 10.1038/nrm2755 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi: 10.1016/j.devcel.2011.05.015 . [DOI] [PubMed] [Google Scholar]
- 18.Hupalowska A, Miaczynska M. The new faces of endocytosis in signaling. Traffic. 2011;13(1):9–18. doi: 10.1111/j.1600-0854.2011.01249.x . [DOI] [PubMed] [Google Scholar]
- 19.Kelly BT, Owen DJ. Endocytic sorting of transmembrane protein cargo. Curr Opin Cell Biol. 2011;23(4):404–12. doi: 10.1016/j.ceb.2011.03.004 . [DOI] [PubMed] [Google Scholar]
- 20.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30(17):3481–500. doi: 10.1038/emboj.2011.286 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458(7237):445–52. doi: 10.1038/nature07961 . [DOI] [PubMed] [Google Scholar]
- 22.Gleason RJ, Akintobi AM, Grant BD, Padgett RW. BMP signaling requires retromer-dependent recycling of the type I receptor. Proc Natl Acad Sci U S A. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Brien CE, Bonanno L, Zhang H, Wyss-Coray T. Beclin 1 regulates neuronal transforming growth factor-β signaling by mediating recycling of the type I receptor ALK5. Mol Neurodegener. 2015;10:69 doi: 10.1186/s13024-015-0065-0 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, et al. A global analysis of SNX27—retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat Cell Biol. 2013;15(5):461–71. doi: 10.1038/ncb2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heldin CH, Moustakas A. Signaling Receptors for TGF-β Family Members. Cold Spring Harb Perspect Biol. 2016;8(8). doi: 10.1101/cshperspect.a022053 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu P, Lin X, Feng X-H. Posttranslatonal Regulation of Smads. Cold Spring Harb Perspect Biol. 2016. doi: 10.1101/cshperspect.a022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gumienny TL, Macneil L, Zimmerman CM, Wang H, Chin L, Wrana JL, et al. Caenorhabditis elegans SMA-10/LRIG is a conserved transmembrane protein that enhances bone morphogenetic protein signaling. PLoS Genet. 2010;6(5):e1000963 doi: 10.1371/journal.pgen.1000963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo D, Holmlund C, Henriksson R, Hedman H. The LRIG gene family has three vertebrate paralogs widely expressed in human and mouse tissues and a homolog in Ascidiacea. Genomics. 2004;84(1):157–65. doi: 10.1016/j.ygeno.2004.01.013 . [DOI] [PubMed] [Google Scholar]
- 29.Simion C, Cedano-Prieto ME, Sweeney C. The LRIG family: enigmatic regulators of growth factor receptor signaling. Endocr Relat Cancer. 2014;21(6):R431–R43. doi: 10.1530/ERC-14-0179 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11(6):725–32. . [DOI] [PubMed] [Google Scholar]
- 31.Hedman H, Henriksson R. LRIG inhibitors of growth factor signalling—double-edged swords in human cancer? Eur J Cancer. 2007;43(4):676–82. doi: 10.1016/j.ejca.2006.10.021 . [DOI] [PubMed] [Google Scholar]
- 32.Herhaus L, Sapkota GP. The emerging roles of deubiquitylating enzymes (DUBs) in the TGFβ and BMP pathways. Cell Signal. 2014;26(10):2186–92. doi: 10.1016/j.cellsig.2014.06.012 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imamura T, Oshima Y, Hikita A. Regulation of TGF-β family signalling by ubiquitination and deubiquitination. J Biochem. 2013;154(6):481–9. doi: 10.1093/jb/mvt097 . [DOI] [PubMed] [Google Scholar]
- 34.Mu Y, Sundar R, Thakur N, Ekman M, Gudey SK, Yakymovych M, et al. TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer. Nat Commun. 2011;2:330 doi: 10.1038/ncomms1332 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395(6705):854 doi: 10.1038/27579 . [DOI] [PubMed] [Google Scholar]
- 37.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–7. doi: 10.1038/nature01278 . [DOI] [PubMed] [Google Scholar]
- 38.Schweinsberg PJ, Grant BD. C. elegans gene transformation by microparticle bombardment. WormBook. 2013:1–10. doi: 10.1895/wormbook.1.166.1 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157(3):1217–26. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Praitis V. Creation of transgenic lines using microparticle bombardment methods. Methods Mol Biol. 2006;351:93–107. doi: 10.1385/1-59745-151-7:93 . [DOI] [PubMed] [Google Scholar]
- 41.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. doi: 10.1038/nmeth.2019 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elliott DA, Brand AH. The GAL4 system: a versatile system for the expression of genes. Methods Mol Biol. 2008;420:79–95. Epub 2008/07/22. doi: 10.1007/978-1-59745-583-1_5 . [DOI] [PubMed] [Google Scholar]
- 43.Phelps CB, Brand AH. Ectopic gene expression in Drosophila using GAL4 system. Methods. 1998;14(4):367–79. Epub 1998/06/03. doi: 10.1006/meth.1998.0592 . [DOI] [PubMed] [Google Scholar]
- 44.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–6. doi: 10.1038/nature05954 . [DOI] [PubMed] [Google Scholar]
- 45.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24(5):251–4. . [DOI] [PubMed] [Google Scholar]
- 46.Yakoby N, Lembong J, Schupbach T, Shvartsman SY. Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development. 2008;135(2):343–51. doi: 10.1242/dev.008920 . [DOI] [PubMed] [Google Scholar]
- 47.Sato K, Norris A, Sato M, Grant BD. C. elegans as a model for membrane traffic. WormBook. 2014:1–47. doi: 10.1895/wormbook.1.77.2 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. Embo J. 2004;23(16):3270–81. doi: 10.1038/sj.emboj.7600342 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Leon N, Valdivieso MH. The long life of an endocytic patch that misses AP-2. Curr Genet. 2016;62(4):765–70. doi: 10.1007/s00294-016-0605-3 . [DOI] [PubMed] [Google Scholar]
- 50.Pan CL, Baum PD, Gu M, Jorgensen EM, Clark SG, Garriga G. C. elegans AP-2 and retromer control Wnt signaling by regulating mig-14/Wntless. Dev Cell. 2008;14(1):132–9. doi: 10.1016/j.devcel.2007.12.001 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park SY, Guo X. Adaptor protein complexes and intracellular transport. Biosci Rep. 2014;34(4). doi: 10.1042/BSR20140069 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moghal N, Sternberg PW. The epidermal growth factor system in Caenorhabditis elegans. Exp Cell Res. 2003;284(1):150–9. . [DOI] [PubMed] [Google Scholar]
- 53.Raftery LA, Sutherland DJ. TGF-β family signal transduction in Drosophila development: from Mad to Smads. Dev Biol. 1999;210(2):251–68. doi: 10.1006/dbio.1999.9282 . [DOI] [PubMed] [Google Scholar]
- 54.Upadhyay A, Moss-Taylor L, Kim MJ, Ghosh AC, O'Connor MB. TGF-β Family Signaling in Drosophila. Cold Spring Harb Perspect Biol. 2017. doi: 10.1101/cshperspect.a022152 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315(9):1610–8. doi: 10.1016/j.yexcr.2008.10.014 . [DOI] [PubMed] [Google Scholar]
- 56.Temkin P, Lauffer B, Jager S, Cimermancic P, Krogan NJ, von Zastrow M. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13(6):715–21. doi: 10.1038/ncb2252 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Naslavsky N, Caplan S. EHDs meet the retromer: Complex regulation of retrograde transport. Cell Logist. 2012;2(3):161–5. doi: 10.4161/cl.20582 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gokool S, Tattersall D, Seaman MN. EHD1 interacts with retromer to stabilize SNX1 tubules and facilitate endosome-to-Golgi retrieval. Traffic. 2007;8(12):1873–86. doi: 10.1111/j.1600-0854.2007.00652.x . [DOI] [PubMed] [Google Scholar]
- 59.Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol. 2001;3(6):567–72. doi: 10.1038/35078543 . [DOI] [PubMed] [Google Scholar]
- 60.Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463(7280):464–73. doi: 10.1038/nature08910 . [DOI] [PubMed] [Google Scholar]
- 61.Georgi LL, Albert PS, Riddle DL. daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell. 1990;61(4):635–45. . [DOI] [PubMed] [Google Scholar]
- 62.Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci. 2008;28(1):39–49. doi: 10.1523/JNEUROSCI.2196-07.2008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL 3rd, et al. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279(45):47050–6. doi: 10.1074/jbc.M409703200 . [DOI] [PubMed] [Google Scholar]
- 64.Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol. 2012;14(4):401–8. doi: 10.1038/ncb2464 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Erp S, van den Heuvel DM, Fujita Y, Robinson RA, Hellemons AJ, Adolfs Y, et al. Lrig2 Negatively Regulates Ectodomain Shedding of Axon Guidance Receptors by ADAM Proteases. Dev Cell. 2015;35(5):537–52. doi: 10.1016/j.devcel.2015.11.008 . [DOI] [PubMed] [Google Scholar]
- 66.Foot N, Henshall T, Kumar S. Ubiquitination and the Regulation of Membrane Proteins. Physiol Rev. 2017;97(1):253–81. doi: 10.1152/physrev.00012.2016 . [DOI] [PubMed] [Google Scholar]
- 67.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12(5):295–307. doi: 10.1038/nrm3099 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLean S, Di Guglielmo GM. TGF-β (transforming growth factor-β) receptor type III directs clathrin-mediated endocytosis of TGF-β receptor types I and II. Biochem J. 2010;429(1):137–45. doi: 10.1042/BJ20091598 . [DOI] [PubMed] [Google Scholar]
- 69.Chen CL, Chen CY, Chen YP, Huang YB, Lin MW, Wu DC, et al. Betulinic acid enhances TGF-β signaling by altering TGF-β receptors partitioning between lipid-raft/caveolae and non-caveolae membrane microdomains in mink lung epithelial cells. J Biomed Sci. 2016;23:30 doi: 10.1186/s12929-016-0229-4 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Z, Shi H, Szymczak LC, Aydin T, Yun S, Constas K, et al. Promotion of bone morphogenetic protein signaling by tetraspanins and glycosphingolipids. PLoS Genet. 2015;11(5):e1005221 doi: 10.1371/journal.pgen.1005221 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elderbroom JL, Huang JJ, Gatza CE, Chen J, How T, Starr M, et al. Ectodomain shedding of TβRIII is required for TβRIII-mediated suppression of TGF-β signaling and breast cancer migration and invasion. Mol Biol Cell. 2014;25(16):2320–32. doi: 10.1091/mbc.E13-09-0524 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gudey SK, Sundar R, Mu Y, Wallenius A, Zang G, Bergh A, et al. TRAF6 stimulates the tumor-promoting effects of TGFβ type I receptor through polyubiquitination and activation of presenilin 1. Sci Signal. 2014;7(307):ra2 doi: 10.1126/scisignal.2004207 . [DOI] [PubMed] [Google Scholar]
- 73.Wang L, Liu Z, Shi H, Liu J. Two Paralogous Tetraspanins TSP-12 and TSP-14 Function with the ADAM10 Metalloprotease SUP-17 to Promote BMP Signaling in Caenorhabditis elegans. PLoS Genet. 2017;13(1):e1006568 doi: 10.1371/journal.pgen.1006568 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tian C, Sen D, Shi H, Foehr ML, Plavskin Y, Vatamaniuk OK, et al. The RGM protein DRAG-1 positively regulates a BMP-like signaling pathway in Caenorhabditis elegans. Development. 2010;137(14):2375–84. doi: 10.1242/dev.051615 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gupta BP, Hanna-Rose W, Sternberg PW. Morphogenesis of the vulva and the vulval-uterine connection. WormBook. 2012:1–20. doi: 10.1895/wormbook.1.152.1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmid T, Hajnal A. Signal transduction during C. elegans vulval development: a NeverEnding story. Curr Opin Genet Dev. 2015;32:1–9. doi: 10.1016/j.gde.2015.01.006 . [DOI] [PubMed] [Google Scholar]
- 77.Skorobogata O, Meng J, Gauthier K, Rocheleau CE. Dynein-mediated trafficking negatively regulates LET-23 EGFR signaling. Mol Biol Cell. 2016. doi: 10.1091/mbc.E15-11-0757 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Skorobogata O, Rocheleau CE. RAB-7 antagonizes LET-23 EGFR signaling during vulva development in Caenorhabditis elegans. PLoS One. 2012;7(4):e36489 doi: 10.1371/journal.pone.0036489 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(A) Loss of sma-10 leads to intracellular accumulation of DAF-4. (B) Total GFP intensity measured from worm lysates from either DAF-4::GFP or DAF-4::GFP,sma-10(wk88) mutant background. (C) The colocalization between DAF-4 and SMA-10 is minimal.
(TIF)
(A). hTAC::GFP and hTfr::GFP [16] were expressed in the intestinal cells of wild-type and sma-10 mutant animals. Fluorescence confocal microscopy was performed and intensity measured using Fiji software in the same manner as in Fig 1. As observed, there is no significant decrease in the total fluorescence of GFP in the sma-10 animals. Further, there is not gross change in the localization patterns of the GFP-tagged cargos. (B) Western blots of animals from (A).
(TIF)
Colocalization data in early endosomes (A), recycling endosomes (B), or lysosomes (C) as detected by the colocalization of the endocytic markers tagRFP::RAB-5, RME-1::tagRFP and LMP-1::tagRFP with SMA-6::GFP respectively.
(TIF)
(A) The cytoplasmic tails vary greatly in length among the three organisms. Green boxes indicate the regions which show most conservation with the small 20 aa tail of C. elegans. (B) Alignments of the amino acids from the green region from panel A.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.