Abstract
Investigating the interactions of RNA-binding proteins (RBPs) with RNAs is a complex task for molecular and computational biologists. The molecular biology techniques and the computational approaches to understand RBP–RNA (or ribonucleoprotein, RNP) interactions have advanced considerably over the past few years and numerous and diverse software tools have been developed to analyze these data. Accordingly, laboratories interested in RNP biology face the challenge of choosing adequately among the available software tools those that best address the biological problem they are studying. Here, we focus on state-of-the-art molecular biology techniques that employ crosslinking and immunoprecipitation (CLIP) of an RBP to study and map RNP interactions. We review the different software tools and databases available to analyze the most widely used CLIP methods, HITS-CLIP, PAR-CLIP, and iCLIP.
INTRODUCTION
In mammalian cells, gene expression programs are robustly influenced by post-transcriptional processes affecting pre-messenger RNA (pre-mRNA) splicing and maturation, as well as mRNA transport, editing, translation, and degradation.1–3 These post-transcriptional events are mainly controlled by RNA-binding proteins (RBPs) and noncoding (nc)RNAs.4,5 Through rich and dynamic interactions with subsets of mRNAs, RBPs and ncRNAs can govern the patterns of expressed proteins and hence a broad range of cellular processes (e.g., proliferation, apoptosis, differentiation, immune response) and consequently the function and dysfunction of tissues, organs, and systems.6–9
To investigate RNP interactions on a transcriptome-wide level, several biochemical methods have been developed over the past ~15 years. Some methods assess native RNP associations without crosslinking or RNase digestion and identify the native RNP complex by immunoprecipitation (IP). Since this method, termed RNP IP or RIP, selects full-length or otherwise long RNAs, bound transcripts are most often identified by microarray analysis (RIP-chip)10 or by next-generation sequencing (RIP-Seq) analysis after fragmentation of RNA by sonication.11,12 Recently, some variations of RIP have been reported that include crosslinking with formaldehyde,13,14 but they have not been used widely to identify precise RBP-binding sites as they do not include an RNase digestion step. Therefore, RIP-derived methods have not been included in this review.
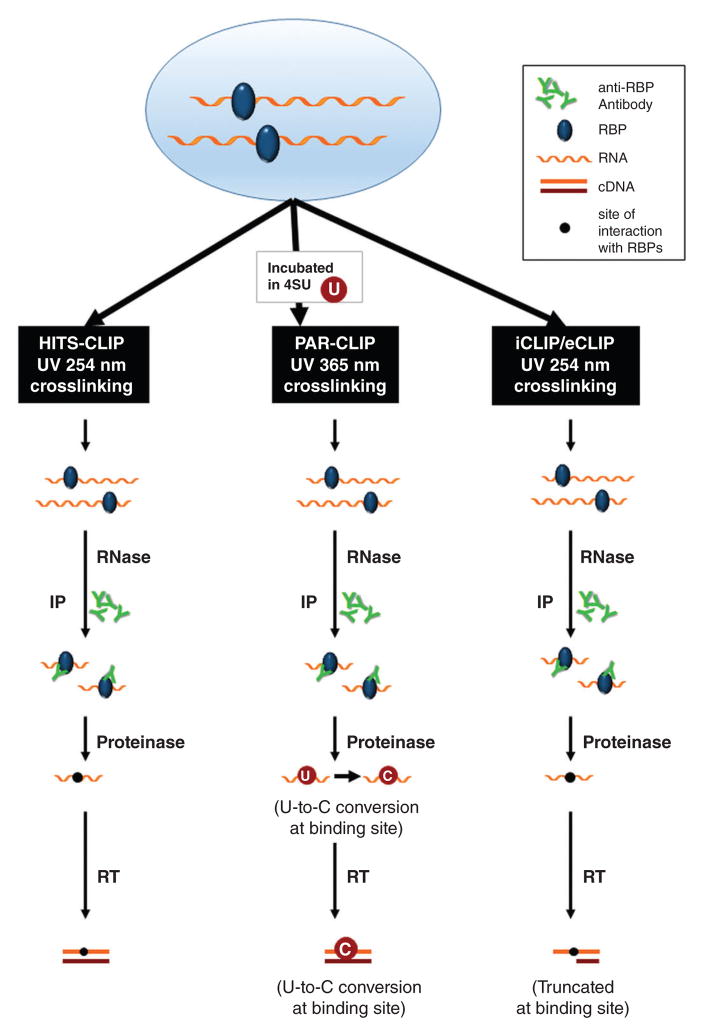
For high-resolution analysis of the RNP interactions, several methodologies have been devised in which the RNP complex is first subjected to crosslinking by irradiation with ultraviolet (UV) light. Following RNase digestion of the crosslinked RNP and IP using a specific anti-RBP antibody, the RNA site where the specific RBP binds can be identified with high precision.15 A number of such CLIP (crosslinking with immunoprecipitation) methods have been developed, including HITS-CLIP (high-throughput sequencing CLIP, also known as CLIP-Seq), PAR-CLIP (photoactivatable-ribonucleoside-enhanced CLIP), and iCLIP/eCLIP (individual-nucleotide/enhanced resolution CLIP) (Figure 1). All of these technologies use high-throughput RNA sequencing (RNA-Seq) to identify en masse the RNA component of the RNP complexes. There are excellent reviews that compare the advantages and limitations of each of these methodologies at the levels of biochemistry and molecular biology.15–18 In this article, we focus instead on the software tools and databases freely available to analyze the data generated by each of these CLIP methods.
FIGURE 1.
Schematic overview of the strategies to identify the sites of RBP-RNA interactions: HITS-CLIP, PAR-CLIP, and iCLIP/eCLIP. All strategies initially involve crosslinking with ultraviolet light (UV) at 254 nm or 365 nm and RNP immunoprecipitation. After RNase digestion and IP, the complexes are digested with protease to release the bound RNA segments, which are used to generate a library for sequencing. PAR-CLIP analysis includes the pre-incubation of cells with 4SU to gain further confidence that the binding sites identified are regions of bona fide interaction with RBPs, and iCLIP/eCLIP analysis truncates fragments at the precise site of interaction with the RBP.
SOFTWARE TOOLS
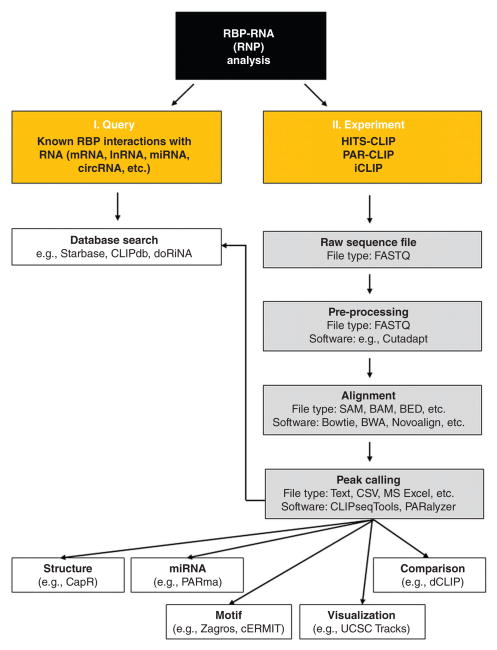
The overall workflow for analyzing different types of CLIP data is very similar. The sequence reads are first pre-processed (adapter sequences are cleaned and low-quality bases removed using software such as Cutadapt), the processed sequence reads are aligned to the genome sequences, and the alignments are used to identify the RNA segments associated with the RBP of interest. The mapped interaction sites can be further used to find signature motifs and structures (Figure 2). In this section, we focus on analysis strategies and specifications for the three most widely used sequencing methods that identify discrete RBP-interaction sites: HITS-CLIP, PAR-CLIP, and iCLIP. The different analysis tools and their salient features are summarized in Table 1. As commercial software tools tend to be costly and restrictive to user customization, they have been excluded from this review (Table 2).
FIGURE 2.
Overview of the general analysis workflow for CLIP-seq data. (I) The user begins by querying whether an RNP complex of interest has already been reported by searching databases like Starbase, CLIPdb, and doRiNA. (II) If the RNP has not been reported, then the user analyzes or generates a dataset that detects these interactions; the ensuing analysis (gray boxes) begins with preparation of the raw sequencing file, preprocessing and alignment of the data, and peak calling (identification of the binding site). At that point (white boxes), the binding sites may be further analyzed (comparison, visualization, and identification of motifs, microRNA interaction sites, structural elements, etc). Once the analyses have been completed, they can be made available for other users.
TABLE 1.
Summary of Different Software Tools and Their Applications
From left to right: software names are followed by the three major CLIP analysis methods and further analyses/comparisons enabled by various tools. Whether the tools are databases or have web-interfaces and the links to each website are also indicated.
TABLE 2.
List of Abbreviations
| Abbreviation | Name |
|---|---|
| API | Application program interface |
| ASPeak | Abundance sensitive peak detection algorithm |
| AURA | Atlas of UTR regulatory activity |
| BAM | Binary alignment/map |
| BLAST | Basic local alignment search tool |
| BLAT | BLAST-like alignment tool |
| cERMIT | Evidence-ranked motif identification tool |
| ChIP | Chromatin immunoPrecipitation |
| CIMS | Crosslinking induced mutation site |
| CircInteractome | Circular RNA interactome |
| CircRNA | Circular RNA |
| CLIP | UV crosslinking with immunoprecipitation |
| CRAC | Crosslinking and cDNA analysis |
| DB | Database |
| doRiNA | Database of RNA interactions |
| FDR | False discovery rate |
| GO | Gene ontology |
| HITS-CLIP | UV crosslinking with immunoprecipitation and high-throughput sequencing |
| HMM | Hidden Markov model |
| HPeak | HMM-based peak calling |
| iCLIP | Individual nucleotide-resolution crosslinking and immunoprecipitation |
| IP | Immunoprecipitation |
| IRES | Internal ribosome entry site |
| mEAT | MicroRNA enrichment analysis tool |
| miRNA | MicroRNA |
| mRNA | Messenger RNA |
| MUMMIE | Multivariate Markov modeling inference engine |
| ncRNA | Non-coding RNA |
| PARalyzer | PAR-CLIP data analyzer |
| PARCLIP | Photoactivatable-ribonucleoside-enhanced CLIP |
| PARma | PAR-CLIP miRNA assignment |
| RBP | RNA binding protein |
| RIP | RNA immunoprecipitation |
| RNA | Ribonucleic acid |
| RNP | Ribonucleoprotein |
| RT | Reverse transcription |
| SAM | Sequence alignment/map |
| Seq | Sequencing |
| UCSC | University of California, Santa Cruz |
| UTR | Untranslated region |
| UV | Ultraviolet |
| ZTNB | Zero-truncated negative binomial |
HITS-CLIP/CLIP-Seq
The advantage of this method over previous versions of RIP, including RIP followed by formaldehyde crosslinking, is that HITS-CLIP (also named CLIP-Seq) uses irradiation with UV light for crosslinking,57,58 enabling subsequent RNase digestion to eliminate unprotected RNA segments and detect direct and specific contacts between RNA and protein. This method takes advantage of the natural photoreactivity of nucleic acid bases, particularly pyrimidines (C and U), at UV light wavelengths of 254 nm.59 Numerous tools are available to analyze HITS-CLIP, including many that can be used for analysis of PAR-CLIP and iCLIP:
CLIPSeqTools
This compendium of tools for analyzing HITS-CLIP/CLIP-Seq data is written in Perl and can be installed and run on a Linux server. CLIPSeqTools can pre-process FASTQ files, remove the adapters, mask the repeats, and align the reads to the genome using a fast and accurate aligner named STAR (spliced transcripts alignment to a reference).60 It can be customized to generate tables for the identified regions, annotations for the positional binding preferences [e.g., 5′-untranslated region (UTR), 3′UTR, coding region (CR), exons or introns], and plots for different types of distributions. After processing, CLIPSeqTools supports the export of data to BED (browser extensible data) files for visualization, motif analysis, evaluation of evolutionary conservation, and generation of tables. For sample comparison CLIPSeqTools uses upper-quartile normalization. Pre-mRNAs, introns, and exons can also be exported in DESeq61 format for further analysis of binding differences. CLIPSeqTools uses SQLite3 as the default database engine, increasing the flexibility of retrieving information from the sequenced data and extending annotations.19 Website: http://mourelatos.med.upenn.edu/clipseqtools/.
ASPeak
The ASPeak (abundance sensitive peak) detection algorithm, as the name suggests, corrects for the abundance of a transcript in the cell with consideration of background levels. ASPeak is implemented in Perl and uses BedGraph files as input, so the alignment needs to be done outside of the ASPeak pipeline (on personal computers, servers, or computer clusters). The read counts for any genomic interval (coding, UTRs, introns, intergenic, or user-defined) are modeled as negative binomial distributions.20 Website: https://sourceforge.net/projects/aspeak/.
CLIPZ
A unique feature of CLIPZ is that it is both a database of RBP–RNA interactions (HITS-CLIP and PAR-CLIP) and a web tool to analyze user-specified data.21 The web tool uses the Burrows-Wheeler aligner (BWA)62 and permits the user to find ‘clusters’ based on alignment to the genome or transcriptome and to create ‘super-clusters’ from multiple samples. It also allows the generation of enriched mRNA sites relative to mRNA expression abundance63 and the visualization of data using genome or transcriptome browsers. CLIPZ can also be used for functional annotation of other types of short-read data such as for mRNA-Seq, DGE (digital gene expression), and small RNA cloning. CLIPZ also includes several tools specific to miRNA analysis such as miRNA expression profiling, principal component analysis (PCA), clustering, target site prediction, and motif analysis. However, as with most online NGS analysis tools, data transfer and analysis are slow. Website: http://www.clipz.unibas.ch/.
PIPE-CLIP
PIPE-CLIP is a comprehensive online tool for analyzing different types of CLIP data, including HITS-CLIP, PAR-CLIP, and iCLIP.22 The PIPE-CLIP pipeline uses SAM (sequence alignment/map) or BAM (the compressed, binary form of SAM) alignment files and the web interface uses the GALAXY platform,64 and therefore many web-based ata analysis tools and genomes are also available. PIPE-CLIP allows flexible removal of PCR duplicates and uses a zero-truncated negative binomial (ZTNB) model for identifying the enriched peaks. It has the same data transfer issues and slow analysis common to web-based tools, but the source code and command line tools are available from github (https://github.com/QBRC/PIPE-CLIP), which makes fast analysis possible. Website: https://pipeclip.qbrc.org/root.
Piranha
Piranha can also be used for all variations of CLIP (iCLIP, PAR-CLIP, HITS-CLIP) by modeling count distributions and external covariates such as transcript abundance. When the conversion of thymidine to cytidine (T-to-C transition) is used as a covariate, this software can process PAR-CLIP data in a way similar to how the PARalyzer software (below) processes them. For finding differentially used binding sites between samples, Piranha uses read counts in the first tissue or condition as a covariate of the second. It also uses a ZTNBR regression model when covariates are provided and otherwise uses a ZTNB model for finding the enriched peaks. BED or BAM alignment files are used for data input.23 Binding motifs are identified using the distributed mutual exclusion (DME) algorithm.65 Website: http://smithlabresearch.org/software/piranha/.
MiClip
MiClip is a novel method to identify high-confidence protein-RNA binding sites from HITS-CLIP and PAR-CLIP datasets by assigning a probability for each potential binding site. It is available as an R package (https://cran.r-project.org/src/contrib/Archive/MiClip/) and as a web tool on GALAXY. Using aligned SAM files as input, MiClip first removes PCR duplicates and uses two rounds of HMM: the first finds enriched regions and the second finds binding sites of RBPs in the enriched regions. It generates tables and creates BedGraph files for easy visualization in the UCSC Genome browser.66 A Perl script is also provided for 7-mer seed motifs.24 Website: http://galaxy.qbrc.org/root?tool_id=mi_clip
HITS-CLIP Analysis
This MATLAB toolbox detects RNA-protein binding sites in HITS-CLIP datasets following a two-stage analysis: the first HMM round identifies enriched locations and the second assesses the reliability of mutations and determines the binding sites. This toolbox aligns reads and provides essential MATLAB functions to identify binding sites using semi-supervised learning. Website: https://qbrc.swmed.edu/softwares.php.
Pyicoclip
Pyicoclip is a part of Pyicoteo package which can call peaks from HITS-CLIP data without a control sample by creating background frequencies through randomization of reads within the same gene. It implements a modified false discovery rate (FDR) algorithm proposed by Yeo et al.67 The input files are aligned files in ELAND, SAM, BAM, or BED format and the output is compatible BED format. The associated scripts can help analyze large and diverse set of NGS data in an efficient manner.25 Website: http://regulatorygenomics.upf.edu/Software/Pyicoteo/index.html.
GraphProt
This program uses HITS-CLIP datasets and secondary structure from the RNA Shapes program68 to generate binding profiles at nucleotide resolution, high-affinity target sites, and binding motifs.26 GraphProt provides a flexible machine learning framework to identify RBP-binding models by using a graph-kernel approach for RNA secondary structure based on Support Vector Machine (SVM) algorithms. The starting file format is FASTA, but it can also work with data generated using ‘RNAcompete.’69 Website: http://www.bioinf.uni-freiburg.de/Software/GraphProt/.
CIMS
Crosslinking-induced mutation site (CIMS) is a novel tool to analyze HITS-CLIP data for determining the exact RBP-RNA crosslink site at single-nucleotide resolution. This analysis method is based on the observation that UV-crosslinked aminoacid-RNA adducts introduce reverse transcription (RT) errors in cDNAs at certain frequencies which are captured by HITS. These mutations could be deletions, insertions, or substitutions (usually discarded in standard HITS-CLIP data analysis) that can be identified with statistical significance using FDR. CIMS is written in Perl and runs on Linux/Unix systems. The input files are two BED files: one is a list of reads uniquely aligned to the genome and the other is a list of coordinates of all the mutations. For alignment, Novoalign (Novocraft Technologies) is recommended for easy conversion of data format using the scripts supplied, but BWA62 can also be used for alignment with custom scripts.27,28 Website: http://zhanglab.c2b2.columbia.edu/index.php/CIMS_Documentation.
CLIPper
CLIPper is a Python tool that can be used to find peaks in HITS-CLIP data. The input file is a BAM file generated after aligning the reads to the genome. It uses a three-stage filter to reduce the false positives: first an FDR or a read coverage cut-off, then a poisson p-value cut-off, and finally a cubic spline to approximate the shape of the peak. The output is a table in BED (bed8) format.29 Website: http://yeolab.github.io/software/.
CLIP-PyL
The CLIP-PyL package, with scripts written in Python, can detect peaks in HITS-CLIP data and generate base-specific coverage metrics. The software needs aligned BAM files as input files along with a BED file containing the gene or transcript in which the user is interested. The program creates pile-ups of mapped reads and can generate a PDF file with coverage plots or BedGraph files that can be uploaded to any genome browsers for visualization. Website: https://github.com/lb3/CLIP-PyL.
PyCRAC
PyCRAC is a suite of Python scripts which can be used to analyze HITS-CLIP, PAR-CLIP or CRAC (CRross-linking and Analysis of cDNA) data70 and many other types of data. Novoalign is recommended for alignment; the software tool then counts the reads, clusters them and calculates FDR. The tool provides utilities to remove adapter contamination and PCR duplicates.30 The output files can be used to find motifs using a supplied script called pymotif or by using MEME.71 Website: http://sandergranneman.bio.ed.ac.uk/Granneman_Lab/pyCRAC_software.html.
TEPeaks
TEPeaks is still under development, but is designed to identify enriched regions of RNA or DNA bound to proteins. It can be used for analysis of HITS-CLIP data and enhances the method implemented by MACS software72 by identifying ‘narrow’ peaks. It is written in Python and the input files are BAM or SAM containing multiple (ideally ~100) alignments and GTF file for gene or transcript models.31 Website: http://hammelllab.labsites.cshl.edu/software/#TEToolkit.
RNAseqlib
RNAseqlib provides a simplified pipeline for analyzing RNA-Seq data from sources including mRNA, HITS-CLIP, Ribo-Seq and SELEX-Seq.32 It is written in Python and uses FASTQ files as input and bowtie/tophat as the aligner. Website: https://rnaseqlib.readthedocs.io/en/clip/.
ProfileSeq
ProfileSeq is a broad-use software written to test profiles of a test sample relative to a control sample and generates a nonparametric probability comparison of the signal density. This software can be used for HITS-CLIP, chromatin IP (ChIP)-Seq, GRO-Seq,73 and other types of HITS following nucleic acid enrichment. The input files are BED files or ‘position files’ (specific to this program).33 Web-site: https://bitbucket.org/regulatorygenomicsupf/profileseq/.
miRTarCLIP
miRTarCLIP identifies miRNA target sites from HITS-CLIP and PAR-CLIP data. Using FASTQ files for data input, the integrated workflow of miRTarCLIP includes automatic removal of adapter sequences, filtering of low-quality reads, alignment of reads to 3′UTRs, T-to-C conversions of PAR-CLIP data, and identification of high-confidence miRNA target sites.74 The program has an intuitive web-based workflow, but the users need to install it in their own servers.34 Website: http://mirtarclip.mbc.nctu.edu.tw/index.php.
Zagros
Zagros is a motif-discovery software for HITS-CLIP data. It can be used for finding motifs using just the binding sequences provided as a FASTA file (input), but it can also use structure information or crosslink modification events (or ‘diagnostic events’) extracted from the sequenced data using extractDEs program and can be used with iCLIP or PAR-CLIP data. Website: http://smithlabresearch.org/software/zagros/.
RNAcontext, RBPmotif
RNAcontext identifies RBP-binding motifs and structural preferences with high accuracy. It is used for finding motifs de novo and analyzing secondary structure to assess if the binding sequence motif is enriched in the context of a particular structure relative to unbound sequences. It requires a list of sequences along with their structure annotation profiles estimated from SFOLD, and RNA-binding affinity estimates for any RBP as the input. It generates a list of RNA-binding domains and the predicted motifs. RBPmotif is a web server implementation of the RNAcontext.35 Website: http://rnamotif.org/.
mCarts
mCarts is an HMM-based method for improved identification of RNA motifs by considering the number of motifs, the distance between individual motif sites, their accessibility, and their conservation. mCarts requires two BED files, a positive dataset comprising HITS-CLIP read clusters, and a negative dataset which typically includes gene regions without HITS-CLIP reads. It also needs ‘library’ files (included in the tool for human and mouse) of information about the genes, UTRs, 10-kb sequences upstream and downstream of genes, and phylogenetic trees for 20 mammalian species.36 Website: http://zhanglab.c2b2.columbia.edu/index.php/MCarts_Documentation.
CapR
CapR is a software that uniquely identifies secondary structures in CLIP data. Given that RNA motifs interacting with RBPs are very short and usually degenerate, and hence it is difficult to elucidate the specificity of RBP–RNA interactions, CapR uses computational methods to assess secondary structures of RNAs (stems, hairpin loops, bulge loops, internal loops, multi-branch loops, and exterior loops) in regions near binding sites. The required input file is a FASTA or multi-FASTA file containing sequences of the RBP-bound RNAs.37 Website: https://github.com/fukunagatsu/CapR.
Deepnet-rbp
RNA molecules in the cell have secondary and tertiary structures. Deepnet-rbp is a flexible software framework written in Python for modeling structural binding preferences and predicting RBP-binding sites by taking into account secondary and tertiary RNA structure using RNAshapes and JAR3D.75,76 Briefly, it uses restricted Boltzmann machines (RBMs)77 as fundamental building blocks of the deep learning and performs maximum likelihood estimation (MLE) to train the model, after which the softmax function predicts the sequence and/or structural features of RBP-binding sites.38 Website: https://github.com/thucombio/deepnet-rbp.
PAR-CLIP
PAR-CLIP (photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation) is based on the HITS-CLIP technology, but cells are first cultured with a photoreactive ribonucleoside analogue, typically 4-thiouridine (4SU), which helps to identify specific sites of RNA-protein crosslinking.78 The precise binding site can be identified by scoring T-to-C transitions in the sequenced cDNA. Besides many software tools that can also be used to analyze HITS-CLIP data (section above), there are several tools specific to PAR-CLIP:
PARalyzer
PARalyzer (PAR-CLIP data analyzer) is pioneering software for PAR-CLIP analysis. It uses alignments generated from bowtie aligner79 (bowtie or SAM/BAM format) and generates two kernel density estimates, one for T-to-C conversion, and one for non-conversion events. If the likelihood of T-to-C conversion is found to be higher than non-conversion after a filtering (usually 5 events or more), it is considered to be an interaction site. It can use very short read lengths (13 bases or higher) after adapter trimming, and thus it can identify accurately RBP-binding sites.39 Website: https://ohlerlab.mdc-berlin.de/software/PARalyzer.
PAR-CLIP HMM
The PAR-CLIP HMM tool integrates reads distribution and mutation counts. Since it is an R package, it needs Rcpp to be installed for seamless integration of R and C++. The input is aligned reads and it uses Bayesian HMM.40 Website: https://qbrc.swmed.edu/softwares.php.
wavClusteR
wavClusteR is a pipeline for analyzing PAR-CLIP data written in R and available as a part of the Bioconductor package. wavClusteR first identifies true PAR-CLIP (T-to-C) conversions in aligned sorted BAM files different from sequencing errors or single-nucleotide polymorphisms (SNPs) and then identifies RBP-binding sites (clusters) at high resolution using a Bayesian framework. The pipeline also helps export the data in UCSC genome browser formats for visualization and for motif search, and uses multiple computational cores in parallel, which accelerates data analysis. This pipeline can also be used for analysis of bisulfite sequencing and DNA methylation.41,42 Website: https://bioconductor.org/packages/release/bioc/html/wavClusteR.html.
Bmix
BMix addresses the possible presence of high-frequency false positive mutations by considering multiple sources of noise in PAR-CLIP data (e.g., SNPs, sequencing errors, misalignments) though probabilistic modeling of the substituted bases. It is written in MATLAB or R and uses aligned BAM files generated by bowtie as input.43 Website: https://github.com/cbg-ethz/BMix.
BackCLIP
This tool identifies the common background that might be present in PAR-CLIP data and gives the user the option of removing it. BackCLIP is written in Python and the background file and the PAR-CLIP clusters must be in BED format.44 Website: https://github.com/phrh/BackCLIP.
PARma
The PAR-CLIP miRNA assignment (PARma) tool is specific for Argonaute (AGO) PAR-CLIP analyses. It identifies target sites of microRNAs from AGO PAR-CLIP data in two steps: first, a PAR-CLIP-specific analysis computes the likely seed sites for microRNAs, and second, a novel pattern-discovery tool (KmerExplain) estimates seed probabilities. Both of these steps are iteratively applied until a convergence is reached. The input is one or more BED files containing aligned PAR-CLIP reads with the name field containing the number of reads mapped at that genomic location obtained from the aligned SAM file.45 Website: https://www.bio.ifi.lmu.de/PARma.
MicroMUMMIE
The computational framework MicroMUMMIE integrates sequence and binding information from a PAR-CLIP experiment and identifies microRNA target sites. The tool uses the AGO PAR-CLIP analysis generated using PARalyzer and searches the complete UTR and not just the clusters of bound RNA.46 Website: https://ohlerlab.mdc-berlin.de/software/microMUMMIE_99/.
cERMIT/mEAT
The evidence-ranked motif identification tool (cERMIT) can be used effectively for identification of motifs in PAR-CLIP data. In the case of AGO PAR-CLIP, the motif analysis evaluates a pre-defined set of microRNAs based on complementary or canonical seed matches (miRNA enrichment analysis approach or mEAT).47 Website: https://ohlerlab.mdc-berlin.de/software/PAR-CLIP_motif_analysis_tool_87/.
iCLIP and eCLIP
Individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) is another improvement of the HITS-CLIP technologies to identify RBP-binding sites even more precisely—down to the nucleotide level. The method includes a cDNA circularization step that truncates the cDNAs at the protein-RNA crosslink sites and thereby generates data with high resolution and specificity.80 Enhanced CLIP (eCLIP) represents an improvement over the current iCLIP protocol by maintaining the single-nucleotide resolution of iCLIP but decreasing the need for PCR amplification by ~1000-fold, thus reducing the numbers of PCR duplicates.81 For eCLIP, normalization to sample input is preferred over normalization to IgG control IP samples. Many of the software tools for HITS-CLIP analysis are able to analyze iCLIP/eCLIP data, but there is a need to refine iCLIP/eCLIP analyses in many cases:
iCLIPpro
iCLIPro is a robust tool that corrects for previously unrecognized effects of iCLIP fragment length on the identification of binding sites for some RBPs, thus improving the identification of binding sites. iCLIPro first visualizes coinciding and non-coinciding fragment start sites in an aligned BAM file and then identifies the best way to analyze iCLIP data by generating overlap heatmaps.48 Website: http://www.biolab.si/iCLIPro/doc/.
COMPARATIVE ANALYSIS
Most of the HITS-CLIP/PAR-CLIP/iCLIP analysis tools do not compare between samples. It is relatively simple to compare binary events (i.e., whether a binding site is shared or not between samples), but it is problematic to compare binding strengths for which specialized software are necessary.
dCLIP
This software tool finds differentially bound regions in two HITS-CLIP, PAR-CLIP or iCLIP experiments. dCLIP is unique in that it finds regions with differences in binding strengths rather than just the binary event of whether a binding site is shared or not. It uses a MA-plot normalization followed by HMM to identify shared and differentially bound regions. The dCLIP software is written in Perl, needs SAM alignment files (from both single-end or paired-end sequencing) for input, and generates tables and BED files for visualization in the UCSC genome browser.49 Website: https://qbrc.swmed.edu/softwares.php.
Databases
Searchable public databases of RBP–RNA interactions are the easiest and most readily available sources of curated RNP data. They usually provide a simple web interface and do not need specific computational skills to mine vast amounts of published RNP data. Many databases also provide additional features such as filters, overlaps of datasets, generation of networks, and primer design.
StarBase
StarBase contains RNA–RNA and protein–RNA interaction data from 108 CLIP-Seq datasets (including PAR-CLIP, HITS-CLIP, and iCLIP) from 37 independent studies. It contains 285,000 protein–RNA interactions and several thousand miRNA–RNA interactions. All of these datasets can be accessed through this interactive web tool.50 Website: http://starbase.sysu.edu.cn/.
CLIPdb
CLIPdb is a database of RBP–RNA interactions created based on 395 publicly available HITS-CLIP datasets for 111 RBPs from four organisms—human, mouse, worm, and yeast. This unified computational database can be used to compare transcriptome-wide binding sites among species. The binding sites can be visualized on the genome browser or can be downloaded.51 Website: http://lulab.life.tsinghua.edu.cn/clipdb/.
AURA2
AURA2 (Atlas of UTR Regulatory Activity) is a meta-database of interaction of trans-binding factors with human and mouse UTRs. It includes experimentally validated binding sites for RBPs, ncRNAs, cis-elements, and RNA epigenetics. It has a user-friendly interface which offers numerous data-mining features including coregulation search, network generation, and regulatory enrichment testing. Gene expression profiles can also be combined with these analyses.52 Website: http://aura.science.unitn.it/.
doRiNA 2.0
doRiNA 2.0 is an upgraded version of the original database which now contains 100 RBP datasets for human (hg19), 30 for mouse (mm9), and 6 for Caenorhabditis elegans (ce6). Unlike other databases, doRiNA uses published RBP target sites, provides APIs (application program interfaces) for incorporating third-party analysis tools and pipelines, and allows local installations of this database.53 Website: http://dorina.mdc-berlin.de.
RBPmap
RBPmap is a database and a web tool for searching experimentally validated RBP-binding motifs and for predicting RBP-binding sites on a given sequence for motif of interest. RBPmap is recommended for mapping RBPs in human, mouse, and fruitfly genomes but it can be used for other genomes as well. The program uses a position-specific background model to map splice sites, 5′ and 3′ UTRs, noncoding RNA and intergenic regions. The program and the database can be installed locally in a Linux server.54 Website: http://rbpmap.technion.ac.il/index.html.
RBPDB
RBPDB (RNA-binding protein database) contains experimentally validated RNA-binding sites, both in vitro and in vivo. The database currently contains information for about 1171 proteins from human, mouse, fruitfly, and worm. Users can search this database using a web interface which allows searching for genes or motifs or browsing by binding domain or by organism. It also allows bulk data downloads.55 Website: http://rbpdb.ccbr.utoronto.ca/.
CircInteractome
CircInteractome (circRNA interactome) is a new database and web tool that identifies RBP- and miRNA-binding sites on human circular RNAs (circRNAs). It uses publicly available circRNA, miRNA, and RBP databases and provides computational binding sites on circRNAs using BLAT82 for RBPs and TargetScan83 for miRNAs. It allows the user to design junction-spanning primers to detect circRNAs, design siRNAs for circRNA silencing, and identify potential internal ribosomal entry sites (IRES).56 Website: http://circinteractome.nia.nih.gov/.
CONCLUDING REMARKS
Different molecular biological methods are available for interrogating RBP–RNA interactions and some three dozen software tools and databases can guide the user in analyzing RBP–RNA interactions from published and unpublished data. Typically, web-based tools such as those reviewed here are easy to use, but they tend to be slow and generally cannot be customized. Nonetheless, they offer a first step for navigating this field and provide quick and generally useful results requiring a relatively short learning period. On the other hand, some open-source script-based tools may be more useful for certain analyses, even though there is a steeper learning curve. The purpose of this review is not to evaluate which is the best software, but to assist the readers with choosing the tool that best suits their needs, depending on the computational resources available and the specific RNP-relevant question they are investigating.
Acknowledgments
This work was supported entirely by the Intramural Research Program of the NIA and NIH.
Footnotes
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
- 1.Yoon JH, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, Kim KM, Noh JH, White EJ, Martindale JL, et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. doi: 10.1038/ncomms6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 6.Abdelmohsen K, Gorospe M. RNA-binding protein nucleolin in disease. RNA Biol. 2012;9:799–808. doi: 10.4161/rna.19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. WIREs RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimmeler S, Nicotera P. MicroRNAs in age-related diseases. EMBO Mol Med. 2013;5:180–190. doi: 10.1002/emmm.201201986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 11.Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium EP. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh G, Ricci EP, Moore MJ. RIPiT-Seq: a high-throughput approach for footprinting RNA:protein complexes. Methods. 2014;65:320–332. doi: 10.1016/j.ymeth.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrickson DG, Kelley DR, Tenen D, Bernstein B, Rinn JL. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016;17:28. doi: 10.1186/s13059-016-0878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley KJ, Steitz JA. The “Observer Effect” in genome-wide surveys of protein-RNA interactions. Mol Cell. 2013;49:601–604. doi: 10.1016/j.molcel.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 17.Milek M, Wyler E, Landthaler M. Transcriptome-wide analysis of protein-RNA interactions using high-throughput sequencing. Semin Cell Dev Biol. 2012;23:206–212. doi: 10.1016/j.semcdb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 18.McHugh CA, Russell P, Guttman M. Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 2014;15:203. doi: 10.1186/gb4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maragkakis M, Alexiou P, Nakaya T, Mourelatos Z. CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite. RNA. 2016;22:1–9. doi: 10.1261/rna.052167.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kucukural A, Ozadam H, Singh G, Moore MJ, Cenik C. ASPeak: an abundance sensitive peak detection algorithm for RIP-Seq. Bioinformatics. 2013;29:2485–2486. doi: 10.1093/bioinformatics/btt428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khorshid M, Rodak C, Zavolan M. CLIPZ: a database and analysis environment for experimentally determined binding sites of RNA-binding proteins. Nucleic Acids Res. 2011;39:D245–D252. doi: 10.1093/nar/gkq940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen B, Yun J, Kim MS, Mendell JT, Xie Y. PIPE-CLIP: a comprehensive online tool for CLIP-seq data analysis. Genome Biol. 2014;15:R18. doi: 10.1186/gb-2014-15-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, Hannon GJ, Sanford JR, Penalva LO, Smith AD. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012;28:3013–3020. doi: 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Chen B, Kim M, Xie Y, Xiao G. A model-based approach to identify binding sites in CLIP-Seq data. PLoS One. 2014;9:e93248. doi: 10.1371/journal.pone.0093248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Althammer S, Gonzalez-Vallinas J, Ballare C, Beato M, Eyras E. Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics. 2011;27:3333–3340. doi: 10.1093/bioinformatics/btr570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maticzka D, Lange SJ, Costa F, Backofen R. Graph-Prot: modeling binding preferences of RNA-binding proteins. Genome Biol. 2014;15:R17. doi: 10.1186/gb-2014-15-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc. 2014;9:263–293. doi: 10.1038/nprot.2014.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb S, Hector RD, Kudla G, Granneman S. PAR-CLIP data indicate that Nrd1-Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast. Genome Biol. 2014;15:R8. doi: 10.1186/gb-2014-15-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Y, Tam OH, Paniagua E, Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015;31:3593–3599. doi: 10.1093/bioinformatics/btv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riley TR, Slattery M, Abe N, Rastogi C, Liu D, Mann RS, Bussemaker HJ. SELEX-seq: a method for characterizing the complete repertoire of binding site preferences for transcription factor complexes. Methods Mol Biol. 2014;1196:255–278. doi: 10.1007/978-1-4939-1242-1_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kremsky I, Bellora N, Eyras E. A quantitative profiling tool for diverse genomic data types reveals potential associations between chromatin and pre-mRNA processing. PLoS One. 2015;10:e0132448. doi: 10.1371/journal.pone.0132448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou CH, Lin FM, Chou MT, Hsu SD, Chang TH, Weng SL, Shrestha S, Hsiao CC, Hung JH, Huang HD. A computational approach for identifying micro-RNA-target interactions using high-throughput CLIP and PAR-CLIP sequencing. BMC Genomics. 2013;14(Suppl 1):S2. doi: 10.1186/1471-2164-14-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS Comput Biol. 2010;6:e1000832. doi: 10.1371/journal.pcbi.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, Lee KY, Swanson MS, Darnell RB. Prediction of clustered RNA-binding protein motif sites in the mammalian genome. Nucleic Acids Res. 2013;41:6793–6807. doi: 10.1093/nar/gkt421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukunaga T, Ozaki H, Terai G, Asai K, Iwasaki W, Kiryu H. CapR: revealing structural specificities of RNA-binding protein target recognition using CLIP-seq data. Genome Biol. 2014;15:R16. doi: 10.1186/gb-2014-15-1-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Zhou J, Hu H, Gong H, Chen L, Cheng C, Zeng J. A deep learning framework for modeling structural features of RNA-binding protein targets. Nucleic Acids Res. 2016;44:e32. doi: 10.1093/nar/gkv1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun J, Wang T, Xiao G. Bayesian hidden Markov models to identify RNA-protein interaction sites in PAR-CLIP. Biometrics. 2014;70:430–440. doi: 10.1111/biom.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comoglio F, Sievers C, Paro R. Sensitive and highly resolved identification of RNA-protein interaction sites in PAR-CLIP data. BMC Bioinformatics. 2015;16:32. doi: 10.1186/s12859-015-0470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sievers C, Schlumpf T, Sawarkar R, Comoglio F, Paro R. Mixture models and wavelet transforms reveal high confidence RNA-protein interaction sites in MOV10 PAR-CLIP data. Nucleic Acids Res. 2012;40:e160. doi: 10.1093/nar/gks697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golumbeanu M, Mohammadi P, Beerenwinkel N. BMix: probabilistic modeling of occurring substitutions in PAR-CLIP data. Bioinformatics. 2016;32:976–983. doi: 10.1093/bioinformatics/btv520. [DOI] [PubMed] [Google Scholar]
- 44.Reyes-Herrera PH, Speck-Hernandez CA, Sierra CA, Herrera S. BackCLIP: a tool to identify common background presence in PAR-CLIP datasets. Bioinformatics. 2015;31:3703–3705. doi: 10.1093/bioinformatics/btv442. [DOI] [PubMed] [Google Scholar]
- 45.Erhard F, Dolken L, Jaskiewicz L, Zimmer R. PARma: identification of microRNA target sites in AGO-PAR-CLIP data. Genome Biol. 2013;14:R79. doi: 10.1186/gb-2013-14-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majoros WH, Lekprasert P, Mukherjee N, Skalsky RL, Corcoran DL, Cullen BR, Ohler U. MicroRNA target site identification by integrating sequence and binding information. Nat Methods. 2013;10:630–633. doi: 10.1038/nmeth.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Georgiev S, Boyle AP, Jayasurya K, Ding X, Mukherjee S, Ohler U. Evidence-ranked motif identification. Genome Biol. 2010;11:R19. doi: 10.1186/gb-2010-11-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hauer C, Curk T, Anders S, Schwarzl T, Alleaume AM, Sieber J, Hollerer I, Bhuvanagiri M, Huber W, Hentze MW, et al. Improved binding site assignment by high-resolution mapping of RNA-protein interactions using iCLIP. Nat Commun. 2015;6:7921. doi: 10.1038/ncomms8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang T, Xie Y, Xiao G. dCLIP: a computational approach for comparative CLIP-seq analyses. Genome Biol. 2014;15:R11. doi: 10.1186/gb-2014-15-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang YC, Di C, Hu B, Zhou M, Liu Y, Song N, Li Y, Umetsu J, Lu ZJ. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genomics. 2015;16:51. doi: 10.1186/s12864-015-1273-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dassi E, Re A, Leo S, Tebaldi T, Pasini L, Peroni D, Quattrone A. AURA 2: Empowering discovery of post-transcriptional networks. Translation (Austin) 2014;2:e27738. doi: 10.4161/trla.27738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blin K, Dieterich C, Wurmus R, Rajewsky N, Landthaler M, Akalin A. DoRiNA 2.0--upgrading the doRiNA database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2015;43:D160–D167. doi: 10.1093/nar/gku1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paz I, Kosti I, Ares M, Jr, Cline M, Mandel-Gutfreund Y. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014;42:W361–W367. doi: 10.1093/nar/gku406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2011;39:D301–D308. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 58.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 59.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 60.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jaskiewicz L, Bilen B, Hausser J, Zavolan M. Argonaute CLIP--a method to identify in vivo targets of miRNAs. Methods. 2012;58:106–112. doi: 10.1016/j.ymeth.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith AD, Sumazin P, Xuan Z, Zhang MQ. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc Natl Acad Sci USA. 2006;103:6275–6280. doi: 10.1073/pnas.0508169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janssen S, Giegerich R. The RNA shapes studio. Bioinformatics. 2015;31:423–425. doi: 10.1093/bioinformatics/btu649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ray D, Kazan H, Chan ET, Pena Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 70.Bohnsack MT, Tollervey D, Granneman S. Identification of RNA helicase target sites by UV cross-linking and analysis of cDNA. Methods Enzymol. 2012;511:275–288. doi: 10.1016/B978-0-12-396546-2.00013-9. [DOI] [PubMed] [Google Scholar]
- 71.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roll J, Zirbel CL, Sweeney B, Petrov AI, Leontis N. JAR3D Webserver: Scoring and aligning RNA loop sequences to known 3D motifs. Nucleic Acids Res. 2016;44:W320–W327. doi: 10.1093/nar/gkw453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steffen P, Voss B, Rehmsmeier M, Reeder J, Giegerich R. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22:500–503. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- 77.Salakhutdinov R, Hinton G. An efficient learning procedure for deep Boltzmann machines. Neural Comput. 2012;24:1967–2006. doi: 10.1162/NECO_a_00311. [DOI] [PubMed] [Google Scholar]
- 78.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and micro-RNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultra-fast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huppertz I, Attig J, D’Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, Konig J, Ule J. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]