Abstract
IMPORTANCE
Uveal melanoma (UM) is an intraocular primary malignant neoplasm that often gives rise to metastatic disease for which there are no effective therapies. A substantial proportion of UMs express the cancer-testis antigen PRAME (preferentially expressed antigen in melanoma), which can potentially be targeted by adoptive T-cell therapy.
OBJECTIVE
To determine whether there may be a rationale for PRAME-directed T-cell therapy for metastatic UM.
DESIGN, SETTING, AND PARTICIPANTS
An experimental study using a retrospective cohort of 64 patients with UM (median follow-up, 62 months) was conducted from January 8, 2015, to November 20, 2016, at the Leiden University Medical Center. Clinical, histopathologic, and genetic parameters were compared between 64 PRAME-positive and PRAME-negative UMs. HLA class I restricted, PRAME-specific T cells were stimulated with UM cell lines to measure their antigen-specific reactivity against these cell lines, which were analyzed for PRAME expression by real-time quantitative polymerase chain reaction. Uveal melanoma metastases from 16 unrelated patients were assessed for PRAME expression by messenger RNA fluorescence in situ hybridization and for HLA class I expression by immunofluorescence staining.
MAIN OUTCOMES AND MEASURES
Interferon γ production for antigen-specific reactivity and detection of PRAME and HLA class I expression in primary and metastatic UM.
RESULTS
Of the 64 patients in the study (31 women and 33 men; mean [SD] age at the time of enucleation, 60.6 [15.6] years), PRAME expression was negative in 35 primary UMs and positive in 29 primary UMs. Positive PRAME expression was associated with a high largest basal diameter (15.0 vs 12.0 mm; P = .005), ciliary body involvement (59% vs 26%; P = .008), and amplification of chromosome 8q (66% vs 23%; P = .002). PRAME-specific T cells reacted against 4 of 7 UM cell lines, demonstrating that T-cell reactivity correlated with PRAME expression. Metastatic UM samples were positive for PRAME messenger RNA in 11 of 16 patients and for HLA class I in 10 of 16 patients, with 8 of 16 patients demonstrating coexpression of both PRAME and HLA class I.
CONCLUSIONS AND RELEVANCE
PRAME is expressed in many primary and metastatic UMs, and about half of the metastatic UMs coexpress PRAME and HLA class I. The finding that PRAME-specific T cells in this study reacted against PRAME-positive UM cell lines suggests a potential role for PRAME-directed immunotherapy for selected patients with metastatic UM.
Uveal melanoma (UM) is a pigment cell–derived malignant neoplasm that occurs in the eye and can lead to metastases, usually affecting the liver. Several characteristics are associated with the development of metastases, such as large tumor size, the presence of epithelioid cells, the loss of 1 chromosome 3 and the presence of additional copies of chromosome 8q,1,2 and a specific gene expression profile known as class 2.3,4 In addition, high-risk tumors are characterized by an inflammatory phenotype, with high numbers of infiltrating CD4+ and CD8+ T cells, macrophages, and increased expression of HLA class I and II.5–9
Treatment options currently used to treat metastatic UM include liver-directed chemotherapy and systemic targeted and immune therapies.10 However, these treatments have resulted in durable responses in very few patients.11 The potential efficacy of immunotherapy has been limited, presumably owing to the small number of mutations leading to neoantigen expression in UM; immunotherapy with checkpoint inhibitors has shown very low response rates in metastatic UM.12–14
Another possible treatment option for patients with metastasized UM is targeted therapy with T cells directed against tumor-associated antigens. Recently, Field et al15 reported that the cancer-testis antigen PRAME (preferentially expressed antigen of melanoma) is expressed in many primary UMs and is a marker for increased risk of metastasis in Class 1 and disomy 3 UMs. PRAME was first identified as a tumor-associated antigen through analysis of the specificity of tumor-reactive T-cell clones derived from a patient with metastatic cutaneous melanoma.16 Subsequently, it has been shown that PRAME (Ensembl: ENST00000398741.5) is expressed in many malignant neoplasms, including cutaneous melanoma, breast carcinoma, non–small cell lung cancer, and leukemia,16,17 whereas normal healthy tissues express minimal or no PRAME, with the exception of the testis and endometrium.16 Griffioen et al18 have shown that PRAME-specific CD8+ T cells isolated from healthy individuals and patients with advanced melanoma were able to recognize and lyse cells expressing HLA class I and high levels of PRAME. However, these T cells had a low avidity for PRAME-expressing tumor cells, and their T-cell receptors (TCRs) will therefore unlikely be useful for therapeutic applications. A previous study isolated PRAME-specific HLA-A2 restricted T-cell clones, which exhibited a high specificity and reactivity for PRAME, derived from a patient who underwent an HLA-A2 mismatched stem cell transplant.19 These PRAME-specific T cells were able to lyse multiple PRAME-positive malignant cell lines, including cell lines derived from cutaneous melanoma, lung cancer, breast cancer, and acute myeloid leukemia. It was demonstrated by TCR gene transfer that the high-affinity, PRAME-specific TCR could potentially be used for PRAME-TCR gene therapy. A clinical trial to evaluate PRAME-TCR gene therapy is being initiated. To determine whether patients with metastatic UM may be potential candidates for PRAME-TCR gene therapy, we expanded our analysis to UM and tested whether PRAME is expressed in primary and metastatic UM and whether high-affinity, PRAME-specific T cells can recognize PRAME-expressing UM cell lines.
Methods
Study Population
Formalin-fixed, paraffin-embedded (FFPE), and fresh-frozen tissue was obtained from 64 patients with primary UM who underwent enucleation at the Leiden University Medical Center (Leiden, the Netherlands) between September 21, 1999, and October 6, 2008, with a median follow-up of 62 months (range, 5–181 months). Patient DNA and RNA were isolated from fresh-frozen tissues for chromosome and gene expression analyses. Each tumor sample was processed for histopathologic evaluation. Patient medical records were scrutinized for clinical and histopathologic features, including age at enucleation, largest basal diameter (in millimeters), thickness (in millimeters), extraocular extension, ciliary body involvement, cell type, and mitotic count. Survival data and information on cause of death were obtained from patient medical records and from the Integral Cancer Center West, and they were updated in 2015. Follow-up time is indicated as the time from enucleation until death or last follow-up. Formalin-fixed, paraffin-embedded tissue blocks containing UM metastases from 17 unrelated patients were collected from our pathological archives for HLA class I staining and messenger RNA (mRNA) fluorescence in situ hybridization (FISH) because metastatic samples were not available from the original cohort. Because positive controls demonstrated a poor quality of the FFPE tissue sample from 1 patient, 16 cases were analyzed further. Tumor material was handled according to the Dutch National Ethical Guidelines (Code for Proper Secondary Use of Human Tissue) and the tenets of the Declaration of Helsinki.20 The Medical Ethics Committee of the Leiden University Medical Center approved this study and waived the need for informed consent, following the regulations laid down for use of patient material according to the Federation of Medical Scientific Societies (FEDERA).
DNA and Gene Expression Analysis
From 64 fresh-frozen specimens, DNA for single-nucleotide polymorphism and copy number analyses was extracted with the QIAmp DNA Mini kit (Qiagen). Single-nucleotide polymorphism analysis was performed with the Affymetrix 250K_NSP-chip and Affymetrix Cytosc an HD chip (Affymetrix) to assess aberrations in chromosomes 3 and 6. Information on the copy number of 8q was obtained by droplet digital polymerase chain reaction. The RNA for gene expression profiling was isolated with the RNeasy mini Kit (Qiagen). PRAME RNA expression was measured on the Illumina HT-12v4 chip (Illumina) using probe ILMN_1700031.21 Expression data for disomy 3 UMs were included in a previous publication.15
Recognition of UM Cell Lines By PRAME-Specific T Cells
Uveal melanoma cell lines were cultured in Roswell Park Memorial Institute medium 1640 supplemented with fetal calf serum, 10%, glutamine, 1%, penicillin, 2%, and streptomycin (Gibco; Thermo Fisher Scientific Inc) at 37°C and 5% carbon dioxide. We used the following primary tumor-derived cell lines: 92.1,22 Mel202, Mel270, Mel285, and Mel290,23 as well as cell lines OMM2.3 and OMM2.5, which were derived from metastases of the same patient from which cell line Mel270 was derived.24 T-cell clones and UM cell lines were coincubated at a responder to stimulator ratio of 1:4. We incubated 5000 T cells with 20 000 tumor cells in a round-bottomed, 96-well plate for 18 hours. Two PRAME-specific T-cell clones (HSS1 and HSS3) recognizing the SLLQHLIGL epitope of PRAME in the context of HLA-A*02:01 were used.19 If HLA-A2 was not present in the UM cell lines, we introduced HLA-A2 (European Nucleotide Archive: AF055066.1) using retroviral vectors.25 To confirm HLA-A2 expression on the UM cell lines, we used the HLA-A2 restricted T-cell clone HSS12, which recognizes peptide FTWEGLYNV from the ubiquitously expressed gene USP11 (Ensembl: ENST00000377107.6).26 Our negative control clone was pp65-A2, which is also HLA-A2 restricted but recognizes a peptide from the pp65 (European Nucleotide Archive: EF531301.1) gene of cytomegalovirus that is not expressed on the UM cell lines.27 After 18 hours of coincubation, supernatants were harvested and interferon γ (IFN-γ) secretion was measured by enzyme-linked immunosorbent assay (Sanquin Reagents).
Detection and Scoring of PRAME mRNA in UM Cell Lines and FFPE Tissue Sections
Detailed information about detection of PRAME expression by real-time quantitative polymerase chain reaction on UM cell lines and by mRNA FISH in primary as well as metastasized FFPE tissue sections is described in the eAppendix in the Supplement.
HLA Class I Staining and Scoring
Immunofluorescence staining for expression of human HLA-A, HLA-B/C, and β2-microglobulin (β2M) was performed on paraffin-embedded samples of metastases as previously described,28 and we used the scoring system of Ruiter et al29 and other studies30,31 (eAppendix in the Supplement).
Statistical Analysis
Statistical analysis was performed with SPSS, version 20.0.0 (IBM Corp). PRAME gene expression was dichotomized as negative and positive. Clinical, histopathologic, and genetic parameters were compared between both groups using the Pearson χ2 test for categorical prognostic parameters and the Mann-Whitney U test for continuous prognostic parameters. The Kaplan-Meier curve and log-rank test were used to perform disease-specific survival analysis for patients with primary UM with negative and positive PRAME expression. Death due to metastasis was considered an event. Patients who died owing to another cause or an unknown cause were censored.
Results
Distribution of PRAME Expression in Primary UM
PRAME gene expression was analyzed in 64 primary UMs using an RNA expression microarray (Figure 1A). The mean (SD) age of the patients at enucleation was 60.6 (15.6) years, and 33 of the 64 patients (52%) were men. When expression values were plotted from lowest to highest, an inflection point in the slope was noted at the sample with expression of 7.23 Illumina units, so we took this point as the threshold for positive PRAME expression. Tumors with expression less than 7.23 Illumina units were categorized as PRAME-negative UM (n = 35), and those with expression of 7.23 Illumina units or more were categorized as PRAME-positive UM (n = 29).
Figure 1. PRAME Expression in Primary Uveal Melanoma (UM) and Correlation With Survival.
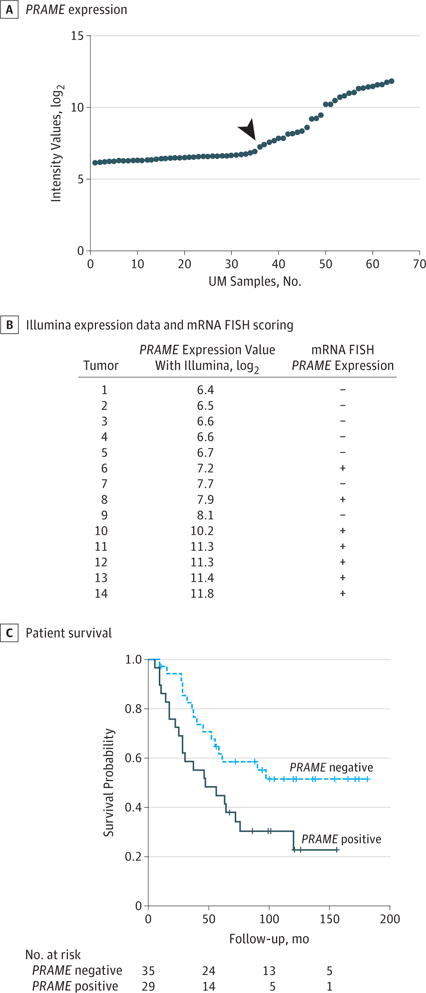
A, PRAME expression determined using 2 different probes in 64 cases of UM using an Illumina HT-12v4 microarray. Using probe ILMN_1700031, tumors are dichotomized into negative and positive. The samples on the right of the arrow are categorized as positive PRAME expression and the samples on the left of the arrow are categorized as negative PRAME expression. B, Illumina expression data and messenger RNA (mRNA) fluorescence in situ hybridization (FISH) scoring for 14 primary UMs demonstrating specificity and sensitivity of the PRAME probe sets. The κ value for mRNA FISH between both observers was 0.857. Plus sign indicates positive expression; minus sign, negative expression. C, Survival curve of patients with negative and positive PRAME-expressing primary UM.
We compared the clinical, histopathologic, and chromosome data between the 2 groups of patients with tumors (Table 1). Both groups did not differ in age or sex. PRAME expression was associated with prognostically poor tumor characteristics; PRAME-positive tumors had the largest median basal diameter (15.0 vs 12.0 mm; P = .005) and a more frequent involvement of the ciliary body (59% vs 26%; P = .008) compared with PRAME-negative tumors. Of the 29 PRAME-positive tumors, 21 (72%) showed monosomy of chromosome 3, whereas this was the case in 20 (57%) of the 35 PRAME-negative tumors (P = .21). PRAME expression correlated with amplification of chromosome 8q (66% vs 23%; P = .002). Tumors with chromosome 8q copies were categorized as follows: a copy number between 1.9 and 2.1 was categorized as normal, between 2.2 and 3.1 as gain, and more than 3.1 as amplification of chromosome 8q. When comparing the Kaplan-Meier survival curves, we found that patients with PRAME-positive tumors had a shorter disease-specific survival than patients with PRAME-negative tumors (median survival, 47 vs 88 months; P = .02) (Figure 1C).
Table 1.
Comparison of Clinical, Histopathologic, and Genetic Features Between PRAME-Negative and PRAME-Positive Primary Uveal Melanoma
| Clinical and Histopathologic Feature | Patients, No. (%)a | P Value | |
|---|---|---|---|
|
PRAME-Negative Tumors (n = 35) |
PRAME-Positive Tumors (n = 29) |
||
| Sex | |||
| Male | 19 (54) | 14 (48) | .63b |
| Female | 16 (46) | 15 (52) | |
| Age at enucleation, median (range), y | 58.9 (12.8–84.8) | 62.7 (33.4–88.4) | .77c |
| Largest basal diameter, median (range), mm | 12.0 (8–20) | 15.0 (9–30) | .005c |
| Thickness, median (range), mm | 7.0 (2–12) | 9.0 (2–12) | .30c |
| Mitotic count, median (range) | 5.0 (1–33) | 5.5 (0–20) | .66c |
| Ciliary body involvement | |||
| No | 26 (74) | 12 (41) | .008b |
| Yes | 9 (26) | 17 (59) | |
| Cell type | |||
| Spindle | 14 (40) | 8 (28) | .30b |
| Mixed or epithelioid | 21 (60) | 21 (72) | |
| Extraocular extension | |||
| None or superficial | 23 (66) | 17 (59) | .56b |
| Deep, total, or episcleral | 12 (34) | 12 (41) | |
| TNM staging | |||
| I–IIB | 26 (74) | 15 (52) | .06b |
| IIIA–IIIC | 9 (26) | 14 (48) | |
| Metastases | |||
| No | 19 (54) | 8 (28) | .03b |
| Yes | 16 (46) | 21 (72) | |
| Chromosome | |||
| 3 status | |||
| Disomy | 15 (43) | 8 (28) | .21b |
| Monosomy | 20 (57) | 21 (72) | |
| 8q status | |||
| Normal | 11 (31) | 3 (10) | |
| Gain of 8q | 16 (46) | 7 (24) | .002b |
| Amplification of 8q | 8 (23) | 19 (66) | |
| 6p status | |||
| Normal | 23 (66) | 20 (69) | .78b |
| Gain of 6p | 12 (34) | 9 (31) | |
Percentages are rounded and may not total 100.
Pearson χ2 test.
Mann-Whitney U test.
Recognizing UM Cell Lines by PRAME-Specific T-Cell Clones
Two previously identified PRAME-specific T-cell clones (HSS1 and HSS3) were used to determine whether UM cell lines can potentially be recognized by PRAME-specific T cells.19 First, PRAME-dependent recognition of the PRAME-specific T-cell clones was demonstrated by introducing full-length PRAME into HLA-A2–positive cell line SW480. As demonstrated in Figure 2A, the PRAME-specific T-cell clones do not recognize the HLA-A2–negative, PRAME-negative SW480 cells. However, retroviral introduction of PRAME into SW480 resulted in efficient recognition, comparable to HLA-A2–positive, PRAME-positive LCL-JY and K562+A2. Before the UM cell lines were tested for recognition by the PRAME-specific T-cell clones, the HLA-A2 expression of the UM cell lines was measured by fluorescence-activated cell sorting analysis using specific antibodies directed against HLA class I (W632) and HLA-A2 (BB7.2). We confirmed that only UM cell line Mel290 was HLA-A2 positive. The remaining UM cell lines were HLA-A2 negative and were therefore retrovirally transduced with HLA-A*02:01. After retroviral transduction, all 7 UM cell lines were recognized by the USP11-specific T-cell clone HSS12, confirming that HLA-A*0201 was now expressed on all UM cell lines. The results shown in Figure 2B demonstrate that 4 of 7 UM cell lines were efficiently recognized by the PRAME-specific T-cell clones HSS1 and HSS3, as coincubation led to production of IFN-γ by both of the PRAME-specific T-cell clones. Using real-time quantitative polymerase chain reaction, we confirmed that the 4 UM cell lines recognized by the PRAME-specific T-cell clones expressed PRAME (Figure 2C). These data demonstrate that PRAME-positive UM cell lines can be recognized by PRAME-specific T cells and that the recognition potential of the T cells correlated with PRAME expression in these cell lines.
Figure 2. Recognition of PRAME-Positive Uveal Melanoma (UM) Cell Lines by PRAME-Specific T Cells.
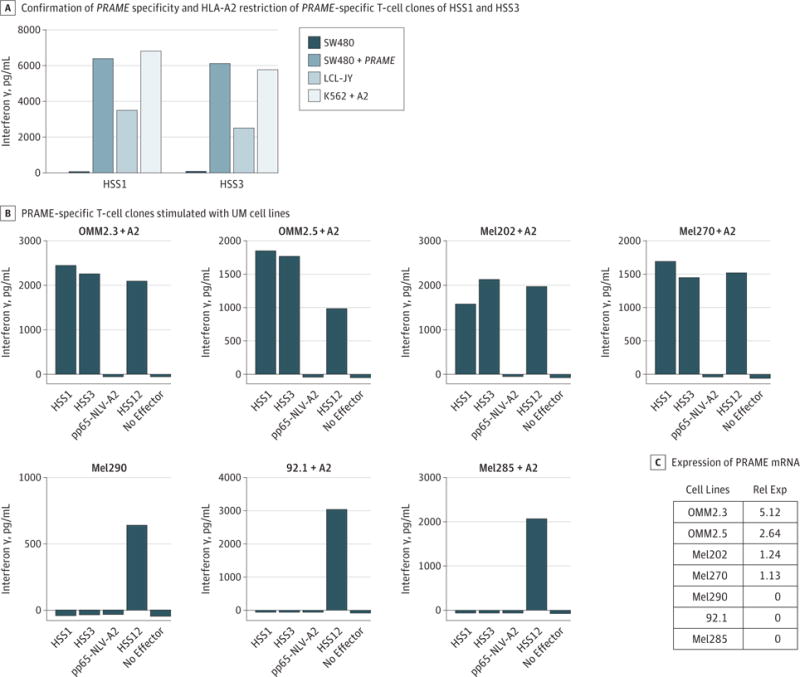
A, PRAME-specific T-cell clones HSS1 and HSS3 were stimulated with the PRAME-negative HLA-A2–positive cell line SW480, SW480 retrovirally transduced with PRAME (SW480+PRAME), HLA-A2–positive LCL-JY cell line expressing low levels of PRAME, and HLA-A2 transduced K562 cell line (K562+A2). B, PRAME-specific T-cell clones HSS1 and HSS3 were stimulated with different UM cell lines. Uveal melanoma cell lines negative for HLA-A2 were retrovirally transduced with HLA-A2 (+ A2). Clone pp65-A2 recognizes a peptide of pp65 in the context of HLA-A2 and serves as a negative control. Clone HSS12 is reactive against the household gene USP11, and was used to confirm that the UM cell lines are HLA-A2 positive. Interferon γ production was measured after 18 hours of co-culture by standard enzyme-linked immunosorbent assay. Experiments were carried out in duplicate. C, The expression of PRAME messenger RNA (mRNA) in different cells was measured by real-time quantitative polymerase chain reaction and is shown as fold expression of the PRAME-positive cutaneous melanoma cell line Mel1.14, which was set to 1. Rel Exp indicates relative expression of PRAME.
PRAME Expression in UM Metastases
Next, we analyzed PRAME expression in 16 UM metastatic samples using an mRNA FISH assay to stain PRAME mRNA in FFPE tissue sections. We validated this technique on primary UM with known PRAME expression levels. Two consecutive FFPE tissue sections from 14 primary tumors were hybridized with both GAPDH (Ensembl: ENST00000229239.9) and PRAME in a dye-mirrored way. One section of each tumor was stained using the GAPDH probe set labeled with the dye Quasar 570 together with the PRAME probe set labeled with Quasar 670, while the other section was stained with the same probe sets but labeled the other way around. The presence of PRAME was determined as described in the eAppendix in the Supplement by 2 independent observers (M.H.M.H. and S.J.L.), with a κ score of 0.857 (Figure 1B). All tumors that were PRAME negative according to the Illumina data (<7.23) were also scored as PRAME negative by the mRNA FISH technique. All tumors with a PRAME expression level greater than 10 according to the Illumina data were scored positive by the mRNA FISH technique. We subsequently tested PRAME by mRNA FISH in metastases from 16 patients with UM. GAPDH but not PRAME expression was seen in tumors from 5 patients, whereas GAPDH as well as PRAME expression was present in metastases from 11 patients. An example of negative and positive PRAME expression by mRNA FISH is shown in Figure 3. In 2 patients, multiple metastases were available (2 in 1 patient and 4 in the second patient), and PRAME expression was identical between the different metastases of each patient. The results of all patients together demonstrate expression of PRAME in metastatic UM in 11 of 16 patients.
Figure 3. PRAME Expression by Messenger RNA (mRNA) Fluorescence In Situ Hybridization (FISH) in Uveal Melanoma (UM) Metastases.
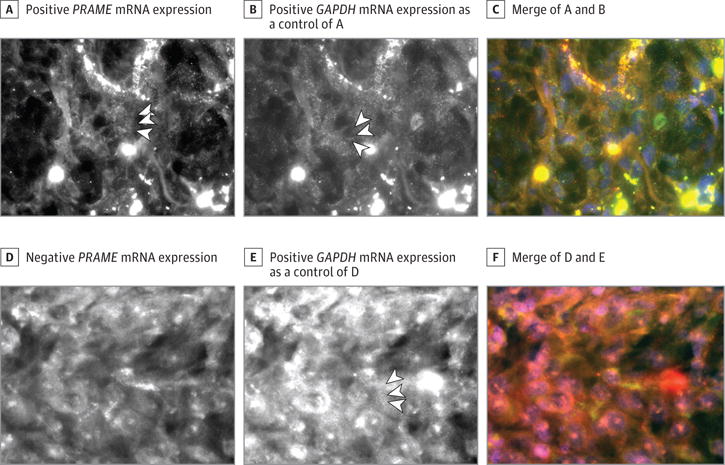
A, PRAME mRNA expression of PRAME-positive UM metastasis; PRAME mRNA is shown as small white speckles (arrowheads). B, GAPDH mRNA expression of UM metastasis shown in A; GAPDH mRNA is shown as small white speckles (arrowheads). C, Merged images with 4′,6-diamidino-2-phenylindole dilactate (DAPI) (blue) of A and B; PRAME is shown in small green dots and GAPDH in small red dots. D, PRAME-negative UM metastasis. E, GAPDH mRNA expression of UM metastasis shown in D; GAPDH mRNA is shown as small white speckles (arrowheads). F, Merged images with DAPI (blue) of D and E. GAPDH is shown in small red dots.
HLA Class I Expression in UM Metastases
For TCR-mediated immunotherapeutic approaches, target antigens are recognized in the context of HLA class I molecules. It is therefore essential that not only PRAME but also HLA class I is expressed on UM metastases to induce a potent immune reactivity of TCR-modified, PRAME-specific T cells. We therefore analyzed HLA class I expression by triple immunofluorescence HLA class I staining on the FFPE tissue sections of the 16 patients with UM metastases. When multiple metastases from a single patient were available, HLA class I expression patterns were similar between different metastases. For 10 of the 16 patients with UM metastases, expression of HLA-A and HLA-B/C, as well as β2M expression, was high and homogeneous. In 2 patients, the staining for HLA-A and HLA-B/C, as well as β2M, was weak but positive, whereas in 3 patients, expression of HLA-A and HLA-B/C, as well as of β2M, was negative. Overall, UM metastases from 10 of 16 patients (63%) demonstrated a high HLA class I expression.
As mentioned previously, PRAME expression was observed in 11 of 16 patients (69%). In 8 of the 16 patients, concomitant expression of PRAME and HLA class I was observed in the metastases, suggesting that half of metastatic UMs could be positive for both PRAME and HLA class I (Table 2) and, therefore, can potentially be candidates for treatment with PRAME-TCR gene therapy.
Table 2.
PRAME mRNA FISH Expression and HLA Class I Expression in UM Metastases
| Patient No. | Location of Metastases | PRAME mRNA FISH Expression | Staining Intensity/% Positive Tumor Cellsa | HLA Class I Expression | ||
|---|---|---|---|---|---|---|
| β2M | HLA-A | HLA-B/C | ||||
| 1 | Liver | Positive | Unknown | Unknown | Unknown | Unknown |
| 2 | Subcutaneous in breast | Positive | 3/5 | 3/5 | 3/5 | Positive |
| 3 | Liver | Positive | 3/5 | 3/5 | 3/5 | Positive |
| 4 | Cutaneous | Negative | 3/5 | 3/5 | 3/5 | Positive |
| Cutaneous | Negative | 3/5 | 3/5 | 3/5 | Positive | |
| Cutaneous | Negative | 3/5 | 3/5 | 3/5 | Positive | |
| Cutaneous | Negative | 3/5 | 3/5 | 3/5 | Positive | |
| Liver | Negative | 3/5 | 3/5 | 3/5 | Positive | |
| 5 | Liver | Positive | 3/5 | 3/5 | 3/5 | Positive |
| 6 | Liver | Positive | 3/5 | 3/5 | 3/5 | Positive |
| 7 | Liver | Positive | 3/5 | 3/5 | 3/5 | Positive |
| 8 | Liver | Negative | 3/5 | 3/5 | 3/4 | Positive |
| 9 | Bone | Positive | 3/5 | 3/5 | 3/5 | Positive |
| Bone | Positive | 3/5 | 3/5 | 3/5 | Positive | |
| 10 | Liver | Negative | 0 | Unknown | 0 | Negative |
| 11 | Liver | Negative | 1/4 | 1/4 | 1/4 | Weak |
| 12 | Mesentery of transverse colon | Negative | 0 | 0 | 0 | Negative |
| 13 | Liver | Positive | 0 | 0 | 0 | Negative |
| 14 | Liver | Positive | 3/4 | 3/4 | 3/4 | Positive |
| 15 | Lung | Positive | 2/1 | 2/1 | 2/1 | Weak |
| 16 | Liver | Positive | 3/5 | 3/5 | 3/5 | Positive |
Abbreviations: β2M, β2-microglobulin; FISH, fluorescence in situ hybridization; mRNA, messenger RNA; UM, uveal melanoma.
The staining intensity scoring system is described in the eAppendix in the Supplement.
Discussion
In the present study, we found that PRAME was expressed at the RNA level in 45% of primary UMs (29 of 64), and PRAME expression was associated with ciliary body involvement, largest basal diameter, and amplification of chromosome 8q. PRAME expression occurred in disomy 3 as well as in monosomy 3 tumors, which confirms the findings reported by Field et al.32 Among all primary UMs, PRAME expression was associated with a poorer disease-specific survival.
We have demonstrated that the high-affinity, PRAME-specific T cells are able to efficiently recognize PRAME-positive UM cell lines. Furthermore, we demonstrated that 50% of the 16 patients with UM have metastases that express both PRAME and HLA class I. In 2 patients from whom multiple metastases were available, we were able to demonstrate that PRAME expression was identical between the different metastases of each patient.
PRAME-TCR–specific T-cell therapy is directed against all patients with metastatic UM with PRAME-positive metastases, irrespective of the size of the primary tumor. The PRAME-TCR–engineered T cells used in the upcoming gene therapy study to treat acute myeloid leukemia and metastatic sarcoma recognize and lyse tumor cells when PRAME is processed and presented on the tumor cell surface in the context of HLA-A*02:01.19 Therefore, this gene therapy study will be available for all HLA-A*02:01–positive patients, which comprise 50% of the Western European and North American population. In patients with primary UM, the frequency of HLA-A*02:01 was also around 50%.33 We are currently also searching for other high-affinity, PRAME-specific TCRs restricted to other HLA class I molecules to broaden the PRAME-TCR gene therapy options. If the clinical studies using this specific TCR demonstrate promising results, we will initiate clinical studies with these newly identified PRAME-TCRs. For patients with metastatic UM, we will perform a biopsy of the metastases and analyze whether PRAME is present to determine whether to treat them with PRAME-TCR gene therapy. Any effect of this treatment on the primary tumor is unlikely because most of these patients have already undergone enucleation or radiation, and therefore less or no primary tissue should remain.
The advantage of PRAME-TCR gene therapy is that a broad group of patients with different malignant neoplasms can be treated with this TCR therapy as long as the patient is HLA-A2 positive and the tumor expresses PRAME. The PRAME-specific T cells, however, may exhibit some reactivity against mature dendritic cells and kidney epithelial cells, which express PRAME at a lower level.19 This reactivity could lead to nephrotoxicity and long-term depletion of mature dendritic cells when the PRAME-TCR therapy is applied in a clinical setting. Therefore, the PRAME-TCR–engineered T cells will be equipped with the suicide switch iCasp9 to eliminate reactive T cells in case of adverse effects. Successful elimination of iCasp9-transduced T cells has been reported in patients with graft-vs-host disease after a haploidentical stem cell transplant, leading to resolution of the graft-vs-host disease by elimination of alloreactive T cells in peripheral blood and the central nervous system.34
Limitations
This study has several limitations. First, we tested PRAME expression in a small cohort of patients with UM metastases because no other adequate FFPE tissues were available. However, we were still able to detect PRAME in 11 of 16 samples and in 8 of 16 patients together with HLA class I, thereby showing that concomitant expression of PRAME and HLA class I is present in UM metastases. Second, 2 of the 4 tumors with intermediate PRAME values (7.23–8.13) were positive with mRNA FISH, and the other 2 were scored negative (Figure 1B), thereby showing that the sensitivity of our PRAME probes might not be high enough to demonstrate PRAME gene expression at low levels with 100% sensitivity. The detection of PRAME might be degraded in these tissues because we used FFPE samples. Most important, however, no tumors with a negative expression in the microarray were scored by the mRNA FISH as PRAME positive, thereby providing high specificity.
Conclusions
We found that 45% of the tested primary UMs express PRAME and that PRAME expression in these patients is associated with known risk factors for metastasis and with poorer overall survival. We found that 50% of the analyzed UM metastases expressed both PRAME and HLA class I and that high-affinity, PRAME-specific T cells were efficiently recognizing UM cell lines expressing PRAME. These findings provide supportive evidence for including patients with metastatic UM in clinical trials using PRAME-TCR gene therapy.
Supplementary Material
Key Points.
Question
Is there a rationale for preferentially expressed antigen in melanoma (PRAME)–specific T-cell therapy for patients with metastatic uveal melanoma?
Findings
This experimental cohort study found that PRAME is expressed in primary uveal melanoma and its metastases, as is HLA class I. Also, PRAME-specific T cells showed reactivity against PRAME-positive uveal melanoma cell lines.
Meaning
For patients with metastatic uveal melanoma, the use of PRAME-specific T-cell therapy may be helpful.
Acknowledgments
Funding/Support: This project was supported by Algemene Nederlandse Vereniging Ter Voorkoming van Blindheid (Dr Jager), Stichting Blinden Penning (Dr Jager), Landelijke Stichting voor Blinden en Slechtzienden (Dr Jager), Novartis Foundation (Dr Jager), Rotterdamse Stichting Blindenbelangen (Dr Jager), a Minerva fellowship of the Leiden University Medical Center Kankerfonds (Dr Heemskerk), Horizon2020 CURE UM 667787 (Dr Jager), grant R01 CA125970 from the National Cancer Institute (Dr Harbour), grant F30 CA206430 from Research to Prevent Blindness, Inc (Mr Field), a Senior Scientific Investigator Award (Dr Harbour), and the Alcon Research Institute (Dr Harbour). The Bascom Palmer Eye Institute also received funding from core grant P30EY014801 from the National Institutes of Health, grant W81XWH-13-1-0048 from the Department of Defense Grant, and an unrestricted grant from Research to Prevent Blindness.
Role of the Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and the decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Gezgin and Jager had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Dr Gezgin and Mr Luk are joint first authors. Drs Heemskerk and Jager are joint senior authors.
Study concept and design: Gezgin, Luk, Field, Luyten, Szuhai, Harbour, Heemskerk, Jager.
Acquisition, analysis, or interpretation of data: Gezgin, Luk, Cao, Dogrusöz, van der Steen, Hagedoorn, Krijgsman, van der Velden, Field, Szuhai, Jordanova, Heemskerk, Jager.
Drafting of the manuscript: Gezgin, Luk, van der Steen, Hagedoorn, Field, Szuhai, Jordanova, Heemskerk, Jager.
Critical revision of the manuscript for important intellectual content: Gezgin, Luk, Cao, Dogrusöz, Krijgsman, van der Velden, Luyten, Harbour, Heemskerk, Jager.
Statistical analysis: Gezgin, Dogrusöz, Krijgsman.
Obtained funding: Heemskerk, Jager.
Administrative, technical, or material support: Gezgin, Luk, Cao, van der Steen, Hagedoorn, van der Velden, Jordanova.
Study supervision: Luyten, Szuhai, Harbour, Jordanova, Heemskerk, Jager.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Dr Harbour reported being the inventor of intellectual property related to gene expression profiling and BAP1 mutations in uveal melanoma and receiving royalties from its commercialization and serving as a paid consultant for Castle Biosciences, licensee of this technology. No other disclosures were reported.
References
- 1.Damato B, Duke C, Coupland SE, et al. Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology. 2007;114(10):1925–1931. doi: 10.1016/j.ophtha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Versluis M, de Lange MJ, van Pelt SI, et al. Digital PCR validates 8q dosage as prognostic tool in uveal melanoma. PLoS One. 2015;10(3):e0116371. doi: 10.1371/journal.pone.0116371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64(20):7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onken MD, Worley LA, Char DH, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119(8):1596–1603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blom DJ, Luyten GP, Mooy C, Kerkvliet S, Zwinderman AH, Jager MJ. Human leukocyte antigen class I expression: marker of poor prognosis in uveal melanoma. Invest Ophthalmol Vis Sci. 1997;38(9):1865–1872. [PubMed] [Google Scholar]
- 6.Maat W, Ly LV, Jordanova ES, de Wolff-Rouendaal D, Schalij-Delfos NE, Jager MJ. Monosomy of chromosome 3 and an inflammatory phenotype occur together in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49(2):505–510. doi: 10.1167/iovs.07-0786. [DOI] [PubMed] [Google Scholar]
- 7.Mäkitie T, Summanen P, Tarkkanen A, Kivelä T. Tumor-infiltrating macrophages (CD68+ cells) and prognosis in malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2001;42(7):1414–1421. [PubMed] [Google Scholar]
- 8.Bronkhorst IH, Ly LV, Jordanova ES, et al. Detection of M2-macrophages in uveal melanoma and relation with survival. Invest Ophthalmol Vis Sci. 2011;52(2):643–650. doi: 10.1167/iovs.10-5979. [DOI] [PubMed] [Google Scholar]
- 9.Bronkhorst IH, Vu TH, Jordanova ES, Luyten GP, Burg SH, Jager MJ. Different subsets of tumor-infiltrating lymphocytes correlate with macrophage influx and monosomy 3 in uveal melanoma. Invest Ophthalmol Vis Sci. 2012;53(9):5370–5378. doi: 10.1167/iovs.11-9280. [DOI] [PubMed] [Google Scholar]
- 10.Blum ES, Yang J, Komatsubara KM, Carvajal RD. Clinical management of uveal and conjunctival melanoma. Oncology (Williston Park) 2016;30(1):29–32. 34–43, 48. [PubMed] [Google Scholar]
- 11.Pereira PR, Odashiro AN, Lim LA, et al. Current and emerging treatment options for uveal melanoma. Clin Ophthalmol. 2013;7:1669–1682. doi: 10.2147/OPTH.S28863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luke JJ, Callahan MK, Postow MA, et al. Clinical activity of ipilimumab for metastatic uveal melanoma: a retrospective review of the Dana-Farber Cancer Institute, Massachusetts General Hospital, Memorial Sloan-Kettering Cancer Center, and University Hospital of Lausanne experience. Cancer. 2013;119(20):3687–3695. doi: 10.1002/cncr.28282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maio M, Danielli R, Chiarion-Sileni V, et al. Efficacy and safety of ipilimumab in patients with pre-treated, uveal melanoma. Ann Oncol. 2013;24(11):2911–2915. doi: 10.1093/annonc/mdt376. [DOI] [PubMed] [Google Scholar]
- 14.Zimmer L, Vaubel J, Mohr P, et al. Phase II DeCOG-study of ipilimumab in pretreated and treatment-naïve patients with metastatic uveal melanoma. PLoS One. 2015;10(3):e0118564. doi: 10.1371/journal.pone.0118564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field MG, Decatur CL, Kurtenbach S, et al. PRAME as an independent biomarker for metastasis in uveal melanoma. Clin Cancer Res. 2016;22(5):1234–1242. doi: 10.1158/1078-0432.CCR-15-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda H, Lethé B, Lehmann F, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6(2):199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 17.Epping MT, Bernards R. A causal role for the human tumor antigen preferentially expressed antigen of melanoma in cancer. Cancer Res. 2006;66(22):10639–10642. doi: 10.1158/0008-5472.CAN-06-2522. [DOI] [PubMed] [Google Scholar]
- 18.Griffioen M, Kessler JH, Borghi M, et al. Detection and functional analysis of CD8+ T cells specific for PRAME: a target for T-cell therapy. Clin Cancer Res. 2006;12(10):3130–3136. doi: 10.1158/1078-0432.CCR-05-2578. [DOI] [PubMed] [Google Scholar]
- 19.Amir AL, van der Steen DM, van Loenen MM, et al. PRAME-specific allo-HLA-restricted T cells with potent antitumor reactivity useful for therapeutic T-cell receptor gene transfer. Clin Cancer Res. 2011;17(17):5615–5625. doi: 10.1158/1078-0432.CCR-11-1066. [DOI] [PubMed] [Google Scholar]
- 20.World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 21.de Lange MJ, van Pelt SI, Versluis M, et al. Heterogeneity revealed by integrated genomic analysis uncovers a molecular switch in malignant uveal melanoma. Oncotarget. 2015;6(35):37824–37835. doi: 10.18632/oncotarget.5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Waard-Siebinga I, Blom DJ, Griffioen M, et al. Establishment and characterization of an uveal-melanoma cell line. Int J Cancer. 1995;62(2):155–161. doi: 10.1002/ijc.2910620208. [DOI] [PubMed] [Google Scholar]
- 23.Verbik DJ, Murray TG, Tran JM, Ksander BR. Melanomas that develop within the eye inhibit lymphocyte proliferation. Int J Cancer. 1997;73(4):470–478. doi: 10.1002/(sici)1097-0215(19971114)73:4<470::aid-ijc3>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 24.Chen PW, Murray TG, Uno T, Salgaller ML, Reddy R, Ksander BR. Expression of MAGE genes in ocular melanoma during progression from primary to metastatic disease. Clin Exp Metastasis. 1997;15(5):509–518. doi: 10.1023/a:1018479011340. [DOI] [PubMed] [Google Scholar]
- 25.Heemskerk MH, de Paus RA, Lurvink EG, et al. Dual HLA class I and class II restricted recognition of alloreactive T lymphocytes mediated by a single T cell receptor complex. Proc Natl Acad Sci U S A. 2001;98(12):6806–6811. doi: 10.1073/pnas.111162298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amir AL, van der Steen DM, Hagedoorn RS, et al. Allo-HLA-reactive T cells inducing graft-versus-host disease are single peptide specific. Blood. 2011;118(26):6733–6742. doi: 10.1182/blood-2011-05-354787. [DOI] [PubMed] [Google Scholar]
- 27.van Loenen MM, de Boer R, van Liempt E, et al. A Good Manufacturing Practice procedure to engineer donor virus–specific T cells into potent anti-leukemic effector cells. Haematologica. 2014;99(4):759–768. doi: 10.3324/haematol.2013.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Esch EM, Tummers B, Baartmans V, et al. Alterations in classical and nonclassical HLA expression in recurrent and progressive HPV-induced usual vulvar intraepithelial neoplasia and implications for immunotherapy. Int J Cancer. 2014;135(4):830–842. doi: 10.1002/ijc.28713. [DOI] [PubMed] [Google Scholar]
- 29.Ruiter DJ, Ferrier CM, van Muijen GN, et al. European BIOMED-1 Concerted Action on Clinical Relevance of Proteases in Tumour Invasion and Metastasis Quality control of immunohistochemical evaluation of tumour-associated plasminogen activators and related components. Eur J Cancer. 1998;34(9):1334–1340. doi: 10.1016/s0959-8049(98)00151-8. [DOI] [PubMed] [Google Scholar]
- 30.Mehta AM, Jordanova ES, Kenter GG, Ferrone S, Fleuren GJ. Association of antigen processing machinery and HLA class I defects with clinicopathological outcome in cervical carcinoma. Cancer Immunol Immunother. 2008;57(2):197–206. doi: 10.1007/s00262-007-0362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordanova ES, Gorter A, Ayachi O, et al. Human leukocyte antigen class I, MHC class I chain–related molecule A, and CD8+/regulatory T-cell ratio: which variable determines survival of cervical cancer patients? Clin Cancer Res. 2008;14(7):2028–2035. doi: 10.1158/1078-0432.CCR-07-4554. [DOI] [PubMed] [Google Scholar]
- 32.Field MG, Durante MA, Decatur CL, et al. Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas. Oncotarget. 2016;7(37):59209–59219. doi: 10.18632/oncotarget.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maat W, Haasnoot GW, Claas FH, Schalij-Delfos NE, Schreuder GM, Jager MJ. HLA class I and II genotype in uveal melanoma: relation to occurrence and prognosis. Invest Ophthalmol Vis Sci. 2006;47(1):3–6. doi: 10.1167/iovs.05-1122. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Dotti G, Krance RA, et al. Inducible caspase-9 suicide gene controls adverse effects from alloreplete T cells after haploidentical stem cell transplantation. Blood. 2015;125(26):4103–4113. doi: 10.1182/blood-2015-02-628354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


