Abstract
We have previously shown that the DNA replication licensing factor ORC4 forms a cage around the chromosomes that are extruded in both polar bodies during murine oogenesis, but not around the chromosomes that are retained in the oocyte or around the sperm chromatin. We termed this structure the ORC4 cage. Here, we tested whether the formation of the ORC4 cage is necessary for polar body extrusion (PBE). We first experimentally forced oocytes to extrude sperm chromatin as a pseudo-polar body and found that under these conditions the sperm chromatin did become enclosed in an ORC4 cage. Next, we attempted to prevent the formation of the ORC4 cage by injecting peptides that contained sequences of different domains of the ORC4 protein into metaphase II oocytes just before the cage normally forms. Our rationale was that the ORC4 peptides would block protein-protein interactions required for cage formation. Two out of six tested peptides prevented the ORC4 cage formation and simultaneously inhibited polar body extrusion (PBE), resulting in the formation of two pronuclei that were retained in the oocyte. Together, these data demonstrate that ORC4 oligomerization is required to form the ORC4 cage and that it is required for PBE.
Keywords: Meiosis, oocyte, ORC4, polar body extrusion
Introduction
During mammalian female meiosis, developing oocytes reduce the number of chromosomes to 1N by ejecting them in two polar bodies in a process called polar body extrusion (PBE) (Ma et al., 2016; Maro et al., 1986). Polar bodies arise from asymmetric divisions in which most of the cytoplasm is retained in the oocyte (Fig. 1-1). To achieve the first unequal division, actin filaments associated with Rab11 containing vesicles reposition the germinal vesicle chromosome plate to the cortex of the oocyte before cytokinesis (Holubcova et al., 2013; Schuh and Ellenberg, 2007). In the second unequal division, the metaphase plate is already near the oolema. It is not clear how the set of chromosomes that will be ejected is chosen. In the first meiotic division, it could simply be the chromosomes that are more proximal to the cortex, but in the second division the metaphase plate is not originally aligned perpendicular to the oolema (Maro and Verlhac, 2002). Also, the oocyte is not simply discarding damaged DNA since it appears that the chromosomes in both the first or second polar body are intact because they can both be used to substitute for oocyte DNA and produce live pups in mice (Wakayama et al., 1997; Wakayama and Yanagimachi, 1998).
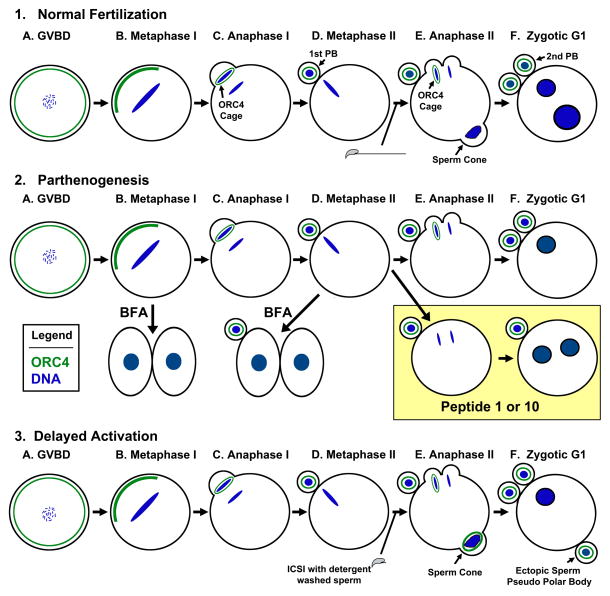
Figure 1. Diagram of ORC4 in Mouse Early Embryonic Development. (1) Normal Fertilization.
ORC4 is present in the cortex of GVBD oocytes (A), and coalesces near the chromosomes in metaphase I (B). ORC4 forms a cage around the chromosomes that are ejected in the first (C, D) and second (E, F) polar bodies. (E) The sperm chromatin forms a “sperm cone” which can be thought of as an abortive polar body that is reabsorbed by the oocyte by G1. (2) Parthenogenesis: ORC4 behaves the same way in parthenogenetically activated oocytes. BFA treatment of metaphase I (B) and metaphase II (D) oocytes results in the oocytes dividing into two equal sized cells and no polar body. The effects of BFA and Peptide injection are also shown. (3) Delayed Activation: Detergent washed sperm do not activate oocytes when injected. If activation by SrCl2 is delayed, the sperm chromatin is surrounded by ORC4 and ejected as a pseudo-polar body (F).
We recently discovered an unexpected link between PBE and the DNA licensing protein ORC4 (Nguyen et al., 2015). ORC4 is one of six proteins that bind to DNA replication origins during the G1 stage of the cell cycle to establish the start sites for DNA synthesis (Takeda et al., 2005). While the primary roles for the ORC proteins are well established to be in DNA replication, recent evidence suggests that many of them have additional functions that are related to the cell cycle, possibly to help coordinate the timing of DNA synthesis with other cellular processes (Sasaki and Gilbert, 2007; Scholefield et al., 2011). While studying the roles of the six ORC proteins in early mammalian embryogenesis, we found that ORC4 forms a unique structure that surrounds the set of chromosomes that will be extruded during anaphase in both meiotic divisions of murine oogenesis, but not around the set of chromosomes that remains in the oocyte (Nguyen et al., 2015) (Fig. 1–1C & E). We termed this structure the ORC4 cage. Here, we tested whether this ORC4 cage was required for PBE. We found that when the ORC4 cage formation was experimentally inhibited prior to second meiotic division, the oocytes did not extrude a second polar body.
Methods
Animals
B6D2F1 mice obtained from National Cancer Institute (Raleigh, NC) were used as sperm and oocyte donors. Animal care and experimental protocols for handling and the treatment procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Hawai’i.
Preparation of Spermatozoa and Oocytes
Swim-up spermatozoa were prepared from the cauda epididymis as described (Mohar et al., 2002). The spermatozoa suspension was used for intracytoplasmic sperm injection (ICSI). Metaphase II (MII) oocytes were prepared from superovulated mice as described previously (Ortega et al., 2012). Briefly, oviducts were removed 14–15 hours after the injections of hCG and the cumulus cells were released from the oviducts into 0.1% bovine testicular hyaluronidase in CZB (Szczygiel and Yanagimachi, 2003) medium for 10 minutes to disperse cumulus cells. The cumulus-free oocytes were washed and kept in CZB under a controlled environment (37ºC and 5% CO2).
To collect immature metaphase I and anaphase I oocytes, females were induced to superovulate by injections of 5 IU of hCG, as described (Yamauchi et al., 2012). Oviducts were removed 48 hours after hCG injection and placed in HEPES-CZB (Kimura and Yanagimachi, 1995) medium in a petri dish at room temperature. Germinal vesicle (GV) oocytes with surrounding cumulus cells were placed in CZB drops under mineral oil and cultured for 2 hours. The oocytes that underwent germinal vesicle breakdown were collected and cultured for 4 and 9 hours to reach metaphase I and anaphase I, respectively.
Intracytoplasmic Sperm Injection (ICSI)
ICSI was carried out as described (Kimura and Yanagimachi, 1995). Briefly, ICSI was performed using Eppendorf Micromanipulators (Micromanipulator TransferMan, Eppendorf, Germany) with a piezoelectric actuator (PMM Controller, model PMAS-CT150; Prime Tech, Tsukuba, Japan). After ICSI, oocytes were cultured in CZB at 37°C under 5% CO2.
Immunocytochemistry (ICC)
ICC was performed as described previously (Ortega et al., 2012). Embryos were cultured in CZB until they reached the desired stage after ICSI or parthenogenesis activation, and fixed in 2% paraformaldehyde for 30 minutes at room temperature. After fixing, cells were washed, blocked with 5% BSA for 1 h at room temperature, then incubated in primary antibody [Polyclonal goat anti-ORC4 (L-15, catalog no. sc-19725 and C-15, catalog no. sc-19726; Santa Cruz Biotechnology, Santa Cruz, CA ), monoclonal mouse anti-His (catalog no. CGAB-HIS-0050; Genecopoeia)] at 1:50 dilution overnight at 4°C. The cells were washed again, then incubated in secondary antibody at 1:1000 dilutions at room temperature for 1 h. Cells were washed again, and mounted with ProLong Gold anti-fade reagent with DAPI (catalog no. P-36931; Invitrogen). Images were collected on an Olympus FV1000 confocal microscope using Fluoview v. 2.1 software.
mORC4-His6 tagged protein expression
Recombinant plasmid containing hexaHistidine (His6) tagged mORC4 (Genecopiea), with a T7 promoter, was transformed into E. coli (BL21(DE3) pLysS; Invitrogen) to induce protein overexpression. An individual colony of transformed E. coli plated on LB (containing 150 μg/mL ampicillin and 35 μg/mL chloramphenicol) was collected to inoculate a 50 mL overnight culture (TB + Amp and chloramphenicol). After 18 hours 5 mL of the overnight culture were added to 1 L of Terrific Broth (+150 μg/mL ampicillin and 35 μg/mL chloramphenicol). Induction of ORC4-His6 expression was started when the OD600 reaches 0.8 – 1.0. To induce protein expression, bacteria was cooled to ~18°C, induced with 0.5 mM IPTG and incubated at room temp with 300 RPM shaking for 18h. Coomassie staining and Western blot was used to monitor mORC4 expression and track purity of the sample following nickel-nitrilotriacetic acid (NiNTA, GE Healthcare) chromatography and size exclusion chromatography purification.
Microinjection of mORC4-His6 tagged protein into oocyte
Recombinant mORC4-His6 protein was microinjected into the cytoplasm of MII oocytes, as described (Nishioka et al., 2009). In all experiments 10 pl of 1.8 μg/ml of peptide was injected into each oocyte. Following the microinjection, the oocytes were incubated for varying time points in CZB free calcium medium containing 10 mM of SrCl2 to induce activation. ICC was used to visualize ORC4 cage formation and overlap between His6 and endogenous ORC4 antibody staining.
Induction of ectopic sperm polar body formation
Sperm were washed with 0.5% SDS for 1 min, then heated at 95°C for 5 minutes, and finally incubated with TritonX-100 for 10 minutes to remove the SDS. A single SDS-washed, heated sperm head was injected into the oocyte. Following ICSI, injected oocytes were cultured in CZB 37°C under 5% CO2 for three hours and then activated by transferring them to media containing SrCl2. At 1h, 2h, and 3h after activation, the oocytes were analyzed by phase microscopy and ICC to evaluate polar body extrusion and ORC4 cage formation.
Brefeldin treatment of oocytes and embryos
GV and MII oocytes were incubated with BFA (10 mM/ml) supplemented with 10 mM of SrCl2 at desired time points (12h and 2h or 5h, respectively) to inhibit polar body formation. All oocytes and embryos were incubated at 37°C in a humidified atmosphere of 5% CO2 in air.
Microinjection of Peptides into oocytes
Six different peptides were injected into the cytoplasm of MII oocytes 1 hr before activation. In all experiments, 10 pl of 2.0 μg/μl RNA was injected into each oocyte. Following microinjection, the oocytes were incubated in CZB free calcium medium containing 10 mM of SrCl2 for one hour. Phase microscopy and ICC were used to evaluate polar body extrusion.
mORC4-His6 fragment deletions
mORC4-His6 plasmids were used to create assembly vectors by using NdeI and AvaI enzymes. Fragmented inserts with desired sequences were amplified by PCR reactions. The primers used to amplify the desired sequences included the homology arms at the beginning and at the end of the assembly vectors. Gibson assay was then used to join vector and fragmented inserts together and the Gibson products were electrophoresed into bacteria. Fragments were confirmed by single PCR colony and sequencing.
mRNA synthesis
Plasmids containing the mORC4-His6 and ORC4-His6 fragment mRNAs were linearized by an enzyme downstream of the ORC4 gene. mMESSAGE mMACHINE T7 transcription kit (Thermo Scientific) was used to synthesize mRNA ORC4-His6 constructs. Agarose (1%) electrophoresis was used to confirm the presence of mRNA product, and isolate mRNA by Direct-zol RNA miniprep (Zymo Research). ORC4-His6 mRNA constructs were stored at −80°C prior to microinjection.
Injection of mRNA
Each ORC4-His6 mRNA constructs was used for microinjection into the cytoplasm of MII oocytes. A microinjection volume of 10 pl/oocyte was used in all experiments. Following microinjection, the oocytes were incubated for 6 hours before parthenogenesis activation by using CZB free calcium medium contained 10 mM of SrCl2 for 3 h. ICC and Phase microscopy were used to visualize polar body extrusion.
Results
ORC4-His6Integrates into the ORC4 Cage
We wished to first ascertain that the ORC4 cage, observed previously by our lab using ICC, truly contained ORC4 and was not an off-target affect of the antibodies (Nguyen et al., 2015). We used two different commercial antibodies to identity ORC4 as a major component of the chromosome cage. We made mouse ORC4 constructs with histidine tags for two variants of ORC4: variant 1 that represents the full length ORC4, 433 amino acids in length (v1-mORC4-His6) and variant 2 (Diez-Roux et al., 2011) that encodes the first 121 amino acids of the full length ORC4 (v2-mORC4-His6). Both mORC4-His transcripts were expressed in bacteria and the proteins isolated on a nickel column. The purified protein, v1-mORC4-His6, was electrophoresed and assayed with the ORC4 antibody we used for ICC and with an antibody to the His6-tag (SFig. 1). The antibody we used for ORC4 ICC previously (Nguyen et al., 2015) recognized a protein at an identical molecular weight as the anti-histidine antibody, verifying that our prior ICC experiments identified ORC4 (Nguyen et al., 2015).
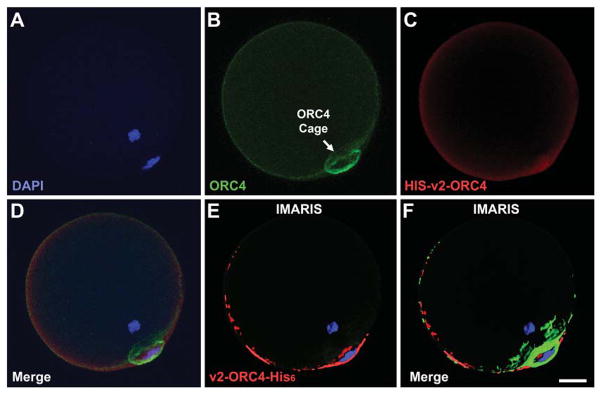
We next attempted to inject each ORC4 construct variant into MII oocytes to determine if they become incorporated into the ORC4 cage as it forms after MII oocytes are activated Fig. 1–1E). Our previous results demonstrated that the ORC4 cage is formed after metaphase is completed, during anaphase of both meiotic divisions (Nguyen et al., 2015). Oocytes were injected with the recombinant ORC4-His6 protein and stimulated to MII to examine if the His6 protein integrates with native ORC4 in the cage structure. The full length, v1-ORC4-His6 proved to be too difficult to solubilize for injections, but the smaller variant 2, v2-ORC4-His6, could be injected. When oocytes were subsequently activated and then stained with an antibody to the His6, we found that v2-ORC4-His6 did incorporate into the ORC4 cage (Fig. 2E & F). This supported the conclusion that ORC4 was part of the ORC4 cage that surrounded the chromosomes to be extruded from the cytoplasm.
Figure 2. Incorporation of v2-ORC4-HIS6 into the ORC4 Cage.
Purified v2-ORC4-His6 protein was injected into MII oocytes prior to activation. The oocytes were developed to G1 with the second polar body, then fixed and immunostained for ORC4 (green) and for the Histidine tag (red). The figure show confocal sections of one oocytes with filters for DAPI (A), ORC4 (B), His-tag (C), and merged (D). IMARIS software was used to identify where the v2-ORC4-His6 was localized (E and F). Bar = 10 μm.
A New ORC4 Cage is formed when the Oocyte Expels Sperm Chromatin
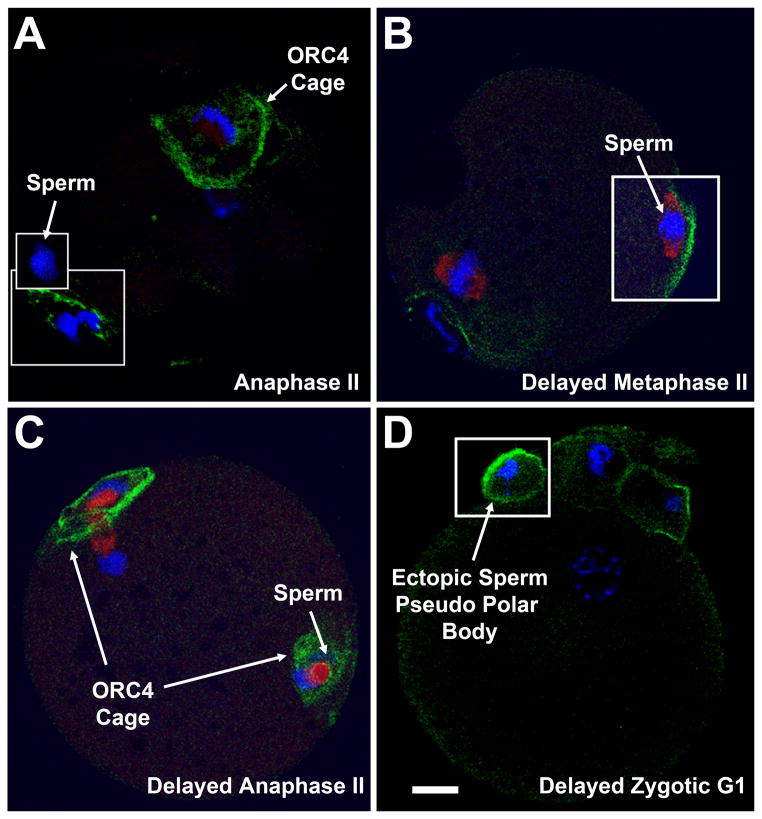
The recruitment of ORC4-His6 to chromosomes during anaphase in meiosis suggests that ORC4 has a novel function associated with PBE. If so, the ORC4 cage should not be formed when PBE was inhibited, and, conversely, a new ORC4 cage should form if chromatin that is normally retained in the zygote was extruded. When we inhibited PBE with Brefeldin A (Wang et al., 2008) (SFig. 2) or Cytochalasin B (SFig. 3), parthenogenetically activated oocytes underwent DNA synthesis but either divided into equal sized cells or did not undergo cytokinesis, and no ORC4 cage was formed. This suggested that the ORC4 cage formation is associated with PBE. We then tested whether an ORC4 cage formed if sperm chromatin was induced to be expelled at metaphase II. During normal fertilization the decondensing sperm nucleus forms a “sperm cone” in which the oolemma around the sperm nucleus blebs outward in a manner similar to the initial stages of PBE (Jedrusik et al., 2007; Longo, 1988) (Fig. 1-1E). This bleb is normally resolved and the sperm chromatin is reabsorbed into the zygote. We never found an ORC4 cage around the sperm chromatin in normal fertilization (Nguyen et al., 2015) (Fig. 3A). The sperm cone, however, can be experimentally induced to form a complete vesicle that is ejected from the oocyte as an ectopic sperm pseudo polar body, similar to PBE (Fig. 1–3E & F). This occurs when sperm are washed with an ionic detergent and then injected into oocytes. These sperm fail to activate the oocyte preventing it from completing its second meiotic division, but do begin to decondense. When these fertilized oocytes were incubated for 2 h, then activated with SrCl2, the sperm chromatin is expelled as in a vesicle that resembles a polar body (Deng and Li, 2009). We performed this experiment and found that a new ORC4 cage formed around the extruded sperm chromatin. ORC4 first accumulated at the oolemma near the decondensing sperm chromatin (Fig. 3B), then an ORC4 cage formed around the sperm chromatin (Fig. 3C). At G1, the sperm chromatin has been ejected as a pseudo polar body with an ORC4 cage (Fig. 3D). The ORC4 cage formed normally around the maternal chromatin that was extruded in the second polar body in these oocytes. These data suggests that an ORC4 cage forms around sperm chromatin when it is induced to be expelled by experimental manipulation.
Figure 3. A New ORC4 Cage Forms with Ectopic Sperm Pseudo Polar Bodies.
(A) Confocal image of a normal ICSI fertilized oocyte at anaphase II. The ORC4 cage surrounds the maternal chromosomes that will be extruded, and the DNA in the first polar body (lower inset). The sperm chromatin is devoid of ORC4 cage (upper inset). (B–D) An embryo injected with a detergent washed spermatozoa and activated 2 hrs later. (B) The delayed embryo in metaphase II before activation. ORC4 begins to accumulate at the oolemma new the decondensing nucleus. Tubulin (red) accumulates around the poles of the chromatin mass. (C) By anaphase II, ORC4 has completely surrounded the sperm chromatin. (D) At G1, the sperm of the delayed activated zygote has been ejected into an ectopic pseudo polar body in which the chromatin is surrounded by an ORC4 cage. Bar = 10 μm.
Disruption of the ORC4 Cage by ORC4 Peptides Prevents PBE
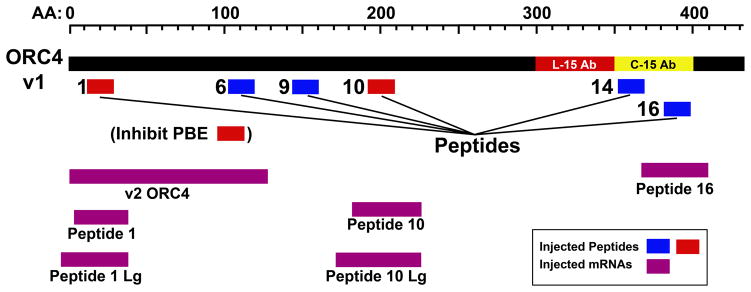
The experiments above suggested that ORC4 cage formation is disrupted when PBE is inhibited, and that an ORC4 cage forms around sperm chromatin when it is extruded from the oocyte. This indicates that the ORC4 cage is associated with PBE, but does not prove it is necessary for PBE. We first attempted to test whether the ORC4 cage was required for PBE by disrupting it with siRNA to Orc4 but this had no effect, presumably because there was already a sufficient amount of ORC4 protein present in the GV oocyte to construct the cage (Nguyen et al., 2015). Therefore, we disrupted the cage more directly using ORC4 peptides. We reasoned that the ORC4 cage required oligomerization, either ORC4 self-association or ORC4 interacting with another protein. We generated a putative three-dimensional structure for the full-length mouse ORC4 protein using Phyre II software (SFig. 4) and identified six, 18 amino acid long peptides that represented sequences of the protein that faced external surfaces that could be involved in protein-protein interactions to form polymers. The six peptides are shown in relation to the full length, v1-ORC4 protein in Fig. 4. We note that no soluble peptides could be identified for one section of the ORC4 protein, between amino acids 210 and 340.
Figure 4. ORC4 Peptides and Fragments.
The full length version 1 ORC4 with 433 amino acids is shown in relation to the six peptides and five ORC4 fragments used in this study. The peptides are shown in blue (peptides with no effect on PBE) and red (the two peptides that prevented PBE). The positions of the five ORC4 fragments that the mRNA sequences used in this study coded for are shown in purple.
Individual peptides were synthesized and injected into MII oocytes 1 h before parthenogenetic activation. Four of the peptides that we injected, 6, 9, 14 and 16, had no effect on either the ORC4 cage or the extrusion of the polar body (Table 1, Fig. 5F), and were used as negative control samples in progressive experiments. Peptides 1 and 10, however, disrupted the ORC4 cage in anaphase II (Fig. 5B) or prevented its formation (Fig. 5C) in many of the oocytes injected. Injection of peptides 1 and 10 caused the parthenogenetically activated oocytes to produce two pronuclei (2 PN) within the oocyte rather than ejecting half of the chromosomes in a polar body in 22.1% and 11.5% of the oocytes, respectively (Table 1, Fig. 1–2E & F box, Fig. 5D). Peptide 1 proved to be much less soluble than peptide 10, so we were not able to inject as many oocytes with peptide 1, even though it appeared to be more effective. In all cases of 2 PN oocytes, there was no evidence of the ORC4 cage (Fig. 5D). When the ORC4 cage was disrupted by peptide 1 or peptide 10 injection, DNA synthesis still occurred, demonstrating that only one function of ORC4 was inhibited (SFig. 5B). Together, the data obtained after injection of ORC4 peptides suggested that the binding sites for ORC4 polymerization are close to the amino acid sequences that were in peptides 1 and 10.
Table 1. Injection of ORC4 Peptides.
Peptides were injected into MII oocytes before parthenogenetic activation. The percentages of oocytes that progressed to having two pronuclei (2 PN) instead of the normal development to one PN and a polar body, are shown.
| Peptide | AA Position in ORC4 | No. Oocytes | 2 PN |
|---|---|---|---|
| 1 | 12 to 29 | 68 | 15 (22.1%)* |
| 6 | 102 to 119 | 122 | 0 |
| 9 | 138 to 155 | 96 | 0 |
| 10 | 191 to 208 | 139 | 16 (11.5%) |
| 14 | 352 to 369 | 123 | 2 (1.6%) |
| 16 | 382 to 399 | 88 | 2 (2.3%) |
For each peptide, at least two independent experiments were performed. There was no statistically significant difference between the individual experiments within each peptide, or between peptides 6, 9, 14 and 16 using, using the Fisher’s Exact Test (p < 0.05).
There was no statistical difference between peptides 1 and 10, but both were statistically different from all other peptides. All p values are listed in STable 1.
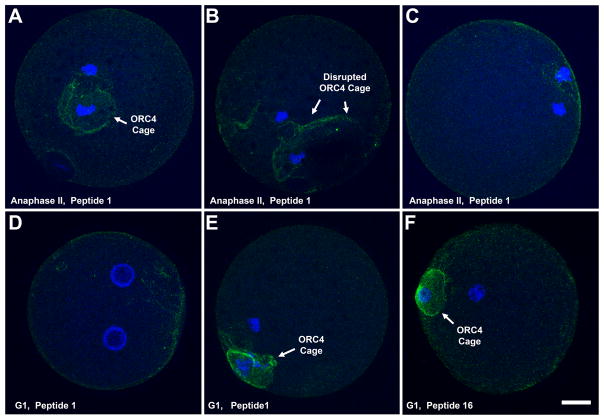
Figure 5. Injection of ORC4 Peptides Disrupts ORC4 Cage and Inhibits PBE.
MII oocytes were injected with ORC4 peptides one hour before parthenogenetic activation. The oocytes were stained at anaphase II or G1 for ORC4. (A) An oocyte with peptide 1 that formed a normal ORC4 cage at anaphase II. (B) An oocyte with the ORC4 cage disrupted at anaphase II. (C) An oocyte with peptide 1 with no visible ORC4 cage. (D) An oocyte injected with peptide 1 that did not extrude a polar body. Both maternal sets of chromosomes formed pronuclei. Some ORC4 staining was visible at the cortex. (E) An oocyte injected with peptide 1 that formed a normal polar body with an ORC4 cage. (F) An oocyte injected with control peptide 16 that progressed to PBE. Oocytes were stained with an antibody for ORC4 (green) and DAPI (blue). Bar = 10 μm.
Disruption of the ORC4 Cage with ORC4 Fragments
Peptide injection proved to be a limiting factor because these small protein fragments had low solubility under conditions which proved optimal for oocyte survival post injection. The solutions were viscous and sticky causing many of the injected oocytes to die. Furthermore, it was possible that the sites on the ORC4 protein that were responsible for polymerization to form the ORC4 cage were larger than 18 amino acids, and larger peptides would have been more difficult to inject. We therefore created five mRNAs encoding for ORC4 fragments that contained the gene sequences for ORC4 fragments that were 36 to 55 amino acids in length that included those of peptides 1, 10 and 16 (Table 2, Fig 4). This was done by deletion mutation of the original murine v1-ORC4 plasmid. The ORC4 fragments were inserted into expression plasmids and mRNA for each was synthesized. We also created mRNA for the v2-ORC4. To test our ability to inject intact mRNA into oocytes and whether oocytes would generate protein from injected mRNA, we first prepared and injected mRNA for GFP into MII oocytes and examined the oocytes by fluorescent microscopy before and after parthenogenetic activation. All oocytes fluoresced green (SFig. 6) indicating that MII oocytes are capable of synthesizing protein from mRNA. Protein synthesis was evident before and after activation, and continued for up to 3 hrs after activation.
Table 2. Injection of mRNA for ORC4 Fragments.
mRNA for five different ORC4 fragments were injected into oocytes before parthenogenetic activation, and the progression to 2 PN was assessed.
| ORC4 Frag | AA Position in ORC4 | No. AAs | No. Oocytes | 2 PN |
|---|---|---|---|---|
| v2-ORC4 | 1 to 121 | 121 | 128 | 32 (25%)* |
| Peptide 1 | 12 to 47 | 36 | 120 | 32 (26.7%) |
| Peptide 1 Lg | 2 to 47 | 46 | 152 | 40 (26.3%) |
| Peptide 10 | 190 to 235 | 46 | 158 | 29 (18.4%) |
| Peptide 10 Lg | 180 to 235 | 55 | 140 | 35 (25.0%) |
| Peptide 16 | 374 to 413 | 40 | 127 | 5 (3.9%) |
For each ORC4 fragment mRNA that was injected, two experiments were performed. There was no statistical significantly difference between experiments for each peptide, or between v2-ORC4, Peptide 1, Peptide 1 Lg, Peptide 10, or Peptide 10 Lg. Peptide 16 was highly statistically significant for all other peptides. All p values are listed in STable 2.
We next injected mRNA for the five ORC4 fragments we had created, and for the v2-ORC4 variant into MII oocytes, and then parthenogenetically activated them. We found that v2-ORC4, which was found to be incorporated into the ORC4 cage when injected as recombinant protein, caused the oocyte to produce two pronuclei instead of a polar body in 25.1% of oocytes (Table 2). Similar percentages of 2 PN oocytes resulted from the injection of the four ORC4 fragment mRNAs that contained peptides 1 and 10. From the oocytes injected with the mRNA encoding for peptide 16 only 3.9% progressed to 2PN, which was significantly different compared to other mRNAs. (STables 1-3). As with the peptide injections, in all cases of 2PN oocytes, there was no visible ORC4 cage.
Discussion
Our previous work demonstrated that ORC4 forms a cage around the ejected chromosomes in both meiotic divisions (Nguyen et al., 2015). Here, we provide several lines of evidence to support our hypothesis that the ORC4 cage is required for polar body extrusion (PBE): When PBE is inhibited, the ORC4 cage is no longer formed; when the oocyte is induced to extrude the sperm chromatin, an ORC4 cage is formed; and when ORC4 cage formation is specifically inhibited by target peptides, PBE is also inhibited. Cage formation was specifically targeted by in the injection of both ORC4 peptides and mRNA that encoded for larger ORC4 fragments, producing the most convincing evidence that the ORC4 cage is required for PBE.
Two areas of ORC4 appeared to be involved in oligomerization, as judged by their ability to prevent ORC4 cage formation, the sequences defined by peptides 1 and 10. Neither peptide, nor the larger versions of these sequences that were injected as mRNA, were able to inhibit PBE all of the time, but the percentages of 2PN parthenogenetically activated oocytes that were formed (11.5% to 26.7%) were all clearly statistically different from controls. There are many possible reasons for the inability of the peptides to inhibit cage formation all the time, including the instability of the peptides injected, the limit on the amount of peptide or mRNA that could be injected, and the efficiency of the peptides and ORC4 fragments to inhibit the cage. When we attempted to lengthen the ORC4 domain that inhibited cage formation with mRNA, we did not increase the percentage of oocytes that developed to 2 PN for peptide 1. For peptide 10, however, increasing the number of amino acids from 18 to 46 increased the percentage of 2 PN oocytes from 11.5% to 18.4%, a statistically significant increase. This suggests that at least one of the putative polymerization domains for ORC4 is larger than 18 AA. It is clear that of the six areas of the ORC4 protein that were injected, only two of them were able to prevent ORC4 cage formation and in every instance that the cage was inhibited, so was PBE.
One interesting apparent paradox in the data may provide some insight into how the ORC4 cage is formed. One would expect that the ORC4 cage which is a three dimensional sphere (Nguyen et al., 2015), would include not only simple polymerization but some kind of branching to link the polymers. We showed that v2-ORC4-His6 becomes part of the ORC4 cage (Fig. 2) but excessive amounts of this ORC4 variant can also prevent its formation (Table 2). One explanation for these results is that v2-ORC4 plays a role in branching to form the cage, so that too much will prevent the cage from forming by recruiting all the available full length v1-ORC4 for branching instead of polymerization.
The more usual function of ORC4 is to participate in the ORC complex that identifies origins of replication (Quintana et al., 1997; Takeda and Dutta, 2005). ORC4 is part of the core that forms the ORC (Vashee et al., 2001), and it binds directly to DNA and can form non-canonical structures with DNA (Kusic et al., 2010; Stefanovic et al., 2008). ORC4 exists in the cytoplasm before it forms the ORC (Vashee et al., 2001), so its presence in the cytoplasm of the oocyte (Nguyen et al., 2015) is consistent with its behavior in somatic cells. It is possible that the sequestration of ORC4 in the ORC4 cage also functions to delay DNA replication during the two divisions of the meiotic oocyte. There is also an accrual of ORC1, ORC2, ORC3 and ORC5 in the area between the separating chromosomes at anaphase I and anaphase II in oogenesis (Nguyen et al., 2015; Ortega et al., 2012) that may serve a similar function. The ability of ORC4 to bind directly to DNA, itself, may contribute to the formation of the ORC4 cage that coats the extruded chromatin. It is also possible that ORC4 cage formation participates in asymmetric divisions of other cell types and in the extrusion of chromatin from developing erythroblasts (Ji et al., 2008; Keerthivasan et al., 2010).
This work demonstrates that the DNA licensing protein ORC4 forms a large cytoplasmic structure, the ORC4 cage that is required for PBE. This is the first example of a functional structure formed by the ORC proteins. Current work is focused on understanding the mechanisms of how the ORC4 cage coordinates PBE and DNA replication.
Supplementary Material
References
- Deng M, Li R. Sperm chromatin-induced ectopic polar body extrusion in mouse eggs after ICSI and delayed egg activation. PLoS One. 2009;4:e7171. doi: 10.1371/journal.pone.0007171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nurnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dolle P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS biology. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holubcova Z, Howard G, Schuh M. Vesicles modulate an actin network for asymmetric spindle positioning. Nat Cell Biol. 2013;15:937–947. doi: 10.1038/ncb2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A, Ajduk A, Pomorski P, Maleszewski M. Mouse oocytes fertilised by ICSI during in vitro maturation retain the ability to be activated after refertilisation in metaphase II and can generate Ca2+ oscillations. BMC Dev Biol. 2007;7:72. doi: 10.1186/1471-213X-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Jayapal SR, Lodish HF. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol. 2008;10:314–321. doi: 10.1038/ncb1693. [DOI] [PubMed] [Google Scholar]
- Keerthivasan G, Small S, Liu H, Wickrema A, Crispino JD. Vesicle trafficking plays a novel role in erythroblast enucleation. Blood. 2010;116:3331–3340. doi: 10.1182/blood-2010-03-277426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biology of Reproduction. 1995;52:709–720. doi: 10.1095/biolreprod52.4.709. [DOI] [PubMed] [Google Scholar]
- Kusic J, Tomic B, Divac A, Kojic S. Human initiation protein Orc4 prefers triple stranded DNA. Mol Biol Rep. 2010;37:2317–2322. doi: 10.1007/s11033-009-9735-8. [DOI] [PubMed] [Google Scholar]
- Longo FJ. Reorganization of the egg surface at fertilization. Int Rev Cytol. 1988;113:233–269. doi: 10.1016/s0074-7696(08)60850-5. [DOI] [PubMed] [Google Scholar]
- Ma H, O’Neil RC, Marti Gutierrez N, Hariharan M, Zhang ZZ, He Y, Cinnioglu C, Kayali R, Kang E, Lee Y, Hayama T, Koski A, Nery J, Castanon R, Tippner-Hedges R, Ahmed R, Van Dyken C, Li Y, Olson S, Battaglia D, Lee DM, Wu DH, Amato P, Wolf DP, Ecker JR, Mitalipov S. Functional Human Oocytes Generated by Transfer of Polar Body Genomes. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro B, Johnson MH, Webb M, Flach G. Mechanism of polar body formation in the mouse oocyte: an interaction between the chromosomes, the cytoskeleton and the plasma membrane. Journal of embryology and experimental morphology. 1986;92:11–32. [PubMed] [Google Scholar]
- Maro B, Verlhac MH. Polar body formation: new rules for asymmetric divisions. Nat Cell Biol. 2002;4:E281–283. doi: 10.1038/ncb1202-e281. [DOI] [PubMed] [Google Scholar]
- Mohar I, Szczygiel MA, Yanagimachi R, Ward WS. Sperm Nuclear Halos Can Transform Into Normal Chromosomes After Injection Into Oocytes. Mol Reprod Dev. 2002;62:416–420. doi: 10.1002/mrd.10147. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Ortega MA, Ko M, Marh J, Ward WS. ORC4 Surrounds Extruded Chromatin in Female Meiosis. J Cell Biochem. 2015;116:778–786. doi: 10.1002/jcb.25033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Ortega MA, Marh J, Alarcon VB, Ward WS. Unique Pattern of ORC2 and MCM7 Localization During DNA Replication Licensing in the Mouse Zygote. Biol Reprod. 2012;87:62, 61–69. doi: 10.1095/biolreprod.112.101774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana DG, Hou Z, Thome KC, Hendricks M, Saha P, Dutta A. Identification of HsORC4, a member of the human origin of replication recognition complex. J Biol Chem. 1997;272:28247–28251. doi: 10.1074/jbc.272.45.28247. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Current opinion in cell biology. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Scholefield G, Veening JW, Murray H. DnaA and ORC: more than DNA replication initiators. Trends Cell Biol. 2011;21:188–194. doi: 10.1016/j.tcb.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Stefanovic D, Kusic J, Divac A, Tomic B. Formation of noncanonical DNA structures mediated by human ORC4, a protein component of the origin recognition complex. Biochemistry. 2008;47:8760–8767. doi: 10.1021/bi800684f. [DOI] [PubMed] [Google Scholar]
- Szczygiel M, Yanagimachi R. Intracytoplasmic sperm injection. In: Nagy A, Gertsenstein M, Vintersten K, Behringer R, editors. Manipulation of the Mouse Embryo - A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 2003. pp. 585–597. [Google Scholar]
- Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J Biol Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Hayashi Y, Ogura A. Participation of the female pronucleus derived from the second polar body in full embryonic development of mice. Journal of reproduction and fertility. 1997;110:263–266. doi: 10.1530/jrf.0.1100263. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Yanagimachi R. The first polar body can be used for the production of normal offspring in mice. Biol Reprod. 1998;59:100–104. doi: 10.1095/biolreprod59.1.100. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang ZB, Zhang X, FitzHarris G, Baltz JM, Sun QY, Liu XJ. Brefeldin A disrupts asymmetric spindle positioning in mouse oocytes. Dev Biol. 2008;313:155–166. doi: 10.1016/j.ydbio.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Riel JM, Ward MA. Paternal DNA damage resulting from various sperm treatments persists after fertilization and is similar before and after DNA replication. J Androl. 2012;33:229–238. doi: 10.2164/jandrol.111.013532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.