Abstract
Purpose
This study aimed to establish a method for testing Staphylococcus aureus in the vitreous of endophthalmitis with matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS), which is simple, fast, and sensitive.
Methods
S. aureus at different numbers was either mixed with homogenized vitreous or inoculated in porcine eyes for culturing, followed by homogenization. The homogenized vitreous samples, with or without centrifugation, were stained with Gram and Coomassie Blue (CBB) dyes and cultured with blood agar. The pellet of the vitreous mixture was analyzed with MALDI-TOF-MS.
Results
The minimum detectable levels of S. aureus in H2O and in the pellet of homogenized vitreous were 9.0 × 103 (positive rate, 22.2%) and 1.0 × 104 CFU/μl (positive rate, 11.1%), respectively. In the vitreous samples inoculated with S. aureus and cultured for 12 h, the number of S. aureus increased in a dose-dependent manner to the number of bacteria in the inoculate. In the supernatant of the homogenized vitreous, there were traces of bacteria identified with Gram staining. On the blood agar plates, the supernatant grew a few colonies, while the pellet grew intensive colonies. The vitreous fragments that were stained with CBB were displayed in the supernatants, in small numbers, and in the pellets. When the inoculated number was 1.0 × 104 CFU/μl or higher, the bacteria in the vitreous pellets could be identified in all samples (100%, n = 9). However, bacteria could be detected in only two out of nine spots of pellets (22.2%) if the number of inoculated S. aureus was 1.0 × 103 CFU/μl.
Conclusions
A method for testing S. aureus directly from vitreous samples of endophthalmitis by the combination of easy extraction methods and a MALDI-TOF- MS assay was provided. This rapid identification method is easily adaptable for use in clinical routine and can help reduce the delay in diagnosis, allowing for earlier therapeutic intervention in patients.
Introduction
Bacterial endophthalmitis is a devastating infection resulting from ocular trauma, postoperative endophthalmitis, and migration of bloodborne organisms into the eye. More than 75% of endophthalmitis cases worldwide are induced by bacteria [1,2]. We retrospectively investigated the bacteria from endophthalmitis samples (aqueous humor and vitreous) and found that the three organisms most commonly responsible are Staphylococcus spp. (43.31%), Bacillus spp. (7.87%) and Streptococcus spp. (6.30%) [3]. Gentile et al. [2] reported that for 988 endophthalmitis isolates, the most prevalent pathogens were coagulase-negative Staphylococcus (39.4%), followed by Streptococcus viridans species (12.1%) and Staphylococcus aureus (11.1%). Diagnosis of the specific pathogen in endophthalmitis is the key to clinically effective treatment, including the administration of the appropriate antibiotics. Identification of pathogens from intraocular samples is often dependent on Gram stains, culturing in broth and on solid agar media, and various biochemical reactions, all of which can take upwards of 2 days [4-6].
Few developments in microbiological diagnostics have had such a significant impact on species-level identification of microorganisms as matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry (MS) [7]. Conventional identification methods rely on biochemical criteria and require additional pretesting and lengthy culture procedures. In comparison, MALDI-TOF-MS can identify bacteria and fungi within minutes directly from colonies grown on culture plates. The reliability and accuracy of the method have been demonstrated in numerous studies, and different systems approved by the U.S. Food and Drug Administration (FDA) are already commercially available [8]. This radically new, methodically simple approach profoundly reduces the turnaround time (TAT) by more than a day by shortening the time necessary for identification of the colonies grown on culture plates [9,10]. In addition, novel applications of the system besides microbial species-level identification, i.e., typing and resistance gene determination, are being explored [7,11,12]. Meanwhile, applications of MALDI-TOF-MS for identification of pathogens from positive blood cultures or directly from patient samples (such as urine) are promising for patients with infectious diseases, although these MS systems are commonly used for identifying bacterial colonies on plates [13-15].
S. aureus is one of the most common pathogens responsible for endophthalmitis, often leading to uniformly poor visual outcomes and the possibility of blindness. Approximately 10% of patient infections with culture-positive aqueous and vitreous samples are due to S. aureus [16,17]. Early identification of S. aureus would greatly improve therapeutic outcomes for this disease. However, complete identification of bacteria using conventional methods usually takes several days.
Rapid identification of the microorganisms causing endophthalmitis is crucial for patient management and treatment. To our knowledge, no study has specifically tested the application of MALDI-TOF-MS directly on vitreous samples of endophthalmitis. We report here the evaluation of an extraction method combined with MALDI-TOF-MS for direct identification of S. aureus using an ex vivo endophthalmitis model of S. aureus inoculated in the porcine vitreous.
Methods
S. aureus strain
The S. aureus strain used to develop the extraction protocol was S. aureus ATCC 25923 (American Type Culture Collection, Manassas, VA). The strain was stored at −80 °C in 10% glycerol and cultured on blood agar plates (bioMérieux, Craponne, France) for 24 h at 37 °C. A colony was picked and suspended in broth for 24 h to increase the bacterial yield. The broth was then centrifuged at 6,000 ×g for 10 min, and the supernatant was discarded. The pellet was washed with 1.0 ml of PBS (1X; 10 mM Na2HPO4, 2 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, pH 7.4). Following further centrifugation (6,000 ×g for 10 min), the supernatant was discarded. The pellet was resuspended in 1.0 ml of PBS and vortexed thoroughly. The suspension was diluted to the proper concentration, and 50 μl was added to the blood agar to quantify the S. aureus present.
Mixing of S. aureus and vitreous suspension
Porcine eyeballs were obtained from a butcher within 8 h of slaughter. The porcine vitreous was taken out and homogenized by ball mill (Scientz, Ningbo China). To determine the minimal bacterial concentration needed for reliable MALDI-TOF-MS identification, sequential dilutions of S. aureus in double-distilled water were mixed with homogenized vitreous to make suspensions. The mixed vitreous was then centrifuged at 6,000 ×g for 10 min. Parts of the supernatant and the resuspended pellets were applied on the blood agar to count the S. aureus. The pellet was washed three times with 1.0 ml of sterile distilled water and then resuspended in 6 μl of sterile distilled water and vortexed thoroughly. Two microliters were then applied in triplicate on the sample plates for MALDI-TOF-MS (VITEK MS, bioMérieux) to identify the organisms present.
Inoculation and identification of S. aureus in vitreous
Fresh porcine eyeballs were washed and sterilized, followed by inoculation with 80 μl suspensions of S. aureus. These 80-μl suspensions were divided into 20-μl aliquots that were injected at four different locations into the eyeball (three before the equator and one at the posterior pole, at 120-degree intervals). Then, the cornea and iris of each eyeball were removed and incubated at 37 °C for 12 h. The experiments consisted of nine eyeballs separated equally into three groups of S. aureus inoculate concentrations (103 CFU/μl, 104 CFU/μl, and 105 CFU/μl).
The vitreous was separated from the cultured porcine eyeball and homogenized, centrifuged, and washed as stated above. The pellet was resuspended in 6 μl of sterile distilled water and vortexed thoroughly, followed by MALDI-TOF-MS analysis. Meanwhile, parts of the homogenates, supernatants, and pellets were aliquoted for staining and incubating on the blood agar plates.
Gram staining vitreous mixtures
The homogenized vitreous, as well as the supernatants and pellets, was subjected to Gram staining after centrifugation. Briefly, 10 μl of the sample was smeared on a sterile glass slide. The smear was stained using a Gram staining kit (Kailin, Jiangmen, China) following the manufacturer’s instructions. Gram-stained slides were viewed and photographed on a microscope with a camera (Olympus DP73, Tokyo, Japan).
Coomassie Blue staining vitreous mixture
Similarly, the homogenized vitreous, as well as its supernatants and pellets, was subjected to Coomassie Blue (CBB) staining after centrifugation. Approximately 10 μl of the sample was smeared on a sterile glass slide. The smear was stained with 0.5% CBB G-250 (MP biomedicals, Santa Ana, CA) [18]. The slides were briefly washed with distilled water, blotted dry, covered with cover glass, and examined with a microscope and photographed.
Culture of infected vitreous on blood agar
The homogenized porcine vitreous was centrifuged at 6,000 ×g for 10 min. Then, 50 μl of supernatant was inoculated on the blood agar plate. The pellet was first resuspended in sterile distilled water (equal to the volume of vitreous), and then 50 μl of the resuspension was spread on a blood agar plate. The plates were then incubated at 37 °C for 24 h.
In addition, two specimens of human vitreous from patients with suspected endophthalmitis were collected with vitrectomy and examined for regular culturing and identification by methods we reported previously [3]. At the same time, a small portion of specimen was subject to centrifugation, and the pellet was analyzed with MALDI-TOF-MS. This study was approved by the Ethics Committee of Zhongshan Ophthalmic Center, Sun Yat-sen University and that the study adhered to the tenets of Declaration of Helsinki and the ARVO statement on human subjects.
MALDI-TOF-MS analysis
Sample preparation for MALDI-TOF-MS analysis was performed as previously described. Species identification was performed using the bioMérieux IVD system (Database version 2.0). Briefly, 1 μl of samples (pellet was resuspended in H2O) were distributed on a MALDI-TOF-MS FlexiMass-DS disposable target slide (Cat. #: TO-430), and then 1-μl matrix solution, a-cyano-4-hydroxycinnamic acid (Vitek® MS-CHCA) was added. As a control, one colony of S. aureus from a subculture on a blood agar plate was also smeared on the target slide. Escherichia coli ATCC 8739 was used for external calibration. Once dry, the target slide was loaded into the mass spectrometer. MALDI-TOF-MS analysis was performed on a VITEK MS instrument with a nitrogen laser (337 nm) operating in positive linear mode with delayed extraction at a 58 kV accelerating voltage. Each spectrum was automatically collected in the positive ion mode as an average of 500 laser shots (five laser shots at 100 profiles per sample). According to the manufacturer’s specifications, S. aureus was identified using VITEK MS automation control and Myla software.
Results
Identification of S. aureus mixed with porcine vitreous
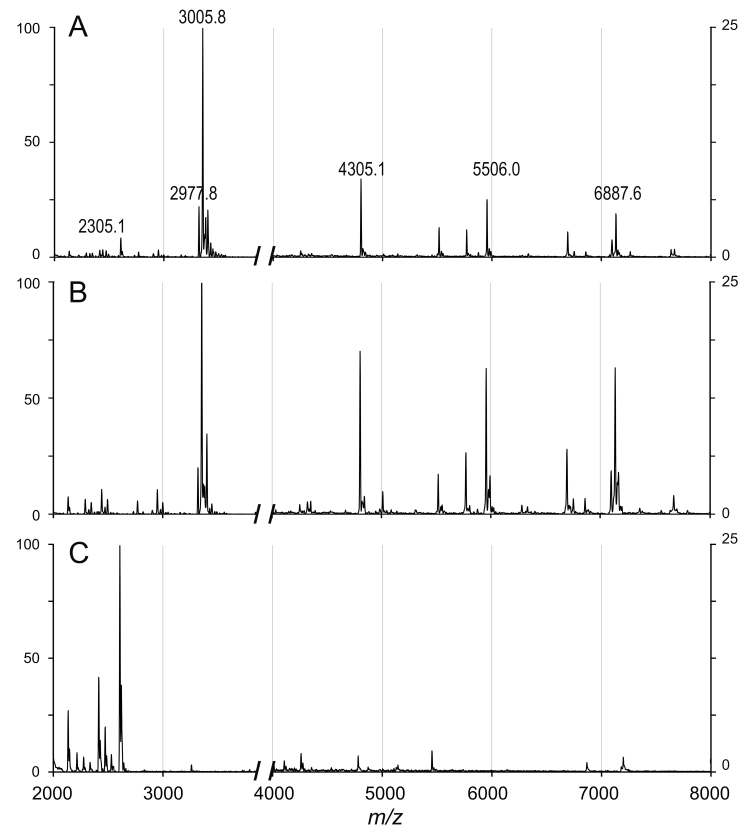
The mixtures of the suspension of S. aureus and homogenized vitreous were subjected to centrifugation, and the pellets were analyzed with MALDI-TOF-MS to determine the bacteria. In general, the mass spectra of S. aureus and its mixture with vitreous displayed similar patterns (Figure 1). Among the 18 total sampled spots, all spots could be identified (100%) when the concentration of the bacterial suspension was 3.0 × 104 CFU/μl or more (Table 1). The minimum detectable level of S. aureus suspension in vitreous was 1.0 × 104 CFU/μl (11.1%). For comparison, S. aureus suspension in H2O was directly determined with VITEK MS. The bacteria were identified in all spots when the concentration was 3.0 × 104 CFU/μl or more, while four out of 18 sampled spots displayed S. aureus at 9.0 × 103 CFU/μl (22.2%).
Figure 1.
Mass spectra of vitreous, Staphylococcus aureus, and the mixture of both. The mixture of porcine vitreous and S. aureus (A), S. aureus (ATCC 25923F; B), and vitreous (C) was analyzed with matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS; VITEK MS). S. aureus is identified in A and B. Details regarding the approach can be found in the Materials and Methods section.
Table 1. S. aureus identified in mixture of vitreous using MALDI-TOF-MS.
| Bacterial concentration in vitreous/water(CFU/μl) | Mixture with vitreous |
Control (with H2O) |
||
|---|---|---|---|---|
| Identified/ 18 spots | % | Identified/ 18 spots | % | |
| 1.0×105 |
18 |
100 |
18 |
100 |
| 5.0×104 |
18 |
100 |
18 |
100 |
| 4.0×104 |
18 |
100 |
18 |
100 |
| 3.0×104 |
18 |
100 |
18 |
100 |
| 2.0×104 |
8 |
44.44 |
15 |
83.33 |
| 1.0×104 |
2 |
11.11 |
10 |
55.56 |
| 9.0×103 |
0 |
|
4 |
22.22 |
| 8.0×103 |
0 |
|
0 |
|
| 7.0×103 | 0 | 0 | ||
Growth of S. aureus in porcine vitreous
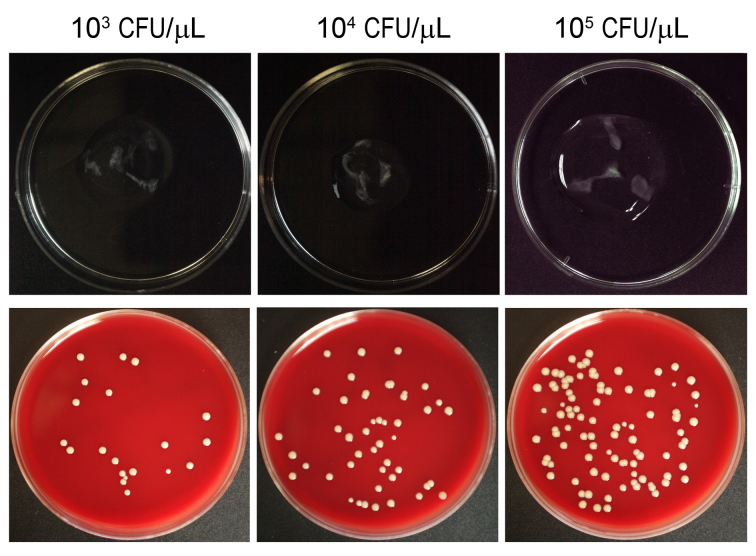
The three different concentrations of S. aureus grew in porcine vitreous in a dose-dependent manner after 12 h of incubation (Figure 2). The clear vitreous became cloudy, and opacity developed along the injection path. Similarly, the homogenized vitreous contained bacteria that developed different densities of colonies after inoculation on the blood agar. The higher the concentration of S. aureus inoculated in the vitreous, the more colonies that appeared on the plate. The total number of bacteria in each vitreous sample was calculated as shown in Table 2.
Figure 2.
Comparison of the growth of Staphylococcus aureus inoculated in porcine vitreous. 103, 104, and 105 CFU/ μl of S. aureus were inoculated (20 μl each injected in four directions, with a total of 80 μl for each eyeball) and incubated for 12 h. Bacteria grown in vitreous can be seen in the upper panel. The vitreous samples were homogenized and diluted (100 times). Fifty microliters of each sample were spread on blood agar plates and cultured for 12 h before counting to examine the number of S. aureus within each eyeball.
Table 2. Identification of S. aureus in pellets of cultured vitreous by Vitek MS.
| Inoculated concentrations(CFU/μl) | Cultured concentrations(CFU/μl) | Total spots* | Vitek MS identification results | Concordant identification [no. (%)] | No identification [no. (%)] | Discordance [no. (%)] |
|---|---|---|---|---|---|---|
| 1.0×105 |
1.0×106 |
9 |
S. aureus |
9 (100) |
0 (0) |
0 (0) |
| 1.0×104 |
0.9×105 |
9 |
S. aureus |
9 (100) |
0 (0) |
0 (0) |
| 1.0×103 | 1.1×104 | 9 | S. aureus | 2(22.2)§ | 7 (77.8) | 0 (0) |
* 3 spots for each eyeball and 3 eyeballs in each group. § 2 spots from the two eyeballs.
Staining evaluation of S. aureus–inoculated vitreous
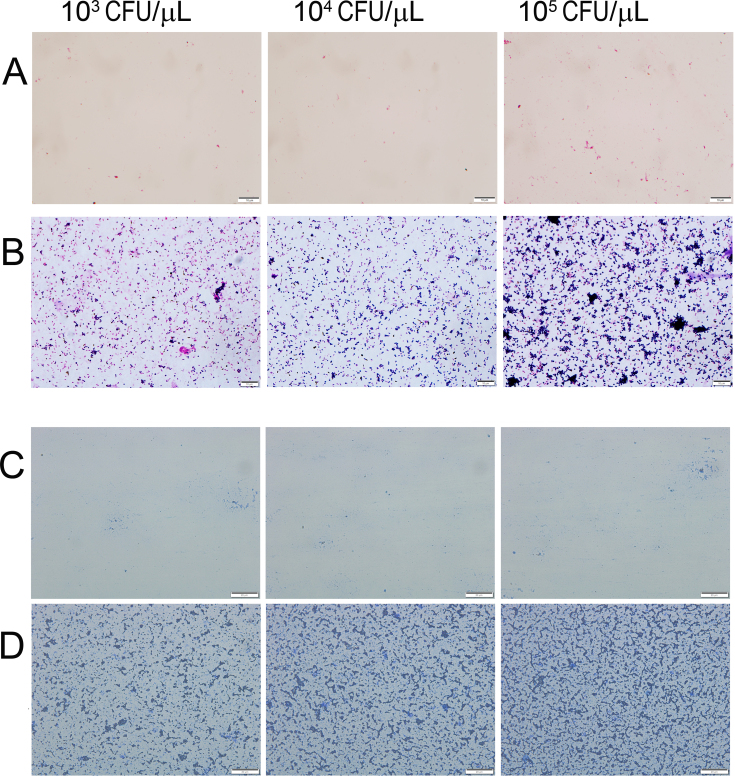
Gram staining was applied to the bacteria in vitreous post-centrifugation (Figure 3A,B). S. aureus can be seen as disperse or aggregate punctuate staining under microscope. There was a high proportion of S. aureus in the pellets, which increased in a dose-dependent manner in the vitreous inoculated with increasing concentrations of bacteria. In contrast, few bacteria were found in the supernatant of the vitreous samples under the microscope.
Figure 3.
Staining of the separations of porcine vitreous inoculated with Staphylococcus aureus over 12 h. After homogenizing and centrifuging, Gram staining (A, B) and Coomassie Blue staining (C, D) were conducted to observe the vitreous supernatants (A, C) and pellets (B, D). In A and B, the scale bar is equal to 10 µm, and in C and D, the scale bar is equal to 20 µm.
CBB staining was used as an indicator to show whether the structure of the vitreous had been disrupted. Figure 3C,D shows that there are small pieces of bulk or strip structures in the pellets post-centrifugation. In the supernatant, few thin filiform structures were observed.
Culture evaluation on blood agar plates
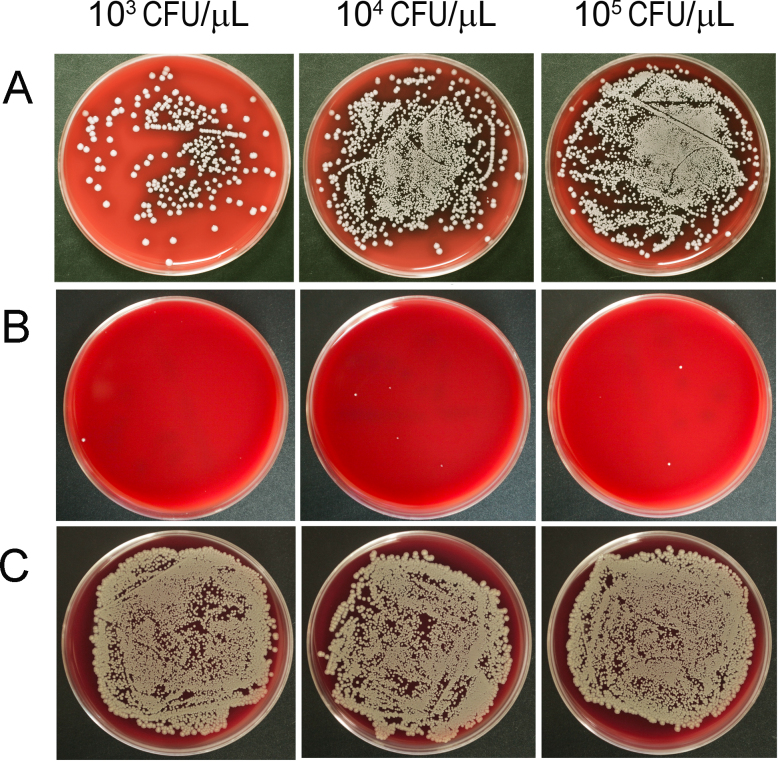
The homogenized vitreous from the inoculated eyeballs, as well as the supernatant and the pellets of vitreous post-centrifugation, was smeared and cultured on blood agar plates (Figure 4). A great number of colonies appeared on the plates smeared with the homogenized vitreous and the resuspended pellets. However, after 24 h of incubation, there were only traces of colonies on the plates inoculated with supernatant.
Figure 4.
Culture plates of the separations of porcine vitreous inoculated with Staphylococcus aureus. After homogenization and centrifugation, the vitreous lysates (A, 50 μl of 100X lysate), supernatant (B, 50 μl of 100X solution), and pellet (C, 50 μl of 10X resuspension) were smeared on blood agar plates and incubated for 24 h. Few colonies were identified in the supernatant samples across all concentrations of S. aureus.
Identification of S. aureus cultured in the porcine eyeball
S. aureus suspensions at 1.0 × 103, 1.0 × 104, and 1.0 × 105 CFU/μl were inoculated separately into porcine eyeballs and incubated for 12 h. The number of the resulting S. aureus in the vitreous after homogenizing and centrifuging was determined by counting the colonies on the blood agar plates (Table 2). After incubation for 12 h, the number of bacteria inoculated in vitreous increased greatly, in a dose-dependent way, in all three groups. The number of bacteria increased approximately ten times for all three concentration groups. After centrifugation, the pellets were used to identify S. aureus present. Table 2 shows that in the two highest concentration groups, S. aureus could be identified in all samples. In the lower concentration 1.0 × 103 vitreous group, two spots out of the nine total spots were positive for S. aureus (22.2%).
Comparison of identification methods for patients’ vitreous
We tested the new method to analyze two vitreous samples from patients with endophthalmitis. Table 3 compared direct MS analysis and traditional methods (culturing + MS analysis, reconfirmed with the VITEK 2 Colorimetric identification Card). The data displays that we can get the same results with direct MS analysis as the traditional approaches for different bacteria in vitreous.
Table 3. Comparison of identification of isolates from two vitreous specimens of patients with endophthalmitis.
| Approaches | Patient A | Patient B |
|---|---|---|
| Surgery |
vitrectomy |
vitrectomy |
| Specimen description |
mixture (certain amount, in an eSwab™ tube)* |
mixture (certain amount, in an eSwab™ tube)* |
| Regular culturing & identification | ||
| Culturing |
nutrient broth, colonies on plate |
nutrient broth, colonies on plate |
| Vitek MS |
Serratia marcescens
(confidence: 99.9%) |
Staphylococcus epidermidis
(confidence: 99.9%) |
| Vitek 2 colorimetry |
Serratia marcescens |
Staphylococcus epidermidis |
| Direct MS analysis | ||
| Centrifuging |
Pellets (from 150 μl mixture) |
Pellets (from 180 μl mixture) |
| Vitek MS |
Serratia marcescens
(confidence: 99.9%) |
Staphylococcus epidermidis
(confidence: 98.3%) |
| Vitek 2 colorimetry | Serratia marcescens | Staphylococcus epidermidis |
* eSwab™ sample collect/transport system contains 1mL of Amies medium. Usually surgeons are required to add 0.5 ml or more vitreous samples to this sample tube.
Discussion
Endophthalmitis, one of the most severe infections in ophthalmology, often leads to a poor visual prognosis. Prompt surgical treatment, sample assays for pathogen identification, and appropriate antibiotic therapy compose the primary therapeutic approach. At present, species determination requires at least 2 days after collection of the surgical vitreous samples from the patient [19]. Therefore, new methods for rapid identification are required. Here, we present a proof-of-concept validation of a method for the direct identification of S. aureus in vitreous, without a subculture of bacteria, based on MALDI-TOF-MS analysis. This approach can be completed within 1 h after the vitreous sample is obtained from the patient. The main advantage of this method is speed, allowing for the initiation of the most appropriate therapy for the patient 1 to 2 days sooner. Furthermore, we tested this method in two vitreous samples from patients with endophthalmitis and obtained the same results as traditional approaches. However, we need more patient vitreous samples to explore this method to substantially improve the clinical management of endophthalmitis via early administration of the appropriate antibiotic agents.
The vitreous humor, composed of 99% water, is a transparent gelatinous mass with a viscosity two to four times that of water. In addition to water, salts, and small organic substances, the vitreous is structured by a network of collagen type II fibrils with glycosaminoglycan, hyaluronan, opticin, and other proteins [20]. When vitreous specimens are obtained from patients with endophthalmitis for examination, the gel-like vitreous and its constituents may interfere with the MS identification of bacteria. The association between the bacterial concentration and the identification ability of MALDI-TOF-MS has been described in previous studies. The minimum bacterial concentrations required for MALDI-TOF-MS identification are >107 CFU/ml in blood culture broth and >106 CFU/ml on culture plates [5,21]. In the current study, mechanical homogenization and centrifugation were used for the S. aureus vitreous samples to separate them into pellets that can be used for smearing and culture, as well as MALDI-TOF-MS assays. The results revealed that the minimum detectable number of S. aureus with MS was 1 × 104 CFU/µl. This suggests that the pretreatment methods had eliminated the vitreous influence on the MS analysis of S. aureus. This extraction protocol is inexpensive and quick and can be used in a clinical laboratory, with the classic configuration of the bioMérieux VITEK MS system without modification of the automat or the database. This work will help to reduce costs, as well as to minimize the time for identification of the pathogen.
The ultimate objective of the present study was to speed up the identification process for the ophthalmologist when deciding the management of patients. The application of MALDI-TOF-MS could be greatly beneficial in the treatment of endophthalmitis, and this technology is already used in clinical practice, such as blood culture [15,22]. Currently, there are no precise assessments of the effect of MALDI-TOF-MS on a vitreous sample of endophthalmitis. In this study, we merely assayed S. aureus as the pathogen in a model of ex vivo endophthalmitis and tested two vitreous samples from patients with endophthalmitis. Thus far, there are limitations for the application of MALDI-TOF-MS to identify bacteria in vitreous pellets. First, for a vitreous sample with low bacterial loads (<1 × 104 CFU/µl), which may be the case at an early stage of endophthalmitis, the MALDI-TOF-MS assay will not work. The second limitation is the insufficient volume of vitreous obtained from a patient (e.g., a diagnostic vitreous sample/biopsy). Fortunately, therapeutic vitrectomy in most cases can obtain a large volume of vitreous sample, i.e., undiluted (500 μl) and diluted vitreous samples (2 ml) [23]. Finally, another consideration of this approach is that it supplements the regular culturing method (culturing combining with MALDI-TOF-MS testing or colorimetric identification card in a clinical laboratory) for human vitreous samples. In general, since there may be possible discrepancies among various specimens, diverse bacteria, and different identification methods, our results should be confirmed by using additional surgical samples of endophthalmitis on a larger scale of investigation in the future.
In conclusion, this rapid identification method, based on the combination of easy extraction methods and a MALDI-TOF-MS assay, constitutes a powerful supplementary method for the identification of bacteria directly from vitreous samples of endophthalmitis. This method is easily adaptable for use in clinical routine and can help reduce the delay in diagnosis, allowing for earlier therapeutic intervention in patients.
Acknowledgments
This work was partially supported by grants from the Guangzhou Science Technology and Innovation Commission (#201607020011), the Innovation Commission and the Fundamental Research Funds of the Key Lab of Ophthalmology and Visual Science, Guangdong (2014B030301040) and the Fundamental Research Funds of State Key Laboratory, China
References
- 1.Duan F, Wu K, Liao J, Zheng Y, Yuan Z, Tan J, Lin X. Causative Microorganisms of Infectious Endophthalmitis: A 5-Year Retrospective Study. J Ophthalmol. 2016;2016:6764192. doi: 10.1155/2016/6764192. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=27413545&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gentile RC, Shukla S, Shah M, Ritterband DC, Engelbert M, Davis A, Hu DN. Microbiological spectrum and antibiotic sensitivity in endophthalmitis: a 25-year review. Ophthalmology. 2014;121:1634–42. doi: 10.1016/j.ophtha.2014.02.001. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24702755&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 3.Wang N, Yang Q, Tan Y, Lin L, Huang Q, Wu K. Bacterial Spectrum and Antibiotic Resistance Patterns of Ocular Infection: Differences between External and Intraocular Diseases. J Ophthalmol. 2015;2015:813979. doi: 10.1155/2015/813979. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26576294&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong BK, Lee CS, Van Gelder RN, Garg SJ. Emerging techniques for pathogen discovery in endophthalmitis. Curr Opin Ophthalmol. 2015;26:221–5. doi: 10.1097/ICU.0000000000000145. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25759963&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christner M, Rohde H, Wolters M, Sobottka I, Wegscheider K, Aepfelbacher M. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J Clin Microbiol. 2010;48:1584–91. doi: 10.1128/JCM.01831-09. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20237093&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson TL, Paraskevopoulos T, Georgalas I. Systematic review of 342 cases of endogenous bacterial endophthalmitis. Surv Ophthalmol. 2014;59:627–35. doi: 10.1016/j.survophthal.2014.06.002. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25113611&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 7.van Belkum A, Chatellier S, Girard V, Pincus D, Deol P, Dunne WM., Jr Progress in proteomics for clinical microbiology: MALDI-TOF MS for microbial species identification and more. Expert Rev Proteomics. 2015;12:595–605. doi: 10.1586/14789450.2015.1091731. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26472137&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 8.Cheng K, Chui H, Domish L, Hernandez D, Wang G. Recent development of mass spectrometry and proteomics applications in identification and typing of bacteria. Proteomics Clin Appl. 2016;10:346–57. doi: 10.1002/prca.201500086. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26751976&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlek AL, Bonten MJ, Boel CH. Direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry improves appropriateness of antibiotic treatment of bacteremia. PLoS One. 2012;7:e32589. doi: 10.1371/journal.pone.0032589. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22438880&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneiderhan W, Grundt A, Worner S, Findeisen P, Neumaier M. Work flow analysis of around-the-clock processing of blood culture samples and integrated MALDI-TOF mass spectrometry analysis for the diagnosis of bloodstream infections. Clin Chem. 2013;59:1649–56. doi: 10.1373/clinchem.2012.198218. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23881934&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 11.Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015;6:791. doi: 10.3389/fmicb.2015.00791. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26300860&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charretier Y, Schrenzel J. Mass spectrometry methods for predicting antibiotic resistance. Proteomics Clin Appl. 2016;10:964–81. doi: 10.1002/prca.201600041. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=27312049&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 13.Ferreira L, Sanchez-Juanes F, Munoz-Bellido JL, Gonzalez-Buitrago JM. Rapid method for direct identification of bacteria in urine and blood culture samples by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: intact cell vs. extraction method. Clin Microbiol Infect. 2011;17:1007–12. doi: 10.1111/j.1469-0691.2010.03339.x. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20718803&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 14.Veron L, Mailler S, Girard V, Muller BH, L’Hostis G, Ducruix C, Lesenne A, Richez A, Rostaing H, Lanet V, Ghirardi S, van Belkum A, Mallard F. Rapid urine preparation prior to identification of uropathogens by MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2015;34:1787–95. doi: 10.1007/s10096-015-2413-y. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26054715&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 15.Jamal W, Saleem R, Rotimi VO. Rapid identification of pathogens directly from blood culture bottles by Bruker matrix-assisted laser desorption laser ionization-time of flight mass spectrometry versus routine methods. Diagn Microbiol Infect Dis. 2013;76:404–8. doi: 10.1016/j.diagmicrobio.2013.04.013. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23726652&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 16.Assaad D, Wong D, Mikhail M, Tawfik S, Altomare F, Berger A, Chow D, Giavedoni L. Bacterial endophthalmitis: 10-year review of the culture and sensitivity patterns of bacterial isolates. Can J Ophthalmol. 2015;50:433–7. doi: 10.1016/j.jcjo.2015.07.013. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26651302&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 17.Ren Z, Wang ZQ, Li R, Luo SY, Deng SJ, Sun XG. Etiological analysis on bacterial endophthalmitis. Zhonghua Yan Ke Za Zhi. 2007;43:1106–9. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18331681&dopt=Abstract [PubMed] [Google Scholar]
- 18.Oki K, Miyata Y, Shimada A, Nagase T, Katsura Y, Kosano H. Cell-mediated contraction of vitreous explants from chicken embryo: possibility of screening for therapeutic agents against proliferative vitreoretinal diseases. Mol Vis. 2013;19:2374–84. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24319331&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai T, Kohzaki K, Watanabe A, Tsuneoka H, Shimadzu M. Use of DNA microarray analysis in diagnosis of bacterial and fungal endophthalmitis. Clin Ophthalmol. 2012;6:321–6. doi: 10.2147/OPTH.S29230. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22399844&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kokavec J, Min SH, Tan MH, Gilhotra JS, Newland HS, Durkin SR, Grigg J, Casson RJ. Biochemical analysis of the living human vitreous. Clin Experiment Ophthalmol. 2016;44:597–609. doi: 10.1111/ceo.12732. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26891415&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 21.Wang MC, Lin WH, Yan JJ, Fang HY, Kuo TH, Tseng CC, Wu JJ. Early identification of microorganisms in blood culture prior to the detection of a positive signal in the BACTEC FX system using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. J Microbiol Immunol. 2015;48:419–24. doi: 10.1016/j.jmii.2013.10.006. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24388584&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 22.Moussaoui W, Jaulhac B, Hoffmann AM, Ludes B, Kostrzewa M, Riegel P, Prevost G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry identifies 90% of bacteria directly from blood culture vials. Clin Microbiol Infect. 2010;16:1631–8. doi: 10.1111/j.1469-0691.2010.03356.x. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20825442&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 23.Chiquet C, Maurin M, Thuret G, Benito Y, Cornut PL, Creuzot-Garcher C, Rouberol F, Pechinot A, Lina G, Romanet JP, Bron A, Vandenesch F. Analysis of diluted vitreous samples from vitrectomy is useful in eyes with severe acute postoperative endophthalmitis. Ophthalmology. 2009;116:2437–41e1. doi: 10.1016/j.ophtha.2009.06.007. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19815283&dopt=Abstract [DOI] [PubMed] [Google Scholar]