Abstract
N6-methyladenosine (m6A) and m5C methylation are two major types of RNA methylation, but the impact of joint modifications on the same mRNA is unknown. Here, we show that in p21 3′UTR, NSUN2 catalyzes m5C modification and METTL3/METTL14 catalyzes m6A modification. Interestingly, methylation at m6A by METTL3/METTL14 facilitates the methylation of m5C by NSUN2, and vice versa. NSUN2-mediated m5C and METTL3/METTL14-mediated m6A methylation synergistically enhance p21 expression at the translational level, leading to elevated expression of p21 in oxidative stress-induced cellular senescence. Our findings on p21 mRNA methylation and expression reveal that joint m6A and m5C modification of the same RNA may influence each other, coordinately affecting protein expression patterns.
Keywords: NSUN2, METTL3, METTL14, p21 mRNA Methylation, Translational Regulation
Introduction
Methylation is a widespread post-transcriptional modification of RNAs (Meyer et al, 2012; Dominissini et al, 2012) and determines translation efficiency, mRNA turnover, and RNA processing (Das et al, 2008; Seshadri et al, 2009; Ji and Chen, 2005; Schaefer et al, 2010; Zhang et al, 2012; Hussain et al, 2013; Yuan et al, 2014). Although methylation at all four ribonucleotides (A, U, C and G) has been reported (Björk et al, 1987; Persson et al, 1992), methylation of an mRNA at other than the 5′-cap occurs predominantly as N6-methyladenosine (m6A), with a small amount of 5-methylcytosine (m5C) identified in mRNA isolated from cultured hamster cells (Carroll et al, 1990; Dubin and Taylor, 1975). Apart from m5C and m6A, N(1)-Methyladenosine (m(1)A and pseudouridineare are also prevalent post-transcriptional RNA modification for coding and non-coding RNAs (Li et al, 2016; Zhao et al., 2017). However, the functional interconnection between different types of RNA methylation, especially m6A and m5C, has not been studied.
The tRNA methyltransferase NSun2 (Misu) is a typical RNA methyltransferase catalyzing the formation of m5C in coding and no-coding RNAs (Hussain et al, 2013; Xing et al, 2015, Tang et al, 2015, Cai et al, 2016). Methylation of mRNAs by NSUN2 is found to regulate the translation or turnover of mRNAs encoding CDK1, TP53, SHC, p16 (CDKN2A), and p27 (CDKN1B), thus affecting the expression of these genes in processes such as cell proliferation, oxidative stress, and replicative senescence (Hussain et al, 2013; Xing et al, 2015; Tang et al, 2015; Cai et al, 2016). METTL3 (methyltransferase like 3), METTL14 (methyltransferase-like 14), and WTAP (Wilm’s tumor 1-associated protein) form a methyltransferase complex, which catalyzes m6A formation (Liu et al, 2014; Ping et al, 2014). METTL3/METTL14-mediated RNA methylation is also involved in the regulation of mRNA stability or translation. For example, METTL3/METTL14 synergistically control the formation of m6A in a large number of mRNAs encoding developmental regulators, thereby inhibiting their expression at the levels of mRNA turnover and RNA processing in embryonic stem cells (Lin et al, 2014; Wang et al, 2014). However, whether NSUN2 and METTL3/METTL14 regulate the expression of an mRNA that is modified both at m5C and m6A has not been reported.
The levels of the broad inhibitor of cyclin-dependent kinases CDKN1A (p21Cip1), increases in cells exposed to a wide variety of stress agents, including genotoxins, oxidants, and metabolic perturbations. The increased p21 expression is believed to contribute to the growth arrest that follows exposure to these insults (Deng et al, 1995; Gorospe et al, 1996a; Waldman et al, 1996) and affects profoundly the outcome of the stressed cells, frequently favoring cell survival (Gorospe et al, 1996b; Gorospe and Holbrook, 1996; Gorospe et al, 1997). Not unexpectedly, p21 expression is tightly regulated at multiple levels. Transcriptional regulation of p21 levels, particularly by the transcription factor TP53, has been intensively reported (Gartel and Tyner, 1999). Beyond transcription, p21 levels are also regulated via stabilization of p21 mRNA and protein as well as by controling translation of p21 mRNA (Gartel and Tyner, 1999; Wang et al, 2000; Lal et al, 2004; Iakova et al, 2004; Timchenko et al, 1996). The RNA-binding protein (RBP) HuR binds to the 3′UTR of p21 mRNA and stabilizes it in cells exposed to short-wavelength UV light (UVC) (Wang et al, 2000). AUF1 and HuR competitively associate with the p21 3′UTR and regulate p21 mRNA stability (Lal et al, 2004). The RBPs CUGBP1 and calreticulin (CRT) competitively interact with the same sequences within the p21 5′UTR and modulate in opposing directions p21 the translation, in turn influencing cell growth arrest and senescence (Iakova et al, 2004). Another mode of post-transcriptional regulation of p21 is through alteration in the stability of the p21 protein, as illustrated by the extended half-life of p21 through interaction with the transcription factor CEBPα (Timchenko et al, 1996).
In the present study, we set out to investigate the possibility that p21 expression might be modulated by changes in p21 mRNA methylation. We report that NSUN2 catalyzes m5C modifications and METTL3/METT14 m6A modifications in the 3′UTR of p21 mRNA. Interestingly, methylation by METTL3/METTL14 facilitates the methylation by NSUN2, and vice versa. In addition, joint methylation at m5C and m6A synergistically enhance p21 expression at the level of translation in a model of oxidative stress-induced cellular senescence. Our findings provide a novel mechanism by which p21 expression is regulated.
Materials and Methods
Cell culture, FACS, transfection, and SA-β-gal staining
TP53-deficient human colon carcinoma cells (HCT116 p53−/−) and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin, at 37°C in 5% CO2. All plasmids were transfected using lipofectamine 2000 (Invitrogen) and cells were collected 48 to 72 h after transfection for further analysis. SA-β-galactosidase staining was performed as described previously (Tang et al, 2015; Cai et al, 2016).
Knockdown of NSUN2, METT3, and METTL14
To silence NSUN2, METT3, and METTL14, cells were transfected with siRNA (10 nM) targeting NSUN2 (GGUGUAGAAAUAACAGCGGUGAAGA), METTL3 (CTGCAAGTATGTTCACTATGA), or METTL14 (AAGGATGAGTTAATAGCTAAA) mRNAs, or a control siRNA (UUGUUCGAACGUG UCACGUTT) using RNAiMAX (Invitrogen).
RNA isolation and real-time qPCR
Total cellular RNA was isolated by using the RNeasy Mini Kit (Qiagen). For reverse-transcription (RT) followed by real-time, quantitative (q)PCR analysis of the levels of p21 and GAPDH mRNAs, primers GACTCTCAGGGTCGAAAACGG and GCGGATTAGGGCTTCCTCTT for p21 mRNA and CGAGTCAACGGATTTGGTGGTAT and AGCCTTCTCCATGGTGAAGAC for GAPDH mRNA were used.
Constructs and reporter gene assays
For construction of the pGL3-5′UTR, pGL3-CR, pGL3-3′UTR reporter vectors, p21 5′UTR fragment was amplified by using primer pairs CCCAAGCTTGTTGTATATCAGGGCCGC and CATGCCATGGGGCGCCTCCTCTGAG and inserted between Hind IIIand Ncol I sites of the pGL3-promoter vector; p21 CR fragment was amplified by using primer pairs GCTCTAGAATGTCAGAACCGGCTGGG and GCTCTAGATTAGGGCTTCCTCTTGGAGA and inserted into Xba I site of the pGL3-promoter vector; p21 3′UTR was amplified by using primer pairs GCTCTAGATCCGCCCACAGGAAGCCTGCAGT and GCTCTAGATACAAGTAAAGTCACTAAGAATC and inserted into Xba I site of the pGL3-promoter vector (Promega).
For reporter gene assays, each of the pGL3-derived vectors was co-transfected with pRL-CMV vector by Lipofectamine 2000 (Invitrogen). Forty eight hours after transfection, cell lysates were collected and the firefly and renilla luciferase activities were measured with a dual luciferase assay system (Promega) following the manufacturer’s instructions. All firefly luciferase measurements were normalized to renilla luciferase measurements from the same sample.
Transcript preparation
The fragments of p21 mRNA were amplified by PCR by using cDNA as a template. All 5′ primers contained the T7 promoter sequence (CCAAGCTTCTAATACGACTCACTATAGGGAGA). To prepare templates for the p21 5′UTR (positions 1–125), CR (positions 126–620), 3′UTR (positions 621–2158), 3′UTR1 (positions 621–800), 3′UTR2 (positions 778–957), 3′UTR3 (positions 935–1114), 3′UTR4 (positions 1092–1271), 3′UTR5 (positions 1249–1428), 3′UTR6 (positions 1046–1585), 3′UTR7 (positions 1563–1742), 3′UTR8 (positions 1720–1899), 3′UTR9 (positions 1877–2056), 3′UTR10 (positions 1979–2258), the following primer pairs were used: (T7) GTTGTATATCAGGGCCGC and GGCGCCTCCTCTGAGT for 5′UTR, (T7)ATGTCAGAACCGGCTGGG and TTAGGGCTTCCTCTTGGAGA for CR, (T7) TCCGCCCACAGGAAGCCTGCAT and TACAAGTAAAGTCACTAAGAATC, (T7) ATGTCAGAACCGGCT and CTCCAGTGGTGTCTCGGTGAC for CR1, (T7) TGTCACCGAGACACCAC and AGGTCCACATGGTCTTCCTCTGC for CR2, (T7) GCAGAGGAAACCAT and TTAGGGCTTCCTCTTGGAGAAGA, (T7) TCCGCCCACAGGAAG and TAAATAATTCTAATGCCAGAGGCTGGGG for 3′UTR1, (T7) GCCTCTGGCATTAGAA and GTGGGGAGGAGGAAGTAGCAGG for 3′UTR2, (T7) GCCAGCTACTTCCTC and CCACTCGGTGAGGCCCCTTCAA for 3′UTR3, (T7) TTTGAAGGGGCCTCA and GGACCCCACCTTCCCfor 3′UTR4, (T7) AAGGCAGGGGGAAGG and CAGCAGAGGGGAATTGCAGAG for 3′UTR5, (T7) GGCTCTGCAATTCCCC and GCTGCTTGAGCTGCCTGAGGTA for 3′UTR6, (T7) CTACCTCAGGCAGCT and ATGAGGAAGGTCGCTGGACGATTTG for 3′UTR7, (T7) AATCGTCCAGCGACC and GGCTCAACGTTAGTGCCAGGAAAGAC for 3′UTR8, (T7) TTTCCTGGCACTAAC and GCCCTCGAGAGGTTTACAGTCTAGG for 3′UTR9, and (T7) GGCCTGGACTGTTTT and TACAAGTAAAGTCACTAAGAATCATT for 3′UTR10.
These PCR products were transcribed in vitro following the manufacturer’s instructions (Thermo). The p21 3′UTR10 mutants [3′UTRCm (bearing C2079G), 3′UTR10m1 (bearing A1986U), 3′UTR10m2 (bearing A2037U), 3′UTR10m3 (bearing A2044U), 3′UTR10m4 (bearing A2061U), 3′UTR10m5 (bearing A2044U and A2061U) were prepared by overlapping PCR. The p27 5′UTR and SHC 5′UTR fragments were described previously (Tang et al, 2015; Cai et al, 2016).
Bisulfite RNA sequencing
Bisulfite RNA sequencing was performed to identify the methylation site (m5C) within an RNA fragment, as previously described. Briefly, 1 μg in vitro methylated p21 3′UTR fragment (methylated by NSUN2 by using cold SAM (Sigma)) or RNA isolated from cells was dissolved in 10 μl of RNase-free water and mixed with 42.5 μl of 5 M sodium bisulfite (Epitect) and 17.5 μl DNA protection buffer (Epitect), incubated in 70°C for 5 min and 60 °C for 60 min, repeating for 3–5 cycles. Samples were desalted by using Micro Bio-spin6 columns and then de-sulfonated by 1 M Tris (pH 9.0, 1/1, V/V) at 37°C for 1 h, followed by ethanol precipitation. The bisulfite-converted fragments (0.2 μg) were reverse-transcribed by RevertAid First-Strand cDNA Synthesis Kit (Thermo) using primer GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCTTAACCTTC CTACCATTCC and subjected to PCR analysis by using primer pairs CCAAGCTTCTAATACGACTCACTATAGGGAGAGTTTGTTGGGAATGGGGCCTGGACTGTTTT and CTTAACCTTCCTACCATTCCTACAAGTAAAGTCACTAAGAATCATT. To test the m5C formation in the endogenous p21 mRNA, The bisulfite-converted cellular RNA were reverse-transcribed by RevertAid First Strand cDNA Synthesis Kit (Thermo) using primer GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTATATTCA and subjected to PCR by using primer pairs GGTTTTTGTTTTTTTATTTAGATTGT and GCAGGGTCCGAGGTATTC. The PCR products were inserted into the pGEM-T Easy Vector System (Promega). The plasmids purified from single clones were sequenced, the sequences aligned with the corresponding p21 mRNA sequence and the cytosines retained were considered to be methylated.
In vitro translation assays
For in vitro translation assays, a cell-free translation system (Promega) in rabbit reticulocyte lysate (RL) was used. Luc-3′UTR fragment was amplified by PCR by using primer pairs (T7) CCATGGAAGACGCCAAAAACATA and TTTTTTTTTTTTTTTTTTTCTAGATACAAGTAAAGTCAC from pGL3-3′UTR reporter vector. The Luc-3′UTR fragment then was in vitro transcribed and further methylated by NSUN2, METTL3/METTL14, or by NSUN2 + METTL3/METTL14 in vitro or kept untreated. The methylated and non-methylated transcripts (0.01 nM) were used for in vitro translation assays. The translation efficiency was determined by measuring the activity of firefly luciferase.
LC-MS analysis
To determine the formation of m5C or m6A in in vitro methylated RNA fragments, in vitro methylated RNA fragments (1 μg) were digested by nuclease P1 (Sigma) and alkaline phosphatase (Thermo). The products were subjected to HPLC-MS analysis at Tsinghua University Mass Spectrum Center (Beijing, China). This method combined High pressure liquid chromatography (HPLC) and mass spectrometry (MS) to measure the mass (by MS) and amount (by HPLC) of m6A or m5C in RNA samples which is digested to single nucleotides. The area value of a peak appeared from a specific nucleotide was calculated by calculous to reflect the amount of methylation.
Measurement of m6A methylation in cells
To measure the levels of m6A methylation in cells, 1 μg of anti-m6A antibody, 20 μg of cellular RNA, and 20 μl (in 50% slurry) protein-G sepharose were incubated in IPP buffer [150 nM NaCl, 0.1% NP-40, 10 mM Tris. HCl (pH. 7.4)] plus 1 U/μl RNasin in 200 μl at 4°C for 2 h. The beads were washed with the IPP buffer for 5 times. RNA isolated from the IP beads were subjected to reverse transcription (RT) followed by real-time, quantitative (q)PCR analysis.
In vitro methylation assays
The in vitro methylation assay by NSUN2 was performed as described previously19. Briefly, 50 μl of reaction mixture containing 0.2 nM purified His-NSun2, 0.01 nM RNA in vitro-transcribed RNA fragments, and 1 μCi of 3H-labeled S-adenosyl-L-methionine (Amersham Bioscience) in reaction buffer (5 mM Tris HCL[pH 7.5], 5 mM EDTA, 10% glycerol, 1.5 mM dithiothreitol, 5 mM MgCl2) supplemented with inhibitors (leupeptin [1 μg/ml], aprotinin [1 μg/ml], 0.5 mM phenylmethylsulfonyl fluoride, and RNasin [5 U/μl]) wasincubated for 45 min at 37°C. Unincorporated 3H S-adenosyl-L-methionine in the reactions were removed by using Qiaquick Spin Columns (Qiagen) and the incorporated radioactivity was measured by liquid scintillation counting.
For in vitro methylation assays by METTL3/METTL14, HeLa cells were co-transfected with vectors expressing flag-METTL3 and flag-METTL14. Forty-eight h later, cell lysates were prepared and subjected to immunoprecipitation assays to enrich flag-METTL3 and flag-METTL14 proteins. The enriched flag-METTL3 and METTL14 in the IP materials (~0.1 nM) were used for in vitro methylation assays in a standard 50 μL of reaction mixture containing 0.1 nM RNA fragment, 1 μCi of 3H-labeled S-adenosyl-L-methionine (Amersham Bioscience), 80 mM KCl, 1.5 mM MgCl2, 0.2 U/μL RNasin, 10 mM DTT, 4% glycerol, and 15 mM HEPES (pH 7.9). Unincorporated 3H S-adenosyl-L-methionine was removed by using Qiaquick Spin Columns (Qiagen) and incorporated radioactivity was measured by liquid scintillation counting. RNA fragment ACGAGUCCUGGACUGAAACGGACUUGU was used as a positive control, as described (Liu et al, 2014).
Antibodies and Western blot analysis
Monoclonal anti-GAPDH, polyclonal anti-p66SHC was from BD Biosciences. Polyclonal anti-METTL3 was from Proteintech, Monoclonal anti-p21 antibody and monoclonal anti-CDK1 were from Santa Cruz. Polyclonal anti-METTL14 was from Abcam. Monoclonal anti-flag antibody was from Sigma. After secondary antibody incubations, signals were detected by SuperSignal WestPico Chemiluminescent Substrate (Pierce) following the manufacturer’s instruction and quantitated by densitometric analysis with ImageMaster VDS software.
Results
NSUN2, METTL3, and METTL14 regulate the expression of p21
To test if p21 expression might be regulated by methylation, we examined if p21 protein levels were altered in HeLa cells in which these methyltransferases (NSUN2, or METTL3, or METTL14) had been silenced by transfection of specific siRNAs. As shown in Fig. 1A (left), Western blot analysis revealed that reducing NSUN2, METTL3, or METTL14 markedly decreased the levels of p21 protein (by ~80% after silencing NSUN2 or METT14 and by ~70% after silencing METTL3). Conversely, overexpression of NSUN2, METTL3, or METTL14 increased p21 protein levels (by ~2.7 fold, ~3.8 fold, ~3.1 fold, respectively) (Fig. 1A, Right). Given that p53-mediated transcriptional regulation is a well-known mechanism controlling p21 expression, we further assessed p21 protein levels in p53-deficient human colon carcinoma cells (HCT116 p53−/− cells) in which NSUN2, METTL3, or METTL14 were individually silenced. As shown in Fig. 1B by Western blot analysis, knockdown of NSUN2, METTL3, or METTL14 in HCT116 p53−/− cells decreased p21 protein levels (by ~80% after NSUN2 silencing, by ~70% after silencing METTL3 or METTL14), indicating that NSUN2, METTL3, and METTL14 regulated p21 expression levels in a p53-independent manner. In addition, knockdown of NSUN2, METTL3, or METTL14 did not alter the mRNA levels of p21 in HCT116 p53−/− cells (Fig. 1C). In sum, NSUN2, METTL3, and METTL14 did not appear to affect p21 expression levels by altering mRNA turnover or transcription and instead may affect p21 translation.
Figure 1. NSUN2, METTL3, and METTL14 regulate p21 expression.

(A) HeLa cells were transfected with siRNAs targeting NSUN2 (siNSUN2), METTL3 (siMET3), or METTL14 (siMET14) (left), or with a vector expressing flag-tagged NSUN2 (pNSUN2), METTL3 (pMET3), or METTL14 (pMET14) (right). Forty-eight hours later, cell lysates were prepared and subjected to Western blot analysis to assess the levels of proteins NSUN2, METTL3, METTL14, p21, and GAPDH. Data are representative from three independent experiments. (B) HCT116 p53−/− cells were transfected with siRNAs targeting NSUN2, METTL3 (siMET3), or METTL14 (siMET14). Forty-eight hours later, cell lysates were prepared and subjected to Western blot analysis to assess the levels of proteins NSUN2, METTL3, METTL14, p21, and GAPDH. Data are representative from three independent experiments. (C) RNA was prepared from cells described in Fig. 1B and subjected to real-time qPCR to assess p21 mRNA levels. Data represent the mean ±SD from 3 independent experiments. (D) Cell lysates were prepared from cells silenced with METTL3 (siMET3), as described in Fig. 1A. CDK1, p27, p53, and SHC protein levels were determined by Western blot analysis.
NSUN2 has been found to regulate the expression of SHC, CDK1, TP53, p16, and p27 (Hussain et al, 2013; Xing et al, 2015, Tang et al, 2015, Cai et al, 2016). To ask whether METTL3/METTL14 and NSUN2 share targets other than p21 mRNA, the levels of CDK1, p27, and SHC were determined by Western blot analysis in cells in which METTL3 was silenced. As shown in Fig. 1D, knockdown of METTL3 did not alter the levels of CDK1, p27, or SHC, suggesting that METTL3/METTL14 and NSUN2 specifically share p21 mRNA as the same target.
NSUN2 and METTL3/METTL14 cooperatively enhance p21 expression
To further investigate the impact of NSUN2, and METTL3/METTL14 on p21 expression, the levels of p21 in cells in which NSUN2 and METTL3/METTL14 were silenced individually or jointly were assessed by Western blot analysis. As shown in Fig. 2A, combined knockdown of NSUN2 and METTL3 (NSUN2+METTL3) was more effective in reducing p21 levels than silencing NSUN2 or METTL3 individually (with reductions by ~50% (p<0.05), ~38% (p<0.05), and ~64% (p<0.05), respectively). Similarly, p21 levels in NSUN2+METTL14 co-silenced cells were much lower than those in cells where only NSUN2 or METTL14 were silenced [with reductions by ~43% (p<0.05), ~42% (p<0.05), and ~66% (p<0.01), respectively) (Fig. 2B). In agreement with the findings shown in Fig. 1C, knockdown of NSUN2, METTL3, METT14, NSUN2 and METTL3 together, or NSUN2 and METTL14 together did not influence the mRNA levls of p21 (Fig. 2C and 2D). These results suggest that NSUN2, METTL3, and METTL14 may cooperatively enhance p21 expression.
Figure 2. NSUN2 and METTL3 or METTL14 synergistically enhance p21 expression.

(A) Upper, HCT116 p53−/− cells were transfected with NSUN2 siRNA, METTL3 siRNA (siMET3), or with both (siNSUN2 + siMET3). Forty-eight h later, cell lysates were prepared and subjected to Western blot analysis to assess the levels of NSUN2, METTL3, p21, and GAPDH. Bottom, data are means±SD from 3 independent experiments; significance is analyzed by Student’s t test (**,p<0.01; *, p<0.05). Data are representatives of 3 independent experiments. (B) Upper, HCT116 p53−/− cells were transfected with NSUN2 siRNA, METTL14 siRNA (siMET14), or with both (siNSUN2+siMET14). Forty-eight h later, cell lysates were prepared and subjected to Western blot analysis to assess the levels of NSUN2, METTL14, p21, and GAPDH. Bottom, data are means±SD from 3 independent experiments; significance is analyzed by Student’s t test (*, p<0.05). (C, D) Real-time qPCR analysis was used for analyzing p21 mRNA levels in cells described in Fig. 2A and Fig. 2B, respectively. Data are means±SD from 3 independent experiments.
To further test this hypothesis, we constructed pGL3-derived reporters bearing p21 5′UTR (pGL3-5′UTR), p21 CR (coding region, pGL3-CR), and p21 3′UTR (pGL3-3′UTR) (Fig. 3A, schematic) and tested the activity of these reporters in HCT116 p53−/− cells after silencing NSUN2, METTL3, and METTL14 separately and in combination. As shown in Fig. 3B–D, individual knockdown of NSUN2, METTL3, and METTL14 strongly decreased the luciferase activity of reporter pGL3-3′UTR (by ~53.5%, ~40.5%, and ~40.2%, METTL14, respectively), but did not alter significantly luciferase activity from pGL3, pGL3-5′UTR, or pGL3-CR. Notably, simultaneous knockdown of NSUN2+METTL3 or NSUN2+METTL14 further decreased the activity of pGL3-3′UTR (by ~64.5% for NSUN2+METTL3, and by ~82.3% for NSUN2+METTL14) (Fig. 3E–F). In sum, NSUN2, METTL3, and METTL14 cooperatively regulate the expression of p21; the NSUN2 and METTL3/METTL14 response region is located in the p21 3′UTR.
Figure 3. NSUN2 and METTL3 or METTL14 synergistically enhance the activity of pGL3-derived reporter bearing p21 3′UTR.

(A) Schematic representation depicting the pGL3-derived reporters used for reporter gene assays. (B, C, D) HCT116 p53−/− cells were transfected with each of the reporters depicted in Fig. 3A together with a pRL-CMV control reporter. Twenty-four h later, cells were further transfected with a siRNA targeting NSUN2 (B), METTL3 (C), or METTL14 (D), or with a control siRNA (Control) and cultured for an additional 48 h. Firefly luciferase activity against Renilla luciferase activity was analyzed. Data represent the means ± SD from 3 independent experiments; significance was analyzed by Student’s t tes (**, p<0.01)t. (E) HCT116 p53−/− cells were co-transfected with a pGL3-3′UTR reporter or a pGL3 vector and a pRL-CMV control reporter. Twenty-four h later, cells were further transfected with NSUN2 siRNA (siNSUN2), METTL3 siRNA (siMET3), or NSUN2 siRNA plus METTL3 siRNA (siNSUN2 + siMET14) and cultured for an additional 48h. Firefly luciferase activity against Renilla luciferase activity was analyzed. Data represent the means ± SD from 3 independent experiments; significance was analyzed by Student’s t test (**, p<0.01). (F) HCT116 p53−/− cells were co-transfected with a pGL3-3′UTR reporter or a pGL3 vector and a pRL-CMV control reporter. Twenty-four h later, cells were further transfected with NSUN2 siRNA (siNSUN2), METTL14 siRNA (siMET14), or NSUN2 siRNA plus METTL14 siRNA (siNSUN2+siMET14) and cultured for an additional 48 h. Firefly luciferase activity against Renilla luciferase activity was analyzed. Data represent the means ± SD from 3 independent experiments; significance was analyzed by Student’s t test (**, p<0.01).
METTL3/METTL14 and NSUN2 methylate the p21 3′UTR
The RNA methyltransferase NSUN2 catalyzes the formation of m5C in RNAs. METTL3 and METTL14 form a stable heterodimer that functions in catalyzing the formation of m6A in RNAs. To test if NSUN2 methylated p21 mRNA, the p21 mRNA fragments described in Fig. 4A were used for in vitro methylation assays by using purified his-NSUN2 and 3H-labeled S-Adenosyl methionine (SAM) (Materials and Methods). To test if METTL3/METTL14 methylated p21 mRNA, HCT116 p53−/− cells were co-transfected with vectors expressing flag-METTL3 and flag-METTL14. Forty-eight h later, cell lysates were prepared and subjected to immunoprecipitation analysis to enrich flag-METTL3 and flag-METTL14. The IP materials and 3H-labeled SAM were used for in vitro methylation assays using as substrates the fragments depicted in Fig. 4A. The p27 3′UTR was included as a negative control and SHC 5′UTR served as a positive control for NSUN2-mediated RNA methylation; p21 cDNA served as a negative control for the METTL3/METTL14-mediated p21 mRNA methylation. As shown in Fig. 4B, NSUN2 was capable of methylating SHC 5′UTR, and a number of p21 mRNA regions (CR, 3′UTR, 3′UTR1, 3′UTR3, and 3′UTR10), but it did not methylate other p21 mRNA segments (5′UTR, 3′UTR2, 3′UTR4, 3′UTR5, 3′UTR6, 3′UTR7, 3′UTR8, 3′UTR9) or p27 3′UTR (Fig. 4B, left and right). METTL3/METTL14 methylated some p21 3′UTR segments (specifically 3′UTR10), but not other areas of p21 mRNA (5′UTR, CR, 3′UTR1, 3′UTR2, 3′UTR3, 3′UTR4, 3′UTR5, 3′UTR6, 3′UTR7, 3′UTR8, 3′UTR9) or p21 cDNA (Fig. 4C, left and right). Therefore, the shared region methylated by both NSUN2 and METTL3/METTL14 is p21 3′UTR10 (positions 1979–2158).
Figure 4. NSUN2 and METTL3/METTL14 methylate p21 mRNA in vitro.
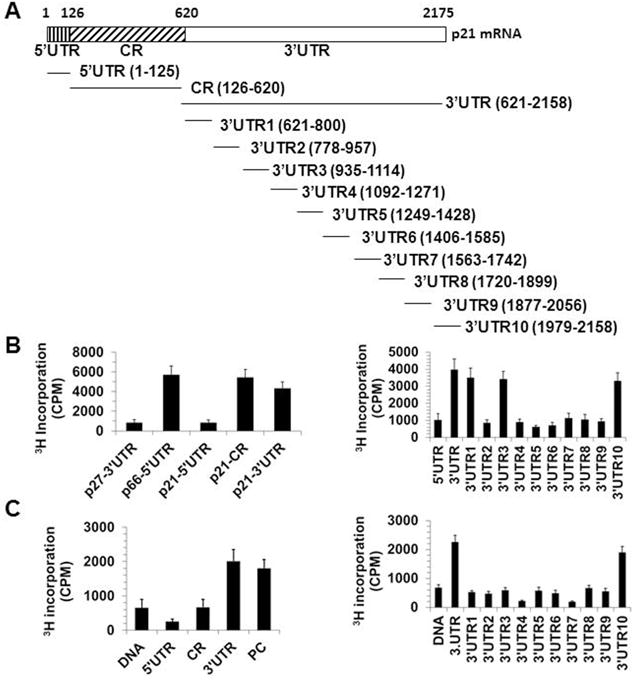
(A) Schematic depiction of the p21 mRNA fragments used for in vitro methylation assays. (B) In vitro methylation assays by using purified his-NSUN2 and the fragments shown in Fig. 4A. Incorporation of 3H-labeled SAM into p21 5′UTR, CR, and 3′UTR fragments (left) as well as 5′UTR, 3′UTR, 3′UTR1, 3′UTR2, 3′UTR3, 3′UTR4, 3′UTR5, 3′UTR6, 3′UTR7, 3′UTR8, 3′UTR9, and 3′UTR10 fragments (right). The incorporation of 3H-labeled SAM into p27 3′UTR and SHC 5′UTR (p66-5′UTR) served as negative control or positive control, respectively. (C) In vitro methylation assays by using immunoprecipitated flag-METTL3 and flag-METTL14 as well as the fragments depicted in Fig. 4A. Incorporation of 3H-labeled SAM into p21 cDNA (DNA), 5′UTR, CR, and 3′UTR fragments (left) as well as p21 cDNA (DNA), 3′UTR, 3′UTR1, 3′UTR2, 3′UTR3, 3′UTR4, 3′UTR5, 3′UTR6, 3′UTR7, 3′UTR8, 3′UTR9, and 3′UTR10 fragments (right). The incorporation of 3H-labeled SAM into p21 cDNA (DNA) and RNA fragment ACGAGUCCUGGACUGAAACGGACUUGU served as a negative control and a positive control (PC), respectively.
METTL3/METTL14 and NSUN2 cooperatively methylate p21 mRNA in vitro
To determine the formation of m5C or m6A in the NSUN2- or METTL3/METTL14-methylated fragments, the CR and 3′UTR fragments of p21 mRNA were methylated in vitro by NSUN2 by using non-isotopic SAM; p21 3′UTR was methylated by flag-METTL3/flag-METTL14 in vitro by using non-isotopic SAM. These methylated fragments or unmethylated fragments (same reaction but without adding NSUN2 or METTL3/METTL14) were subjected to MS-HPLC analysis. As shown in Fig. 5A, NSUN2 catalyzed the formation of m5C at the CR and 3′UTR of p21 mRNA. Given that methylation of p21 3′UTR, but not p21 CR, is involved in the regulation of p21 expression by NSUN2 (Fig. 3B), we did not identify the methylation sites in CR and instead identified the methylation sites in 3′UTR10. As shown in Fig. 5B (left) by bisulfite sequencing assays, C2079 was identified as the major methylation site in 3′UTR10 by NSUN2 in vitro. The observation that mutation of C2079 (C–G) greatly reduced the methylation of 3′UTR10 by NSUN2 further supported this view (Fig. 5B, right).
Figure 5. Measurement of m5C or m6A in NSUN2 or METTL3/METTL14 methylated fragments.

(A) p21 CR and 3′UTR fragments were in vitro-methylated by purified his-NSUN2 and non-isotopic SAM (+NSUN2) or kept untreated (−NSUN2), whereupon these fragments were subjected to HPLC-MS analysis to determine the formation of m5C. Data represent the peak value of m5C. (B) left, p21 3′UTR10 fragment was in vitro methylated by non-isotopic SAM and subjected to bisulfite RNA sequencing analysis to identify the methylation sites, as described in “Materials and Methods”. The percentage of methylation and the number of positive colones from the total colones sequenced are indicated. Right, in vitro methylation assays by using purified his-NSUN2 and fragments 5′UTR, 3′UTR10, and 3′UTR10 mutating C2079 (C-G, 3′UTRCm). (C) left, p21 3′UTR10 fragment was in vitro-methylated by immunoprecipitated flag-METTL3/flag-METTL14 and non-isotopic SAM (+MET3/MET14) or kept untreated (-MET3/MET14), whereupon these fragments were subjected to HPLC-MS analysis to determine the formation of m6A. The peak values of m6A are indicated. Right, in vitro methylation assays by using immunoprecipitated flag-MET3/flag-MET14 as well as 3′UTR10 and its variants m1-m5.
We then examined the formation of m6A in p21 3′UTR10 by METTL3 and METTL14 in vitro. As shown, METTL3/METTL14 catalyzes the formation of m6A in p21 3′UTR (Fig. 5C, left). The A residue in the middle of motifs AAC and GAC was recognized as the site for m6A methylation. Analysis of the sequence of 3′UTR10 revealed that 3′UTR10 contained four potential sites for m6A formation (A1986, A2037, A2044, and A2061). A-to-U mutations at A2044 (m3) or A2061 (m4), but not at A1986 (m1) or A2037 (m2), reduced the methylation levels of 3′UTR10 by METTL3/METTL14; mutation of A2044 and A2061 together (m5) further reduced the methylation levels of 3′UTR10 by METTL3/METTL14 (Fig. 5C, right). These results suggest that A2044 or A2061 are the methylation sites of METTL3/METTL14.
Based on the findings that NSUN2 andMETTL3 or METTL14 cooperatively regulated p21 expression levels (Fig. 2A, 2B, 3E, and 3F) and that p21 3′UTR10 was identified as the common target fragment for NSUN2- and METTL3/METTL14-mediated RNA methylation (Fig. 4B and 4C), we tested the interaction between NSUN2-mediated m5C formation and METTL3/METTL14-mediated m6A formation. To this end, fragment 3′UTR10 methylated by NSUN2 and non-isotopic SAM was further subjected to in vitro methylation assays by using METTL3/METTL14 and 3H-labeled SAM, and vice versa. As shown in Fig. 6A and 6B, pre-methylation by NSUN2 augmented significantly the effect of METTL3/METTL14 in methylating p21 3′UTR10 (from 1967 CPM to 3073 CPM) (Fig. 6A); likewise, pre-methylation by METTL3/METTL14 augmented significantly the effect of NSUN2 in methylating p21 3′UTR10 (2978 CPM compared with 4249 CPM) (Fig. 6B).
Figure 6. NSUN2 and METTL3/METTL14 cooperatively methylate p21 3′UTR and thereby enhancing the translation of p21.

(A) p21 3′UTR fragment was in vitro-methylated by NSUN2 (Pre-m5C) and non-isotopic SAM or kept untreated (Control). These fragments were further in vitro-methylated by flag-METTL3/METTL14 and 3H-labeled SAM. Data represent the means ± SD from 3 independent experiments; significance was analyzed by Student′s t test (**, p<0.01). (B) p21 3′UTR3 or 3′UTR10 fragment was in vitro methylated by METTL3/METTL14 and non-isotopic SAM (Pre-m6A) or kept untreated (Control). These fragments were further in vitro methylated by flag-METTL3/METTL14 and 3H-labeled SAM. Data represent the means ± SD from 3 independent experiments; significance was analyzed by Student′s t test (**, p<0.01). (C) Luc-3′UTR10 chimeric transcript transcribed from pGL3-3′UTR10 reporter was methylated by NSUN2, METTL3/METTL14 (MET3/14), NSUN2 + METTL3/METTL14 (NSUN2 + MET3/14), or kept untreated (Control). These transcripts then were used for in vitro translation assays. Luciferase activity was represented to reflect the efficiency of translation. (D) Luc-3′UTR10 (3′UTR10) or LUC-3′UTR10Cm (3′UTR10Cm, mutating C2079) was methylated in vitro by NSUN2 (Met) or kept untreated (Unmet), whereupon in vitro translation assays were performed. (E) Luc-3′UTR10 or Luc-3′UTR10Am was methylated by METTL3/METTL14 in vitro (Met) or kept untreated (Unmet), whereupon in vitro translation assays were performed. (F) Luc-3′UTR10, Luc-3′UTR10Am, or Luc-3′UTRCm was methylated by NSUN2+METTL3/METTL14 in vitro (Met) or kept untreated (Unmet), whereupon in vitro translation assays were performed. Data in C-F represent the means ± SD from 3 independent experiments; significance was analyzed by Student′s t test (**, p<0.01; *, p<0.05).
METTL3/METTL14 and NSUN2 cooperatively enhance p21 translation
In light of the fact that p21 mRNA levels were not altered in cells silenced with NSUN2, METTL3, or METTL14, we tested whether methylation by NSUN2 or METTL3/METTL14 influenced the translation of p21. To this end, in vitro-transcribed reporter transcripts Luc-3′UTR (transcribed from pGL3-3′UTR) were methylated in vitro individually by NSUN2 or by METTL3/METTL14, or jointly by NSUN2+METTL3/METTL14; unmethylated RNA was included in control reactions. These transcripts were then used for in vitro translation assays and reporter activities were used as readout of the efficiency of translation. As shown in Fig. 6C, methylation by NSUN2 and METTL3/METTL14 led to ~2.5-fold and 2.2-fold luciferase activities, respectively; methylation by NSUN2 and METTL3/METTL14 showed ~3.3-fold the activity level measured in the control group. Furthermore, methylation of the Luc-3′UTR10Am (mutating A2044 and A2061) and Luc-3-UTR10Cm (mutating C2079) by NSUN2, METTL3/METTL14, or NSUN2 and METTL3/METTL14 together was less effective than methylation of Luc-3′UTR10 under same conditions in elevating the translational efficiency (Fig. 6D–F). In conclusion, NSUN2-mediated m5C formation facilitates METTL3/METTL14-mediated m6A formation in p21 3′UTR, and vice versa; both types of methylation cooperate to further enhance p21 translation.
NSUN2 and METTL3/METTL14 cooperatively methylate p21 mRNA in cells
To test if NSUN2 and METTL3/METTL14 methylated p21 mRNA in cells, NSUN2, METTL14, or NSUN2+METTL14 were silenced in HCT116 p53−/− cells. RNA was isolated and subjected to IP-based reverse transcription (RT) followed by real-time, quantitative (q)PCR analysis to assess m6A levels in p21 mRNA, and subjected to bisulfite RNA sequencing analysis to assess the formation of m5C at C2079 of p21 mRNA. As shown in Fig. 7A, the anti-m6A antibody, but not the anti-IgG antibody, effectively immunoprecipitated the p21 mRNA. As anticipated, knockdown of METTL14 reduced m6A methylation in p21 mRNA by ~56%. In keeping with the findings that NSUN2-mediated m5C methylation augmented METTL3/METTL14-mediated m6A methylation of p21 3′UTR, knockdown of NSUN2 reduced singnificantly the levels of p21 mRNA methylation at m6A by ~16%. Knockdown of NSUN2 and METTL14 together was more effective than individually silencing NSUN2 or METTL14 in reducing the levels of m6A methylation of p21 mRNA by ~79%. On the other hand, knockdown of NSUN2 or METTL3/METTL14 reduced m5C levels at C2079 (~26.3% vs. ~11.1% in Control vs. siNSUN2 and ~26.3% vs. ~18.8% in Control vs. siMETTL14) in cells expressing LUC-3′UTR (Fig. 7B). Joint knockdown of NSUN2 and METTL14 together further reduced the formation of m5C at C2079 (Control vs. siNSUN2+siMETTL14, ~26.3% vs. ~7.1%) (Fig. 7B). In sum, METTL3/METTL14 and NSUN2 cooperatively methylate p21 mRNA in cells.
Figure 7. NSUN2 and METTL3/METTL14 cooperatively methylate p21 3′UTR in vivo.

(A) top, RNA isolated from HCT116 p53−/− cells was subjected to IP assays by using an antibody recognizing m6A or a control IgG antibody. The levels of p21 mRNA in the IP materials normalized against were analyzed by RT-qPCR analysis. Data represent the means ± SD from 3 independent experiments. Bottom, HCT116 p53−/− cells were transfected with siRNAs targeting NSUN2 (siNSUN2) or METTL14 (siMET14) or both (siNSUN2+siMET14). Forty-eight h later, RNA was prepared and subjected to IP assays by using m6A antibody. The levels of p21 mRNA were determined by RT-qPCR analysis. Data represent the means ±SD from 3 independent experiments; significance was analyzed by Student’s t test (*, p<0.05; **, p<0.01). (B) HCT116 p53−/− cells were transfected with a pGL3-3′UTR vector. Twenty four h later, cells were further transfected with a NSUN2 siRNA, METTL14 siRNA, NSUN2 and METTL14 siRNA, or kept untreated (Control) and cultured for additional 48 h. RNA was isolated and subjected to bisulfite RNA sequencing to assess the methylation of C2079 (m5C). The percent of methylation and the number of positive colons from the total colons sequenced are indicated.
NSUN2 and METTL3/METTL14 cooperatively enhance p21 expression levels in cellular sensecence induced by oxidative stress
By methylating SHC, p53, and p16 mRNAs, NSUN2 promotes the expression of SHC, p53, and p16 in oxidative stress-induced cellular senescence [15]. Exposure of HCT116 p53−/− cells to H2O2 (50 μM, 48 h) greatly increased p21 levels (~4.2 fold) and NSUN2 (~2.5 fold) as well as the percentage of cells expressing senescence-associated β-galactosidase [(SA)-β-gal] (~7.21% in untreated cells, ~85.43% in H2O2-treated cultures) (Fig. 8A and 8B). Interestingly, cells exposed to oxidative stress also expressed higher levels of METTL3 (~2.9 fold) and METTL14 (~3.3 fold) (Fig. 8A). To test the impact of NSUN2- and METTL3/METTL14-mediated regulation of p21 in oxidative stress-induced cellular senescence, HCT116 p53−/− cells in which NSUN2 and METTL14 were silenced (alone or jointly) were exposed to H2O2 (50 μM, 48 h) and p21 levels were determined by Western blot analysis. As shown in Fig. 8C (left and right), H2O2 treatment induced p21 less robustly in cells with silenced NSUN2 or METTL14, but cells in which NSUN2+METTL14 were jointly silenced showed the weakest expression of p21, in agreement with the results shown in Fig. 2B. Moreover, knockdown of NSUN2 or METTL14 lowered the amount of cells expressing senescence-associated β-galactosidase activity following H2O2 treatment, but joint knockdown of NSUN2 and METTL14 most strongly suppressed the number of cells displaying senescence-associated β-galactosidase activity. Together, our findings indicate that modification of p21 mRNA by NSUN2-mediated m5C and METTL3/METTL14-mediated m6A modification cooperatively enhance p21 translation, in turn promoting oxidative stress-induced cellular senescence.
Figure 8. NSUN2 and METTL3/METTL14 synergistically enhance the expression of p21 in oxidative stress-induced cell senescence.
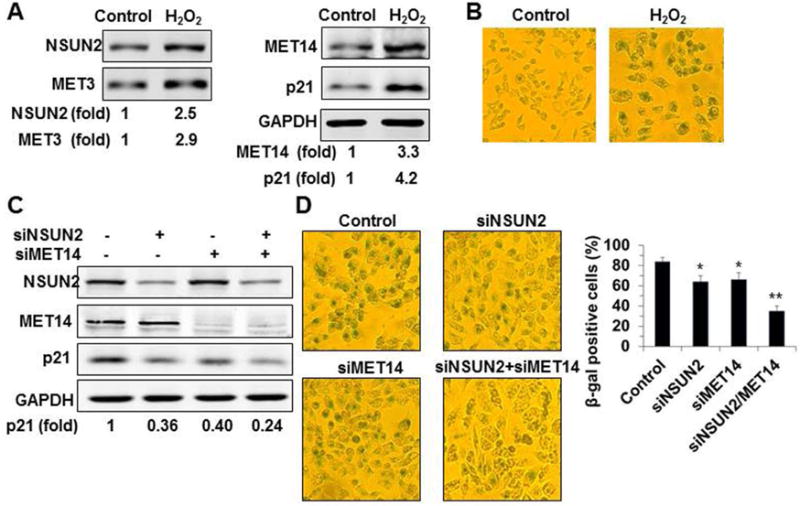
(A) HCT116 p53−/− cells were exposed to H2O2 (50 μM) for 48 h. The levels of NSUN2, METTL3, METTL14, p21, and GAPDH were analyzed by Western blot analysis. Data are representatives from 3 independent experiments. (B) The cells described in Fig. 8A were subjected to SA-β-gal analysis and the percentage of SA-β-gal-positive cells was counted. Data are representatives of 3 independent experiments. (C) HCT116 p53−/− cells were transfected with siRNAs to silence NSUN2 or METTL14 (siMET14) or both proteins. Twenty-four h later, cells were exposed to H2O2 (50 μM) and cultured for additional 48 h. The levels of NSUN2, METTL14, p21, and GAPDH were analyzed by Western blot analysis. Data are representatives of 3 independent experiments. (D) Cells described in Fig. 8C were subjected to SA-β-gal analysis. Data represent the means ± SD from 3 independent experiments and statistical significance was analyzed by Student’s t test (*, p<0.05; **, p<0.01).
Discussion
In the present study, we provide evidence that NSUN2 and METTL3/METTL14 cooperatively methylate p21 mRNA in vitro and in cells, in turn enhancing p21 production at the level of translation (Figs. 1–7). Although the detailed mechanism whereby translation is regulated by these RNA modifications, m5C and m6A methylation mutually promote the expression of p21 and hence cell senescence following oxidative damage (Fig. 8). Notably, methylation by NSUN2 in the coding region (CR) of p21 did not significantly influence the expression of p21 (Figure 3B and Fig. 4B). Our previous studies revealed that methylation of the ICAM1 CR by NSUN2 similarly had no effect on the regulation of ICAM1 expression (Luo et al, 2016), although methylation of the CR of SHC mRNA by NSUN2 did enhance SHC translation (Cai et al, 2016). Therefore, whether or not mRNA methylation influences mRNA fate may depend on the location of the methylated site or on additional factors interacting with the mRNA.
METTL3 and METTL14 form a stable heterodimer core complex that catalyzes the m6A modification of mammalian nuclear RNAs (Liu et al, 2014; Ping et al, 2014). By interacting with METTL3/METTL14, WTAP regulates the activity of METTL3/METTL14-mediated RNA methylation (Liu et al, 2014; Ping et al, 2014). Thus far, METTL3/METTL14-mediated RNA methylation was found to regulate the RNA processing and turnover of mRNAs encoding developmental regulators to help sustain embryonic stem cells (ESCs) (Lin et al, 2014; Wang et al, 2014). Interestingly, NSUN2-mediated RNA methylation is required for balancing stem cell self-renewal and differentiation in skin (Blanco et al, 2011, Blanco et al, 2016). It is plausible to postulate that NSUN2 and METTL3/METTL14 may cooperate to influence stem cell function by regulating p21 or other unidentified targets. In addition, elevation of p21 under stress conditions can promote cell survival. Thus, our findings that H2O2 induce the expression of NSUN2, METTL3, and METTL14 support a function for these methyl transferases to upregulate p21 expression in order to elicit cell survival. The fact that p21 is elevated in senescent cells while NSUN2 levels are reduced (Tang et al, 2015) suggests that NSUN2 and METTL3/METTL14 do not regulate p21 expression in replicative senescence.
Methylation of m6A is a predominant modification of RNAs. In different RNA species, the levels of m6A have been found to be 1 to 3 residues per 1,000 nucleotides (Carroll et al, 1990). In addition to m6A, m5C is also a widespread modification of mRNAs (Xing et al, 2015; Tang et al, 2015; Cai et al, 2016; Luo et al, 2016; Squires et al, 2012; Zhao et al., 2017). The present study indicates that NSUN2-mediated m5C methylation promotes METTL3/METTL14-mediated m6A methylation and reciprocally m6A methylation by METTL3/METTL14 enhances NSUN2-mediated m5C methylation, jointly inducing p21 translation. Because C2079 is adjacent to A2044 and A2061, the reciprocal influence between m5C and m6A methylation may depend on whether m5C and m6A sites are close enough. Given that NSUN2 and METTL3/METTL14 target numerous coding and non-coding RNAs for methylation(Meyer et al, 2012; Dominissini et al, 2012; Hussain et al, 2013; Liu et al, 2014; Ping et al, 2014), their impact on mRNAs jointly modified at m5C and m6A may not be limited to p21 mRNA and affect other mRNAs widely.
Acknowledgments
This work was supported by Grants 81230008, 81420108016, and 91339114 from the National Science Foundation of China; Grant B07001 (111 project) from the Ministry of Education of People’s Republic of China. M.G. was supported by the NIA-IRP, NIH.
Footnotes
Conflicts of Interest
We declare no conflict of interest
References
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortés-Garrido R, Gkatza N, Dietmann S, Frye M. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–40. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011;7(12):e1002403. doi: 10.1371/journal.pgen.1002403. Epub 2011 Dec 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR, Ericson JU, Gustafsson CE, Hagervall TG, Jönsson YH, Wikström PM. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- Cai X, Hu Y, Tang H, Hu H, Pang L, Xing J, Liu Z, Luo Y, Jiang B, Liu T, Gorospe M, Chen C, Wang W. RNA methyltransferase NSUN2 promotes stress-induced HUVEC senescence. Oncotarget. 2016;7:19099–110. doi: 10.18632/oncotarget.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol Cell Biol. 1990;10:4456–4465. doi: 10.1128/mcb.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Thotala DK, Kapoor S, Karunanithi S, Thakur SS, Singh NS, Varshney U. Role of 16S ribosomal RNA methylations in translation initiation in Escherichia coli. EMBO J. 2008;27:840–851. doi: 10.1038/emboj.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Amato NJ, Wang P, McGowan SJ, Niedernhofer LJ, Wang Y. Simultaneous Quantification of Methylated Cytidine and Adenosine in Cellular and Tissue RNA by Nano-Flow Liquid Chromatography-Tandem Mass Spectrometry Coupled with the Stable Isotope-Dilution Method. Anal Chem. 2015;87:7653–9. doi: 10.1021/acs.analchem.5b00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res. 1999;246:280–9. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- Gorospe M, Martindale JM, Sheikh MS, Fornace AJ, Jr, Holbrook NJ. Regulation of p21CIP1/WAF1expression by cellular stress: p53-dependent and -independent mechanisms. Mol Cell Differ. 1996a;4:47–65. [Google Scholar]
- Gorospe M, Wang X, Guyton K, Holbrook NJ. Protective role of p21Waf1/Cip1 against prostaglandin A2-mediated apoptosis of human colorectal carcinoma cells. Mol Cell Biol. 1996b;116:6654–6660. doi: 10.1128/mcb.16.12.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M, Holbrook NJ. Role of p21 in prostaglandin A2-mediated cellular arrest and death. Cancer Res. 1996;156:475–479. [PubMed] [Google Scholar]
- Gorospe M, Cirielli C, Wang X, Seth P, Capogrossi M, Holbrook NJ. p21Waf1/Cip1 protects against p53-mediated apoptosis of human melanoma cells. Oncogene. 1997;114:929–935. doi: 10.1038/sj.onc.1200897. [DOI] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, Frye M. NSun2-Mediated Cytosine-5 Methylation of Vault Noncoding RNA Determines Its Processing into Regulatory Small RNAs. Cell Rep. 2013;4:255–61. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–17. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2005;22:624–36. doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Xiong XS, Wang K, Wang LX, Shu XT, Ma SQ, Yi CQ. TranscrIptome-wide mapping reveals reversible and dynamic N (1)-methyladenosine methylome. Nat Chem Biol. 2016;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- Lin S, Gregory RI. Methyltransferases modulate RNA stability in embryonic stem cells. Nat Cell Biol. 2014;16:129–31. doi: 10.1038/ncb2914. [DOI] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Feng J, Xu Q, Wang W, Wang X. NSun2 Deficiency Protects Endothelium From Inflammation via mRNA Methylation of ICAM-1. Circ Res. 2016;118:944–56. doi: 10.1161/CIRCRESAHA.115.307674. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson BC, Gustafsson C, Berg DE, Björk GR. The gene for a tRNA modifying enzyme, m5U54-methyltransferase, is essential for viability in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:3995–3998. doi: 10.1073/pnas.89.9.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang X, Rendtlew Danielsen JM, Liu F, Yang YG. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri A, Dubey B, Weber MH, Varshney U. Impact of rRNA methylations on ribosome recycling and fidelity of initiation in Escherichia coli. Mol Microbiol. 2009;72:795–808. doi: 10.1111/j.1365-2958.2009.06685.x. [DOI] [PubMed] [Google Scholar]
- Tang H, Fan X, Xing J, Liu Z, Jiang B, Dou Y, Gorospe M, Wang W. NSUN2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging (Albany NY) 2015;7:1143–58. doi: 10.18632/aging.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Wilde M, Nakanishi M, Smith JR, Darlington GJ. CCAAT/enhancer-binding protein α (C/EBP α) inhibits cell proliferation through the p21(WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- Waldman T, Lengauer C, Kinzler KW, Vogelstein B. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature. 1996;831:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–9. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Yi J, Cai X, Tang H, Liu Z, Zhang X, Martindale JL, Yang X, Jiang B, Gorospe M, Wang W. NSUN2 Promotes Cell Growth via Elevating CDK1 Translation. Mol Cell Biol. 2015;35:4043–52. doi: 10.1128/MCB.00742-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Tang H, Xing J, Fan X, Cai X, Li Q, Han P, Luo Y, Zhang Z, Jiang B, Dou Y, Gorospe M, Wang W. Methylation by NSUN2 represses the levels and function of miR-125b. Mol Cell Biol. 2014;34:3630–41. doi: 10.1128/MCB.00243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu Z, Yi J, Tang H, Xing J, Yu M, Tong T, Shang Y, Gorospe M, Wang W. NSUN2 stabilizes p16INK4 mRNA by methylating the p16 3′UTR. Nature Communications. 2012;3:712. doi: 10.1038/ncomms1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications 2017. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]


