Abstract
Native transthyretin (TTR) homotetramer dissociation is the first step of the fibrils formation process in amyloid disease. A large number of specific point mutations that destabilize TTR quaternary structure have shown pro-amyloidogenic effects. Besides, several compounds have been proposed as drugs in the therapy of TTR amyloidosis due to their TTR tetramer binding affinities, and therefore, contribution to its integrity. In the present paper we have explored key positions sustaining TTR tetramer dynamical stability. We have identified positions whose mutations alter the most the TTR tetramer equilibrium dynamics based on normal mode analysis and their response to local perturbations. We have found that these positions are mostly localized at β-strands E and F and EF-loop. The monomer-monomer interface is pointed out as one of the most vulnerable regions to mutations that lead to significant changes in the TTR-tetramer equilibrium dynamics and, therefore, induces TTR amyloidosis. Besides, we have found that mutations on residues localized at the dimer-dimer interface and/or at the T4 hormone binding site destabilize the tetramer more than the average. Finally, we were able to compare several compounds according to their effect on vibrations associated to the ligand binding. Our ligand comparison is discussed and analyzed in terms of parameters and measurements associated to TTR-ligand binding affinities and the stabilization of its native state.
Introduction
Amyloid diseases involve protein misfoldings into cross-β-sheet structures and aggregations, ultimately leading to amyloid fibril formation. Amyloid fibrils deposition on various tissues causes systematic organ dysfunctions like cardiomyopathy, ophthalmopathy, and neuropathy. While most of current treatments are limited to reduce the symptoms, numerous clinical trials deal with other alternative therapies. Among them, transthyretin(TTR) results as a possible molecular target of amyloidosis disruption. TTR misfolds and misassembles are responsible of hereditary amyloidosis like senile systemic amyloidosis[1], familial amyloid polyneuropathy[2], and familial amyloid cardiomyopathy[3].
TTR is an amyloidogenic protein whose native homotetramer is involved in the transport of thyroxine hormone (T4) in plasma. The four TTR monomers are assembled with D2 symmetry. Each monomer is composed of two four-stranded antiparallel β-sheets and a short α-helix. Two monomers form a dimer stabilized by a hydrogen bond network between the two β-strands H and F at the monomer-monomer interface. Two of such dimers form the tetramer with a weaker dimer-dimer interface stabilized mostly by hydrophobic contacts between pairs of AB and GH loops. Two T4 hormone binding sites are located in the long hydrophobic cavities formed by each symmetric half of the dimer-dimer interface.
TTR tetramer dissociation into dimers is a required early step in the amyloid cascade[4],[5],[6]. The resulted pair of dimers quickly dissociates into monomers and, as a consequence, partial monomer denaturation leads to fibril assembly and other aggregates formation. It is therefore expected that TTR tetramer stabilization contributes to inhibit TTR fibrillogenesis[7],[8],[9],[10],[11],[12],[13],[14],[15],[16].
Specific point mutations that destabilize the TTR tetramer increase the propensity of fibril formation[9],[16],[17],[18]. The understanding at the molecular level of TTR tetramer equilibrium dynamics requires identification of the network of residues whose mutations directly affect their relative structural and dynamics stability.
The binding of thyroxine hormone T4 inhibits TTR amyloidogenesis by establishing interactions with residues at the dimer-dimer interface that bridge both subunits. Several drugs that bind to the hydrophobic T4 binding site are known to stabilize the native TTR tetramer conformation, and therefore slow amyloidogenesis[19],[20],[21]. The exploration of new classes of compounds with certain affinity to the TTR tetramer enlighten about promising candidates in the therapy against the disease.
Here in, we present a systematic selection of key positions sustaining TTR tetramer stability. Adapting a previously developed method[22] to identify key position residues whose mutations directly affect the affinity for a ligand, we identify key position residues whose mutations significantly alter the TTR tetramer equilibrium dynamics. The method makes use of normal mode analysis and mode responses to local perturbations. This combination has been previously proved as a useful tool to simulate effect of mutations in a large variety of proteins and, therefore, identify functionally important residues[22].
TTR tetramer ligand-free and ligand-bound conformations co-exist as local minima within the TTR energy landscape. The conformational changes involved in ligand-binding should be achieved by its intramolecular vibrational dynamics. We explore the possibility that the relatively high affinity of certain ligand to TTR can be a consequence of significant changes introduced on the subspace of normal modes of the tetramer involved in the unbound-to-bound conformational changes. If ligand binding modifies these modes, energy barriers that dissociate the TTR-ligand complex will be harder to overcome by thermal fluctuations. In a previous paper[22] we have presented a procedure to define and compare normal mode subspaces associated to ligand-binding. In the present paper we adapt this method to analyze TTR-ligand complex dynamics stability. In order to contribute with new complementary dynamics features to future explorations of potential new drugs, we compare several ligands according to their effect on vibrations associated to ligand binding.
Methods
Normal modes analysis
Normal mode analysis has been calculated using the coarse-grained Elastic Network Model (ENM)[23],[24],[25],[26],[27]. Normal modes and molecular dynamics, with the help of powerful computational tools that allow further structural and dynamics analysis[28],[29], become standard techniques for analyzing and enhancing our understanding of molecular mechanisms. Briefly, ENM represents the N residues of a protein by their α-carbons (nodes), connected by uniform springs to their neighbours within a cut-off distance rc. Here in, a value of rc = 7Å has been used for all the selected TTR structures.
The potential of the network of N nodes is defined as[23],[30], [31]
| (1) |
where rij ≡ ri−rj is the vector connecting nodes i and j, and the zero superscript indicates the equilibrium position that corresponds to the coordinates of the α-carbons in an experimental structure. The type of interaction between nodes i and j defines the value of the force constant kij as[32]:
| (2) |
being γ a scaling constant to match the theoretical B-factors with the experimental ones. RING program[33],[34] has been used to define inter-residue connectivities.
Normal modes are obtained by diagonalizing the 3Nx3N Hessian matrix H, a matrix of second-order partial derivatives of the potential energy
| (3) |
where q is an orthogonal matrix whose columns qk are the eigenvectors of H, that is, the normal modes, and Λ is the diagonal matrix of eigenvalues λk of H.
A. Mode perturbations, comparison and key residues
Single point mutations of a residue i are simulated by introducing perturbations to the local interactions involving the ith residue[35], [36], [37], [38]. Previous works have shown that this can be performed by changing the force constants kij that connect i with other residues j by a small amount δγ = 0.05 [22]. Here in, in order to simulate mutations introduced in the TTR tetramer, we simultaneously modify the force constants of the four ith residue on each monomer. The new set of perturbed normal modes (N being the number of residues of each monomer) is compared with the unperturbed {qk}k = 1,2x3N−6 modes as follows.
Firstly, we calculate the overlap matrix O with elements defined as the dot product
| (4) |
The similarity Riis defined as
| (5) |
with defined as the maximum value of among the projections (k′ = 1, 4x3N) of mode on the {qk}k = 1,4x3N modes.
The lower the value of Ri, the larger effects that mutations in the residue i introduce on the TTR tetramer equilibrium dynamics. The effects of mutations of residue i on the TTR tetramer equilibrium dynamics are finally evaluated by normalizing Ri as
| (6) |
Where and σR are the average and standard deviation of the distribution of Ri over all residues. Key position residues are selected as those presenting relative large negative values of Zi. That is, residues with large negative values of Zi are residues whose mutations alter the most the TTR tetramer equilibrium dynamics.
B. Subspace of modes associated to ligand-binding: Definition and comparison
Dynamics associated to tetramer-ligand assembly is defined as the subspace of normal modes that overlap the most with the direction dictated by structural distortions introduced by ligand binding. The procedure to select these modes follows the procedure previously described elsewhere[22]. Firstly, unbounded TTR tetramer and TTR tetramer-ligand complex structures are superposed minimizing the root-mean-square-deviation (RMSD) between Cα atoms. The normalized difference vector v between them is projected on the basis of the ligand-free tetramer normal modes
| (7) |
with
| (8) |
We calculate the mode participation number[39],[40],[22] as a quantitative measure of the degree of delocalization of v among the different tetramer normal modes
| (9) |
The value of Pq, rounded to the nearest higher integer, indicates the size of the subspace of modes that retains the direction of the ligand-binding. Values of Pq ≈ 4x3N−6 mean that all vibrations of the tetramer are involved in the process, while values of Pq ≈ 1 indicate that the direction of structural changes is dictated by one single normal mode. The first Pq modes ordered by index fk in decreasing values of (ck)2 define the subspace S of modes required to achieve a good description of the conformational change. In this way, S retains normal modes most involved in the tetramer-ligand complex formation.
Once defined S, the corresponding subspace Sligand is obtained by establishing a one-to-one correspondence between ligand-free tetramer’s normal modes {qk}k = 1,4x3N−6 and normal modes calculated using the tetramer-ligand complex structure. This is achieved by maximizing of the trace of the square of the overlap matrix O whose elements are defined as the dot product
| (10) |
This can be done by using a variant of the Min-Cost algorithm[41],[42] that selects those elements of the O matrix, one for each row, and each pertaining to a different column (or vice versa), which maximize the sum of their squared values.
The comparison of ligand-free tetramer and ligand-bound tetramer subspaces of modes, S and Sligand, associated to the conformational change upon ligand-binding is performed through the calculation of the corresponding Gramian matrix[43],[44], [45], [46], [22]. Firstly, each is projected onto the set of modes and the corresponding vector projection is obtained as
| (11) |
The Gramian matrix G of the set of vectors , being Pq the dimension of subspaces, is calculated as the matrix of inner products with elements
| (12) |
By diagonalizing G
| (13) |
we obtain eigenvalues that vary in the [0:1] range. Therefore, the similarity of the two subspaces can be quantify as
| (14) |
The smaller the value of , the stronger the effect of the ligand on tetramer vibrations associated to the conformational changes required in the TTR tetramer-ligand complex dissociation. As a consequence, TTR tetramer-ligand complex dissociation slows down and the complex gain stability.
depends on subspace dimensionalities. Therefore, for each TTR tetramer-ligand complex in the dataset we normalize the values of as:
| (15) |
where and σref are the average and standard deviation of a reference distribution built by performing the comparison of Sligand with subspaces Sref obtained by randomly selection of Pq ligand-free normal modes within windows of ± 10 modes centered on each original qk of subspace S.
Results and discussion
The structure of the tetrameric TTR-T4 complex is depicted in Fig 1. T4 hormones are localized at the hydrophobic cavities of each dimer-dimer interface. Dimers are composed of two monomers (A-A’ and B-B’ respectively). Each monomer is formed by two four antiparallel β-strands and a short α-helix. The monomer-monomer interfaces are stabilized by hydrogen bond networks between the two β-strands H and F generating eight-stranded β-sandwich tertiary structures. The list of structures of TTR tetramer-ligand complexes used in this work is indicated in Table 1, including ligand free TTR tetramer. We applied several filters to TTR structures available in the PDB database in order to obtain a well curated dataset: (i) crystal structures with resolution < 1.7 Å, (ii) structures without missing residues between reside 11 and 124, (iii) structures without mutations, (iv) crystal structures with optimal Spearman rank correlation coefficient between experimental and theoretical B-factors > 0.6 Å, (v) structures with ligands that have been demonstrated to inhibit fiber formation, (vi) structures with ligands whose relative affinities have been proved experimentally using either competition binding assays of isothermal titriation calorimetry (ITC).
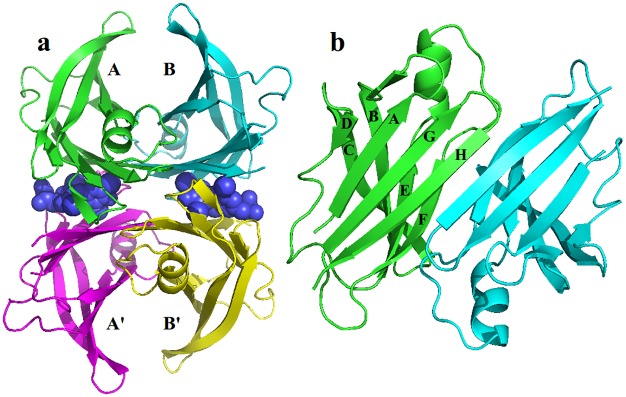
Fig 1. Structure of TTR-T4 complex.
T4 molecules are displayed in the binding cavities as spheres and TTR monomers as ribbons. (a) The two monomers present in the asymmetric unit of the crystal structures used are labelled as A and B, symmetry related monomers as A’ and B’. (b) View of the A-A’ monomer-monomer interface.
Table 1. TTR PDB coordinates used in this paper.
Effect of mutations on dynamics
The intramolecular vibrational dynamics of a protein is intrinsically related to its structural stability. Therefore, changes in the TTR tetramer equilibrium dynamics can be associated to changes in its relative stability. It is expected that mutations with large effects on TTR tetramer dynamics will destabilize it, increasing the propensity of TTR fibril formation and amyloid diseases.
Single point mutations have been simulated on each residue of the ligand-free TTR tetramer structure in order to identify key position residues sustaining TTR tetramer dynamics stability. The value of Zi (see Section Mode perturbations, comparison and key residues) is a measure of the change in the TTR tetramer equilibrium dynamics introduced by mutations on the ith residue. The lower the value of Zi, the larger the effect of mutations of residue ith on the tetramer stability.
Firstly, in Table 2 we analyse key position residues selected as those ranked with the lowest 10% values of Zi. Most of them are localized in the loop segment β-strands E, EF-loop, and β-strands F. For the sake of robustness, it is important to mention that results shown in Table 2 were mostly reproduced using two ligand-free TTR tetramer structures availables (PDB_id: 1f4l and 2qgb). The RMSD between these structures is 0.47Å. Despite changes in the ordering, similar qualitative results have been obtained for both structures. More than 75% of residues ranked with the lowest 20% values of Zi are reproduced on both structures and ~90% match by +/- one residue. The complete list of the lowest 20% values of Zi obtained using both ligand-free structures is provided in S1 and S2 Tables. Fig 2 shows their localization within the TTR structure. Almost half of them are located at the monomer-monomer interface, revealing as one of the most vulnerable regions to mutations that lead to significant changes in the TTR-tetramer equilibrium dynamics. Interestingly, in Table 2 we also show that among top 10% positions with low Zi, most of them correlates with mutations previously reported to induce TTR amyloidogenesis. Particularly, V30, with four different mutations in this position and with Zi = −1.17, has been identified as one of the most prominent familial amyloid polyneuropathy mutation[14],[16],[52].
Table 2. Key position residues.
| Residue | Zi | Secondary Structure Element | Type | Intra- monomer IACs |
Inter-monomer IACs |
Mutation | <ΔΔG> |
|---|---|---|---|---|---|---|---|
| His 88 | -1,87 | EF-loop | * | 7 | 1 | His → Arg[56] | -1,03 |
| Tyr 69 | -1,58 | β-strand E | 13 | Tyr → His; Tyr → Ile | 18.03 | ||
| Thr 75 | -1,52 | α-helix | 9 | 8,18 | |||
| Phe 95 | -1,46 | β-strand F | * | 9 | 2 | 20,56 | |
| Phe 87 | -1,44 | EF-loop | * | 4 | 6 | Phe → Met | 18,53 |
| Ile 73 | -1,38 | β-strand E | 11 | Ile → Val | 17,49 | ||
| Glu 89 | -1,33 | EF-loop | * | 5 | 3 | Glu → Gln; Glu → Lys | 3,55 |
| Val 71 | -1,32 | β-strand E | 9 | Val → Ala | 16,81 | ||
| Val 93 | -1,22 | β-strand F | * | 6 | 2 | Val → Met | 9,74 |
| Val 30 | -1,17 | β-strand B | 7 | Val → Leu; Val → Met Val → Ala; Val → Gly |
23,46 | ||
| Asp 74 | -1,15 | β-strand E | 7 | Asp → His (Non Amyloidogenic) |
7,63 |
Type:* monomer-monomer interface[47]
IAC: Inter Atomic Contact.
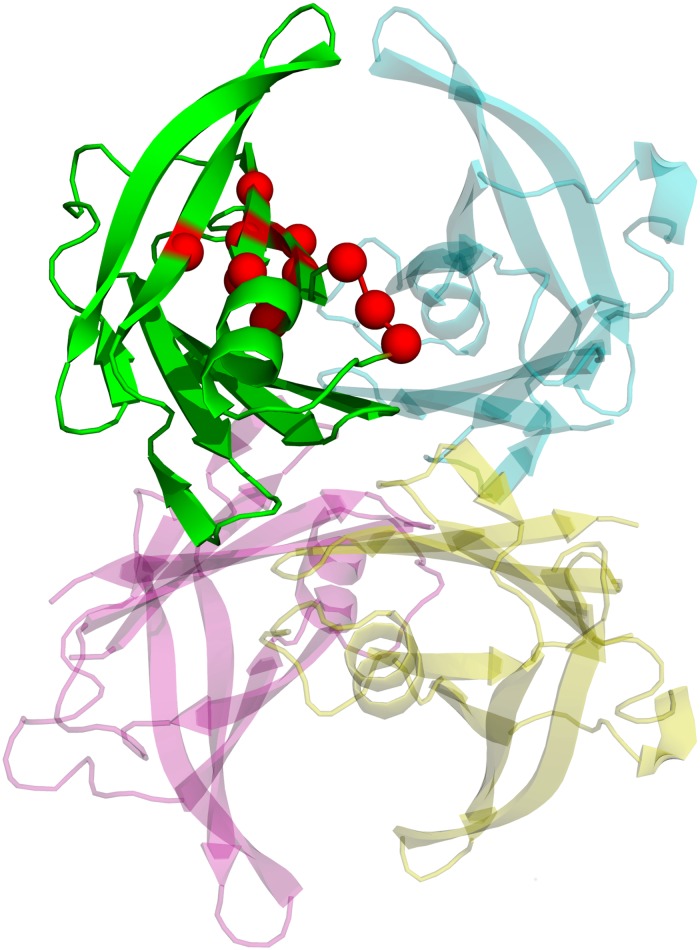
Fig 2. Key position residues shown as spheres.
Their localizations are displayed in only one monomer.
Key position residues have been analyzed using using FoldX algorithm[53],[54] that predicts differences in the free energy changes ΔΔG = ΔGmutant-ΔGwild-type between the unfolding free energy of a mutant (= ΔGmutant) and wild type (ΔGwild-type) protein. The average values ΔΔG over all possible mutation performed on each key position residue are listed in Table 2. In good agreement with our predictions, we found that most key position residues are strongly destabilizing (ΔΔG>2kcal/mol)[55].
Table 3 shows values of Zi for residues previously identified as belonging to the thyroxine hormone binding site (TBS), monomer-monomer, and dimer-dimer interfaces [47], [48], [57], [58]. All of the substrate-binding cavity residues present negative values of Zi, indicating that mutations on these residues destabilize the tetramer more than the average. T4 binding sites are at the weakly dimer-dimer interfaces. We observe negative values of Zi in dimer-dimer interface residues within the large L110-V122 fragment (β-strands G, GH-loop, and β-strands H), and L17-D18 (β-strands A), while positive values are observed for A19-S23 segment (AB-loop) and S85 (EF-loop). Finally, all monomer-monomer interface residues also present negative values of Zi. That is, mutations on any of the residues of the large F87-V122 segment introduce larger perturbations on the tetramer structure than the average. Fig 3 shows the localization of all these residues within the TTR structure.
Table 3. Zi values of residues of substrate-binding cavity, monomer-monomer, and dimer-dimer interfaces.
| Residue | Zi | Secondary Structure Element | type | Intra-monomer IACs | Inter-monomer IACs | Mutation |
|---|---|---|---|---|---|---|
| Met 13 | -0,16 | β-strand A | † | 9 | Met → Ile (Non Amyloidogenic) |
|
| Lys 15 | -0,45 | β-strand A | † | 8 | ||
| Leu 17 | -0,21 | β-strand A | #,† | 8 | 2 | |
| Asp 18 | -0,30 | β-strand A | # | 8 | 1 | Asp → Asn; Asp → Gly; Asp → Glu |
| Ala 19 | 0,15 | AB-loop | # | 7 | 3 | Ala → Asp |
| Val 20 | 0,18 | AB-loop | # | 6 | 2 | Val → Ile |
| Arg 21 | 0,53 | AB-loop | # | 6 | 1 | |
| Gly 22 | 0,19 | AB-loop | # | 4 | 1 | |
| Ser 23 | 0,14 | AB-loop | # | 5 | 1 | Ser → Asn |
| Ser 85 | 0,31 | EF-loop | # | 4 | 1 | |
| Phe 87 | -1,44 | EF-loop | * | 4 | 6 | |
| His 88 | -1,87 | EF-loop | * | 7 | 1 | |
| Glu 89 | -1,33 | EF-loop | * | 5 | 3 | Glu → Gln; Glu → Lys |
| His 90 | -0,92 | EF-loop | * | 5 | 1 | His → Asp; His → Asn (Non Amyloidogenic) |
| Glu 92 | -0,53 | β-strand F | * | 6 | 4 | Gln → Lys |
| Val 93 | -1,22 | β-strand F | * | 6 | 2 | Val → Met |
| Val 94 | -1,08 | β-strand F | * | 6 | 4 | Val → Ala |
| Phe 95 | -1,46 | β-strand F | * | 9 | 2 | |
| Thr 96 | -0,93 | β-strand F | * | 4 | 3 | |
| Tyr 105 | -1,04 | β-strand G | * | 14 | 1 | |
| Thr 106 | -0,37 | β-strand G | † | 4 | ||
| Ile 107 | -1,01 | β-strand G | * | 9 | 1 | Ile → Val; Ile → Phe Ile → Met |
| Ala 108 | -0,43 | β-strand G | † | 4 | ||
| Ala 109 | -0,61 | β-strand G | † | 6 | Ala → Ser (Non Amyloidogenic) Ala → Thr (Non Amyloidogenic) |
|
| Leu 110 | -0,06 | β-strand G | #,† | 6 | 2 | |
| Ser 112 | -0,21 | β-strand G | # | 7 | 1 | Ser → Ile |
| Pro 113 | -0,62 | GH-loop | # | 9 | 1 | |
| Tyr 114 | -1,07 | GH-loop | *,# | 6 | 6 | Tyr → His; Tyr → Cys |
| Ser 115 | -0,18 | β-strand H | *,# | 5 | 4 | |
| Tyr 116 | -0,70 | β-strand H | * | 6 | 6 | Tyr → Ser |
| Ser 117 | 0,44 | β-strand H | #,† | 3 | 2 | |
| Thr 118 | -0,33 | β-strand H | * | 5 | 1 | |
| Thr 119 | -0,58 | β-strand H | *, #, † | 3 | 5 | Thr → Met (Non Amyloidogenic) |
| Ala 120 | -0,91 | β-strand H | *, # | 4 | 2 | Ala → Ser |
| Val 121 | -0,54 | β-strand H | # | 4 | 2 | |
| Val 122 | -0,40 | Loop | *, # | 5 | 3 | Val → Ile; Val → del; Val → Ala |
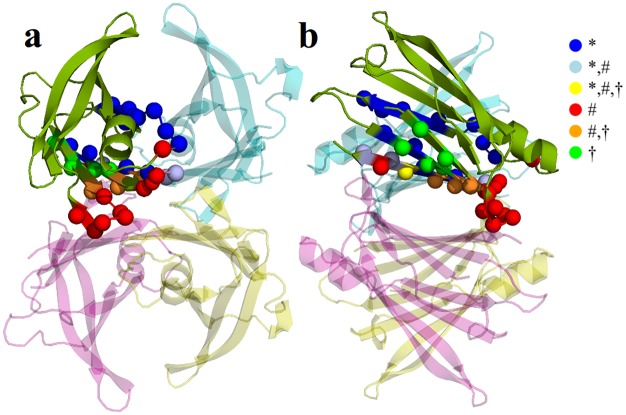
Fig 3. Residues of substrate-binding cavity, monomer-monomer, and dimer-dimer interfaces shown as spheres.
Their localizations are displayed only in one monomer. (* monomer-monomer interface, # dimer-dimer interface, † substrate-binding cavity).
A. Effect of ligand-binding on dynamics
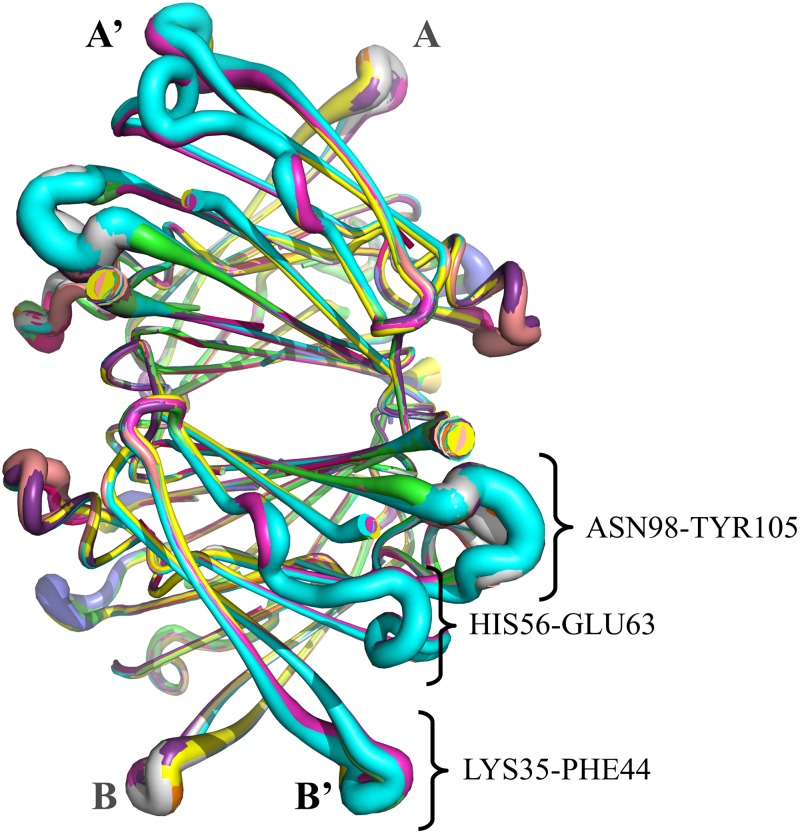
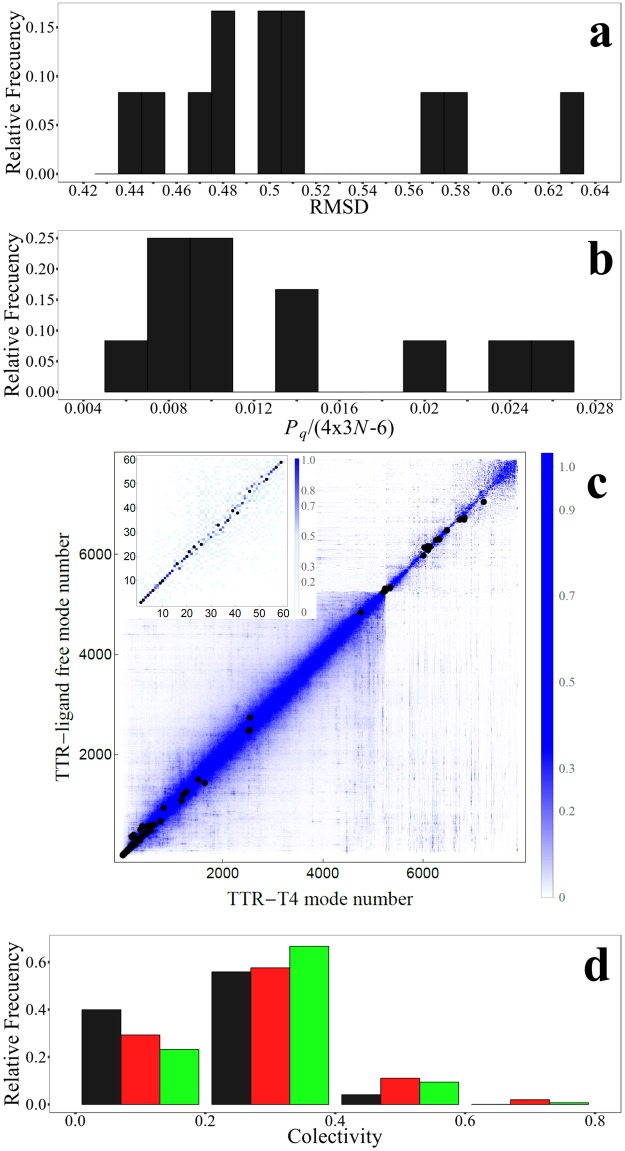
We have performed a comparative analysis of effects of several drugs and compounds (see Table 1) on TTR tetramer ligand-free vibrations involved in the conformational changes associated to TTR tetramer-ligand complex formation/dissociation. Fig 4 shows the superimposed structures listed in Table 1 indicating those regions presenting larger flexibilities. None of the key position residues listed in Table 2 belong to one of these regions. The structural distortions introduced by ligand-binding are analyzed in terms of the RMSD (root mean square difference) between TTR tetramer ligand-free structure and each TTR tetramer-ligand complex structure. Fig 5A displays the corresponding distribution of RMSD values. The low values of RMSD indicate that TTR tetramer ligand-free structure is not largely affected by ligand-bindings. The largest value of 0.63 corresponds to the RMSD with TTR tetramer-T4 complex structure. A detailed information of RMSD values between all structures of our dataset is shown in S1 Fig. TTR tetramer-T4 (pdb: 2ROX) complex structure is the structure that presents the main structural differences with the rest of the structures, mainly due to structural differences between T4 and the other compounds. Despite that, RMSD values between TTR tetramer complexes seem not to follow a clear relation with chemical similarities between ligands, mostly belonging to the large family of polyphenols.
Fig 4. Superimposed putty-style cartoon representations of the 13 structures TTR tetramer-ligand complexes listed in Table 1.
The variable-width corresponds to values of experimental B-factors. The segments of sequence with larger flexibilities are also indicated.
Fig 5.
(a) Distribution of RMSD between TTR tetramer ligand-free structure and each TTR tetramer-ligand complex structure; (b) Distribution of the fraction of normal modes involved in the ligand-induced conformational transition calculated as Pq/4x3N; (c) overlap matrix between TTR tetramer ligand-free and TTR-T4 complex normal modes highlighting modes belonging to the ligand-binding subspace (black dots), the inset displays the overlap matrix for modes within the low-frequency range; (d) Distribution of degree of collectivity, κk, for each normal mode that participates in the conformational change (red), for each normal mode that participate significantly in the flexibility pattern (green), and for all other modes (black).
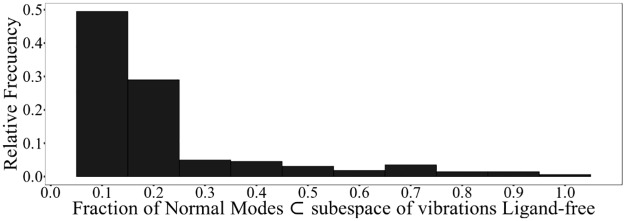
Furthermore, Fig 5B shows the distribution of the fraction of TTR tetramer ligand-free normal modes involved in ligand-induced conformational transitions calculated as values Pq/(4x3N−6) obtained over all TTR tetramer-ligand complexes in our dataset. The average value of Pq = 104 ± 54, and Pq/(4x3N−6) = 0.01± 0.007 indicates that a significantly small fraction of the vibrational space participates in the process of ligand-binding. In order to analyze the composition of subspace of normal modes associated to ligand-binding, Fig 5C depicts the overlap matrix between TTR tetramer ligand-free and TTR-T4 complex normal modes, highlighting modes belonging to the ligand-binding subspace. As it can be seen, normal modes associated to ligand-binding are not uniformly distributed throughout the whole range of modes. Mostly of these modes are localized at the low-frequency range and only a minor contribution corresponds to high frequency modes. This is in good agreement with previous studies that associate low-frequency normal modes with ligand-binding conformational changes[59],[60].
Normal modes can be analyzed in terms of their collectivities, defined as[59]
| (16) |
being , and (j = x, y, z) the components of the ith Cα residue in the k normal mode. κk = N−1 corresponds to normal modes equally distributed throughout all the residues of the protein, and κk = 1 represents normal modes localized on a single residue. Fig 5D points out that modes involved in ligand-induced conformational transition are composed of vibrations within the entire range of frequencies, showing a distribution of collectivities slightly higher than other modes. Besides, normal modes that participate of ligand-binding can be compared with essential normal modes involved in the TTR-tetramer flexibility pattern. In order to do that, vector Blf with elements corresponding to the B-factors associated to each ith residue is expanded on the basis of {qk}k = 1,4x3N−6 modes
| (17) |
with
| (18) |
The mode participation number PB associated to this expansion can be defined as
| (19) |
with an equivalent interpretation as Pq described in Section Subspace of modes associated to ligand-binding: definition and comparison. The first PB modes ordered by index fk in decreasing values of (bk)2 represent the minimum set of modes required to achieve a good representation of the flexibility pattern. The distribution of collectivities of modes related to the flexibility pattern of TTR-tetramer is also displayed in Fig 5D. We can see that subspaces of normal modes associated to ligand-binding do not involve the most collective modes that significantly contribute to protein flexibility.
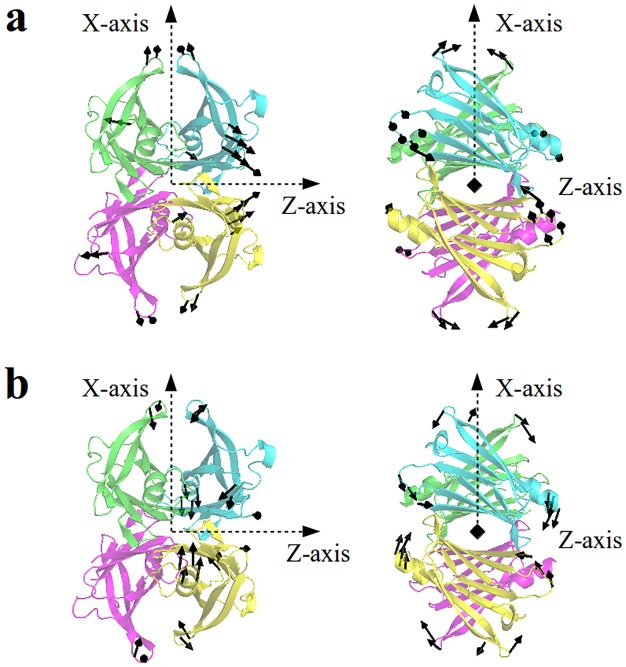
In order to identify common dynamics aspects, we compare ligand-binding subspaces using the subspace for T4-binding as template. Fig 6 shows the distribution of fraction of modes shared between each ligand-binding subspace and T4-binding subspace. While most normal modes change among ligands, two normal modes (normal modes # 22 and 25) are shared by all ligand-binding. Fig 7 depicts these modes. Both modes present 2-fold rotational symmetry with respect to Z axis. They imply a combination of shear (mode #22) and hinge (mode # 25) motions between dimers on only one of the T4-binding cavities. Both modes imply relative motions mainly localized on EF-loop, FG-loop and BC-loop affecting only one of both dimer-dimer interfaces. Previous studies[51] show that TTR ligands present different abilities to saturate the two T4 binding sites, leading to the presence of a main binding site with clear ligand affinity and a second minor site with a much lower ligand affinity. Our results complement these previous structural evidence for asymmetric TTR-ligand binding related to its negative binding cooperativity[61], [62].
Fig 6. Distribution of fraction of normal modes shared between each ligand-binding subspace and T4-binding subspace.
Fig 7. Normal modes 22(a) and 25(b) involved in conformational changes of all ligand-binding of our dataset.
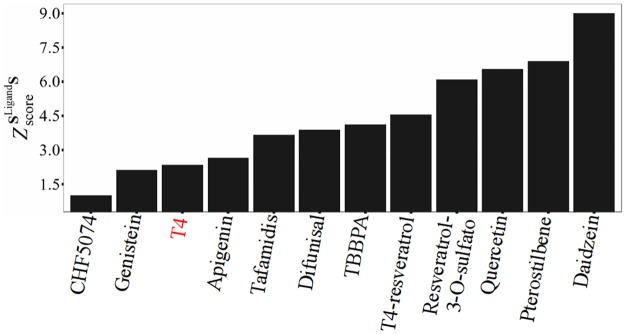
In Fig 8 we display the comparison of our set of ligands and compounds according to their effect on vibrations associated to ligand binding. In order to relate binding affinities to changes introduced on vibrations, we analyze our relative values of in terms of experimental parameters and measurements associated to TTR-ligand binding affinities and the stabilization of its native state. Ligands presenting low values of (see Section Subspace of modes associated to ligand-binding: definition and comparison) introduce significant changes on tetramer vibrations associated to the conformational changes required in the TTR tetramer-ligand complex dissociation. As a consequence, TTR tetramer-ligand complex gain stability and fibrillogenesis is inhibited.
Fig 8. Values of for different ligands.
Performing competition binding assays, it has been shown that the polyphenols pterostilbene and quercetin have different preferential binding sites in TTR than the one of T4[51]. Besides, experiments reveal that, at equal concentrations, quercetin is able to displace pterostilbene[51], in agreement with the lower value of for quercetin with respect to pterostilbene.
While neither pterostilbene nor quercetin is able to displace TTR-bond T4, the polyphenol apigenin displaces it but only at very high concentrations[51]. These results indicate that apigenin presents lower affinity than T4. This is in agreement with our higher value of for apigenin respect to T4. Even more, tafamidis TTR-affinity has shown to be higher than polyphenols and T4, something that can only confirm in the case of pterostilbene and quercetin. Nevertheless, apigenin, T4, and tafamidis are close in order of .
According to competition experiments using resveratrol and radiolabeled T4 as probes, the resveratrol is not able to displace the bound T4. Besides, fluorescence experiments shows that resveratrol metabolites, like resveratrol-3-O-sulfate, have lower affinities for TTR than that of resveratrol[51]. In good agreement with that, we obtain (T4)< (resveratrol)< (resveratrol-3-O-sulfate). Furthermore, fluorometric competitive binding assays, using resveratrol as fluorescent probe, indicates that CHF5074 presents high binding affinity for TTR, in agreement with its low value.
TTR-ligand dissociation constants (Kd) determined by isothermal titriation calorimetry (ITC)[63] indicates that Kd(diflunisal) = 580 nM is higher than Kd(apigenin) = 250 nM, in agreement with values of . Furthermore, ITC experiments also report values of Kd(TBBPA) = 20 nM and Kd(Tafamidis) = 3 nM[50], leading to a final order of affinity to TTR as tafamidis>TBBPA>diflunisal. Values of do not agree with these results, despite both compounds are close in order of .
Relative capacities of genistein, apigenin and daidzen to displace TTR-bound radiolabeled T4 indicates the order of binding affinities for TTR as genistein>apigenin>daidzen, in good agreement with the order given in Fig 8. Among them, only genistein and apigenin are able to displace TTR-bound resveratrol while daidzen results a weak ligand exhibiting rather low binding affinity. That is, TTR-binding affinities varies as genistein> apigenin> resveratrol> daidzen in agreement with the order .
At this point it is interesting to mention that values of do not correlate neither with values of RMSD between ligand-free and ligand-bound structures nor with values of ΔIACs(Inter Atomic Contacts) changes. We obtain Spearman correlation coefficients of -0.22 between and RMSD and -0.07 between and ΔIACs. Therefore, do not depend on these structural features. Ligands that introduce larger structural distortions or changes of inter-residue interactions do not correspond to those that modify the most the TTR-tetramer equilibrium dynamics.
Conclusions
We have analyzed the effects of mutations and ligand-binding pattern on TTR-tetramer equilibrium dynamics. We have explored key positions residues that most affect TTR-tetramer vibrations. Most of them are localized at β-strands E, EF-loop, and β-strands F, with amyloidogenic mutations previously reported. The monomer-monomer interface is pointed out as one of region most vulnerable to mutations that lead to significant changes in the TTR-tetramer equilibrium dynamics and, therefore, induces TTR amyloidogenesis. Besides, we have found that mutations on residues localized at the dimer-dimer interface and/or at the T4 hormone binding site destabilize the tetramer more than the average.
At this point it is important to mention that, in the present work, we explore the effects of mutations on equilibrium dynamics of TTR tetramer. It is expected that this particular aspect, among others structural features, is associated to the stability of the quaternary structure. Considering the diversity of reasons that lead a mutation to be pro-amyloidogenic, a correlation between reported amyloidogenic TTR mutations and Zi, that only quantifies perturbations of TTR tetramer equilibrium dynamics, is not expected. Nevertheless, values of Zi not only represent a complementary contribution to explore new potential mutations but also enlighten about effects of some of the previously reported mutations.
Several drugs and compounds have been compared according to their effect on vibrations associated to the ligand binding. We found that a significantly small fraction of the vibrational space participates of the ligand-induced conformational transitions. We discussed these particular ligand effects in terms of experimental parameters and measurements associated to TTR-ligand binding affinities and stabilization of its native state. In general, the effect of ligands on equilibrium dynamics of ligand-free TTR-tetramer is in good agreement with previous findings obtained with competition binding assays.
Two normal modes have been found to participate in all ligand-bindings of our dataset. Both of them imply motions between dimers on only one of the T4-binding cavities, evidencing the asymmetric TTR-ligand binding related to its negative binding cooperativity.
As resume, in the present paper we focus on the analysis of effects of mutations and ligand-binding on TTR-tetramer equilibrium dynamics. We have found that our analysis of dynamics aspects leads to complementary information that can help for future developments of new drugs against amyloid diseases.
Supporting information
(TIF)
(PDF)
(PDF)
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by Universidad Nacional de Quilmes, grant entitled "Simulación de procesos moleculares de relevancia fisicoquímica y biológica."
References
- 1.Westermark P.; Sletten K.; Johansson B.; Cornwell GG 3rd. Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc Natl Acad Sci USA. 1990;87: 2843–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saraiva MJ., Magalhaes J., Ferreira N., Almeida MR. Transthyretin deposition in familial amyloidotic polyneuropathy. Curr Med Chem. 2012;19: 2304−2311. [DOI] [PubMed] [Google Scholar]
- 3.Ruberg FL., Berk JL. Transthyretin (TTR) cardiac amyloidosis. Circulation. 2012;126: 1286−1300. doi: 10.1161/CIRCULATIONAHA.111.078915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quintas A., Vaz DC., Cardoso I., Saraiva MJ, Brito RM. Tetramer dissociation and monomer partial unfolding precedes protofibril formation in amyloidogenic transthyretin variants. J Biol Chem. 2001;276: 27207−27213. doi: 10.1074/jbc.M101024200 [DOI] [PubMed] [Google Scholar]
- 5.Cardoso I., Goldsbury CS., Muller SA., Olivieri V., Wirtz S., et al. Transthyretin fibrillogenesis entails the assembly of monomers: a molecular model for in vitro assembled transthyretin amyloid-like fibrils. J Mol Biol. 2002;317: 683−695. doi: 10.1006/jmbi.2002.5441 [DOI] [PubMed] [Google Scholar]
- 6.Redondo C., Damas AM., Saraiva MJ. Structural evidence for native state stabilization of a conformationally labile amyloidogenic transthyretin variant by fibrillogenesis inhibitors. Biochem J. 2000;348: 167−172. [DOI] [PubMed] [Google Scholar]
- 7.Johnson SM., Wiseman RL., Sekijima Y., Green NS., Adamski-Werner SL., et al. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: A focus on the transthyretin amyloidoses. Acc Chem Res. 2005;38: 911–921. doi: 10.1021/ar020073i [DOI] [PubMed] [Google Scholar]
- 8.Choi S., Reixach N., Connelly S., Johnson SM., Wilson IA., Kelly JW. A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity. J Am Chem Soc. 2010;132: 1359–1370. doi: 10.1021/ja908562q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connelly S., Choi S., Johnson SM., Kelly JW., Wilson IA. Structure-based design of kinetic stabilizers that ameliorate the transthyretin amyloidoses. Curr Opin Struct Biol. 2010;20: 54–62. doi: 10.1016/j.sbi.2009.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolstoe SE., Wood SP. Drug targets for amyloidosis. Biochem Soc Trans. 2010;38: 466–470. doi: 10.1042/BST0380466 [DOI] [PubMed] [Google Scholar]
- 11.Bulawa CE, Connelly S, DeVit M, Wang L, Weigel C, Fleming J a., et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci. 2012;109: 9629–9634. doi: 10.1073/pnas.1121005109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palaninathan SK. Nearly 200 X-Ray Crystal Structures of Transthyretin: What Do They Tell Us About This Protein and the Design of Drugs for TTR Amyloidoses? Curr Med Chem. 2012;19: 2324–2342. [DOI] [PubMed] [Google Scholar]
- 13.Zanotti G, Cendron L, Folli C, Florio P, Imbimbo B Pietro, Berni R. Structural evidence for native state stabilization of a conformationally labile amyloidogenic transthyretin variant by fibrillogenesis inhibitors. FEBS Lett. Federation of European Biochemical Societies; 2013;587: 2325–2331. doi: 10.1016/j.febslet.2013.06.016 [DOI] [PubMed] [Google Scholar]
- 14.Miroy GJ, Lai Z, Lashuel H a, Peterson S a, Strang C, Kelly JW. Inhibiting transthyretin amyloid fibril formation via protein stabilization. Proc Natl Acad Sci U S A. 1996;93: 15051–15056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller SR, Sekijima Y, Kelly JW. Native state stabilization by NSAIDs inhibits transthyretin amyloidogenesis from the most common familial disease variants. Lab Invest. 2004;84: 545–552. doi: 10.1038/labinvest.3700059 [DOI] [PubMed] [Google Scholar]
- 16.Johnson SM, Wiseman RL, Sekijima Y, Green NS, Adamski-Werner SL, Kelly JW. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: A focus on the transthyretin amyloidoses. Acc Chem Res. 2005;38: 911–921. doi: 10.1021/ar020073i [DOI] [PubMed] [Google Scholar]
- 17.Hurshman Babbes AR., Powers ET., Kelly JW. Quantification of the Thermodynamically Linked Quaternary and Tertiary Structural Stabilities of Transthyretin and Its Disease-Associated Variants: The Relationship between Stability and Amyloidosis. Biochemistry. 2008; 6969–6984. doi: 10.1021/bi800636q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cendron L., Trovato A., Seno F., Folli C., Alfieri B., Zanotti G., et al. Amyloidogenic Potential of Transthyretin Variants INSIGHTS FROM STRUCTURAL AND COMPUTATIONAL ANALYSES. J Biol Chem. 2009;284: 25832–25841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klabunde T., Petrassi HM., Oza VB., Raman P., Kelly JW., Sacchettini JC. Rational design of potent human transthyretin amyloid disease inhibitors. Nat Struct Mol Biol. 2000;7: 312–321. [DOI] [PubMed] [Google Scholar]
- 20.Green NS., Foss TR., Kelly JW. Genistein, a natural product from soy, is a potent inhibitor of transthyretin amyloidosis. Proc Natl Acad Sci USA. 2005;102: 14545–14550. doi: 10.1073/pnas.0501609102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trivella DBB., Bleicher L., de C Palmieri L., Wiggers HJ., Montanari CA., Kelly JW., et al. Conformational differences between the wild type and V30M mutant transthyretin modulate its binding to genistein: implications to tetramer stability and ligand-binding. J Struct Biol. 2010;170: 522–531. doi: 10.1016/j.jsb.2010.03.002 [DOI] [PubMed] [Google Scholar]
- 22.Saldaño T, Monzon A, Parisi G, Fernandez-Alberti S. Evolutionary Conserved Positions Define Protein Conformational Diversity. PLOS Comput Biol. 2016;12: e1004775 doi: 10.1371/journal.pcbi.1004775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tirion MM. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys Rev Lett. 1996;77: 1905–1908. doi: 10.1103/PhysRevLett.77.1905 [DOI] [PubMed] [Google Scholar]
- 24.Bahar I, Erman B, Jernigan RL, Atilgan AR, G CD. Collective motions in HIV-1 reverse transcriptase: examination of flexibility and enzyme function. J Mol Biol. 1999;285: 1023–1037. doi: 10.1006/jmbi.1998.2371 [DOI] [PubMed] [Google Scholar]
- 25.Bahar I, Jernigan RL. Cooperative fluctuations and subunit communication in tryptophan synthase. Biochemistry. 1999;38: 3478–3490. doi: 10.1021/bi982697v [DOI] [PubMed] [Google Scholar]
- 26.Bahar I, Rader a J. Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol. 2005;15: 586–92. doi: 10.1016/j.sbi.2005.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bahar I, Atilgan a R, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2: 173–81. Available: http://www.ncbi.nlm.nih.gov/pubmed/9218955 doi: 10.1016/S1359-0278(97)00024-2 [DOI] [PubMed] [Google Scholar]
- 28.Skjærven L, Yao X-Q, Scarabelli G, Grant B. Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinformatics. 2014;15: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skjærven L; Jariwala S; Yao X-Q; Grant B. Online interactive analysis of protein structure ensembles with Bio3D-web. Bioinformatics. 2016;32: 3510–3512. doi: 10.1093/bioinformatics/btw482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinsen K. Analysis of domain motions by approximate normal mode calculations. Proteins Struct Funct Genet. 1998;33: 417–429. [DOI] [PubMed] [Google Scholar]
- 31.Atilgan a R, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80: 505–15. doi: 10.1016/S0006-3495(01)76033-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong JI, Jang Y, Kim MK. A connection rule for alpha-carbon coarse-grained elastic network models using chemical bond information. J Mol Graph Model. 2006;24: 296–306. doi: 10.1016/j.jmgm.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 33.Martin AJM, Vidotto M, Boscariol F, Walsh I, Tosatto SCE. RING : Networking interacting residues, evolutionary conservation and energetics in protein structures. Bioinformatics. 2010;27: 2003–2005. [DOI] [PubMed] [Google Scholar]
- 34.Piovesan D., Minervini G., Tosatto SC. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016;44(W1): W367–74. doi: 10.1093/nar/gkw315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng W, Brooks BR, Doniach S, Thirumalai D. Network of dynamically important residues in the open/closed transition in polymerases is strongly conserved. Structure. 2005;13: 565–577. doi: 10.1016/j.str.2005.01.017 [DOI] [PubMed] [Google Scholar]
- 36.Zheng W, Brooks BR, Thirumalai D. Low-frequency normal modes that describe allosteric transitions in biological nanomachines are robust to sequence variations. Proc Natl Acad Sci U S A. 2006;103: 7664–9. doi: 10.1073/pnas.0510426103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng W, Thirumalai D. Coupling between normal modes drives protein conformational dynamics: illustrations using allosteric transitions in myosin II. Biophys J. Biophysical Society; 2009;96: 2128–37. doi: 10.1016/j.bpj.2008.12.3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng W, Tekpinar M. Large-scale evaluation of dynamically important residues in proteins predicted by the perturbation analysis of a coarse-grained elastic model. BMC Struct Biol. 2009;9: 45 doi: 10.1186/1472-6807-9-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell RJ, Dean P, Hibbins-Butter DC. Localization of normal modes. 1970;3: 2111–2118. [Google Scholar]
- 40.Taraskin SN, Elliott SR. Anharmonicity and localization of atomic vibrations in vitreous silica. Phys Rev B. 1999;59: 8572–8585. [Google Scholar]
- 41.Carpaneto G, Martello S, Toth P. Algorithms and codes for the assignment problem. Ann Oper Res. 1988; [Google Scholar]
- 42.Kalstein A, Fernández-Alberti S, Bastida A, Soler MA, Farag MH, Zúñiga J, et al. Vibrational dynamics of polyatomic molecules in solution: Assignment, time evolution and mixing of instantaneous normal modes. Theor Chem Acc. 2011;128: 769–782. doi: 10.1007/s00214-010-0832-5 [Google Scholar]
- 43.Krzanowski WJ. Between-Groups Comparison of Principal Components. J Am Stat Assoc. 1979;74: 703–707. [Google Scholar]
- 44.Blows MW, Chenoweth SF, Hine E. Orientation of the genetic variance-covariance matrix and the fitness surface for multiple male sexually selected traits. Am Nat. 2004;163: 329–40. doi: 10.1086/381941 [DOI] [PubMed] [Google Scholar]
- 45.Flury B. Common principal components and related multivariate models. New York: Wiley; 1988. [Google Scholar]
- 46.Cohn RD. Comparisons of multivariate relational structures in serially correlated data. J Biol Environ Stat. 1999;4: 238–257. [Google Scholar]
- 47.Hörnberg a, Eneqvist T, Olofsson a, Lundgren E, Sauer-Eriksson a E. A comparative analysis of 23 structures of the amyloidogenic protein transthyretin. J Mol Biol. 2000;302: 649–669. doi: 10.1006/jmbi.2000.4078 [DOI] [PubMed] [Google Scholar]
- 48.Wojtczak A, Cody V, Luft JR, Pangborn W. Structures of human transthyretin complexed with thyroxine at 2.0 A resolution and 3’,5’-dinitro-N-acetyl-L-thyronine at 2.2 A resolution. Acta Crystallogr D Biol Crystallogr. 1996;52: 758–765. doi: 10.1107/S0907444996003046 [DOI] [PubMed] [Google Scholar]
- 49.Florio P, Folli C, Cianci M, Del Rio D, Zanotti G, Berni R. Transthyretin binding heterogeneity and antiamyloidogenic activity of natural polyphenols and their metabolites. J Biol Chem. 2015;290: 29769–29780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iakovleva I, Begum A, Brannstrom K, Wijsekera A, Nilsson L, Zhang J, et al. Tetrabromobisphenol a is an efficient stabilizer of the transthyretin tetramer. PLoS One. 2016;11: 1–16. doi: 10.1371/journal.pone.0153529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cianci M, Folli C, Zonta F, Florio P, Berni R, Zanotti G. Structural evidence for asymmetric ligand binding to transthyretin. Acta Crystallogr Sect D Biol Crystallogr. International Union of Crystallography; 2015;71: 1582–1592. doi: 10.1107/S1399004715010585 [DOI] [PubMed] [Google Scholar]
- 52.Sipe JD. Amyloidosis. Crit Rev Clin Lab Sci. 1994;31: 325–354. doi: 10.3109/10408369409084679 [DOI] [PubMed] [Google Scholar]
- 53.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: An online force field. Nucleic Acids Res. 2005;33: 382–388. doi: 10.1093/nar/gki387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J Mol Biol. 2002;320: 369–387. doi: 10.1016/S0022-2836(02)00442-4 [DOI] [PubMed] [Google Scholar]
- 55.Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The Stability Effects of Protein Mutations Appear to be Universally Distributed. J Mol Biol. 2007;369: 1318–1332. doi: 10.1016/j.jmb.2007.03.069 [DOI] [PubMed] [Google Scholar]
- 56.Holmgren G, Hellman U, Anan I, Lundgren H, Jonasson J, Stafberg C, et al. Cardiomyopathy in Swedish patients with the Gly53Glu and His88Arg transthyretin variants. Amyloid. 2005;12: 184–188. doi: 10.1080/13506120500223126 [DOI] [PubMed] [Google Scholar]
- 57.Adamski-Werner SL, Palaninathan SK, Sacchettini JC, Kelly JW. Diflunisal Analogues Stabilize the Native State of Transthyretin. Potent Inhibition of Amyloidogenesis. J Med Chem. 2004;47: 355–374. doi: 10.1021/jm030347n [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Begum A, Brännström K, Grundström C, Iakovleva I, Olofsson A, et al. Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin. Environ Sci Technol. 2016;50: 11984–11993. doi: 10.1021/acs.est.6b02771 [DOI] [PubMed] [Google Scholar]
- 59.Tama F, Sanejouand YH. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14: 1–6. Available: http://www.ncbi.nlm.nih.gov/pubmed/11287673 [DOI] [PubMed] [Google Scholar]
- 60.Petrone P, Pande VS. Can Conformational Change Be Described by Only a Few Normal Modes ? Biophys J. Elsevier; 2006;90: 1583–1593. doi: 10.1529/biophysj.105.070045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fergurson K. Active and inactive conformations of the epidermal growth factor receptor. Biochem Soc Trans. 2004;32: 742–745. doi: 10.1042/BST0320742 [DOI] [PubMed] [Google Scholar]
- 62.Cheng S-Y., Pages RA., Saroff HA., Edelhoch H., Robbins J. Analysis of thyroid hormone binding to human serum prealbumin by 8-anilinonaphthalene-1-sulfonate fluorescence. Biochemistry. 1977;16: 3707–3713. [DOI] [PubMed] [Google Scholar]
- 63.Iakovleva I, Brannstrom K, Nilsson L, Gharibyan AL, Begum A, Anan I, et al. Enthalpic Forces Correlate with the Selectivity of Transthyretin-Stabilizing Ligands in Human Plasma. J Med Chem. 2015;58: 6507–6515. doi: 10.1021/acs.jmedchem.5b00544 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper.