Measuring actin assembly in live cells is challenging, because actin tagged with a fluorescent protein is not fully functional1. A fusion of a fluorescent protein to the Lifeact peptide is widely used to localize actin filaments in live cells2. However, we find that these fusion proteins have many concentration-dependent effects on actin assembly in vitro and in fission yeast cells. mEGFP-Lifeact inhibits actin assembly during endocytosis as well as assembly and constriction of the cytokinetic contractile ring. Purified mEGFP-Lifeact and Lifeact-mCherry bind actin filaments with Kds of ~10 μM. Lifeact-mCherry can promote actin filament nucleation and either promote or inhibit filament elongation. Separately and together profilin and formins suppress these effects. Lifeact-mCherry can also promote or inhibit actin filament severing by cofilin. These concentration-dependent effects mean that caution is necessary when overexpressing Lifeact fusion proteins to label actin filaments in cells. Therefore, we used low micro-molar concentrations of tagged Lifeact to follow assembly and disassembly of actin filaments in cells. Careful titrations also gave an estimate of a peak of ~190,000 actin molecules (~500 μm) in the fission yeast contractile ring. These filaments shorten from ~500 to ~100 subunits as the ring constricts.
Quantitative studies of actin assembly are essential to understand the mechanisms of endocytosis, cell motility and cytokinesis. Purified actin labeled with fluorescent dyes was used in assays of bulk samples3 and individual actin filaments4 that defined the parameters of actin assembly. Many studies have used actin tagged with a fluorescent protein, but these fusion proteins cannot replace the native actin gene1. When expressed at low levels, GFP-actin incorporates into cellular structures nucleated by Arp2/3 complex1,5. However, no GFP-actin incorporates into contractile rings in either fission yeast5,6 or many other cells7,8 (with a few exceptions9,10). Even tagging actin with a tetracysteine peptide and FlAsH prevents assembly into contractile rings of fission yeast6, because the Cdc12p formin that polymerizes contractile ring actin filaments11 filters out tagged actin12.
An alternative approach is to express fluorescently-tagged proteins that bind actin filaments such as calponin homology domains13. Riedl et al.2 showed that a peptide consisting of the first 17 residues of the S. cerevisiae protein Abp140 fused to a fluorescent protein labels actin filaments in cells. Advantages of this “Lifeact” peptide include its small size and the lack of homologous genes in organisms other than S. cerevisiae.
Lifeact with fluorescein on its N-terminus binds actin monomers with a Kd of 70 nM and actin filaments with a Kd of 2.2 μM, without affecting the time course of spontaneous polymerization of bulk samples of pyrenyl-actin2. However, more than 1,800 studies have utilized Lifeact fused to a fluorescent protein for expression in cells at unspecified concentrations, and no studies have documented the effects of Lifeact fusion proteins on interactions with actin or actin filament turnover in cells.
We used fission yeast to test the effects of mEGFP-Lifeact on actin assembly during clathrin-mediated endocytosis14,15 and cytokinesis16, both precisely timed events in fission yeast. We expressed a wide range of total mEGFP-Lifeact from the strong repressible P3nmt1 promoter (Fig. 1a, b). These populations of cells grew normally (Fig. 1c). As observed previously17,18, mEGFP-Lifeact concentrated in endocytic actin patches, cytokinetic contractile rings and actin cables (Fig. 1a). The total cellular mEGFP-Lifeact influenced the fluorescence intensities of these structures but not their distributions, sizes or shapes, judging from fixed cells stained by Alexa 568-phalloidin (Fig. 1d), consistent with a previous study17.
Figure 1. Effects of mEGFP-Lifeact on endocytosis in fission yeast cells.
The 3nmt1 promoter controlled the expression of mEGFP-Lifeact. (a) Fluorescence micrograph of a field of cells (pseudo-colored) grown in 5 μg/ml thiamine to induce expression of mEGFP-Lifeact. Lifeact labels actin patches (circle), contractile rings (arrow) and actin cables (arrowhead). This wide range of expression of mEGFP-Lifeact was observed in >10 independent experiments. (b) Distribution of the cytoplasmic mEGFP-Lifeact concentrations expressed by 325 cells across 4 independent experiments. (c) Growth of dilution series of wild type cells and the cells expressing mEGFP-Lifeact at 25°C under either repressing (+ thiamine) or inducing (− thiamine) conditions, repeated in two independent experiments. (d) Fluorescence micrographs of fixed cells stained with Alexa568-phalloidin. (Left) pseudo-colored mEGFP fluorescence. Asterisks: cells expressing low levels of Lifeact. (Middle and Right) Alexa568-phalloidin fluorescence. Staining with rhodamine-phalloidin or Bodipy-phallacidin produced similar results. (e) Fluorescence micrographs of a cell expressing mEGFP-Lifeact. Upper: circles mark seven actin patches used for measuring fluorescence over time. Lower: time course in seconds of the appearance and disappearance of the first patch. Observations were similar in >100 cells across 3 independent experiments. (f) Time courses of average numbers of mEGFP-Lifeact molecules in actin patches of four cells expressing mEGFP-Lifeact at the indicated cytoplasmic concentrations including the cell shown in e (arrowhead in f). Data show the mean of 4 independent experiments. (g) Dependence of the time for mEGFP-Lifeact fluorescence to peak and to disappear (insert) in patches on the cytoplasmic concentration of mEGFP-Lifeact. The gray dots are average times of 5 or more patches in a cell. The blue dots are the average times in bins of 50 μM mEGFP-Lifeact. The red lines are the best linear fits of the binned data points. Data were obtained from n=20 independent experiments.(h) Dependence of the average peak number (blue circles) of mEGFP-Lifeact molecules per actin patch (> 3 patches/cell) on the concentrations of free mEGFP-Lifeact (n = 16 cells). Data were obtained and pooled across 15 independent experiments. The red curve is the best fit of Equation 1 (see Methods) to the data giving a Kd of 20.0 μM. Inset: best fit with constraining the Kd at 9.3 μM (insert). All the error bars = mean ± 1 standard deviation. All scale bars are 5 μm.
However, mEGFP-Lifeact produced concentration-dependent changes in both endocytosis and cytokinesis. The time to assemble and disassemble (Fig. 1f, g) actin patches and to assemble (Fig. 2a, b) and constrict (Fig. 2c, d) contractile rings increased with the concentration of mEGFP-Lifeact.
Figure 2. Effects of mEGFP-Lifeact on cytokinesis.
Cells expressed Rlc1p-tdTomato to mark the contractile ring and a range of concentrations of mEGFP-Lifeact. (a) Time series of fluorescence micrographs of contractile ring assembly in cells expressing (upper cell) 22 μM and (lower cell) 55 μM mEGFP-Lifeact. These cells also expressed Sad1p-mEGFP to mark SPBs. Time zero is defined as the separation of spindle pole bodies (marked with empty arrowheads). Numbers are times in minutes. The scale bar is 2 μm. Contractile ring assembly was measured in n=73 cells pooled across 6 independent experiments. (b) Dependence of time to assemble a contractile ring on the total cytoplasmic concentration of mEGFP-Lifeact. The gray dots are the times to assemble a contractile ring in individual cells. The filled black dots are the average times in bins of 2.75 μM mEGFP-Lifeact. The black line is the best linear fit (R2 = 0.92) of the binned data points. The black circles are the average times in the cells expressing zero or 6 μM mEGFP-Lifeact. (c) Time series of fluorescence micrographs of contractile ring constriction in cells with cytoplasmic concentrations of (upper) 26 μM and (lower) 87 μM mEGFP-Lifeact. Numbers are times in minutes. The scale bar is 2 μm. Contractile ring constriction was measured in n=87 cells pooled across 8 independent experiments. (d) Dependence of the constriction rate of contractile ring on the total cytoplasmic concentration of mEGFP-Lifeact. The gray dots are rates of constriction in individual cells. The black dots are the average times in bins of 21.7 μM mEGFP-Lifeact. The red line is the best linear fit (R2 = 0.93) of the binned data points.
We used purified proteins to investigate possible mechanisms for these effects on cells. Purified mEGFP-Lifeact and Lifeact-mCherry both bound filamentous actin with a 1:1 stoichiometry and Kds of 9.3 ± 1.0 μM (Fig. 3a) and 13.2 ± 0.7 μM (Fig. 3b), ~5-fold weaker than fluorescein-Lifeact (Kd = 2 μM)2. Thus neither the position nor the identity of the fluorescent protein influenced the affinity. Lifeact-mCherry slightly increased the anisotropy of Alexa 488-actin monomers, but unlabeled actin did not affect the anisotropy of Lifeact-mCherry. Thus Lifeact-mCherry interacts very weakly (if at all) with actin monomers.
Figure 3. Lifeact-fluorescent fusion protein binding to actin filaments and effects on barbed end elongation.
(a) Binding of 2 μM actin filaments to a range of mEGFP-Lifeact concentrations. (b) Binding of 2 μM Lifeact-mCherry to a range of actin filament concentrations. The solid line is a fit of the binding equation to the data (Kd = 13.2 ± 0.7 μM). Error bars are standard deviations of the mean of n=3 independent experiments. Inset: Binding of 3 μM actin to a range of Lifeact-mCherry concentrations. The solid line is a fit of the binding equation to the Lifeact-mCherry data (Kd = 5.8 μM). (c) Binding of a range of Lifeact-mCherry concentrations to 3 μM actin filaments with 30 μM Hs cofilin. Open circles: normalized actin in the pellet; squares: normalized cofilin in the pellet and; filled circles: normalized Lifeact-mCherry. (a, c) Data are representative binding curves from three independent experiments. (d, h) Total internal reflection fluorescence (TIRF) microscopy of actin filaments. (d) Micrographs of actin filaments at intervals of 150 s with 0, 1 or 25 μM Lifeact-mCherry. (e, f, g) Dependence of barbed end elongation rates on the concentration of Lifeact-mCherry. Error bars are standard deviations of the mean of n=10 filaments pooled across 3 independent experiments.. (e) Actin alone. Inset shows low Lifeact-mCherry concentrations. (f) Actin with (closed triangles) 1 μM Sc profilin or (open triangles) 5 μM Sc profilin. (g) Actin with (closed squares) 200 nM formin Bni1(FH1FH2)p or with (closed inverted triangles) 50 nM formin Cdc12(FH1FH2)p and 5 μM Sp profilin. (h, i) Evaluation of actin filament nucleation by TIRF microscopy after incubating 0.5 μM actin monomers (20% Alexa 488-labeled) for 600 s. (h) Micrographs of actin with 200 nM Bni1(FH1FH2)p and the indicated concentrations of Lifeact-mCherry. (i) Dependence of the numbers of actin filaments after 600 s on the concentration of Lifeact-mCherry in reactions containing (circles) actin in the absence or presence of (squares) 200 nM Bni1(FH1FH2)p or (triangles) 1 μM Sc profilin. Data are representative of one of 3 sets of independent experiments that each produced samples containing dozens of filaments. Data are normalized separately for each condition to the number of filaments in the absence of Lifeact-mCherry.
Consistent with prior work with fluorescein-Lifeact2, micromolar concentrations of Lifeact-mCherry had no effect on the assembly kinetics of bulk samples of pyrenyl-actin, but decreased the critical concentration for polymerization slightly (Supplementary Fig. S1a–d). Bulk assays measure an ensemble of reactions, including nucleation, elongation, fragmentation, and annealing.
Observations of single actin filaments by total internal reflection fluorescence (TIRF) microscopy (Fig. 3d) revealed that Lifeact-mCherry perturbs many aspects of actin assembly. In spite of low affinities for actin filaments and monomers, submicromolar concentrations of Lifeact-mCherry increased the barbed end elongation rate approximately three-fold, whereas higher concentrations slowed elongation (Fig. 3e). Submicromolar Lifeact-mCherry also increased pointed end elongation from 0.09 ± 0.03 to 0.4 ± 0.1 subunits/s.
Since addition of actin subunits is diffusion limited19, Lifeact-mCherry is unlikely to increase the rate of subunit addition. One possibility is that Lifeact-mCherry promotes elongation by stimulating nucleation, creating short oligomers that grow and anneal to both ends of filaments, very favorable reactions20. We used four approaches to test this hypothesis.
First, fluorescence microscopy showed that submicromolar concentrations of Lifeact-mCherry dramatically increased the number of filaments formed in reactions containing actin with or without 200 nM formin Bni1(FH1FH2)p (Fig. 3h, i). The number of filaments peaked at 1 μM Lifeact-mCherry and decreased at higher concentrations.
Second, we tested profilin, which inhibits nucleation but not barbed end elongation21. Profilin suppressed the effects of Lifeact-mCherry on elongation of barbed ends (Fig. 3f) and nucleation (Fig. 3i).
Third, we tested formin FH2 domains, which nucleate actin filaments and remain processively attached to filament barbed ends during elongation22,23. The Bni1p FH1FH2 construct allows elongation at half the normal rate without profilin. Despite increasing the number of filaments formed (Fig. 3h, i), submicromolar concentrations of Lifeact-mCherry had no effect on the rate of elongation of barbed ends associated with this FH1FH2 construct (Fig. 3g). High concentrations of Lifeact-mCherry slowed the elongation of Bni1p-bound barbed ends up to 30%, but much less than actin alone.
Fourth, we tested the fission yeast cytokinesis formin Cdc12p, which requires profilin to elongate barbed ends24. Barbed ends elongating with Cdc12p and profilin were insensitive to Lifeact-mCherry (Fig. 3g).
Thus independently and together profilin and formins minimize the effects of Lifeact-mCherry on actin filament elongation. Profilin prevents formation of the small oligomers required to make nuclei. Formin FH1FH2 constructs prevent annealing at the barbed end25, so short filaments captured by FH2 domains can only elongate by monomer addition. Analysis of the rates of the reactions showed that nucleation and annealing can account for faster elongation (Supplementary Figure S1e). We speculate that Lifeact-mCherry promotes nucleation by stabilizing actin oligomers by binding to an interface not available in filaments.
Lifeact-mCherry also perturbs interactions of actin filaments with cofilin, a protein important for endocytosis and cytokinesis26,27. Cofilin binds cooperatively to filaments and severs at interfaces between decorated and undecorated segments28. Other actin-binding proteins, including Aip1, myosin, coronin, cortactin and Arg kinase modulate severing activity29–32.
Cofilin reduced the affinity of actin filaments for Lifeact-mCherry. Thirty micromolar Hs cofilin saturated actin filaments 1:1 independent of the concentration of Lifeact-mCherry (Fig. 3c), so the proteins bind independently to actin filaments. However, Lifeact-mCherry binding to cofilin-saturated actin filaments was cooperative (cooperativity factor ω = 8.8) with much lower affinity (Kd = 67 μM) (Supplementary Fig. S2a). Thus cofilin weakens the affinity of actin filaments for Lifeact-mCherry five-fold indirectly without competing for binding sites.
Lifeact-mCherry can promote or inhibit actin filament severing by cofilin. Concentrations of Lifeact-mCherry well below the Kd for binding to actin filaments dramatically reduced severing by 20 nM S. pombe cofilin (Sp cofilin) (Fig. 4a, b). High concentrations of Lifeact-mCherry also inhibited severing by 5, 10 or 20 μM human cofilin 1 (Hs cofilin) (Fig. 4c), which severs filaments optimally at concentrations 1000 x higher than Sp cofilin. On the other hand submicromolar Lifeact-mCherry concentrations stimulated severing (Fig. 4c, red data). Higher concentrations of Hs cofilin required higher concentrations of Lifeact-mCherry to stimulate severing (Fig. 4c, blue and black data).
Figure 4. Nanomolar concentrations of Lifeact-mCherry inhibit filament severing by cofilin.
Conditions: Actin filaments were preformed in the microscopy chamber by incubating 0.5 μM actin monomers (20% Alexa 488-labeled) in microscopy buffer (see Methods) for ~5 min. The buffer containing unpolymerized actin monomers was replaced with the same buffer containing cofilin and Lifeact-mCherry. Images were collected over time by TIRF microscopy. (a) Representative time-series of fluorescent micrographs of preformed actin filaments incubated with 20 nM Sp cofilin or 5 μM Hs cofilin in the absence or presence of 1.5 or 1 μM Lifeact-mCherry. (b, c) Dependence of the rate of actin filament severing on the concentration of Lifeact-mCherry. The units of severing activity are severing events • 10−4 μm−1 s−1. Data represent one of 3 sets of independent experiments that each produced samples containing hundreds of filaments and severing events. (b) Severing by 20 nM Sp cofilin. Inset shows low Lifeact-mCherry concentrations. (c) Severing by (closed circles) 5 μM, (open circles) 10 μM or (squares) 20 μM Hs cofilin. (d) Thermodynamic scheme of cofilin and Lifeact binding to two contiguous filamentous actin sites. Dashed box outlines interactions between a single Lifeact (L) and cofilin (C) with a filamentous actin subunit (F), resulting in Lifeact-bound actin (L_F), cofilin-bound actin (F_C) and Lifeact- and cofilin-bound actin (L_F_C) (no cooperativity). See methods section for a complete description of the model. Asterisks denote binding constants measured in Cao et al.40. Daggers denote binding constants were derived from detailed balance.
A thermodynamic analysis (see Methods) shows that Lifeact-mCherry (i) decreases the affinity of cofilin for an isolated actin subunit 5-fold (Fig. 4d, dashed box), (ii) decreases the number of initial (no neighbor) cofilin binding events and (iii) favors nearest-neighbor binding of cofilin. These effects suggest that these two proteins produce different, incompatible changes in actin filament structure such as twist33 and torsional flexibility34. It follows that Lifeact inhibits severing (Fig. 4b) by reducing the number of bound cofilins. Such conformational changes also likely inhibit filament elongation at saturating Lifeact-mCherry concentrations (Supplementary Figure S2b).
Given the effects of mEGFP-Lifeact on endocytosis and cytokinesis, we explored how to use mEGFP-Lifeact to study actin filaments in live cells. The kinetics of actin patch assembly (Fig. 1g) and cytokinesis (Fig. 2b) in cells expressing low concentrations of mEGFP-Lifeact were indistinguishable from normal cells. However, higher cellular concentrations of mEGFP-Lifeact are required to measure the absolute numbers of polymerized actin in cellular structures.
Therefore, we used endocytic patches to test the feasibility of titrating polymerized actin with mEGFP-Lifeact in live cells. As expected for a bimolecular reaction, the average peak numbers of mEGFP-Lifeact per patch increased with the concentration of free mEGFP-Lifeact (Fig. 1h) measured in parts of the cytoplasm without actin patches or actin cables. The binding curve fit to the data gave a Kd = 20.0 ± 3.8 μM (Fig. 1h), similar to the affinity measured in vitro. Fitting an unconstrained binding reaction to the data extrapolated to a maximum of 7,000 ± 600 mEGFP-Lifeact molecules per patch. If we constrained the Kd at 9.3 μM, the value measured in vitro, the binding curve extrapolated to a maximum of 5,300 ± 180 mEGFP-Lifeact molecules per patch (Fig. 1h insert). Thus the peak number of 5000–7000 mEGFP-Lifeacts per patch is consistent with previous measurements made with either GFP-actin5,15,35 or GFP-cofilin27.
To characterize the assembly and disassembly of actin filaments during cytokinesis, we used low concentrations of mEGFP-Lifeact to measure the time course of actin assembly. Then we benchmarked this data at one time point with an estimate of the number of actin molecules by titration with a range of concentrations of mEGFP-Lifeact.
We measured contractile ring fluorescence in cells expressing 6 μM mEGFP-Lifeact, a concentration that had no effect on the time course of contractile ring assembly (Fig. 2b). We co-expressed Rlc1p-tdTomato to mark rings and used automated segmentation to separate the mEGFP-Lifeact fluorescence in contractile rings from actin patches (Fig. 5a). Manually segmenting a sample of cells confirmed that automated segmentation included in rings only 2–3 actin patches per cell, a small fraction of the peak number of polymerized actin molecules in rings.
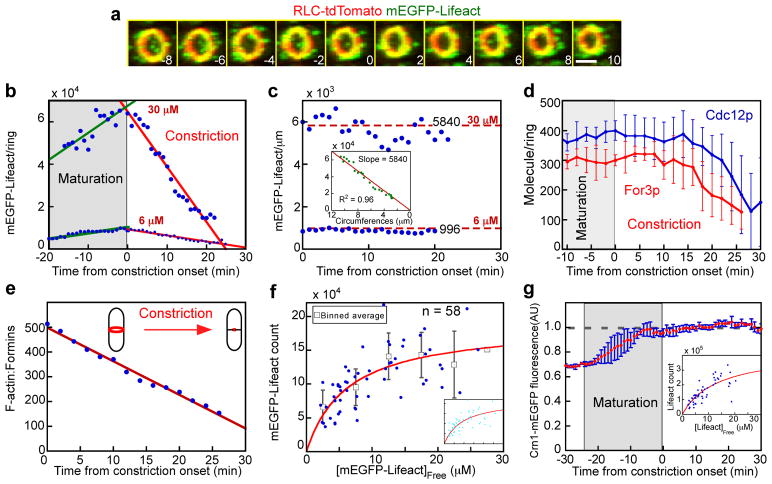
Figure 5. Quantitative analysis of actin filaments during maturation and constriction of contractile rings.
Times are in minutes with zero defined as the onset of constriction. (a–c) Numbers of mEGFP-Lifeact molecules in contractile rings counted with the segmentation method. (a) Time series of fluorescence micrographs of a cell expressing mEGFP-Lifeact (green) and Rlc1p-tdTomato (red). Merged images reconstructed from the z-series were tilted 45° to show the ring surrounded by actin patches. More than 50 cells were analyzed. The scale bar is 2 μm. (b, c, d, e, g) Time courses. (b–c) Lifeact molecules in contractile rings of three cells expressing 6 μM mEGFP-Lifeact and one cell expressing 30 μM mEGFP-Lifeact. These concentrations do not saturate actin in the ring. (b) Numbers of mEGFP-Lifeact molecules in rings. Solid lines are the linear best fits of the data points (green) before and (red) after time zero. (c) Densities (molecules per μm) of mEGFP-Lifeact in rings. Red dashed lines are mean densities of 5840 and 996 molecules per μm. Insert: numbers of Lifeact molecules vs. ring circumference in a cell expressing 30 μM mEGFP-Lifeact. Solid red line is the linear best fit (R2 = 0.97). (d) Numbers of formin molecules in contractile rings: (blue line) Cdc12p-3GFP (n = 4 cells) and (red line) For3p-3GFP (n = 4 cells). (e) Ratio of actin molecules to formin molecules in contractile rings during constriction, assuming a peak number of 190,000 actin molecules that decreased linearly. (f) Dependence of the peak numbers of mEGFP-Lifeact molecules (blue dots) in contractile rings (n = 58 cells) on the concentration of free mEGFP-Lifeact. The smooth red curves are the best fits of Equation 1 either without constraint (R2 = 0.48) or with the Kd constrained to 9.3 μM (insert) (R2 = 0.46). Squares are average numbers of Lifeact in rings in bins of 5 μM mEGFP-Lifeact. Data are from 6 independent experiments. (g) The normalized average fluorescence intensity of Crn1-mEGFP in the cleavage furrows of n=7 cells pooled across 3 independent experiments. Inset: Dependence of peak numbers of Lifeact molecules in contractile rings (blue dots), measured by the subtraction method, on concentration of free mEGFP-Lifeact. The smooth curve (red line) is the best fit of Equation 1. Error bars show mean ± 1 standard deviation.
The fluorescence of mEGFP-Lifeact in contractile rings increased steadily during the 25 min maturation stage (Fig. 5b), peaking just prior to the onset of constriction (designated as time zero). The mEGFP-Lifeact fluorescence decreased linearly over time as the contractile ring constricted (Fig. 5a, b), so the density of actin filaments was constant throughout constriction (Fig. 5c). For comparison, we show that the time course was similar and the fluorescence ~6-fold higher in a cell expressing 30 μM mEGFP-Lifeact (Fig. 5b, c).
We used triple GFP tags to count the small numbers of formins Cdc12p and For3p in rings (Fig. 5d). Their numbers were constant during the maturation phase and most of ring constriction, so the ratio of actin to formins declined linearly as rings constricted (Fig. 5e). The numbers of both formins declined during the last part of constriction.
We used titration to count the number of mEGFP-Lifeact molecules in contractile rings (Fig. 5f). The maximum number was about 150,000 in cells with the highest concentrations (> 10 μM) of free cytoplasmic mEGFP-Lifeact (binned averages, Fig. 5f). Fitting the binding equation to the dependence of contractile ring mEGFP-Lifeact on the free mEGFP-Lifeact concentrations gave a Kd of 6.2 ± 1.8 μM and extrapolated to a maximum of 190,000 ± 20,000 mEGFP-Lifeact per ring. If we constrained the Kd to 9.3 μM (measured in vitro), the binding curve extrapolated to a maximum of 221,000 ± 8,000 mEGFP-Lifeact per ring (Fig. 5f, inset). Assuming 1:1 binding, contractile rings contain ~190,000 polymerized actin molecules. An alternative approach subtracted the contributions from actin patches (Fig. 5g) from the total fluorescence around the equator to measure actin numbers (Fig. 5g, insert). This method indicates that the ring is flanked by another ~94,000 polymerized actins including short-lived whiskers (Supplementary Fig. S3)26.
Electron microscopy of thin sections has provided information about the size and shape of contractile rings since the 1970s36, but preserving, resolving and counting individual actin filaments is challenging. Electron micrographs of intact fission yeast cells showed actin filaments in contractile rings37, but the data were too limited to estimate the numbers of filaments. Kamasaki et al.38 digested the fission yeast cell wall and permeabilized the plasma membrane to allow decoration of actin filaments with myosin subfragment-1. Decorating with myosin stabilized filaments during fixation and revealed their polarity. From electron micrographs of serial sections of six cells they estimated that fission yeast contractile rings contained 1100–2100 filaments between 450 to 610 nm long for a total of about 920,000 actin molecules per ring. This larger number may arise from the cell cycle arrest and release used in this work or challenges of measuring actin filaments by electron microscopy.
Knowing the peak number of 190,000 actin subunits in the contractile ring, we calculated that a cross section of the ring has ~50 filaments. Since the ring is ~125 nm wide37, the center to center spacing of the filaments is ~15 nm. During ring constriction this density of polymerized actin is constant, but the number of actin molecules declines in proportion to the circumference, as first shown by electron microscopy of echinoderm embryos36. Since formins Cdc12 and For3p nucleate these filaments5,11,39, the ratio of actin to formin molecules gives average filament lengths, ~1.4 μm at the onset of constriction and declining linearly as the actin departs before the formins. The peak of 9,000 mEGFP-Lifeact molecules bound in the time course experiment (Fig. 5b, 6 μM) are unlikely to interfere with the 2900 Myo2p molecules, 2000 Myp2 molecules, 500 α-actinin molecules and 1300 Rng2 IQGAP molecules associated with the ring5.
Like any probe in any physical system, Lifeact fused to a fluorescent protein is bound to influence actin assembly in cells. One must be aware of the hazards and use probe concentrations that limit its effects. Fortunately, the negative effects on actin patches and cytokinesis depend on the concentration of mEGFP-Lifeact, so minding the concentration allows for its use. Although formins and profilin temper the negative effects, we caution users to measure the concentration of Lifeact fusion proteins in their cells and keep the concentration of Lifeact in the low micromolar range unless they need to measure the total numbers of polymerized actin molecules.
Materials and Methods
Expression of mEGFP-Lifeact in fission yeast
Fission yeast cells were cultured by standard protocols at 25°C. The gene for the fusion protein mEGFP-glycine-serine-Lifeact was incorporated into the leu locus and controlled by the strong 3nmt1 promoter18. Cells were grown in EMM5S medium without thiamine for 15–20 h to induce the expression of mEGFP-Lifeact. Heterogeneous expression of mEGFP-Lifeact (Fig. 1a) was observed in more than 10 independent experiments. Growth comparisons between the wild type and the cells expressing P3nmt1-mEGFP-Lifeact (Fig. 1c) were consistent in repeated experiments. To drive expression of 6 μM mEGFP-Lifeact, we sub-cloned the constitutive adf1 (cofilin) promoter and made a pFA6a-Padf1-mEGFP-Lifeact-KanMX6 vector that was integrated into the leu locus through the standard lithium-acetate transformation.
List of yeast strains
QC319: kanMX6-P3nmt1-mEGFP-lifeact ade6-M210 leu1-32 ura4-D18 (from Jianqiu Wu)
QC501: kanMX6-P3nm51-mEGFP-lifeact rlc1-TdTomato-NatMX6 (made for this study)
QC568: for3-3GFP-ura4+ rlc1-TDTomato-NatMX6 (made for this study)
KV344: cdc12-3XGFP::kanMX6 leu1-32 his3-D1 ura4-D18 ade6-M216 (Pollard lab stock)
QC256: crn1-mEGFP-KanMX6 rlc1-TdTomato-NatMX6 (made for this study)
QC500: sad1-mEGFP-kanMX6 kanMX6-P3nmt1-mEGFP-lifeact rlc1-TdTomato-NatMX6 (made for this study)
QC503: rlc1-TdTomato-NatMX6 sad1-mEGFP-KanMX6 (made for this study)
QC601: kanMX-Pcof1-mEGFP-Lifeact rlc1-tdTomato-NatMX6 (made for this study)
QC602: kanMX-Pcof1-mEGFP-Lifeact rlc1-tdTomato-NatMX6 sad11-mEGFP-KanMX (made for this study)
Fluorescence imaging
We imaged live yeast cells on a pad of 25% gelatin in EMM5S with an Olympus IX71 microscope with a spinning disk confocal unit (Yokogawa, CSU10) and an EMCCD camera (Andor, X1) calibrated to count the numbers of fluorescent protein molecules in each pixel.
We stained the actin filaments in fixed cells by fixing with 5% formaldehyde for 5 min in TEM buffer (0.1 M Tris-HCl pH 7.4, 1 mM EGTA, 1 mM MgCl2), washing with TEM buffer 3 times for 1 min each, permeabilizing with 1% Triton X-100 in TEM for 5 min and washing with TEM buffer before treatment with 8 μM Alexa568-phalloidin (Invitrogen, CA) in TEM buffer for 10 min. Cells were mounted on 25% gelatin pads for microscopy. Cells were also stained with either rhodamine-phalloidin or Bodipy-phallacidin with similar results.
We counted actin molecules in cellular structures by titration with mEGFP-Lifeact, which binds polymerized actin stoichiometrically (Fig. 3a). The concept41 is to vary the concentration of the fluorescent ligand in the cell and measure the numbers of ligand molecules associated with cellular structures, in this case endocytic actin patches and contractile rings. The bimolecular reaction is:
We verified this reaction by measuring bound mEGFP-Lifeact as a function of LAfree and calculating the Kd representing the dissociation equilibrium constant.
| (Equation 1) |
Fitting Equation 1 to the cellular binding data allowed us to estimate LA-FActin when we could not saturate the Lifeact binding site directly.
Image analysis
We analyzed fluorescence images with Image J (NIH) and Matlab (Mathworks). We measured the total number of mEGFP-Lifeact molecules in each cell from the total fluorescence in a stack of confocal Z-sections and a calibration curve. The total volume of a cell was calculated from its length as the volume of a cylinder plus a sphere (assuming diameter R = 3.74 μm):
| (Equation 2) |
We estimated the length (L) of each cell by measuring the total area of its longitudinal cross section.
We calculated the concentration of LA in the cytoplasm from the total number of mEGFP-Lifeact molecules divided by the cytoplasmic volume of cells, which is 29% of total cellular volume5. We calculated the free mEGFP-Lifeact concentration in the cytoplasm from measurements of fluorescence in an area (S) free of actin cytoskeletal structures on the medial slice of Z-series fluorescence micrographs (interval = 0.36 μm) of a cell. The free Lifeact concentration equals the total number of mEGFP-Lifeact molecules in the area divided by the volume S * 0.36 μm3.
We used a computer assisted semi-automated Image J plug-in to track actin patches in >100 cells in 3 independent experiments. To measure the number of actin patches flanking the cleavage furrow, we made 3D reconstructions from stacks of fluorescence micrographs of a region of interest 1 μm on both sides the contractile ring. We counted the numbers of actin patches from two different viewing angles of the furrow and averaged. We segmented contractile rings to count mEGFP-Lifeact with custom NIH Image J macros and Matlab scripts using the fluorescence of Rlc1p-tdTomatoto mark the contractile ring. We measured contractile ring assembly in 73 cells and ring constriction in 87 cells. Localization of mEGFP-Lifeact in the cleavage furrow was analyzed in more than 50 cells.
An alternative approach to count the number of actin molecules in the contractile ring
The total mEGFP-Lifeact in a band 2 μm wide centered on the cleavage furrow was much higher than in the segmented ring owing to the presence of actin patches, whiskers of actin on the sides of contractile rings and actin cables. We measured the number of actin patches near contractile rings from the fluorescence of mEGFP-Crn1p (coronin). The number of patches was constant at 19 ± 2 (n = 5) during ring constriction (Fig. 5g). We estimated the numbers of mEGFP-Lifeact bound to 19 actin patches at each concentration of cytoplasmic mEGFP-Lifeact from the average number of actin molecules per patch of 3000 (= 6000/2). Subtracting this number from the total mEGFP-Lifeact at the cleavage site gave the numbers of mEGFP-Lifeact in the contractile ring and other structures flanking the ring at each concentration of mEGFP-Lifeact (Fig. 5g insert). These counts were about 50% higher than numbers counted by segmenting rings (Fig. S3), so additional actin filaments flank the ring.
Protein purification
Lifeact-mCherry
Lifeact (sequence: MGVADLIKKFESISKEE), followed by a linker (sequence: RIPGLIN) and the sequence for mCherry was cloned into a pGEX-4T3 vector, which added an N-terminal GST fusion and a thrombin cleavage site. After cleavage with thrombin, the protein began with Gly-Ser, followed by the Lifeact sequence. The construct was expressed in BL21 DE3 cells with 0.5 mM IPTG (added once the cells reached an optical density of 0.8 at 595 nm) and incubation at 16 °C overnight. After harvesting, cell pellets were resuspended in 10 mL of 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 1 mM DTT per gram of cells. The cells were lysed by sonication, and ~100 ml of clarified supernatant was incubated with 6 mL of glutathione-Sepharose resin (GE Healthcare) for 1 h with rotation at 4 °C. The lysate and the resin were then poured into a glass column and the resin was washed with 100 mL of lysis buffer. Fifty units of thrombin (<1% of the concentration of Lifeact-mCherry) were added to the column and incubated at 4 °C overnight. The protein was then eluted with lysis buffer, dialyzed into KMEI buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole (pH 7.0), 1 mM DTT), flash-frozen and stored at −80 °C. Purified Lifeact-mCherry (up to 50 μM) was incubated with monomeric and filamentous actin for 30 min at room temperature. Analysis of the proteins by SDS-PAGE, Coomassie blue staining and densitometry of bands with ImageJ confirmed that no actin was degraded in the presence of the remaining thrombin.
mEGFP-Lifeact
We sub-cloned the coding sequence of mEGFP- glycine-serine-Lifeact into the pGEX6P-1 vector (GE Healthcare) with a Precision Protease cleavage site engineered between the sequences of GST and mEGFP-Lifeact. We induced the expression of recombinant GST-mEGFP-Lifeact in 2 liter cultures of BL21-CordonPlus-RIL cells (Stratagene) with 0.25 mM IPTG overnight at 20°C. Bacteria were harvested and lysed by sonication in 50 mL of lysis buffer (20 mM Tris-HCl pH 8.0, 400 mM NaCl, 1 mM EDTA, 1 mM DTT) with 0.5 mM PMSF and 1 tablet of Protease Inhibitor Cocktail (Roche, eComplete)). We incubated 3 mL of glutathione Sepharose 4B resin (GE Healthcare) with 40 mL of clarified supernatant of the bacterial lysis at 4°C for 1 h before packing the resin in a 1×10 cm column. After washing the column with 10 column volumes of lysis buffer, we incubated the column overnight at 4°C with 1 mL of lysis buffer containing 50 units of Precision Protease (GST tagged, GE Healthcare) and eluted mEGFP-Lifeact with 9 ml of column buffer.
We measured fusion protein concentrations by absorbance at 280 nm using theoretical extinction coefficients of 34,380 M−1cm−1 for Lifeact-mCherry and of 21,890 M−1 cm−1 for mEGFP-Lifeact.
Muscle actin
Actin was purified from an acetone powder of frozen chicken skeletal muscle (Trader Joe’s) using one cycle of polymerization and depolymerization followed by gel filtration on Sephacryl S-300 and storage in Ca-Buffer-G (2 mM Tris-HCl, pH 8.0, 2 mM ATP, 0.1 mM CaCl2, 1 mM NaN3, 0.5 mM DTT). The concentration of actin was measured by absorbance at 290 nm with an extinction coefficient of 26,600 M−1cm−1. Actin was polymerized in 50 mM PIPES, pH 6.8, 50 mM KCl, 0.2 mM CaCl2, 0.2 mM ATP and labeled on lysines by incubating overnight at 4°C with a 13:1 molar ratio of Alexa Fluor 488 carboxylic acid succinimidyl ester (A-20000, Invitrogen, Carlsbad, CA) to actin. After depolymerization, clarification and gel-filtration on Sephacryl S-300, purified actin monomers were typically ~30–50% labeled.
Cofilins
S. pombe (Sp) and human (Hs) cofilin were expressed in BL21(DE3)pLysS cells with 1 mM IPTG at 37 °C for 4 hours. About 8 g of cells expressing Sp cofilin were resuspended in 70 mL of 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 2 mM DTT, 2 mM PMSF, 1 tablet Roche Protease inhibitor cocktail and lysed by sonication. Following clarification of the lysate, Sp cofilin was precipitated with 70% ammonium sulfate, resuspended in 20 mL of 10 mM Tris-HCl pH 8.0, 1 mM NaN3, 1 mM EDTA, 250 mM NaCl, and 2 mM DTT and gel filtered on a 400-ml column of Sephacryl S-200. Peak cofilin fractions were dialyzed in 25 mM Tris-HCl, pH 8.4, and 2 mM DTT, loaded on a 20 mL DEAE-Sepharose column and eluted with a 500-ml gradient of 0–500 mM NaCl in DEAE buffer. About 8 g of cells expressing Hs cofilin were resuspended in 70 mL of 10 mM Tris-HCl (pH 7.5), 1 mM EGTA, and 2 mM DTT and lysed by sonication. Following clarification of the lysate by centrifugation at 21,200 × g for 45 min, the supernatant was applied to a 20 mL DEAE-Sepharose column, and the flow-through of ~70 mL was dialyzed vs. 2 L of 150 mM NaCl, 10 mM Tris-HCl (pH 7.5), 2 mM DTT. The protein was then gel-filtered on a 400-mL column of Sephacryl S-200 resin in the dialysis buffer. Following purification, both Hs and Sp cofilin were dialyzed in KMEI buffer, concentrated in centrifuge filter devices (Corning Glass, 10-kDa molecular mass cutoff), flash-frozen and stored at −80 °C.
Co-sedimentation assays
We measured affinity and stoichiometry of mEGFP-Lifeact and Lifeact-mCherry binding to chicken skeletal muscle actin filaments with a pelleting assay. Actin was polymerized in KMEI buffer and incubated with a range of concentrations of mEGFP-Lifeact or Lifeact-mCherry for 30–60 min at room temperature. Some reactions contained 30 μM Hs cofilin. Reactions were centrifuged at 100,000 × g for 30 min. Supernatants or pellets from 3 independent experiments were analyzed by SDS-PAGE and staining with Coomassie blue. Band intensities were measured with ImageJ software.
TIRF Microscopy
Glass flow chambers were incubated for 1 min each with 0.5% Tween 80 in high-salt Tris-buffered saline (HS-TBS) (50 mM Tris-HCl, pH 7.5, 600 mM KCl), 250 nM N-ethylmaleimide-inactivated skeletal muscle myosin in HS-TBS, and 10% BSA (w/v) in HS-TBS, with washes of HS-TBS after each incubation step. Polymerization was initiated by mixing 0.5 μM actin monomers (20% labeled with Alexa 488) with or without formin or profilin and a range of concentrations of Lifeact-mCherry in standard microscopy buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, 0.3 mM ATP, 15 mM glucose, 50 mM dithiothreitol [DTT], 0.02 μM CaCl2, 20 μg/ml catalase, 100 μg/ml glucose oxidase, and 0.5% methylcellulose [4,000 cP at 2% (w/v)]). Formin-mediated elongation experiments were performed with 200 nM Bni1(FH1FH2)p or 50 nM Cdc12(FH1FH2)p. Bni1p can elongate barbed ends in the absence of profilin, whereas Cdc12p cannot. Therefore, experiments with Bni1p were performed in the absence of profilin, whereas all experiments performed with Cdc12p included profilin. The concentrations of formin used are a reflection of the amount of formin required to nucleate and elongate a sufficient number of filaments for data acquisition and analysis, and do not affect the measured rates. For severing experiments, 0.5 μM actin was polymerized in the chamber until filaments were visible. Unpolymerized monomers were then exchanged with a fresh sample of proteins (i.e. Lifeact-mCherry and cofilin) in microscopy buffer, and imaging was continued. Severing data were obtained from 2 to 3 independent experiments that each produced samples containing hundreds of filaments and severing events.
Fluorescence micrographs were collected every 5–10 s using prism-style total internal reflection microscopy on an Olympus IX-70 inverted microscope and a Hamamatsu C4747-95 CCD (Orca-ER) camera controlled by MetaMorph software (Molecular Devices, Union City, CA). Changes in filament length were measured using ImageJ software (National Institutes of Health) for at least 10 filaments in 2 to 4 independent experiments, typically over a span of at least 300 s. We assumed 370 actin subunits per μm of filament42.
Thermodynamic analysis of Lifeact-mCherry and cofilin binding to actin filaments
Cofilin binds actin filaments cooperatively, with greater affinity for sites with cofilin bound to neighboring subunits. These interactions are best described by a model for cooperative binding to singly contiguous (one nearest neighbor) sites with a binding affinity Kappw, where Kapp is the apparent association equilibrium constant and w is the cooperativity coefficient. We found that even though Lifeact-mCherry and cofilin can bind simultaneously to actin filaments, saturating filaments with cofilin decreases their affinity for Lifeact-mCherry (Fig. 3c). The best fit of a singly contiguous cooperative model to Lifeact-mCherry-actin binding data had a cooperativity factor ω of 8.8, which is similar to the value of ω (8.5) for cofilin binding actin filaments28.
Using our measured actin-binding constants for Lifeact-mCherry and published parameters for cofilin40, we built a thermodynamic model that describes Lifeact-mCherry and cofilin binding to two contiguous filamentous actin subunits (Fig. 4d). This minimal model describes both an initial binding event (no nearest neighbors) and a subsequent binding event to a site with a bound neighbor. We determined values for unknown binding constants by applying a detailed balance.
The top row of the model defines reactions for cofilin binding to two contiguous actin subunits. The first cofilin binds with a Kd of 23 μM to form F_C. Owing to nearest-neighbor cooperativity, the second cofilin binds next to the first with a Kd of 1.4 μM. The left column defines reactions for Lifeact binding to two actin subunits. Lifeact does not bind cooperatively to actin in the absence of cofilin, so the binding constants are the same for both binding reactions (13 μM). Other reactions in the scheme define binding of Lifeact to partially and fully cofilin-decorated actin, and vice versa. The affinity of Lifeact for a cofilin-bound actin subunit was determined from the fit to the binding data in Fig. 3c, which produced a Kd of 67 μM and a cooperativity factor (ω) of 8.8. Because this value of ω is nearly identical to that measured for cofilin, we used the same values of ω+ and ω− as reported for cofilin in Cao et al.40 to determine the affinity of Lifeact to a cofilin-bound actin subunit with a nearest neighbor bound to Lifeact and cofilin (Kd = 4.0).
Lifeact-mCherry decreases the affinity of cofilin for an isolated actin subunit 5-fold (Fig. 4d, dashed box). However, the affinity of cofilin for a binding site with a cofilin-bound neighbor is roughly independent of Lifeact-mCherry. Thus, for a given concentration of cofilin, the number of initial (no neighbor) cofilin binding events decreases in the presence of Lifeact-mCherry, so nearest-neighbor binding of cofilin becomes an increasingly predominant mode of filament saturation.
Cofilin is known to alter actin filament twist33 and increase torsional flexibility34. Given non-overlapping sites on actin the lower affinities of cofilin and Lifeact-mCherry for actin in each other’s presence suggests that these two proteins produce different and incompatible changes in actin filament structure.
Cofilin severs optimally when the number of interfaces between decorated and undecorated filament segments is high. For Sp cofilin, this occurs at nanomolar concentrations, which decorate filaments with a low density of singly bound cofilins28. At these concentrations, a five-fold decrease in the affinity of cofilin for binding sites without a neighboring cofilin will decrease the number of binding events, resulting in fewer decorated segments. This effect ultimately reduces the number of severing events (Fig. 4b).
On the other hand, reducing the affinity of cofilin for actin sites without a neighboring cofilin will not affect the extent of filament decoration by concentrations of cofilin that normally saturate actin filaments (e.g. 5–20 μM) at equilibrium. However, it will likely slow the kinetics of cofilin binding to sites without a neighboring cofilin and therefore the initiation of new decorated segments. This will increase the lifetimes of interfaces between decorated and undecorated segments and increase filament severing in the kinetic regime (Fig. 4c).
Higher concentrations of Lifeact-mCherry will slow cofilin binding to sites without a neighboring cofilin, causing an increasing proportion of filament decoration to occur through nearest-neighbor binding of cofilin. This will ultimately result in longer, but fewer decorated segments, and therefore decreased filament severing (Fig. 4c).
The extent of filament saturation by Lifeact-mCherry in turn depends on the cofilin concentration. Because the affinity of Lifeact-mCherry for actin is inversely proportional to cofilin concentration, the concentration of Lifeact-mCherry that produces maximal severing increases with cofilin concentration (Fig. 4c).
Statistics and Reproducibility
We performed a number of independent experiments to ensure repeatability of our results. Sample sizes and the number of independent experiments are indicated in each figure legend in the manuscript.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM026338. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Cameron Lamoureux, Jeremiah Johnson and Catherine McGuinness for carrying out preliminary experiments for this project.
Footnotes
Author Contributions
N.C., Q.C. and T.D.P. designed the experiments. N.C. performed the biochemistry experiments. Q.C. performed the experiments in yeast. N.C., Q.C. and T.D.P. analysed the data. N.C., Q.C. and T.D.P. wrote the paper.
Competing Financial Interest
The authors declare no competing financial interests.
References
- 1.Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci U S A. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nature methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. Journal of muscle research and cell motility. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 4.Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci U S A. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 6.Chen Q, Nag S, Pollard TD. Formins filter modified actin subunits during processive elongation. J Struct Biol. 2012;177:32–39. doi: 10.1016/j.jsb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerisch G, Weber I. Cytokinesis without myosin II. Curr Opin Cell Biol. 2000;12:126–132. doi: 10.1016/s0955-0674(99)00066-6. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho A, Desai A, Oegema K. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell. 2009;137:926–937. doi: 10.1016/j.cell.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 9.Murthy K, Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- 10.Zhou M, Wang YL. Distinct pathways for the early recruitment of myosin II and actin to the cytokinetic furrow. Mol Biol Cell. 2008;19:318–326. doi: 10.1091/mbc.E07-08-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. The Journal of cell biology. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aizawa H, Sameshima M, Yahara I. A green fluorescent protein-actin fusion protein dominantly inhibits cytokinesis, cell spreading, and locomotion in Dictyostelium. Cell structure and function. 1997;22:335–345. doi: 10.1247/csf.22.335. [DOI] [PubMed] [Google Scholar]
- 13.Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 15.Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol Biol Cell. 2010;21:2894–2904. doi: 10.1091/mbc.E10-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, et al. Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J Cell Biol. 2012;199:831–847. doi: 10.1083/jcb.201209044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coffman VC, Nile AH, Lee IJ, Liu H, Wu JQ. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drenckhahn D, Pollard TD. Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. The Journal of biological chemistry. 1986;261:12754–12758. [PubMed] [Google Scholar]
- 20.Andrianantoandro E, Blanchoin L, Sept D, McCammon JA, Pollard TD. Kinetic mechanism of end-to-end annealing of actin filaments. Journal of molecular biology. 2001;312:721–730. doi: 10.1006/jmbi.2001.5005. [DOI] [PubMed] [Google Scholar]
- 21.Courtemanche N, Pollard TD. Interaction of profilin with the barbed end of actin filaments. Biochemistry. 2013;52:6456–6466. doi: 10.1021/bi400682n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annual review of biochemistry. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 23.Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell motility and the cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. The Journal of cell biology. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q, Pollard TD. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. The Journal of cell biology. 2011;195:485–498. doi: 10.1083/jcb.201103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q, Pollard TD. Actin filament severing by cofilin dismantles actin patches and produces mother filaments for new patches. Curr Biol. 2013;23:1154–1162. doi: 10.1016/j.cub.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Molecular cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Chen Q, Courtemanche N, Pollard TD. Aip1 promotes actin filament severing by cofilin and regulates constriction of the cytokinetic contractile ring. The Journal of biological chemistry. 2015;290:2289–2300. doi: 10.1074/jbc.M114.612978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courtemanche N, Gifford SM, Simpson MA, Pollard TD, Koleske AJ. Abl2/Abl-related gene stabilizes actin filaments, stimulates actin branching by actin-related protein 2/3 complex, and promotes actin filament severing by cofilin. The Journal of biological chemistry. 2015;290:4038–4046. doi: 10.1074/jbc.M114.608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elam WA, Kang H, De La Cruz EM. Competitive displacement of cofilin can promote actin filament severing. Biochemical and biophysical research communications. 2013;438:728–731. doi: 10.1016/j.bbrc.2013.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gandhi M, Achard V, Blanchoin L, Goode BL. Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Molecular cell. 2009;34:364–374. doi: 10.1016/j.molcel.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. The Journal of cell biology. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prochniewicz E, Janson N, Thomas DD, De la Cruz EM. Cofilin increases the torsional flexibility and dynamics of actin filaments. Journal of molecular biology. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 35.Picco A, Mund M, Ries J, Nedelec F, Kaksonen M. Visualizing the functional architecture of the endocytic machinery. Elife. 2015;4 doi: 10.7554/eLife.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder TE. The contractile ring. II. Determining its brief existence, volumetric changes, and vital role in cleaving Arbacia eggs. J Cell Biol. 1972;53:419–434. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanbe T, Kobayashi I, Tanaka K. Dynamics of cytoplasmic organelles in the cell cycle of the fission yeast Schizosaccharomyces pombe: three-dimensional reconstruction from serial sections. J Cell Sci. 1989;94(Pt 4):647–656. doi: 10.1242/jcs.94.4.647. [DOI] [PubMed] [Google Scholar]
- 38.Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arasada R, Pollard TD. Contractile ring stability in S. pombe depends on F-BAR protein Cdc15p and Bgs1p transport from the Golgi complex. Cell reports. 2014;8:1533–1544. doi: 10.1016/j.celrep.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. Journal of molecular biology. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 41.McCormick CD, Akamatsu MS, Ti SC, Pollard TD. Measuring affinities of fission yeast spindle pole body proteins in live cells across the cell cycle. Biophys J. 2013;105:1324–1335. doi: 10.1016/j.bpj.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huxley HE, Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behavior during contraction and rigor. Journal of molecular biology. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.