Abstract
MicroRNAs (miRNAs) are functional RNA molecules which play important roles in the post-transcriptional regulation. miRNAs regulate their target genes by repressing translation or inducing degradation of the target genes’ mRNAs. Many databases have been constructed to provide computationally predicted miRNA targets. However, they cannot provide the miRNA targets expressed in a specific tissue and related to a specific disease at the same time. Moreover, they cannot provide the common targets of multiple miRNAs and the common miRNAs of multiple genes at the same time. To solve these two problems, we construct a database called CSmiRTar (Condition-Specific miRNA Targets). CSmiRTar collects computationally predicted targets of 2588 human miRNAs and 1945 mouse miRNAs from four most widely used miRNA target prediction databases (miRDB, TargetScan, microRNA.org and DIANA-microT) and implements functional filters which allows users to search (i) a miRNA’s targets expressed in a specific tissue or/and related to a specific disease, (ii) multiple miRNAs’ common targets expressed in a specific tissue or/and related to a specific disease, (iii) a gene’s miRNAs related to a specific disease, and (iv) multiple genes’ common miRNAs related to a specific disease. We believe that CSmiRTar will be a useful database for biologists to study the molecular mechanisms of post-transcriptional regulation in human or mouse. CSmiRTar is available at http://cosbi.ee.ncku.edu.tw/CSmiRTar/ or http://cosbi4.ee.ncku.edu.tw/CSmiRTar/.
Introduction
MicroRNAs (miRNAs), 20–25 nucleotides non-coding RNAs, play important roles in the post-transcriptional regulation of gene expression [1–3]. Via binding to the complementary sites within the 3’ untranslated regions (3’ UTRs) of their target genes’ mRNAs, miRNAs induce mRNA degradation or lead to translational inhibition [1,4–6]. miRNAs are known to be involved in a wide range of biological processes including cell development, differentiation, cell-cycle control and apoptosis [7–9].
Understanding the miRNA-target interactions is the crucial step to discern the roles of miRNAs in different biological processes [10]. Many databases have been constructed to provide miRNA targets information. For example, TarBase [11] and miRTarBase [12] collect manually curated miRNA targets with experimental evidence from the literature but they are far from complete. The other databases such as TargetScan [10], miRDB [13], microRNA.org [14], DIANA-microT [15], miRecords [16], MAGIA [17], mirDIP [18], miRSystem [19] and miRGator [20] collect computationally predicted miRNA targets generated from various algorithms. However, these databases usually return thousands of predicted targets of a query miRNA. Researchers have to put extra efforts to extract the interested miRNA targets from a large number of uninterested ones. Since miRNAs regulate their targets in specific tissues, cell types and disease states, it is advantageous to have a database which can return miRNA targets in a specific physiological condition. Three existing databases attempted to meet this need. miTALOS [21] can provide miRNA targets of a specific tissue or cell line. miRWalk [22] can provide miRNA targets related to a specific OMIM disorder. starBase [23] can provide miRNA targets whose expressions are anti-correlated with miRNA’s expression in specific cancer types. However, none of them can provide the miRNA targets expressed in a specific tissue and related to a specific disease at the same time. Therefore, there is still a need for a database which implements both the tissue and disease filters.
The complex circuitry of miRNA-mRNA interactions show the dynamic regulation of gene expression. Recent study showed that overexpressed MRE (miRNA response element)-containing transcripts can soak up the miRNA and upregulate its target genes [24]. Moreover, the competing endogenous RNAs (ceRNAs), transcripts that cross-regulate each other by competing for shared miRNAs, were proposed to describe the new layer of post-transcriptional regulation and linked the functions of coding and non-coding RNAs [25]. Several studies indicated the deregulation of ceRNA network in cancer development [26]. Because most existing databases do not provide the common miRNAs of a set of genes, they cannot be used to find out the shared miRNAs of ceRNAs. Therefore, it is advantageous to have a database which provides researchers the common miRNAs of multiple genes and the common targets of multiple miRNAs.
To meet these two needs, we develop a database called CSmiRTar (Condition-Specific miRNA Targets). CSmiRTar collects computationally predicted targets of 2588 human miRNAs and 1945 mouse miRNAs from four most widely used miRNA target prediction databases (miRDB, TargetScan, microRNA.org and DIANA-microT). CSmiRTar implements (i) a tissue filter for users to search the miRNA targets expressed in a specific tissue, (ii) a disease filter for users to search the miRNA targets related to a specific disease, and (iii) a database filter for users to search the miRNA targets supported by multiple existing miRNA target prediction databases. Moreover, CSmiRTar allows users to search the common targets of a set of input miRNAs under a specific physiological condition and the common miRNAs of a set of input genes under a specific physiological condition. We believe that CSmiRTar will be a useful database for biologists to study the molecular mechanisms of post-transcriptional regulation in human or mouse.
Construction and contents
Data collection and processing
Five data sources were used to construct CSmiRTar. First, the experimentally validated human and mouse miRNA targets were retrieved from miRTarBase [12], which manually collected miRNA-target interactions with experimental evidence from the literature. Second, the computationally predicted human and mouse miRNA targets were retrieved from four most widely used miRNA target prediction databases (TargetScan [10], miRDB [13], microRNA.org [14] and DIANA-microT [15]). The miRNA-target interactions in these four databases were predicted by TargetScan algorithm, MirTarget algorithm, miRanda algorithm and DIANA microT-CDS algorithm, respectively. Since the collected miRNA-target interactions from different databases may use different identifiers (IDs), we have to do ID conversion in order to integrate data from different databases. In CSmiRTar, we used miRBase ID as the miRNA identifier and NCBI gene ID as the gene identifier. That is, all miRNA-target interactions were recorded as miRBase ID-NCBI gene ID pairs in CSmiRTar. Third, the tissues in which a human or a mouse gene is expressed were retrieved from Expression Atlas [27]. Expression Atlas, maintained by EMBL-EBI, provided the genes expressed in a specific tissue by analysing microarray and RNA-seq data in ArrayExpress [28]. Fourth, the diseases to which a human gene is related were retrieved from DisGeNET [29], which manually collected gene-disease associations from the literature and other expert curated databases. Fifth, the diseases to which a human miRNA is related were retrieved from PhenomiR [30], which manually collected miRNA-disease associations from the literature. The statistics of CSmiRTar could be found in Table 1. The collected dataset is already very big. On average, a human gene has 572 predicted miRNAs and a mouse gene has 231 predicted miRNAs. Therefore, biologists already have troubles to find out the functional miRNAs (among so many predicted miRNAs) for a gene of interest.
Table 1. The statistics of CSmirTar.
| Organism | # of collected miRNA-target pairs | # of miRNAs which can be queried | # of genes which can be queried | # of collected tissues in which a gene may be expressed | # of collected diseases to which a gene may be related | # of collected diseases to which a miRNA may be related |
|---|---|---|---|---|---|---|
| Human | 19,006,454 | 2588 | 21137 | 81 | 3696 | 81 |
| Mouse | 6,949,317 | 1945 | 21049 | 51 | X | X |
Implementation of CSmiRTar website
CSmiRTar was built using the scripting language PHP and Codelgniter framework. Crawler was used to retrieve raw data from other databases and Python was used to process the raw data. The processed data was stored in MySQL. The Interactive bar chart was generated by Highcharts.
Utility and discussion
Database interface
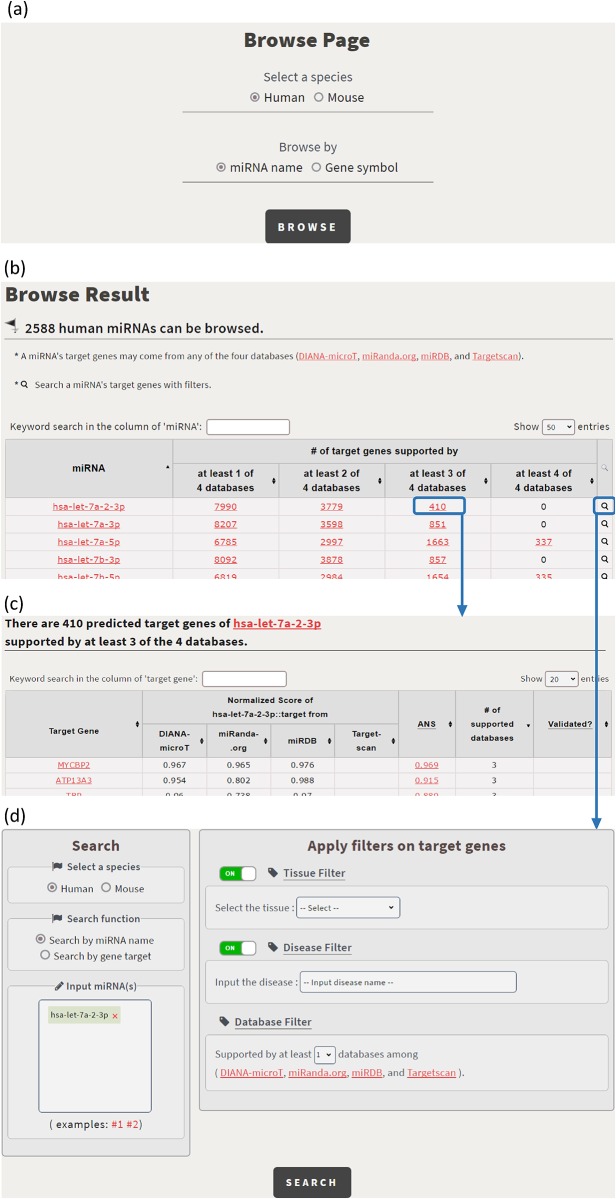
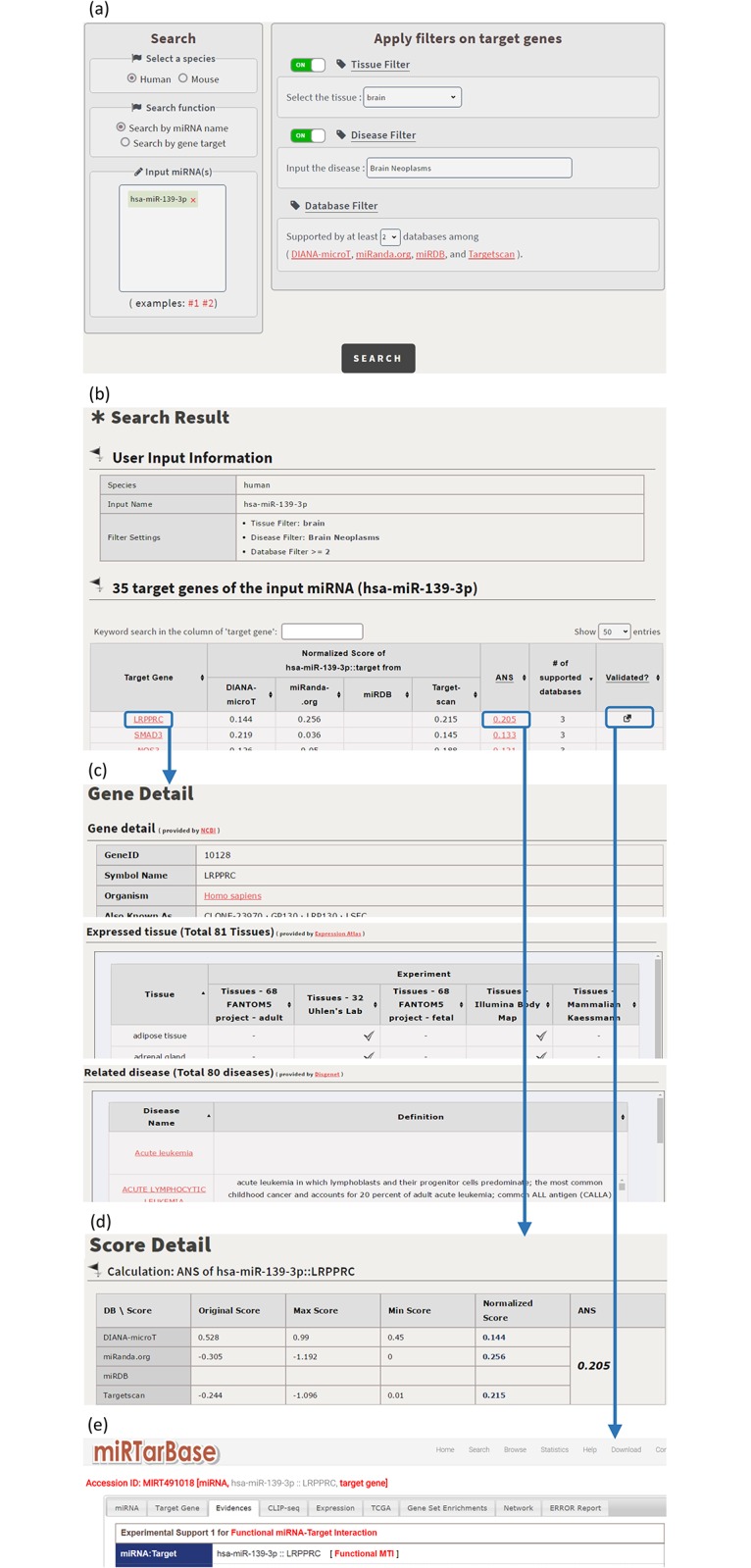
CSmiRTar provides both a search mode and a browse mode. In the search mode, users have four possible ways to search CSmiRTar. First, users can input a miRNA and search its targets which are (i) expressed in a specific tissue, (ii) related to a specific disease, or/and (iii) supported by multiple existing miRNA target prediction databases. After submission, users will see the search results sorted by the number of supported databases or the average normalized score (see Fig 1). Second, users can input a set of miRNAs and search their common targets which are (i) expressed in a specific tissue, (ii) related to a specific disease, or/and (iii) supported by multiple existing miRNA target prediction databases. After submission, users will see the search results sorted by the number of supported databases or the mean average normalized score (see Fig 2). Third, users can input a gene and search its miRNAs which are (i) related to a specific disease or/and (ii) supported by multiple existing miRNA target prediction databases. After submission, users will see the search results sorted by the number of supported databases or the average normalized score (see Fig 3). Fourth, users can input a set of genes and search their common miRNAs which are (i) related to a specific disease or/and (ii) supported by multiple existing miRNA target prediction databases. After submission, users will see the search results sorted by the number of supported databases or the mean average normalized score (see Fig 4).
Fig 1. Search the target genes of an input miRNA.
(a) Search human miR-139-3p’s target genes which are expressed in the brain tissue, related to the brain neoplasms disease and supported by at least two existing databases. (b) CSmirTar returns 35 target genes sorted by the average normalized score (ANS). (c) When clicking on a gene name in the “Target Gene” column, it opens a webpage showing the basic information of this gene, the tissues in which this gene is expressed and the diseases to which this gene is related. (d) When clicking on a score in the “ANS” column, it opens a webpage showing how the ANS is calculated. (e) When clicking on the icon in the “Validated?” column, it links to miRTarBase to show the experimental evidence of the selected miRNA-target pair.
Fig 2. Search the common targets of a set of input miRNAs.
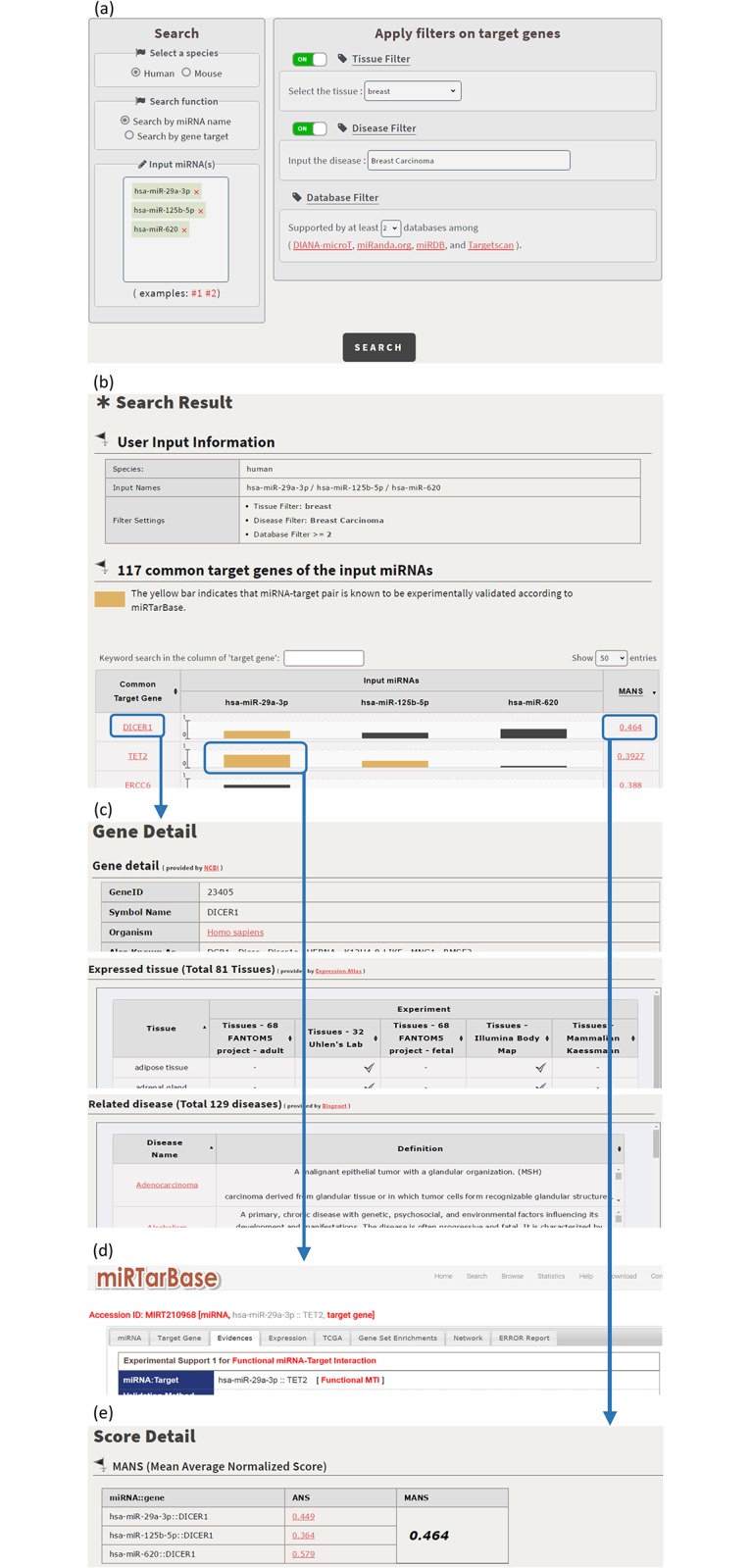
(a) For a set of human miRNAs (miR-29a-3p, miR-125b-5p and miR-620), search their common target genes which are expressed in the breast tissue, related to the breast carcinoma disease and supported by at least two existing databases. (b) CSmirTar returns 117 common target genes sorted by the mean average normalized score (MANS). (c) When clicking on a gene name in the “Common Target Gene” column, it opens a webpage showing the basic information of this gene, the tissues in which this gene is expressed and the diseases to which this gene is related. (d) An orange bar means that the miRNA-target pair has been experimentally validated. When clicking on the orange bar, it links to miRTarBase to show the experimental evidence of the selected miRNA-target pair. (e) When clicking on a score in the “MANS” column, it opens a webpage showing how the MANS is calculated.
Fig 3. Search the miRNAs of an input gene.
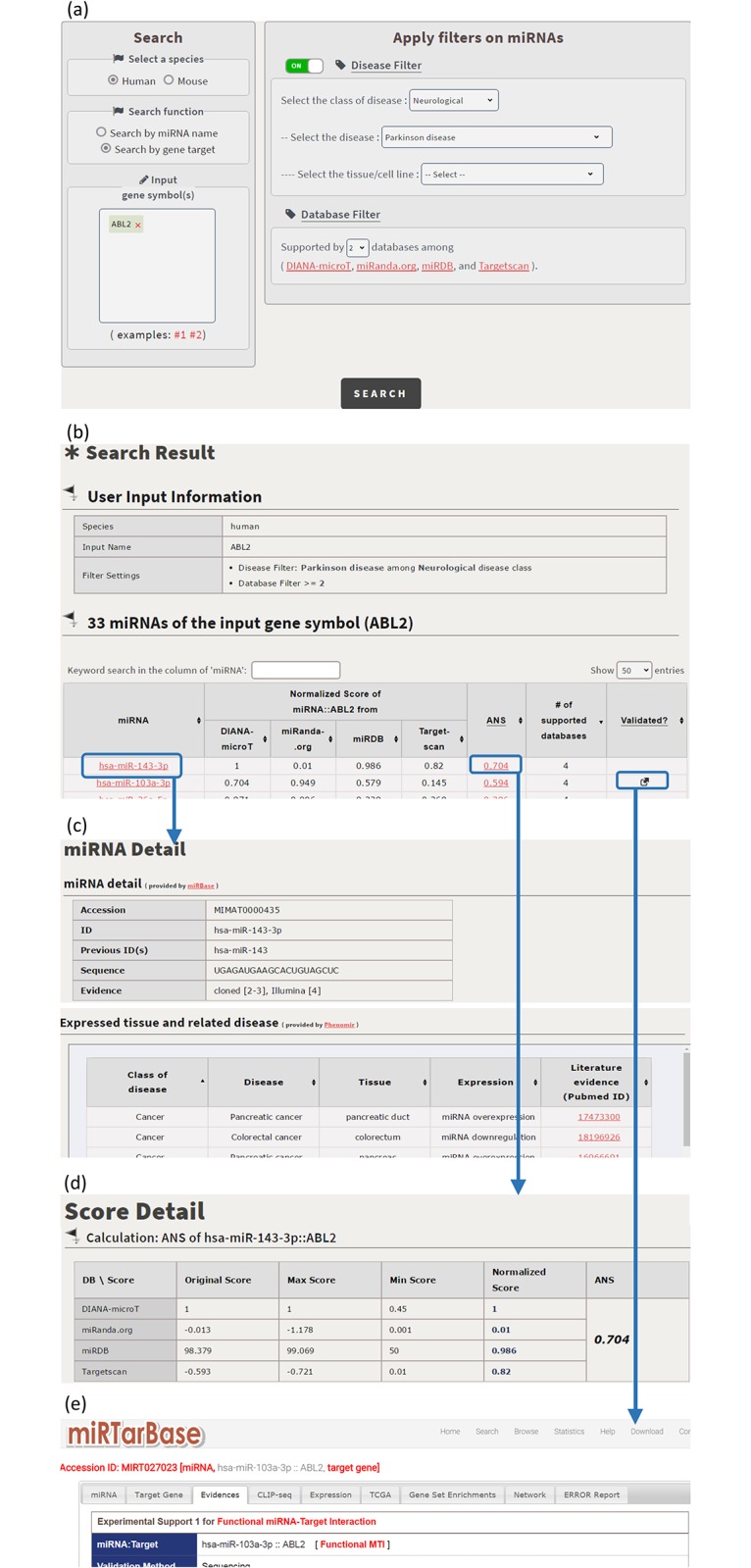
(a) For human gene ABL2, search its miRNAs which are related to Parkinson disease and supported by at least two existing databases. (b) CSmirTar returns 33 miRNAs sorted by the average normalized score (ANS). (c) When clicking on a miRNA name in the “miRNA” column, it opens a webpage showing the basic information of this miRNA and the diseases to which this miRNA is related. (d) When clicking on a score in the “ANS” column, it opens a webpage showing how the ANS is calculated. (e) When clicking on the icon in the “Validated?” column, it links to miRTarBase to show the experimental evidence of the selected miRNA-target pair.
Fig 4. Search the common miRNAs of a set of input genes.
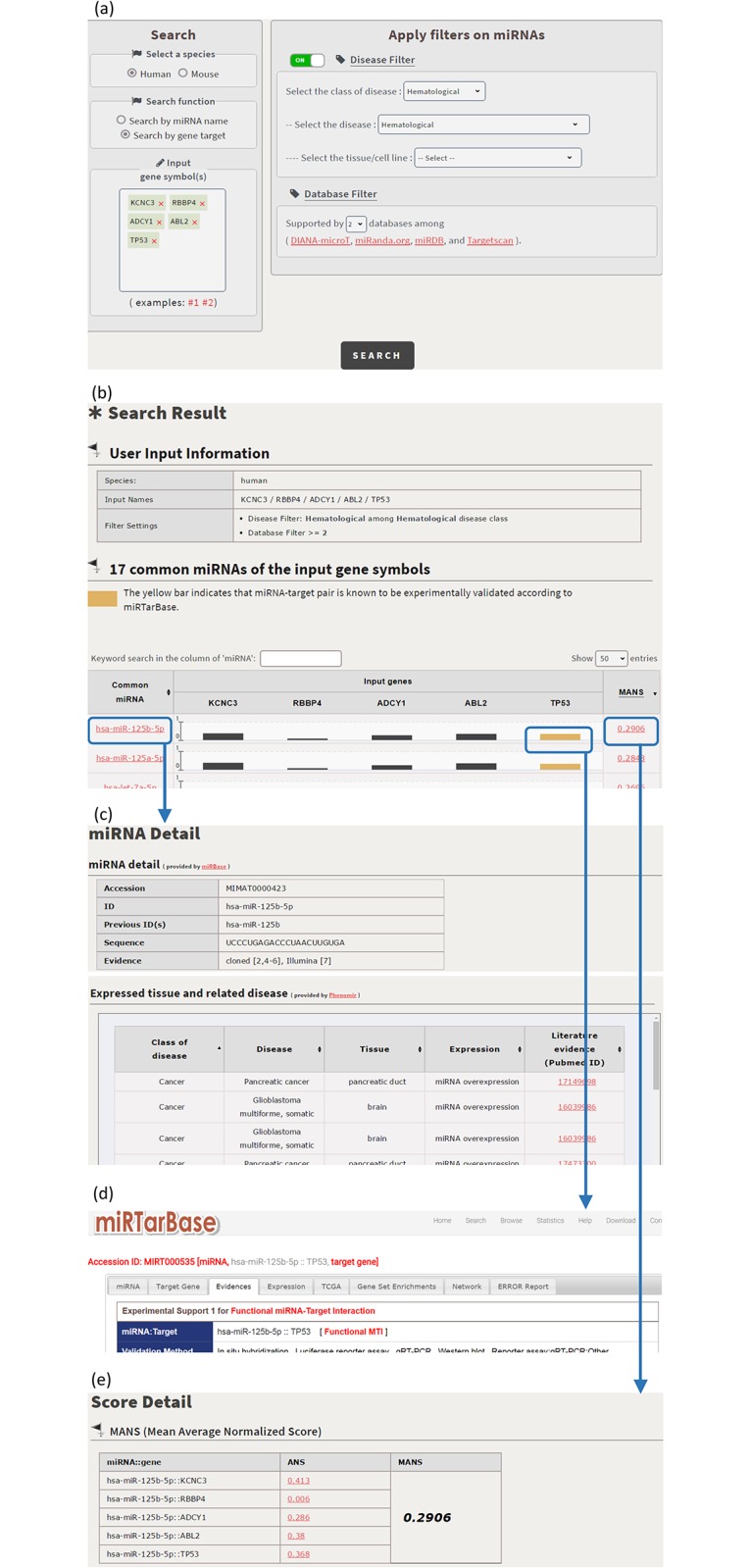
(a) For a set of human genes (KCNC3, RBBP4, ADCY1, ABL2 and TP53), search their common miRNAs which are related to the hematological disease and supported by at least two existing databases. (b) CSmirTar returns 17 common miRNAs sorted by the mean average normalized score (MANS). (c) When clicking on a miRNA name in the “Common miRNA” column, it opens a webpage showing the basic information of this miRNA and the diseases to which this miRNA is related. (d) An orange bar means that the miRNA-target pair has been experimentally validated. When clicking on the orange bar, it links to miRTarBase to show the experimental evidence of the selected miRNA-target pair. (e) When clicking on a score in the “MANS” column, it opens a webpage showing how the MANS is calculated.
In the browse mode, users have two possible ways to browse CSmiRTar. First, users can click on a human/mouse miRNA name and get the miRNA’s targets supported by one or multiple existing miRNA target prediction databases (see Fig 5). Second, users can click on a human/mouse gene name and get the miRNAs, which regulate the gene, supported by one or multiple existing miRNA target prediction databases (see Fig 6).
Fig 5. Browse CSmiRTar by a miRNA name.
(a) The browse page in shown. (b) The 2588 human miRNAs which can be browsed are shown. (c) By clicking on the number 410, it opens a webpage showing the 410 predicted target genes of hsa-let-7a-2-3p supported by at least 3 of the 4 existing miRNA target prediction databases. (d) By clicking on the icon in the rightmost column, it directs users to the search page.
Fig 6. Browse CSmiRTar by a gene name.
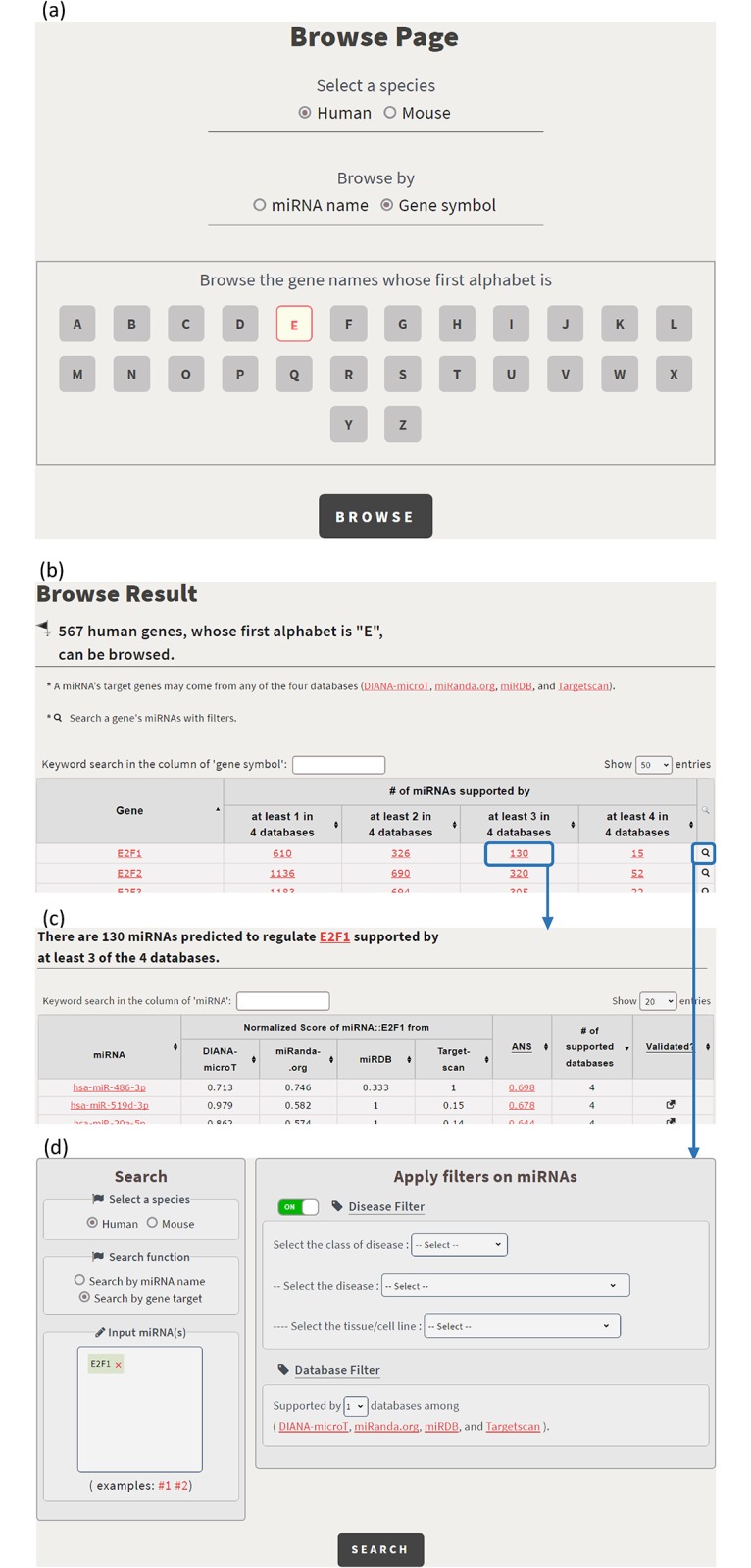
(a) The browse page is shown. (b) The 567 human genes whose first alphabet is “E” are shown. (c) By clicking on the number 130, it opens a webpage showing the 130 miRNAs predicted to regulate E2F1 supported by at least 3 of the 4 existing miRNA target prediction databases. (d) By clicking on the icon in the rightmost column, it directs users to the search page.
Our database/tissue/disease filters can significantly reduce the number of predicted miRNA targets but still keep the functional ones
Identifying the functional targets is a crucial step to dissect the function of miRNAs. Using existing miRNA target prediction databases usually returns thousands of predicted targets per miRNA. Therefore, it is very hard for researchers to choose the biologically plausible candidates for further experimental validation. Besides, it can be expected that many of the predicted targets are non-functional since miRNAs only regulate their target genes in specific tissues, cell types and disease states.
To solve this problem, we implement three different kinds of filters (a database filter, a tissue filter and a disease filter) to efficiently reduce the number of predicted miRNA targets but still keep the functional ones. To show the effectiveness of our filters, we prepare a benchmark set by randomly selecting several experimentally validated miRNA-target pairs in specific tissues and cancers from OncomiRDB [31]. As shown in Table 2 for 10 case studies, our filters can significantly reduce the number of predicted miRNA targets by more than 90% but still keep the experimentally validated miRNA targets. For example, human miR-16-5p is known to regulate the gene PPM1D in breast cancer cells [32]. Even by considering the common predicted targets from four existing miRNA target prediction databases (i.e. setting the database filter equal to four), PPM1D is still hidden in 883 predicted targets of miR-16-5p, suggesting that applying the database filter alone is not an efficient way to reduce the non-functional miRNA targets. If we further apply the tissue filter (selecting breast) and disease filter (selecting invasive breast cancer), PPM1D is now hidden in only 16 predicted targets. Researchers then have a high chance to pick out the functional targets (e.g. PPM1D) of miR-16-5p for further experimental investigation. On the contrary, if using existing miRNA target prediction databases (e.g. miRecords [16], miRWalk [22], miRSystem [19] and starBase [23]), researchers will have difficulty to pick out PPM1D among hundreds or even thousands of predicted targets of miR-16-5p (see Table 3).
Table 2. Our database/tissue/disease filters can significantly reduce the number of predicted miRNA targets but still keep the functional one.
| Human miRNA | Functional target | Reference Pubmed ID | Filter settings: (database/tissue/disease) | # of predicted targets when using only the database filter | # of predicted targets when using all three (database/tissue/disease) filters | Reduced ratio |
|---|---|---|---|---|---|---|
| miR-16-5p | PPM1D | 20668064 | 4 / breast tissue / invasive breast cancer | 883 | 16 | 98.19% = (883–16)/883 |
| miR-301a-3p | PTEN | 21393507 | 4 / breast tissue / invasive breast cancer | 533 | 14 | 97.37% |
| miR-195-5p | SLC2A3 | 22265971 | 3 / bladder tissue / carcinoma of bladder | 3008 | 125 | 95.84% |
| miR-615-5p | IGF2 | 22819824 | 3 / liver tissue / liver neoplasms | 1238 | 57 | 95.40% |
| miR-519d-3p | CDKN1A | 22262409 | 4 / liver tissue / liver neoplasms | 717 | 45 | 93.72% |
| miR-135a-5p | APC | 18632633 | 4 / colon / colorectal neoplasms | 374 | 27 | 92.78% |
| miR-153-3p | MCL1 | 19676043 | 3 / brain / glioblastoma | 1816 | 156 | 91.41% |
| miR-204-5p | EZR | 21416062 | 3 / stomach / stomach carcinoma | 1152 | 104 | 90.97% |
| miR-103a-3p | KLF4 | 22593189 | 3 / colon / colorectal carcinoma | 2095 | 236 | 88.74% |
| miR-497-5p | RAF1 | 21350001 | 4 / breast tissue / breast neoplasms | 887 | 106 | 88.05% |
Table 3. The number of predicted miRNA targets, which include the functional one being examined, in different databases.
| Human miRNA | Functional target | miRecords | miRWalk | miRSystem | starBase | CSmiRTar |
|---|---|---|---|---|---|---|
| miR-16-5p | PPM1D | 151 (6)a | 916 (8) | 151 (6) | 4239 (1) | 16 |
| miR-301a-3p | PTEN | 352 (5) | 715 (8) | 561 (4) | 144 (4) | 14 |
| miR-195-5p | SLC2A3 | 479 (5) | 942 (8) | 404 (5) | 338 (4) | 125 |
| miR-615-5p | IGF2 | 66 (4) | 196 (8) | 3036 (1) | N/A | 57 |
| miR-519d-3p | CDKN1A | 217 (5) | 806 (8) | 19 (6) | 4 (5) | 45 |
| miR-135a-5p | APC | 687 (4) | 1336 (7) | 59 (6) | 2262 (1) | 27 |
| miR-153-3p | MCL1 | 42 (6) | 111 (9) | 177 (5) | 246 (3) | 156 |
| miR-204-5p | EZR | 224 (5) | 9 (11) | 14 (7) | 172 (3) | 104 |
| miR-103a-3p | KLF4 | 3799 (3) | 1751 (7) | 1983 (2) | 299 (3) | 236 |
| miR-497-5p | RAF1 | 11043 (2) | 1303 (8) | 224 (5) | 342 (4) | 106 |
miRecords, miRWalk, miRSystem and starBase were all run with the setting of their algorithm filter as stringent as possible to minimize the number of predicted mRNA targets while still include the functional target being examined. CSmiRTar was run with the settings shown in Table 2.
- a151 (6) means that in miRecords, miR-16-5p has 151 target genes (including PPM1D) predicted by at least 6 different algorithms. Note that in miRecords, 6 is the most stringent setting of the algorithm filter for still reporting PPM1D as a predicted target gene of miR-16-5p.
Our database/disease filters can significantly reduce the number of predicted miRNAs of a gene but still keep the functional ones
As shown in Table 4 for 10 case studies, our filters can significantly reduce the number of predicted miRNAs of a gene by 63% to 95% but still keep the experimentally validated miRNAs which really regulate the gene. For example, human gene MECP2 is known to be regulated by miR-212-3p in gastric cancer cells [33]. Even by considering the common predicted miRNAs from three existing miRNA target prediction databases (i.e. setting the database filter equal to three), miR-212-3p is still hidden in 537 predicted miRNAs of the gene MECP2, suggesting that applying the database filter alone is not an efficient way to reduce the non-functional miRNAs of MECP2. If we further apply the disease filter (selecting gastric cancer in the stomach), miR-212-3p is now hidden in only 22 predicted miRNA of MECP2. Researchers then have a high chance to pick out the functional miRNAs (e.g. miR-212-3p) of MECP2 for further experimental investigation.
Table 4. The database/disease filters can significantly reduce the number of predicted miRNAs but still keep the functional miRNA which regulates the target gene.
| Human target gene | Functional miRNA which regulates the target gene | Reference Pubmed ID | Filter setting: (database/disease) | # of predicted miRNAs when using only the database filter | # of predicted miRNAs when using both (database/disease) filters | Reduced ratio |
|---|---|---|---|---|---|---|
| MECP2 | miR-212-3p | 20020497 | 3 / gastric cancer in stomach | 537 | 22 | 95.90% = (537–22)/537 |
| MCL1 | miR-153-3p | 19676043 | 3 / glioblastoma multiforme, somatic in brain | 253 | 21 | 91.70% |
| CREB1 | miR-181b-5p | 22539488 | 4 / gastric cancer in stomach | 99 | 9 | 90.91% |
| ALCAM | miR-215-5p | 21119604 | 4 / gastric cancer in stomach | 47 | 5 | 89.36% |
| FOXO1 | miR-370-3p | 23029264 | 4 / prostate cancer in prostate gland | 49 | 7 | 85.71% |
| APC | miR-135a-5p | 18632633 | 4 / colorectal cancer in colorectum | 22 | 5 | 77.27% |
| SPRY2 | miR-27a-3p | 20638779 | 4 / pancreatic cancer in pancreas | 26 | 6 | 76.92% |
| KLF4 | miR-103a-3p | 22593189 | 3 / colorectal cancer in colorectum | 128 | 35 | 72.66% |
| ID1 | miR-381-3p | 22592211 | 3 / lung cancer in lung cancer cell line | 56 | 16 | 71.43% |
| PBX3 | let-7c-5p | 21984339 | 4 / colorectal cancer in colorectum | 60 | 22 | 63.33% |
Identifying the shared miRNAs of ceRNAs
An important step to reconstruct the ceRNA network is to identify the shared miRNAs of ceRNAs. CSmiRTar allows users to input a set of genes (e.g. ceRNAs) to search the shared miRNAs which regulate these genes. As shown in Table 5 for five case studies, CSmiRTar can identify the experimentally validated shared miRNAs of ceRNAs. For example, it is known that human ceRNAs (PTEN, VAPA and CNOT6L) are all regulated by miR-17-5p, miR-19a-3p, miR-20a-5p and miR-106b-5p in human prostate cancer cells [34]. By considering the common predicted miRNAs from three existing databases (i.e. setting the database filter equal to three) and applying the disease filter (selecting prostate cancer), CSmiRTar returns 13 predicted shared miRNAs which contain all the four experimentally validated shared miRNAs of the input ceRNAs.
Table 5. The database/disease filters can significantly reduce the number of predicted shared miRNAs but still keep the functional shared miRNAs of ceRNAs.
| Human ceRNAs | Functional shared miRNAs | Reference Pubmed ID | Filter setting: (database/disease) | # of predicted shared miRNAs when using the filters |
|---|---|---|---|---|
| PTEN, VAPA, CNOT6L | miR-17-5p, miR-19a-3p, miR-20a-5p, miR-106b-5p | 22000013 | 3 / prostate cancer | 13 |
| CD44, CDC42 | miR-216a-5p, miR-330-3p | 21149267 | 1 / breast cancer | 192 |
| PTENP1, PTEN | miR-19b-3p, miR-20a-5p | 20577206 | 1 / colorectal cancer | 114 |
| CDK6, ABL1, SRC | miR-203-3p | 23462723 | 2 / breast cancer | 29 |
| FOXF2, RECK, MTSS1 | miR-182-5p | 23383207 | 3 / prostate cancer | 2 |
Identifying the common target genes of a set of miRNAs
In CSmiRTar, users can input a set of miRNAs to search their common target genes. As shown in Table 6 for five case studies, CSmiRTar can successfully identify the experimentally validated common target genes of multiple miRNAs. For example, it is known that human miR-186-5p, miR-216b-5p, miR-337-3p, and miR-760 cooperatively induce cellular senescence by targeting the gene CSNK2A1 in human colorectal cancer cells [35]. By considering the predicted target genes supported by three existing miRNA target prediction databases (i.e. setting the database filter equal to three) and applying the tissue/disease filter (selecting colon/colorectal carcinoma), CSmiRTar returns 23 predicted common target genes which contain the experimentally validated common target gene CSNK2A1 of the input set of miRNAs.
Table 6. The database/tissue/disease filters can significantly reduce the number of predicted common targets but still keep the functional common target of a set of miRNAs.
| A set of human miRNAs | Common target gene | Reference Pubmed ID | Filter settings: (database/tissue/disease) | # of predicted targets when using the filters |
|---|---|---|---|---|
| miR-17-5p, miR-19a-3p, miR-20a-5p, miR-20b-5p, miR-26b-5p, miR-106a-5p, miR-106b-5p | PTEN | 22000013 | 3 / prostate tissue / prostate carcinoma | 20 |
| miR-186-5p, miR-216b-5p, miR-337-3p, miR-760 | CSNAK2A1 | 23137536 | 3 / colon / colorectal carcinoma | 23 |
| miR-330-3p, miR-608, miR-216a-5p | CD44 | 21149267 | 2 / breast tissue / breast neoplasms | 78 |
| miR-130a-3p, miR-301a-3p, miR-454-3p | SMAD4 | 23393589 | 3 / colon / colorectal neoplasms | 116 |
| miR-19b-3p, miR-20a-5p | PTEN | 20577206 | 3 / colon tissue / colon carcinoma | 91 |
Conclusions
In this article, we present CSmiRTar which provide computationally predicted targets of 2588 human miRNAs and 1945 mouse miRNAs. CSmiRTar implements (i) a tissue filter for users to search the miRNA targets expressed in a specific tissue, (ii) a disease filter for users to search the miRNA targets related to a specific disease, and (iii) a database filter for users to search the predicted miRNA targets supported by multiple existing databases,. Moreover, CSmiRTar allows users to search the common targets of a set of input miRNAs under a specific physiological condition and the common miRNAs of a set of input genes under a specific physiological condition. We provide many case studies to show the effectiveness of our filters in reducing the number of predicted miRNA targets but still keep the functional ones. However, users should note that some functional miRNA targets may not be kept when applying both the tissue and disease filters if they are not expressed in normal tissues but are abnormally expressed in disease states. Nevertheless, we believe that CSmiRTar will be a useful database for biologists to study the molecular mechanisms of post-transcriptional regulation in human and mouse.
Acknowledgments
This work was supported by National Cheng Kung University and Ministry of Science and Technology of Taiwan [MOST-105-2221-E-006-203-MY2]. Funding for open access charge: Ministry of Science and Technology of Taiwan.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by National Cheng Kung University and Ministry of Science and Technology of Taiwan [MOST-105-2221-E-006-203-MY2]. Funding for open access charge: National Cheng Kung University and Ministry of Science and Technology of Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 2.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol 2005; 15(3):331–341. doi: 10.1016/j.sbi.2005.05.006 [DOI] [PubMed] [Google Scholar]
- 3.Sontheimer EJ, Carthew RW. Silence from within: endogenous siRNAs and miRNAs. Cell 2005; 122(1):9–12. doi: 10.1016/j.cell.2005.06.030 [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120(1):15–20. doi: 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136(2):215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011, 12(2):99–110. doi: 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- 7.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005; 438(7068):685–689. doi: 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- 8.Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol 2007; 23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406 [DOI] [PubMed] [Google Scholar]
- 9.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 2008; 9(3):219–230. doi: 10.1038/nrm2347 [DOI] [PubMed] [Google Scholar]
- 10.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res 2015; 43:D153–D159. doi: 10.1093/nar/gku1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 2014; 42:D78–D85. doi: 10.1093/nar/gkt1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015; 43:D146–D152. doi: 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res 2008; 36:D149–D153. doi: 10.1093/nar/gkm995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013; 41:W169–W173. doi: 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 2009; 37:D105–D110. doi: 10.1093/nar/gkn851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sales G, Coppe A, Bisognin A, Biasiolo M, Bortoluzzi S, Romualdi C. MAGIA, a web-based tool for miRNA and genes integrated analysis. Nucleic Acids Res 2010; 38:W352–W359. doi: 10.1093/nar/gkq423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirdel EA, Xie W, Mak TW. NAViGaTing the micronome—using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One 2011; 6(2):e17429 doi: 10.1371/journal.pone.0017429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK, Lai LC, et al. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One 2012; 7(8):e42390 doi: 10.1371/journal.pone.0042390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho S, Jang I, Jun Y. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res 2013; 41:D252–D257. doi: 10.1093/nar/gks1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preusse M, Theis FJ, Mueller NS. miTALOS v2: analyzing tissue specific microRNA function. PLoS One 2016; 11(3):e0151771 doi: 10.1371/journal.pone.0151771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015; 12(8):697 doi: 10.1038/nmeth.3485 [DOI] [PubMed] [Google Scholar]
- 23.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014; 42:D92–D97. doi: 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007, 4(9):721–726. doi: 10.1038/nmeth1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011; 147(2):382–395. doi: 10.1016/j.cell.2011.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet 2015; 52(10):710–718. doi: 10.1136/jmedgenet-2015-103334 [DOI] [PubMed] [Google Scholar]
- 27.Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, et al. Expression Atlas update—a database of gene and transcript expression from microarray-and sequencing-based functional genomics experiments. Nucleic Acids Res 2014; 42:D926–D932. doi: 10.1093/nar/gkt1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, et al. ArrayExpress update—simplifying data submissions. Nucleic Acids Res 2015; 43(Database issue):D1113–D1116. doi: 10.1093/nar/gku1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database 2015; bav028 doi: 10.1093/database/bav028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruepp A, Kowarsch A, Schmidl D, Buggenthin F, Brauner B, Dunger I, et al. PhenomiR: a knowledgebase for microRNA expression in diseases and biological processes. Genome Biol 2010, 11(1):R6 doi: 10.1186/gb-2010-11-1-r6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Gu J, Wang T, Ding Z. OncomiRDB: a database for the experimentally verified oncogenic and tumor-suppressive microRNAs. Bioinformatics 2014; 30(15):2237–2238. doi: 10.1093/bioinformatics/btu155 [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, et al. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res 2010, 70(18):7176–7186. doi: 10.1158/0008-5472.CAN-10-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada R, Akiyama Y, Hashimoto Y, Fukamachi H, Yuasa Y. miR-212 is downregulated and suppresses methyl-CpG-binding protein MeCP2 in human gastric cancer. Int J Cancer 2010; 127(5):1106–1114. doi: 10.1002/ijc.25126 [DOI] [PubMed] [Google Scholar]
- 34.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011; 147(2):344–357. doi: 10.1016/j.cell.2011.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SY, Lee YH, Bae YS. MiR-186, miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular senescence by targeting α subunit of protein kinase CKII in human colorectal cancer cells. Biochem Biophys Res Commun 2012; 429(3–4):173–179. doi: 10.1016/j.bbrc.2012.10.117 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.