Abstract
Blimp-1 expression in T cells extinguishes the fate of T follicular helper cells, drives terminal differentiation, and limits autoimmunity. Although various factors have been described to control Blimp-1 expression in T cells, little is known about what regulates Blimp-1 expression in T helper 2 (TH2) cells and the molecular basis of its actions. We report that signal transducer and activator of transcription 3 (STAT3) unexpectedly played a critical role in regulating Blimp-1 in TH2 cells. Furthermore, we found that the cytokine interleukin-10 (IL-10) acted directly on TH2 cells and was necessary and sufficient to induce optimal Blimp-1 expression through STAT3. Together, Blimp-1 and STAT3 amplified IL-10 production in TH2 cells, creating a strong autoregulatory loop that enhanced Blimp-1 expression. Increased Blimp-1 in T cells antagonized STAT5-regulated cell cycle and antiapoptotic genes to limit cell expansion. These data elucidate the signals required for Blimp-1 expression in TH2 cells and reveal an unexpected mechanism of action of IL-10 in T cells, providing insights into the molecular underpinning by which Blimp-1 constrains T cell expansion to limit autoimmunity.
INTRODUCTION
Blimp-1 is a transcriptional repressor with global roles in regulating cellular differentiation (1). First identified as the master regulator associated with B cell differentiation into plasma cells, Blimp-1 has now been described as a critical regulator of several other cell types (2, 3). In T cells, Blimp-1 has been shown to antagonize T follicular helper cell (TFH) differentiation, control interleukin-10 (IL-10) expression in regulatory T (Treg) cells and T helper 1 (TH1) cells, and promote differentiation and function of cytotoxic T lymphocytes (4–8). Furthermore, recent studies have found a critical role for Blimp-1 in driving the inflammatory phenotype associated with IL-23–induced TH17 cells (9).
In CD8 T cells, Blimp-1 is required for the differentiation of shortlived effector cells after viral infection and highly expressed in exhausted T cells induced in response to chronic viral infection (10). Consistent with this, its absence in CD8 effector T cells causes expansion of memory cells, suggesting that Blimp-1 is important for effector cell homeostasis (4, 11). Paradoxically, though, conditional deletion of Blimp-1 in all T cells causes accumulation of effector T cells and associated systemic, fatal autoimmunity, arguing that Blimp-1 limits effector T cell function (12, 13). Polymorphisms of PRDM1 are linked to multiple autoimmune diseases, including Crohn’s disease, ulcerative colitis, and systemic lupus erythematosus (14–18). Together, it appears that Blimp-1 is important for the development of terminally differentiated effector cells, while simultaneously preventing autoimmunity. How Blimp-1 regulates these processes remains poorly understood, and limited mechanistic studies have explored the molecular basis of Blimp-1’s actions.
Although Blimp-1 in T cells has been described in several T cell subsets, including TH1, TH2, TH17, Treg, and T follicular regulatory cells, the signals that regulate the expression of Blimp-1 within each T cell subset remain unclear. In immune cells, transcription factors are frequently regulated by exogenous signals, especially cytokines. Many cytokines exert their effect through members of the signal transducer and activator of transcription (STAT) family. This is certainly the case for T-bet, GATA3, Rorγt, and Bcl6, all of which are important STAT target genes (19, 20). Therefore, several studies have explored which cytokines and STATs are responsible for Blimp-1 induction. In TH1 cells, IL-12 via STAT4 is critical to TH1 differentiation and has also been shown to drive Blimp-1 expression in TH1 cells in an in vivo model (8). In a similar manner, the cytokine IL-23, which is known to promote inflammatory TH17 cells, can drive Blimp-1 in TH17 cells through STAT3 (9). Last, because IL-2 via STAT5 can suppress differentiation of TFH cells, some evidence suggests that the IL-2/STAT5 pathway can drive Blimp-1 expression, which subsequently represses TFH cell development (6, 7). In summary, several cytokines and STAT pathways have been described to promote Blimp-1 expression in various T cell subsets; however, the signals that regulate Blimp-1 expression in TH2 cells are unknown.
In this study, we set out to determine how Blimp-1 is regulated and functions in CD4 T cells. We uncovered a role for STAT3 downstream of IL-10 stimulation in regulating Blimp-1 in TH2 cells. In addition, we found that Blimp-1 expression antagonized STAT5 induction of key T cell survival genes in CD4 T cells, suggesting a previously unappreciated link between IL-10 and Blimp-1 and providing new mechanisms by which Blimp-1 limits autoimmunity.
RESULTS
Blimp-1 expression in TH2 cells requires STAT3
To identify factors important in controlling Blimp-1 expression in TH2 cells, we first determined optimal conditions that drive expression of Blimp-1 in in vitro–generated TH2 cells. We activated naïve CD4 T cells isolated from Blimp-1 YFP (yellow fluorescent protein) reporter mice and determined YFP levels over time as a surrogate for the induction of Blimp-1 itself. Both TH1- and TH2-inducing conditions led to the potent expression of Blimp-1 after 7 to 8 days in culture, consistent with previous reports that Blimp-1 induction occurs late in differentiation after induction of lineage-specific cytokines and transcription factors (fig. S1) (21). No Blimp-1 was evident under conditions in which only IL-2 was added, despite previous reports suggesting that IL-2 is a major inducer of Blimp-1 (7, 21, 22).
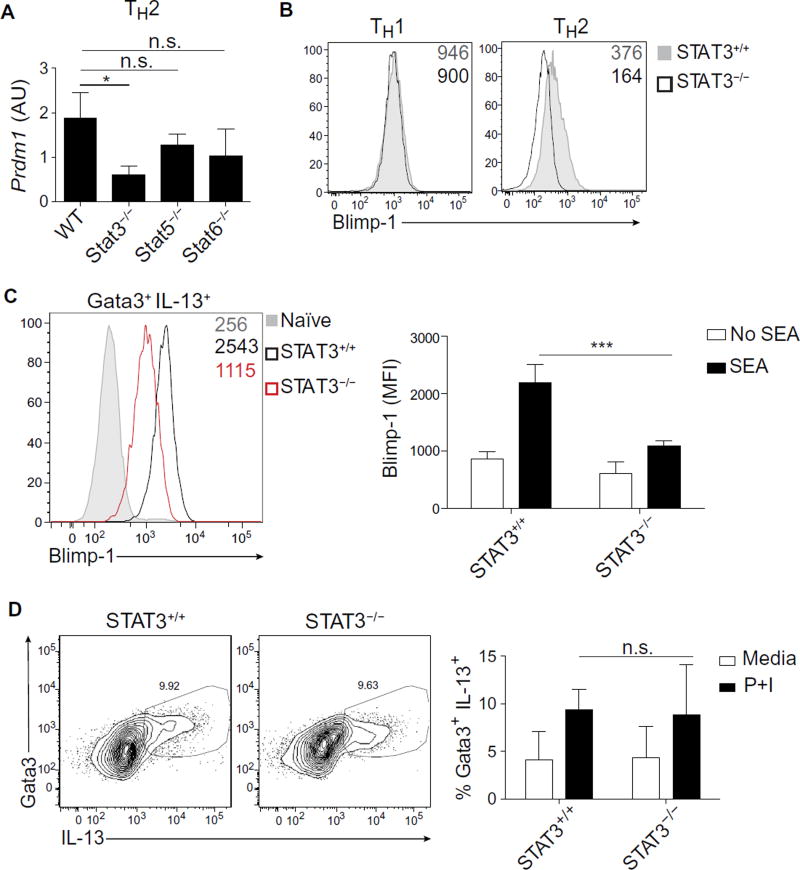
We next explored which of the major factors contributing to TH2 cell differentiation were required for Blimp-1 induction. We first measured Blimp-1 transcripts in the absence of STAT5 or STAT6, which are transcription factors known to be critical for TH2 cell development (20). Neither STAT5 nor STAT6 deficiency had a major effect on Blimp-1 induction (Fig. 1A). We next considered a possible role for STAT3, given that it is also activated in TH2 cells and contributes to TH2 cell differentiation (23). We found that the absence of STAT3 substantially decreased Blimp-1 expression at the level of both mRNA and protein (Fig. 1, A and B). These data were intriguing in light of recent reports that STAT3 can also drive Blimp-1 expression in TH17 cells stimulated with IL-23 (9). Contrary to our expectations, Blimp-1 expression was unaltered in the absence of STAT3 in TH1 cells (Fig. 1B). In contrast to TH2 cells, TH1 cells relied on the STAT1–STAT4–T-bet axis for the induction of Blimp-1, in agreement with previously published reports, suggesting that TH1 cells up-regulate Blimp-1 as part of a global TH1 differentiation program (fig. S2) (8). The fact that TH1 cells rely predominantly on STAT4 whereas TH2 and TH17 cells rely on STAT3 suggests that the Blimp-1 locus (Prdm1) is regulated in a cell type–specific or context-dependent manner.
Fig. 1. STAT3 is required for Blimp-1 induction in TH2 cells.
(A) Expression of Blimp-1 measured by qPCR in STAT+/+ or STAT−/− TH2 cells at day 8. Data are means ± SD. n = 2; *P = 0.0238. WT, wild type; n.s., not significant. AU, arbitrary units. (B) Expression of Blimp-1 protein measured by intracellular flow cytometry in STAT3+/+ or STAT3−/− under indicated conditions at day 7 after activation. Geometric mean fluorescence intensity (MFI) of YFP was shown on the top right of each flow plot. Data are representative of three independent experiments. (C) Expression of Blimp-1 protein measured by intracellular flow cytometry in TH2 cells (Gata3+ IL-13+) isolated from STAT3+/+ or STAT3−/− hosts 9 days after infection with SEA. MFI of Blimp-1 is shown on the top right of the flow plot. Quantification of Blimp-1 MFI is shown on the right. Data are means ± SD (n = 4 to 7) and are representative of two independent experiments. ***P = 0.0002. (D) Percentage of TH2 cells (Gata3+ IL-13+) isolated from STAT3+/+ or STAT3−/− hosts 9 days after infection with SEA. Cells were restimulated with PMA and ionomycin (P+I) for 6 hours in vitro. Quantification of percent TH2 cells is presented as means ± SD (n = 4 to 7) and is representative of two independent experiments.
To establish the idea that STAT3 promotes Blimp-1 expression in TH2 cells in vivo, we immunized mice with soluble egg antigen (SEA) from Schistosoma mansoni, which is known to generate a robust TH2 response (24, 25). Blimp-1 was highly induced in Gata3+ IL-13+ T cells present in the draining lymph node 9 days after SEA immunization (Fig. 1C). STAT3-deficient T cells exhibited substantially lower levels of Blimp-1 compared with STAT3-intact T cells, suggesting that STAT3 is critically important for Blimp-1 expression in TH2 cells both in vitro and in vivo. Despite substantial reduction in Blimp-1 levels in the absence of STAT3, we observed normal TH2 differentiation as measured by Gata3 and IL-13 expression, suggesting that STAT3 and subsequent Blimp-1 expression are not required for TH2 differentiation per se (Fig. 1D). Together, these data suggest that STAT3 is a critical regulator of Blimp-1 in TH2 cells.
IL-10 acting via STAT3 is necessary and sufficient for maximal Blimp-1 expression in TH2 cells
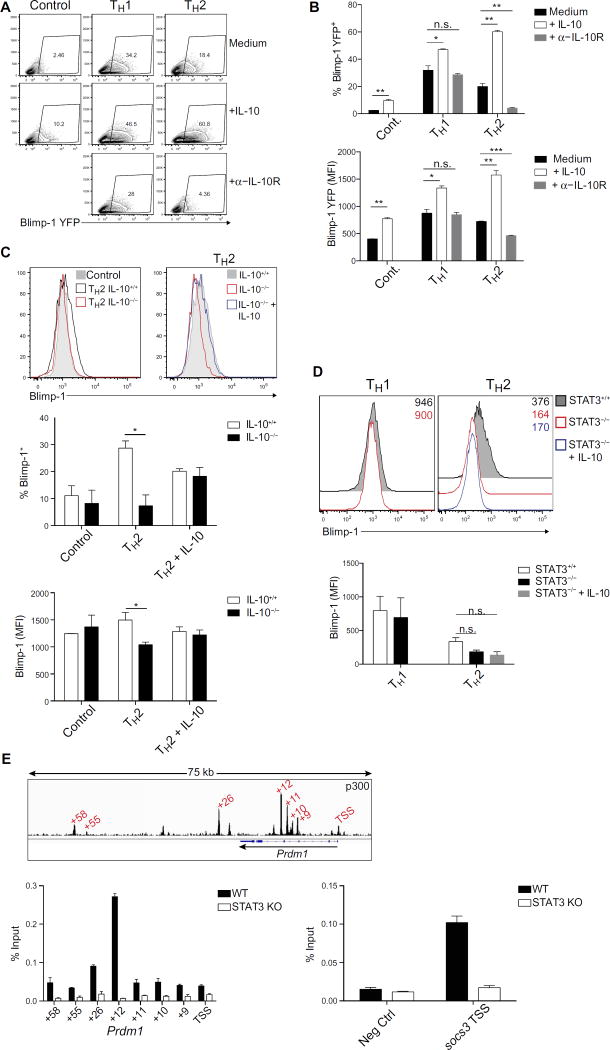
One of the major cytokines produced by TH2 cells is IL-10, which is also a potent STAT3-activating cytokine (26, 27). Therefore, we next explored whether IL-10 was one of the relevant factors responsible for Blimp-1 induction in TH2 cells. Addition of exogenous IL-10 increased Blimp-1 expression in TH1 and TH2 cells early during differentiation when Blimp-1 levels are submaximal (Fig. 2, A and B). Moreover, blockade of the IL-10 receptor had the most pronounced effect on TH2 cells, suggesting a role for autocrine/paracrine IL-10 signaling. Exogenous IL-10 had little effect on Blimp-1 later in differentiation when expression was maximal. However, blockade of the IL-10 receptor still profoundly affected Blimp-1 expression in TH2 cells, substantiating the importance of this cytokine (fig. S3A). In TH1 cells, addition of IL-10 also resulted in modest increases in Blimp-1. However, in contrast to TH2 cells, blockade of IL-10 signaling had no effect on Blimp-1 expression, arguing that the IL-12/STAT4 axis plays a predominant role in regulating Blimp-1 expression in TH1 cells. Hence, IL-10 can support Blimp-1 up-regulation in TH1 cells but is not necessary for its induction; however, it is both necessary and sufficient for maximal Blimp-1 induction in TH2 cells (Fig. 2, A and B). Differences in IL-10 receptor expression or the ability to signal downstream of IL-10 through phosphorylated STAT3 (phospho-STAT3) activation did not explain why IL-10 more profoundly affected Blimp-1 induction in TH2 cells compared to TH1 cells (fig. S3, B to D). Together, these data support a model where TH1 cells use STAT4 to drive Blimp-1 induction, and additional STAT3 activation can modestly support expression; however, TH2 cells exclusively rely on STAT3 to induce Blimp-1.
Fig. 2. IL-10, acting via STAT3, is necessary and sufficient for Blimp-1 induction in TH2 cells.
(A) Expression of YFP in T cells at day 5 under indicated conditions with or without the addition of IL-10 (10 ng/ml) or α–IL-10R (10 µg/ml) throughout the culture. Data are representative of three independent experiments. (B) Quantification of percent YFP and MFI of (A). Data are means ± SD; n = 2. Percent YFP: Control, **P = 0.0063; TH1, *P = 0.0242; TH2 + IL-10, **P = 0.0017; TH2 + α–IL-10R, **P = 0.0097. MFI YFP: Control, **P = 0.0019; TH1, *P = 0.0131; TH2 + IL-10, **P = 0.0050; TH2 + α–IL-10R, ***P = 0.0008. (C) Expression of Blimp-1 protein measured by intracellular flow cytometry in T cells isolated from IL-10+/+ or IL-10−/− mice cultured for 7 days under indicated conditions with or without exogenous IL-10 (10 ng/ml). Quantification of percent YFP and MFI is presented as means ± SD (n = 2) and is representative of two independent experiments. Percent YFP: *P = 0.0248. MFI YFP: *P = 0.0493. (D) Expression of Blimp-1 protein measured by intracellular flow cytometry in STAT3+/+ or STAT3−/− TH2 cells cultured for 7 days with or without exogenous IL-10 (10 ng/ml). MFI of Blimp-1 is shown on the top right of the flow plot. Graph quantifies Blimp-1 MFI across multiple samples. Data are representative of three independent experiments. (E) ChIP-qPCR of STAT3 binding to the Prdm1 locus. Top: p300 binding at Prdm1 in TH2 cells. Sites tested for STAT3 are marked in red. Middle: STAT3 ChIP-qPCR at Prdm1 locus. Data are representative of three independent experiments. Bottom: STAT3 ChIP-qPCR at the negative control site and the positive control site, the Socs3 promoter. Data are representative of three independent experiments. TSS, transcription start site; KO, knockout.
To confirm that IL-10 production by TH2 cells was essential for their ability to acquire optimal Blimp-1 expression, we cultured naïve CD4+ T cells from wild-type and IL-10–deficient mice and measured Blimp-1 expression. IL-10–deficient TH2 cells failed to up-regulate Blimp-1 normally (Fig. 2C, left), and addition of exogenous IL-10 to IL-10–deficient TH2 cells restored Blimp-1 protein expression (Fig. 2C, right). Last, although STAT3 is thought to be the main STAT downstream of IL-10, IL-10 can also activate other STATs (28). We therefore confirmed that STAT3 was solely required for IL-10–dependent induction of Blimp-1 in TH2 cells by demonstrating that, in STAT3-deficient cells, Blimp-1 was not induced by exogenous IL-10 (Fig. 2D).
Because STAT3 was critical for Blimp-1 induction in TH2 cells, we examined whether this effect was direct by assessing whether STAT3 bound the Prdm1 locus. We found that STAT3 bound to multiple sites within the Prdm1 locus, including the promoter and several enhancers also bound by p300 (Fig. 2E). STAT3 binding was strongest at locations where p300, a mark of active enhancers, was most strongly bound in TH2 cells (+12, +26). Therefore, STAT3 appears to be a direct regulator of Blimp-1 expression in TH2 cells after stimulation with IL-10. Collectively, these data show that IL-10 acting via STAT3 is requisite for in vitro control of Blimp-1 expression in TH2 cells.
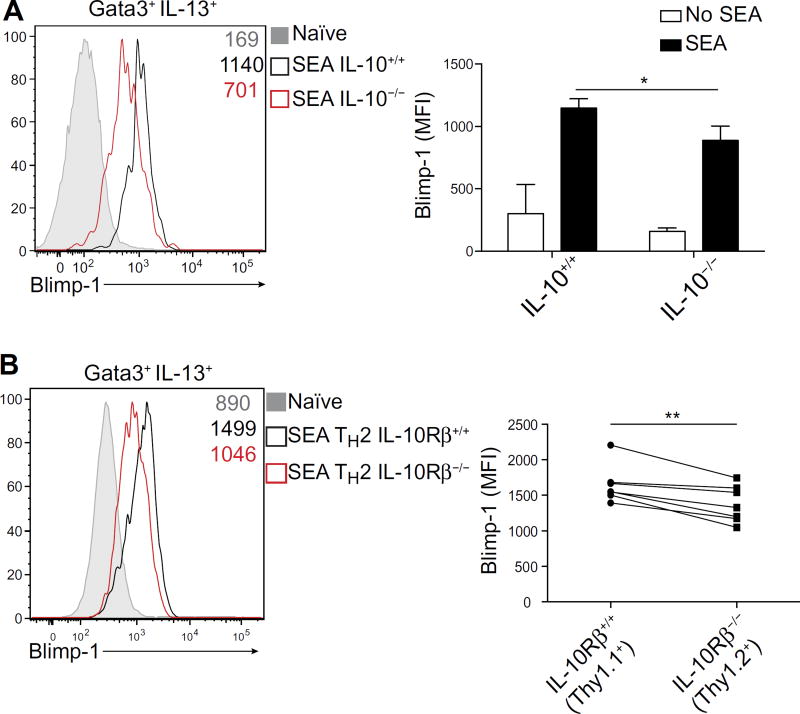
We next sought to determine whether these aspects of Blimp-1 regulation were operative in vivo. We therefore immunized IL-10–intact or IL-10–deficient mice with SEA and found that Blimp-1 was robustly induced in Gata3+ IL-13+ cells from control mice. In contrast, TH2 cells from IL-10–deficient mice had substantially reduced Blimp-1 expression (Fig. 3A). Blimp-1 expression was not as severely affected in the absence of IL-10 compared with cells lacking STAT3, suggesting that additional STAT3-activating cytokines may contribute to Blimp-1 induction in TH2 cells in vivo. Nonetheless, the data indicate that IL-10 and STAT3 are required for optimal Blimp-1 expression.
Fig. 3. IL-10 induces optimal Blimp-1 expression in TH2 cells in vivo.
(A) Expression of Blimp-1 protein in TH2 cells (Gata3+ IL-13+) isolated from Il10+/+ or Il10−/− hosts 9 days after infection with SEA. Quantification of Blimp-1 MFI is shown on the right. Data are means ±SD(n=5)and are representative of two independent experiments. *P = 0.032. (B) Expression of Blimp-1 protein in TH2 cells (Gata3+ IL-13+) isolated from bone marrow chimeras reconstituted with a 50:50 mix of Il10Rβ+/+ (Thy1.1+) or Il10Rβ−/−(Thy1.2+) cells 9 days after infection with SEA. Quantification of Blimp-1 MFI is shown on the right; data are means ± SD (n = 7), using paired t test. Data are representative of two independent experiments. **P = 0.0032.
Our findings demonstrated that IL-10 was a relevant regulator of Blimp-1 expression; however, the approach did not establish whether IL-10 was exerting its effect directly or indirectly on T cells. To answer this question, we generated chimeric mice by transplanting immunodeficient recipients with congenically marked control and IL-10Rβ (IL-10 receptor β)–deficient (Thy1.2+) bone marrow, which we then immunized with SEA. Nine days after immunization, we found that Gata3+ IL-13+ control T cells (Thy1.1+) robustly expressed Blimp-1, consistent with our previous in vivo experiments. However, T cells lacking IL-10Rβ (Thy1.2+) expressed substantially less Blimp-1 (Fig. 3B). Together, our results indicate that IL-10, acting directly on effector T cells, activates STAT3 to drive optimal Blimp-1 expression in TH2 cells both in vitro and in vivo.
IL-10 drives its own expression through STAT3
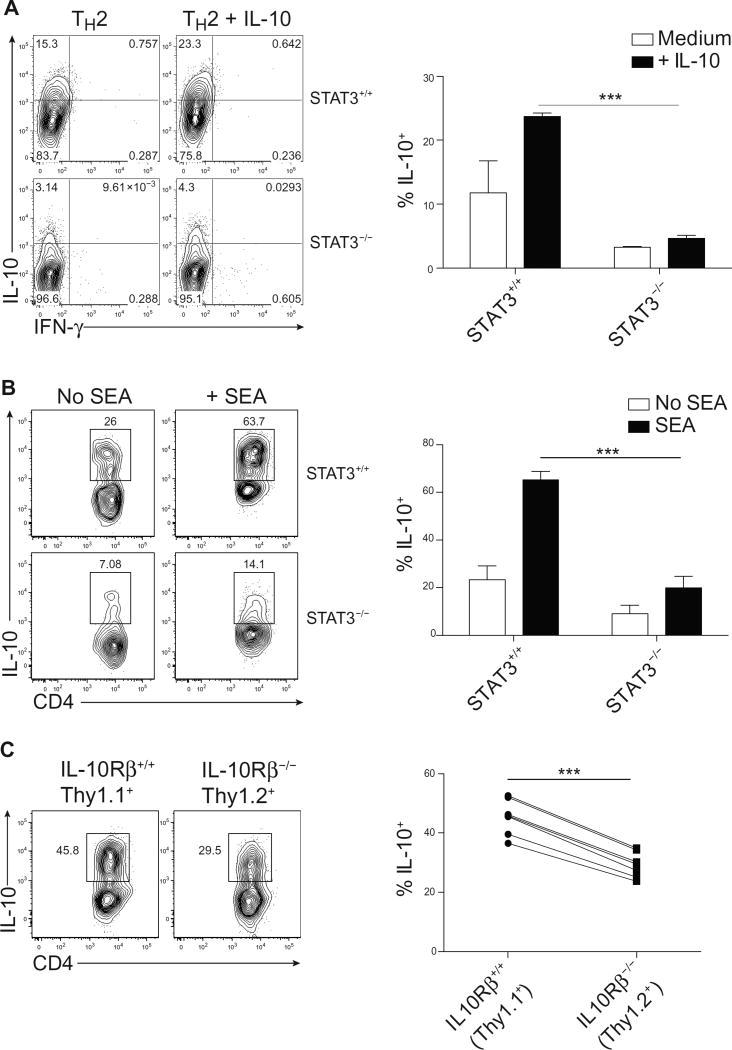
IL-10 is known to be an important product of TH2 cells (26, 28), and having defined IL-10 as an activator of STAT3 and an inducer of Blimp1, we next examined the effect of STAT3 on IL-10 expression itself in TH2 cells. STAT3 is known to control IL-10 expression in T cells downstream of IL-27 and IL-6; thus, we wondered whether STAT3 might also be responsible for controlling IL-10 expression in TH2 cells (29). Therefore, we compared IL-10 production in in vitro–generated control and STAT3-deficient TH2 cells. Although STAT3-deficient TH2 cells up-regulated Gata3 and normally produced IL-13 and IL-4 (fig. S4), IL-10 production was markedly reduced (Fig. 4A). The addition of exogenous IL-10 to control TH2 cells slightly increased IL-10 and IL-4 expression. However, addition of IL-10 to STAT3-deficient T cells failed to promote IL-10 production, establishing the requirement of STAT3 for the effects of exogenous IL-10 in TH2 cells.
Fig. 4. STAT3 promotes IL-10 expression in TH2 cells.
(A) Expression of IL-10 in STAT3+/+ or STAT3−/− TH2 cells cultured for 7 days with or without IL-10 (10 ng/ml). Percent quantified is shown on the right; data are means ± SD (n = 2) and are representative of three independent experiments. ***P = 0.0007. (B) Expression of IL-10 in TH2 cells (Gata3+ IL-13+) isolated from STAT3+/+ or STAT3−/− hosts 9 days after infection with SEA and restimulated in vitro with cognate antigen for 48 hours. Cells were additionally stimulated with PMA and ionomycin for 2 hours for IL-10 staining. Quantification of IL-10 is shown on the right. Data are means±SD (n = 4 to 7) and are representative of two independent experiments. ***P <0.0001. (C) Expression of IL-10 in TH2 cells isolated from bone marrow chimeras reconstituted with a 50:50 mix of Il10Rβ+/+ (Thy1.1+) or Il10Rβ−/−(Thy1.2+) cells 9 days after infection with SEA and restimulated in vitro with cognate antigen for 48 hours. Cells were additionally stimulated with PMA and ionomycin for 2 hours for IL-10 staining. Quantification of IL-10 is shown on the right; data are means ± SD (n = 7), using a paired t test. Data are representative of two independent experiments. ***P < 0.0001.
We next sought to assess the importance of STAT3 in regulating IL-10 expression in TH2 cells in vivo. IL-10 production by TH2 cells generated in response to SEA immunization also required STAT3, because STAT3-deficient TH2 cells had markedly reduced expression of this cytokine (Fig. 4B). These data argue that, in TH2 cells, STAT3 is required for T cell IL-10 production, which in turn reinforces its own expression. To determine whether IL-10 acts in an autocrine/paracrine manner to drive expression of itself in effector T cells, we compared IL-10 production in TH2 cells in transplanted chimeric mice bearing IL-10Rβ–intact (Thy1.1+) and IL-10Rβ–deficient (Thy1.2+) T cells that were immunized with SEA. As expected, Gata3+ IL-13+ IL-10Rβ–expressing cells (Thy1.1+) robustly produced IL-10 (Fig. 4C). However, IL-10 expression was substantially reduced, though not completely absent, in T cells lacking IL-10Rβ (Thy1.2+). This indicated that other STAT3-activating cytokines may contribute to Blimp-1 and IL-10 production in vivo but that IL-10 is critical for the optimal production of itself.
Blimp-1 is required for IL-10 expression but not TH2 differentiation
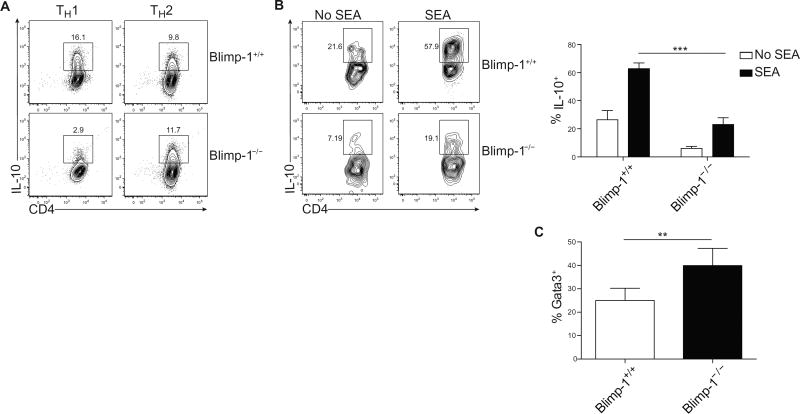
In light of the finding that STAT3 could induce both IL-10 and Blimp-1 expression, we postulated that IL-10 and Blimp-1 could create an autoregulatory loop (5, 8, 13, 30). We first determined the role of Blimp-1 in regulating IL-10 production in TH1 and TH2 cells in vitro by using mice in which Blimp-1 was specifically deleted in T cells (Blimp-1−/−). Although TH1 cells do not require IL-10 for the expression of Blimp-1 (Fig. 2A), the converse is the case: Blimp-1 is necessary for IL-10 expression, consistent with evidence for TH1 cells in vivo (Fig. 5A) (8). In contrast, although TH2 cells require IL-10 for Blimp-1 expression, Blimp-1 is not necessary for IL-10 expression in in vitro–generated TH2 cells. These data suggested that TH1 and TH2 cells differentially regulate IL-10 and Blimp-1 expression, again suggesting that the control of these genes is context-dependent. Given the data indicating a requirement for Blimp-1 in controlling IL-10 expression in TH1 cells, and our data in TH2 cells arguing against this paradigm, we next wanted to determine whether Blimp-1 was entirely dispensable in TH2 cells for the production of IL-10 both in vitro and in vivo. To this end, we immunized Blimp-1−/− mice with SEA and found, in contrast to our in vitro results (Fig. 5A), a substantial reduction, but not a complete absence, of IL-10 production in Blimp-1−/− TH2 cells. This suggests that Blimp-1 can regulate IL-10 expression in TH2 cells but is not an absolute requirement (Fig. 5B). In addition, we noted that Blimp-1−/− effector TH2 cells were slightly increased at the peak of the SEA response (Fig. 5C), consistent with studies on Blimp-1 in CD8 T cells (4, 10, 11). Together, our data argue that IL-10 can act directly on T cells to activate STAT3 and drive both Blimp-1 and further IL-10 production. IL-10 and Blimp-1, in turn, can promote additional IL-10 expression, creating a feed-forward autoregulatory loop. However, the IL-10–STAT3–Blimp-1 axis is also context-dependent, because TH1 cells rely more on Blimp-1 for the expression of IL-10, whereas TH2 cells can partially maintain IL-10 expression in the absence of Blimp-1 expression most likely by using STAT3.
Fig. 5. An autoregulatory loop controls Blimp-1 and IL-10 expression in TH2 cells.
(A) Expression of IL-10 in Blimp-1+/+ or Blimp-1−/− TH1 and TH2 cells cultured for 7 days in vitro. Data are representative of three independent experiments. (B) Expression of IL-10 in TH2 cells (Gata3+ IL-13+) isolated 9 days after infection with SEA from Blimp-1+/+ or Blimp-1−/− and subsequently cultured for 2 days in vitro with soluble antigen. Cells were additionally stimulated with PMA and ionomycin for 2 hours before staining for IL-10. Quantification of percent expressing IL-10 is shown on the right; data are means ± SD (n = 5 to 6). Data are representative of two independent experiments. ***P < 0.0001. (C) Percentage of TH2 cells (CD4+ CD44+ Gata3+) isolated directly ex vivo from Blimp-1+/+ mice or mice where Blimp-1 is conditionally deleted from T cells (Blimp-1 flox/flox CD4-cre+, called Blimp-1−/−) 9 days after infection with SEA. Quantification of TH2 cells. Data are means ± SD (n = 5 to 6) and are representative of three independent experiments. **P = 0.0092.
Blimp-1 antagonizes STAT5-driven T cell proliferation and survival
A potential role for Blimp-1 in executing a program driven by IL-10 was intriguing, given the autoimmune phenotype associated with Blimp-1 deficiency and the role of IL-10 in tolerance (31). However, although the absence of Blimp-1 in T cells led to increased TH2 cell responses after SEA immunization (Fig. 5C), the absence of STAT3, global IL-10, or IL-10 signaling in T cells did not result in altered TH2 cell differentiation (Fig. 1D and fig. S5). These data suggest that Blimp-1 has a role in controlling the immune response independent of driving IL-10 expression. To better understand the actions of Blimp-1 in T cells, we explored the transcriptomic program regulated by Blimp-1 in T cells. However, we reasoned that up-regulating Blimp-1 by inducing TH2 differentiation would likely not clarify genes directly regulated by Blimp-1 per se. We therefore used a simpler, albeit artificial, system, namely, retroviral overexpression of Blimp-1 in nonpolarized T cells (which lack Blimp-1 expression). Despite efficient transduction [marked by green fluorescent protein (GFP)] in control cells, we consistently found that generation of Blimp-1–expressing cells was extremely inefficient (fig. S6). We explored whether Blimp-1 expression was inducing cell death and found a substantial increase in nonviable cells compared with control cells (Fig. 6A, top). However, despite the paucity of cells, we were able to analyze the Blimp-1–dependent transcriptome. Consistent with an increase in cell death, we found that Blimp-1 expression was associated with the down-regulation of cell cycle and cell survival genes (Fig. 6B, top).
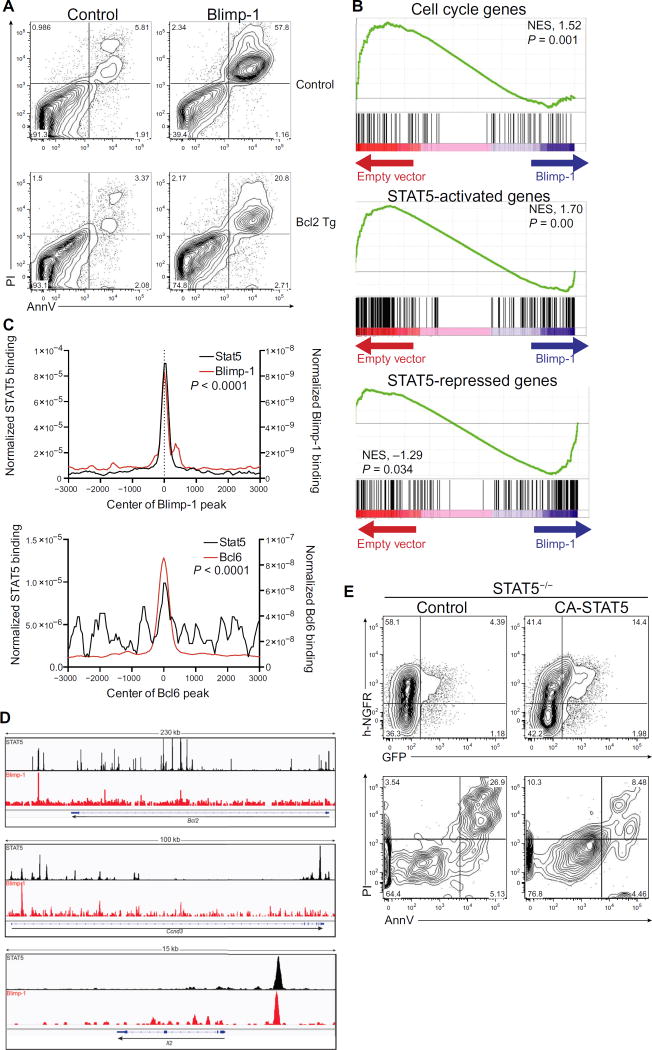
Fig. 6. Blimp-1 antagonizes STAT5 to induce cell death.
(A) Cell viability was measured by Annexin V and PI in CD4+ T cells isolated from control (top) or Bcl2 transgenic (bottom) mice and retrovirally transduced with GFP-expressing control or Blimp-1 virus. Cells are gated on GFP+; data are representative of three independent experiments. (B) GSEA analysis of genes controlling cell cycle or regulated by STAT5 compared to genes regulated by Blimp-1. Each plot shows nonrandom distribution of cell cycle genes or STAT5-regulated genes (33) versus genes regulated by Blimp-1. NES, normalized enrichment score. (C) Genome-wide co-occupancy of Blimp-1 and STAT5 (top) or Bcl6 and STAT5 (bottom). Blimp-1 and Bcl6 peaks were annotated using HOMER to STAT5 peaks found in promoters or enhancers (based on the presence of p300 peaks) in activated T cells. The y axis depicts the coverage normalized by tag count. The P value was calculated using a two-sample Kolmogorov-Smirnov test. (D) Binding of STAT5 and Blimp-1 mapped by ChIP-seq at the Bcl2, Ccnd3, and Il2 loci. The genome browser for Bcl2 (chr1:108,384,057–108,615,038), Ccnd3 (chr17:47,639,714–47,740,290), and Il2 (chr3:37,015,478–37,031,415) is shown. The y axis depicts the normalized tag number, defined as tag count per 1 million. (E) STAT5-deficient T cells overexpressing Blimp-1 (marked by GFP) were transduced with control or retrovirus expressing CA-STAT5, marked by surface expression of h-NGFR. Expression of GFP and h-NGFR is shown 48 hours after retroviral transduction (top). Cell viability [Annexin V (AnnV) and PI] is shown in GFP+ h-NGFR+ cells (bottom). Data are representative of two independent experiments.
Many genes that regulate cell cycle entry and promote cell survival in T cells are targets of STAT5 (32). Therefore, we next examined the intersection between STAT5- and Blimp-1–regulated genes. We found that the expression of STAT5-activated genes (33) was enriched in control cells and was poor in Blimp-1–expressing cells (Fig. 6B, middle and bottom). We therefore investigated whether the cell death phenotype induced by Blimp-1 could be reversed by transgenic, constitutive expression of Bcl2, the canonical STAT5-regulated prosurvival gene. Coexpression of Bcl2 substantially rescued Blimp-1–mediated cell death (Fig. 6A, bottom).
Blimp-1 and STAT5 had notable overlapping genomic binding in T cells (Fig. 6C), suggesting that Blimp-1 might be directly antagonizing STAT5 target genes. STAT5 binding was highly enriched in proximity to Blimp-1 binding sites, whereas Bcl6 binding had limited overlap with STAT5 sites (Fig. 6C). This suggested that Blimp-1 was capable of binding to genes directly activated by STAT5 and potentially repressing expression. To examine this further, we next looked at specific genes known to be involved in cell cycle and regulated by STAT5, such as Bcl2, Ccnd3 (which encodes cyclin D3), and Il2. We found several regions where Blimp-1 and STAT5 both bound, including promoters and potential enhancers (Fig. 6D).
Last, to test whether STAT5 could antagonize Blimp-1–mediated cell death in T cells, we overexpressed a constitutively active form of STAT5 (CA-STAT5) together with Blimp-1 into STAT5-deficient T cells. Consistent with our previous results, overexpression of Blimp-1 alone was associated with low cell recovery, whereas co-overexpression of CA-STAT5 in T cells was sufficient to partially rescue the cell loss and Blimp-1–mediated cell death (Fig. 6E). Collectively, these data suggested that Blimp-1 modifies expression of STAT5-regulated cell cycle genes, potentially by repressing STAT5-activated genes.
Together, our results support a model in which Blimp-1 is induced by IL-10 and antagonizes STAT5’s role in driving T cell survival. We believe that these data uncover an unappreciated role for IL-10 acting directly on T cells to mediate control of effector T cell responses and additionally provide a potential explanation for the role of Blimp-1 in protection from T cell–mediated autoimmunity.
DISCUSSION
A critical role for Blimp-1 in T cells was described nearly a decade ago, with the discovery that T cell–specific deletion of Blimp-1 results in multiorgan autoimmunity and fatal colitis (12, 13). Recent genome-wide association studies have identified polymorphisms in proximity to PRDM1, with diverse human autoimmune diseases (14–18) further implicating the regulation of Blimp-1 expression in constraining T cell– driven autoimmunity. Herein, we show that the regulation of Blimp-1 expression in T cells is complex. TH1 cells primarily use IL-12 acting via STAT4 to induce Blimp-1, whereas TH2 cells rely on IL-10 via STAT3. Blimp-1 and STAT3 promote IL-10 production, which acts in an autocrine/paracrine manner to reinforce expression of itself and Blimp-1. The induction of Blimp-1 in T cells antagonizes STAT5’s role in promoting cell cycle and cell survival genes (fig. S7). This provides a mechanism that helps explain the auto-immunity associated with the absence of Blimp-1.
Our study highlights several unexpected aspects about Blimp-1 expression and function in T cells. Some studies have suggested that IL-2 via STAT5 controls Blimp-1 expression in T cells, whereas recent reports have found that both STAT3 and STAT4 can drive Blimp-1 in TH17 and TH1 cells, respectively (8, 9). Because STAT3 is a major regulator of TH17 cell differentiation and, likewise, STAT4 is as a major driver of TH1 cells, we expected STAT5 or STAT6 to be important for regulating Blimp-1 in TH2 cells. Unexpectedly, we found that STAT3 was necessary for Blimp-1 expression in TH2 cells both in vitro and in vivo. Although a role for STAT3 in regulating TH2 cell differentiation has been described (23), STAT3 is not the predominant STAT family member required for TH2 cell development. The fact that Blimp-1 is controlled by multiple STAT family members suggests that the regulation of the locus is complex and is regulated either in a cell type–specific or in a context-dependent manner. Thus, the availability of specific cytokines in different settings and the regulation of cytokine receptor expression can influence which STAT family member is the dominant regulator of the Prdm1 locus. Further study of how STAT3 and STAT4 differentially regulate Blimp-1 would provide some insight into how Blimp-1 expression is controlled in different T cell subsets. In a similar manner, Blimp-1 and STAT3 can cooperate to regulate IL-10 expression; although Blimp-1 may override the need for STAT3 such as in TH1 cells, the relative importance of each may be context-dependent.
Our finding that STAT3 is critical for Blimp-1 expression in TH2 cells may appear counterintuitive, insofar as STAT3 also positively regulates Bcl6 expression, and Bcl6, in turn, suppresses Blimp-1 (34, 35). Regulation of gene expression represents the integration of diverse signals, and Bcl6 is a locus that is targeted by multiple factors. For example, STAT5, activated by IL-2, is a profound, direct inhibitor of Bcl6. In the setting in which Blimp-1 is induced by STAT3, Bcl6 expression would likely be already extinguished because of STAT5 activation. In this setting, Blimp-1 acts in concert with STAT5 to ensure that Bcl6 expression is extinguished.
In addition, this study highlights the multifaceted role that Blimp-1 plays in T cells. Several studies have found that Blimp-1 controls IL-10 expression by directly activating the Il10 gene, suggesting that Blimp-1 has both activating and repressive capacity (30). Because IL-10 has anti-inflammatory activity, it would appear logical that the multiorgan inflammatory disease associated with T cell–restricted Blimp-1 deficiency would be the result of a lack of IL-10 production, much in the way IL-10–deficient or IL-10R–deficient animals succumb to colitis. However, we found that in the absence of STAT3 or IL-10, Blimp-1 was reduced but TH2 cell differentiation was intact. This was in contrast to an increase in TH2 cells found when Blimp-1 expression was deleted from T cells. These data suggested that Blimp-1 had an IL-10–independent function to regulate cell expansion and/or survival. When we explored the role Blimp-1 had in T cells by forcing expression using a retrovirus, we found a substantial increase in cell death, suggesting that Blimp-1 could directly regulate T cell survival. In support of this hypothesis, we found a decrease in genes associated with cell survival when Blimp-1 expression was enforced in T cells and a concomitant overlap of STAT5 binding with Blimp-1 binding, both genome-wide and at places where STAT5 is known to activate critical T cell survival genes such as Bcl2. Together, these data suggest that Blimp-1 directly controls cell expansion by antagonizing prosurvival STAT5 activity, and this is independent of its function in regulating IL-10 expression.
Our study highlights the potentially important contribution of IL-10 signaling acting directly on T cells. Our in vitro studies demonstrate that IL-10 can act in an autocrine/paracrine manner to drive Blimp-1 and further IL-10 expression. However, in vivo, it is likely that IL-10 can come from a number of sources, of which TH2 effector cells are just one component. Because TH2 effector cells robustly produce IL-10, we believe that paracrine signaling would provide some benefit to the induction of Blimp-1 and IL-10 expression, but these questions would need to be carefully worked out using models where only TH2 effector cells were unable to produce IL-10. Along the same lines, a more thorough analysis of the physiological role of Blimp-1 in TH2 cells is still lacking. Because T cell–restricted Blimp-1–deficient hosts develop spontaneous TH1-like inflammation, performing TH2-driven disease models in these animals becomes challenging. A careful analysis of Blimp-1 deficiency in only TH2 cells should be performed when such a genetic model becomes available.
In summary, we have uncovered several findings related to the role of Blimp-1 in CD4 T cell biology. Although we did not see a role for IL-2 or STAT5 in driving Blimp-1 in our experiments, we found an important role for STAT3 in TH2 cells and confirm previous findings that the STAT1–STAT4–T-bet pathway controls Blimp-1 in TH1 cells. We additionally describe a mechanism for IL-10 on T cells by driving Blimp-1 expression. Last, we show that Blimp-1 can limit expansion and survival of T cells by antagonizing STAT5 activity at critical cell cycle/antiapoptotic genes. Collectively, this study has clarified what drives Blimp-1 expression in T cells and the function of Blimp-1 once expressed. This work has important implications for understanding T cell–driven autoimmunity and may establish a previously unappreciated role for Blimp-1 in T cell– mediated inflammatory bowel disease and colitis.
MATERIALS AND METHODS
Study design
C57BL/6J (000664), CD4-Cre+Tg (017336), Stat6−/−(005977), Tbx21−/−(004648), Thy1.1 (000406), Il10Rb−/− (005027), Il10−/− (002250), and Prdm1fl/fl(008100) mice were purchased from the Jackson Laboratory. Rag−/− mice were purchased from Taconic Farms. Stat4−/− mice were provided by M. Kaplan (Indiana University). Vav-Bcl2 Tg mice were provided by A. Nussenzweig. Stat1−/− and Stat3fl/fl mice were provided by D. Levy (36, 37). Stat5fl/fl mice have been described previously (38), and Prdm1-EYFP (Jax 008828) was provided by S. Crotty (La Jolla Institute for Allergy and Immunology) and E. Meffre (Yale University). All floxed mice were bred with CD4-Cre+ Tg mice. All animal studies were performed according to the National Institutes of Health guidelines for the use and care of live animals and were approved by the Institutional Animal Care and Use Committee of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS). All in vitro experiments included two technical replicates per experiment and were performed independently two or three times, as indicated in the figure legends. All in vivo experiments were performed independently two times, with four to seven animals per group. Flow cytometry was individually performed on each animal to provide statistics.
Naïve CD4+ T cell isolation and activation in vitro
CD4+ T cells isolated from spleen and lymph nodes of 6- to 8-week-old animals were purified by negative selection and magnetic separation (Miltenyi Biotec) followed by sorting for naïve population (CD4+ CD62L+ CD44− CD25−) using FACSAria IIIu (BD). Naïve CD4+ T cells were activated by plate-bound α-CD3 and α-CD28 (10 µg/ml each; BioXCell) in complete RPMI [RPMI with 10% fetal bovine serum, 2 mM glutamine, penicillin (100 IU/ml), streptomycin (0.1 mg/ml), 10 mM Hepes, 1 mM sodium pyruvate, and 50 µM β-mercaptoethanol] for 3 days under neutral conditions or with various cytokines as indicated. Cells were removed at day 3, washed, and rested with IL-2 and fresh cytokines for 2 days. Cells were isolated again (day 5) and restimulated by plate-bound α-CD3 and α-CD28 (1 mg/ml) for an additional 2 to 3 days. Conditions were as follows: control: α–interferon-γ (α–IFN-γ) (10 µg/ml) and α–IL-4 (10 µg/ml); IL-2: IL-2 (100 U/ml), α–IFN-γ (10 µg/ml), and α–IL-4 (10 µg/ml); TH1: IL-12 (10 ng/ml) and α–IL-4 (10 µg/ml); and TH2: IL-4 (10 ng/ml) and α–IFN-γ (10 µg/ml).
Flow cytometry
Cells were resuspended in Hanks’ balanced salt solution and stained for live/dead using LIVE/DEAD Fixable Dead Cell Stain (Life Technologies) and cell surface markers (see table S1 for antibodies). Cells were fixed with either FoxP3 kit (eBio) (transcription factors) or Cytofix/Cytoperm (BD) (cytokines). For intracellular cytokine staining, cells were restimulated for 2 to 4 hours with phorbol 12-myristate 13-acetate (PMA) (50 ng/ml) and ionomycin (1 µg/ml), with the addition of brefeldin A (GolgiPlug, BD). For Blimp-1 staining, cells were fixed for 1 hour and then stained for 45 min. Annexin V and propidium iodide (PI) staining was done according to the protocol provided with Annexin V (BD). For phospho-STAT3 staining, cells were collected at day 5 and restimulated with IL-10 (10 ng/ml) or IL-6 (10 ng/ml) or left unstimulated for 30 min at 37°C, followed by immediate fixation (5× Lyse/Fix, BD) and permeabilization (Perm Buffer III, BD) and staining with phospho-STAT3. Cells were collected using FACSVerse (BD), LSR II (BD), or Fortessa (BD) and analyzed with FlowJo (Tree Star).
SEA immunization
Eight- to 10-week-old mice were injected in the footpad with 100 mg of SEA. Nine days after immunization, draining and nondraining lymph nodes were isolated and crushed into complete RPMI. Cells were counted and plated in round-bottom plates and either stimulated with PMA (50 ng/ml) and ionomycin (1 µg/ml), with the addition of brefeldin A (GolgiPlug, BD) for 6 hours, or stimulated with SEA (20 µg/ml) at 37°C for 48 hours, and then additionally stimulated with PMA (50 ng/ml) and ionomycin (1 µg/ml), with the addition of brefeldin A (GolgiPlug, BD) at 37°C for 2 hours. Cells were stained for flow cytometry as described above.
Bone marrow chimeras
Bone marrow cells were isolated from femurs of mice, and T cells were depleted using Thy1.1 or Thy1.2 positive selection and a magnetic separation kit (Miltenyi Biotec). Cells were counted, mixed in a ratio of 50:50, and injected intravenously into Rag2−/−-deficient hosts. A total of 20 × 106 cells were injected per mouse. Four weeks after reconstitution, mice were bled to check reconstitution efficiency, and 6 weeks after reconstitution, they were used for SEA experiments, as described above.
Retroviral transduction
Blimp-1 complementary DNA (cDNA) was cloned into MigR1 expression vector that uses GFP as a selection marker. CA-STAT5 has been previously described (39) and uses human neural growth factor receptor (h-NGFR) as a selection marker. Sorted naïve CD4+ T cells from wild-type or STAT5−/− mice were cultured in the presence of plate-bound α-CD3 and α-CD28 with α–IFN-γ and α–IL-4 for 36 hours, transduced with control (empty vector) or Blimp-1– or CA-STAT5–expressing retrovirus, and cultured for 2 days. Cells were sorted for RNA isolation (GFP+) or analyzed by flow cytometry.
Quantitative polymerase chain reaction
mRNA was isolated from about 1 × 106 to 2 × 106 cells using the mir-Vana miRNA Isolation Kit (AM1560, ABI). RNA (0.5 to 1 µg) was converted into cDNA using the TaqMan Reverse Transcription Kit (4304134, ABI). Quantitative polymerase chain reaction (qPCR) was performed with TaqMan probes [mouse Blimp-1 (Mm00476128_m1), mouse IL-10Rα (Mm00434151_m1), mouse IL-10Rβ (Mm00434157_m1), and mouse actin (Mm02619580_g1, ABI)] and TaqMan Universal Master Mix (4370048, ABI) and detected by a CFX96 Real-Time PCR Detection Machine (Bio-Rad). ΔΔCt was calculated to determine the relative expression normalized to β-actin.
mRNA sequencing
About 0.5 × 106 cells were isolated by sorting GFP+ cells from three independent biological replicates of CD4 T cells transduced with empty vector or Blimp-1–expressing retrovirus. Total RNA was prepared by using the mirVana miRNA Isolation Kit (AM1560, ABI). A fraction of total RNA (2 µg) was processed into mRNA sequencing library by using the TruSeq RNA Sample Prep Kit (FC-122-1001, Illumina). The libraries were sequenced for 50 cycles (single-read) with HiSeq 2000 (Illumina). Raw sequencing data were decoded and demultiplexed with CASAVA 1.8.2 to generate FASTQ files and mapped onto the mouse genome build mm9 using TopHat 2.0.8, and the gene expression values (reads per kilobase exon per million mapped reads) were calculated with Cufflinks 2.2. All downstream statistical analyses were performed with Partek Genomics Suite 6.6. Gene set enrichment analysis (GSEA) from the Broad Institute (www.broad.mit.edu/gsea) was used to calculate the enrichment of genes in each set. Cell cycle gene lists were assembled using QIAGEN’s SABiosciences cell cycle pathway PCR arrays; STAT5-regulated genes were identified from STAT5-regulated transcriptome analysis and chromatin immunoprecipitation sequencing (ChIP-seq) analysis (GSE77656) (33). Gene lists are in table S2.
ChIP sequencing
The following ChIP-seq data sets were acquired from Gene Expression Omnibus: Blimp-1, GSE75724 (9); Bcl6, GSE41317 (40); STAT5, GSE26553 (38); and p300, GSE40463 (41). For co-occupancy of Blimp-1 or Bcl6 with STAT5, Blimp-1/Bcl6 peaks were annotated with HOMER according to the intersection of STAT5 and p300 peaks found in activated CD4+ T cells.
ChIP-qPCR
Naïve T cells from STAT3-intact or STAT3-deficient hosts were cultured for 5 days under TH2 conditions plus IL-10. Cells were pulsed with IL-10 for 30 min before fixation with 1% formaldehyde. A total of 20 × 106 cells per sample were sonicated with Bioruptor Pico in shearing buffer, STAT3 (C-20; sc-482, Santa Cruz Biotechnology) ChIP was performed overnight using Protein A Dynabeads (Thermo Fisher), and SYBR Green–based qPCR was performed using custom-designed primers (table S3).
Statistics
For statistical analysis, unpaired two-tailed Student’s t test was performed using GraphPad Prism (version 6.0, GraphPad), unless otherwise specified to calculate statistical significance (reported as means ± SD). For analysis of IL-10Rβ chimeras, paired Student’s t tests were used. A P value of 0.05 was considered significant.
Supplementary Material
Fig. S1. Expression of Blimp-1 in T cell subsets in vitro.
Fig. S2. The IL-12–STAT4–T-bet axis controls Blimp-1 expression in TH1 cells.
Fig. S3. IL-10 regulation of TH1 and TH2 cells.
Fig. S4. TH2 differentiation is normal in the absence of STAT3.
Fig. S5. The absence of IL-10 signaling on T cells does not affect TH2 cell differentiation in vivo.
Fig. S6. Transduction of Blimp-1 GFP-expressing retrovirus.
Fig. S7. Model of IL-10–driven Blimp-1 expression via STAT3 in TH2 cells to promote cell death.
Fig. S8. Representative gating strategies.
Table S1. Flow cytometry antibodies.
Table S2. Gene lists for GSEA analysis.
Table S3. ChIP-qPCR custom-designed SYBR primers.
Table S4. Excel file containing source data for Figs. 1, 3, 4, and 5.
Acknowledgments
We thank A. Canela-Rodriguez and A. Nussenzweig for the Bcl2Tg mice, S. Oland for technical help with immunizations, and M. McGeachy and T. Hand for helpful suggestions and comments on the manuscript. We thank J. Simone, J. Lay, and K. Tinsley (Flow Cytometry Section, NIAMS); G. Gutierrez-Cruz (NIAMS Deep Sequencing Core); and the NIAMS Laboratory Animal Care and Use Section staff for technical support. This study used the high-performance computational capabilities of the Helix Systems at the NIH.
Funding: This work was supported by the Intramural Research Programs of NIAMS and by the Postdoctoral Research Associate Program of the National Institute of General Medical Sciences.
Footnotes
SUPPLEMENTARY MATERIALS
Author contributions: Conceptualization: A.C.P., D.J., A.V.V., and J.J.O.; methodology: A.C.P., D.J., A.V.V., G.V., and Y.K.; validation: A.C.P. and D.J.; formal analysis: A.C.P., S.R.B., and G.V.; investigation: A.C.P., D.J., A.H., F.P., D.S.S., and Y.K.; resources: D.J., A.V.V., D.S.S., S.B.S., A.S., and S.M.K.; data curation: A.C.P., S.R.B., G.V., and Y.K.; writing: A.C.P. and J.J.O.; supervision: J.J.O.; and funding acquisition: A.C.P. and J.J.O.
Competing interests: J.J.O. holds NIH patents related to the therapeutic targeting of Jak kinases and has a Collaborative Research Agreement and Development Award with Pfizer Inc. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
REFERENCES AND NOTES
- 1.Bikoff EK, Morgan MA, Robertson EJ. An expanding job description for Blimp-1/ PRDM1. Curr. Opin. Genet. Dev. 2009;19:379–385. doi: 10.1016/j.gde.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Semin. Immunol. 2011;23:341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, Kallies A. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 6.Ballesteros-Tato A, León B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann C, Heinrich F, Neumann K, Junghans V, Mashreghi M-F, Ahlers J, Janke M, Rudolph C, Mockel-Tenbrinck N, Kühl AA, Heimesaat MM, Esser C, Im S-H, Radbruch A, Rutz S, Scheffold A. Role of Blimp-1 in programing Th effector cells into IL-10 producers. J. Exp. Med. 2014;211:1807–1819. doi: 10.1084/jem.20131548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain R, Chen Y, Kanno Y, Joyce-Shaikh B, Vahedi G, Hirahara K, Blumenschein WM, Sukumar S, Haines CJ, Sadekova S, McClanahan TK, McGeachy MJ, O’Shea JJ, Cua DJ. Interleukin-23-induced transcription factor Blimp-1 promotes pathogenicity of T helper 17 cells. Immunity. 2016;44:131–142. doi: 10.1016/j.immuni.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 12.Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat. Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 13.Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, Calame K. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 14.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagacé C, Scott R, Amininejad L, Bumpstead S, Baidoo L, Baldassano RN, Barclay M, Bayless TM, Brand S, Büning C, Colombel J-F, Denson LA, De Vos M, Dubinsky M, Edwards C, Ellinghaus D, Fehrmann RSN, Floyd JAB, Florin T, Franchimont D, Franke L, Georges M, Glas J, Glazer NL, Guthery SL, Haritunians T, Hayward NK, Hugot J-P, Jobin G, Laukens D, Lawrance I, Lémann M, Levine A, Libioulle C, Louis E, McGovern DP, Milla M, Montgomery GW, Morley KI, Mowat C, Ng A, Newman W, Ophoff RA, Papi L, Palmieri O, Peyrin-Biroulet L, Panés J, Phillips A, Prescott NJ, Proctor DD, Roberts R, Russell R, Rutgeerts P, Sanderson J, Sans M, Schumm P, Seibold F, Sharma Y, Simms LA, Seielstad M, Steinhart AH, Targan SR, van den Berg LH, Vatn M, Verspaget H, Walters T, Wijmenga C, Wilson DC, Westra H-J, Xavier RJ, Zhao ZZ, Ponsioen CY, Andersen V, Torkvist L, Gazouli M, Anagnou NP, Karlsen TH, Kupcinskas L, Sventoraityte J, Mansfield JC, Kugathasan S, Silverberg MS, Halfvarson J, Rotter JI, Mathew CG, Griffiths AM, Gearry R, Ahmad T, Brant SR, Chamaillard M, Satsangi J, Cho JH, Schreiber S, Daly MJ, Barrett JC, Parkes M, Annese V, Hakonarson H, Radford-Smith G, Duerr RH, Vermeire S, Weersma RK, Rioux JD. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellinghaus D, Zhang H, Zeissig S, Lipinski S, Till A, Jiang T, Stade B, Bromberg Y, Ellinghaus E, Keller A, Rivas MA, Skieceviciene J, Doncheva NT, Liu X, Liu Q, Jiang F, Forster M, Mayr G, Albrecht M, Häsler R, Boehm BO, Goodall J, Berzuini CR, Lee J, Andersen V, Vogel U, Kupcinskas L, Kayser M, Krawczak M, Nikolaus S, Weersma RK, Ponsioen CY, Sans M, Wijmenga C, Strachan DP, McArdle WL, Vermeire S, Rutgeerts P, Sanderson JD, Mathew CG, Vatn MH, Wang J, Nöthen MM, Duerr RH, Büning C, Brand S, Glas J, Winkelmann J, Illig T, Latiano A, Annese V, Halfvarson J, D’Amato M, Daly MJ, Nothnagel M, Karlsen TH, Subramani S, Rosenstiel P, Schreiber S, Parkes M, Franke A. Association between variants of PRDM1 and NDP52 and Crohn’s disease, based on exome sequencing and functional studies. Gastroenterology. 2013;145:339–347. doi: 10.1053/j.gastro.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jönsen A, Bengtsson AA, Rantapää-Dahlqvist S, Baechler EC, Brown EE, Alarcón GS, Edberg JC, Ramsey-Goldman R, McGwin G, Jr, Reveille JD, Vilá LM, Kimberly RP, Manzi S, Petri MA, Lee A, Gregersen PK, Seldin MF, Rönnblom L, Criswell LA, Syvänen A-C, Behrens TW, Graham RR. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, Catanese JJ, Xie G, Stahl EA, Chen R, Alfredsson L, Amos CI, Ardlie KG, BIRAC Consortium. Barton A, Bowes J, Burtt NP, Chang M, Coblyn J, Costenbader KH, Criswell LA, Crusius JBA, Cui J, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TWJ, Kastner DL, Ke X, Kurreeman FAS, Lee AT, Liu X, Li Y, Martin P, Morgan AW, Padyukov L, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AHM, van der Horst-Bruinsma IE, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth P, YEAR Consortium. Altshuler D, Karlson EW, Toes REM, de Vries N, Begovich AB, Siminovitch KA, Worthington J, Klareskog L, Gregersen PK, Daly MJ, Plenge RM. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X-j, Lu X-l, Lv J-c, Yang H-z, Qin L-x, Zhao M-h, Su Y, Li Z-g, Zhang H. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann. Rheum. Dis. 2011;70:1330–1337. doi: 10.1136/ard.2010.140111. [DOI] [PubMed] [Google Scholar]
- 19.Weinmann AS. Regulatory mechanisms that control T-follicular helper and T-helper 1 cell flexibility. Immunol. Cell Biol. 2014;92:34–39. doi: 10.1038/icb.2013.49. [DOI] [PubMed] [Google Scholar]
- 20.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J. Exp. Med. 2008;205:1959–1965. doi: 10.1084/jem.20080526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J. Immunol. 2007;178:242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 23.Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J, Nguyen ET, Levy DE, Kaplan MH. The transcription factor STAT3 is required for T helper 2 cell development. Immunity. 2011;34:39–49. doi: 10.1016/j.immuni.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wynn TA, Eltoum I, Cheever AW, Lewis FA, Gause WC, Sher A. Analysis of cytokine mRNA expression during primary granuloma formation induced by eggs of Schistosoma mansoni. J. Immunol. 1993;151:1430–1440. [PubMed] [Google Scholar]
- 25.Vella AT, Pearce EJ. CD4+ Th2 response induced by Schistosoma mansoni eggs develops rapidly, through an early, transient, Th0-like stage. J. Immunol. 1992;148:2283–2290. [PubMed] [Google Scholar]
- 26.Mosmann TR, Schumacher JH, Fiorentino DF, Leverah J, Moore KW, Bond MW. Isolation of monoclonal antibodies specific for IL-4, IL-5, IL-6, and a new Th2-specific cytokine (IL-10), cytokine synthesis inhibitory factor, by using a solid phase radioimmunoadsorbent assay. J. Immunol. 1990;145:2938–2945. [PubMed] [Google Scholar]
- 27.Weber-Nordt RM, Riley JK, Greenlund AC, Moore KW, Darnell JE, Schreiber RD. Stat3 recruitment by two distinct ligand-induced, tyrosine-phosphorylated docking sites in the interleukin-10 receptor intracellular domain. J. Biol. Chem. 1996;271:27954–27961. doi: 10.1074/jbc.271.44.27954. [DOI] [PubMed] [Google Scholar]
- 28.Wehinger J, Gouilleux F, Groner B, Finke J, Mertelsmann R, Weber-Nordt RM. IL-10 induces DNA binding activity of three STAT proteins (Stat1, Stat3, and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. FEBS Lett. 1996;394:365–370. doi: 10.1016/0014-5793(96)00990-8. [DOI] [PubMed] [Google Scholar]
- 29.Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA, Ernst M, Saris CJM, O’Shea JJ, Hunter CA. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 2007;8:1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 30.Minnich M, Tagoh H, Bönelt P, Axelsson E, Fischer M, Cebolla B, Tarakhovsky A, Nutt SL, Jaritz M, Busslinger M. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat. Immunol. 2016;17:331–343. doi: 10.1038/ni.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shouval DS, Ouahed J, Biswas A, Goettel JA, Horwitz BH, Klein C, Muise AM, Snapper SB. Interleukin 10 receptor signaling: Master regulator of intestinal mucosal homeostasis in mice and humans. Adv. Immunol. 2014;122:177–210. doi: 10.1016/B978-0-12-800267-4.00005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Q, Li WQ, Aiello FB, Mazzucchelli R, Asefa B, Khaled AR, Durum SK. Cell biology of IL-7, a key lymphotrophin. Cytokine Growth Factor Rev. 2005;16:513–533. doi: 10.1016/j.cytogfr.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Villarino A, Laurence A, Robinson GW, Bonelli M, Dema B, Afzali B, Shih H-Y, Sun H-W, Brooks SR, Hennighausen L, Kanno Y, O’Shea JJ. Signal transducer and activator of transcription 5 (STAT5) paralog dose governs T cell effector and regulatory functions. eLife. 2016;5:e08384. doi: 10.7554/eLife.08384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLOS ONE. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray JP, Marshall HD, Laidlaw BJ, Staron MM, Kaech SM, Craft J. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity. 2014;40:367–377. doi: 10.1016/j.immuni.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee C-k, Raz R, Gimeno R, Gertner R, Wistinghausen B, Takeshita K, DePinho RA, Levy DE. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17:63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- 37.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 38.Yang X-P, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun H-W, Wei L, Vahedi G, Kanno Y, O’Shea JJ, Laurence A. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onishi M, Nosaka T, Misawa K, Mui AL-F, Gorman D, McMahon M, Miyajima A, Kitamura T. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao W, Spolski R, Li P, Du N, West EE, Ren M, Mitra S, Leonard WJ. Opposing actions of IL-2 and IL-21 on Th9 differentiation correlate with their differential regulation of BCL6 expression. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3508–3513. doi: 10.1073/pnas.1301138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vahedi G, Takahashi H, Nakayamada S, Sun H-w, Sartorelli V, Kanno Y, O’Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression of Blimp-1 in T cell subsets in vitro.
Fig. S2. The IL-12–STAT4–T-bet axis controls Blimp-1 expression in TH1 cells.
Fig. S3. IL-10 regulation of TH1 and TH2 cells.
Fig. S4. TH2 differentiation is normal in the absence of STAT3.
Fig. S5. The absence of IL-10 signaling on T cells does not affect TH2 cell differentiation in vivo.
Fig. S6. Transduction of Blimp-1 GFP-expressing retrovirus.
Fig. S7. Model of IL-10–driven Blimp-1 expression via STAT3 in TH2 cells to promote cell death.
Fig. S8. Representative gating strategies.
Table S1. Flow cytometry antibodies.
Table S2. Gene lists for GSEA analysis.
Table S3. ChIP-qPCR custom-designed SYBR primers.
Table S4. Excel file containing source data for Figs. 1, 3, 4, and 5.