Abstract
Immunotherapeutic approaches, particularly PD-1/PD-L1 blockade, have improved the treatment of non-small cell lung cancer (NSCLC), supporting the premise that evasion of immune destruction is of importance for NSCLC progression. However, the signals responsible for upregulation of PD-L1 in NSCLC cells and whether they are integrated with the regulation of other immune-related genes are not known. Mucin 1 (MUC1) is aberrantly overexpressed in NSCLC, activates the NF-κB p65→ZEB1 pathway and confers a poor prognosis. The present studies demonstrate that MUC1-C activates PD-L1 expression in NSCLC cells. We show that MUC1-C increases NF-κB p65 occupancy on the CD274/PD-L1 promoter and thereby drives CD274 transcription. Moreover, we demonstrate that MUC1-C-induced activation of NF-κB→ZEB1 signaling represses the TLR9, IFNG, MCP-1 and GM-CSF genes, and that this signature is associated with decreases in overall survival. In concert with these results, targeting MUC1-C in NSCLC tumors suppresses PD-L1 and induces these effectors of innate and adaptive immunity. These findings support a previously unrecognized central role for MUC1-C in integrating PD-L1 activation with suppression of immune effectors and poor clinical outcome.
Keywords: MUC1-C, NSCLC, PD-L1, TLR9, IFN-γ, MCP-1, GM-CSF, TLR7
INTRODUCTION
A hallmark of human cancers is the evasion of immune destruction 1. Cancers are often infiltrated with immune cells that are ineffective in recognizing tumor antigens 2. In this way, immunotherapy has recently changed the landscape of NSCLC treatment 3. Blockade of the programmed death 1 (PD-1)/programmed death ligand 1 (PD-L1) immune checkpoint, in particular, is broadly effective in the treatment of NSCLCs and can extend survival in patients with tumors not responsive to targeted therapy. However, PD-1/PD-L1 blockade is associated with a response rate of about 20% in NSCLC and these responses are often of short duration 3. These findings have supported the premise that evasion of immune recognition and destruction contributes to the pathogenesis of NSCLC and that additional approaches are needed to enhance the effectiveness of PD-1/PD-L1 blockade 4, 5. Studies in genetically engineered mouse models (GEMMs) have demonstrated that NSCLCs driven by mutant EGFR activate the PD-1/PD-L1 pathway and thereby suppress T-cell function 6. KRAS-driven NSCLCs also increase inflammatory cytokine production to suppress T-cell activity in the tumor microenvironment 7. However, the signaling pathways responsible for regulating PD-L1 expression and other immune related genes in NSCLC cells remain largely unknown.
Mucin 1 (MUC1) is a transmembrane glycoprotein that is aberrantly overexpressed in >80% of NSCLCs 8. Moreover, the overexpression of MUC1 in NSCLCs is associated with poor disease-free and overall survival 8–13. The frequency of mutations in the MUC1 gene is low (~1%) in NSCLCs compared to that for EGFR, KRAS and p53 14–16, emphasizing the importance of aberrant MUC1 expression, rather than mutational activation, for NSCLC pathogenesis. MUC1 consists of two subunits: an N-terminal extracellular mucin subunit (MUC1-N) and a transmembrane C-terminal subunit (MUC1-C) that functions as an oncoprotein 17, 18. MUC1-C includes a 58-amino acid extracellular domain, which forms complexes with galectin-3 and thereby cell surface receptor tyrosine kinases, such as EGFR 19. The MUC1-C 72-amino acid cytoplasmic domain is an intrinsically disordered structure 20, which has the plasticity to interact with multiple kinases and effectors that have been linked to transformation 17, 18. In this context, MUC1-C directly activates the TAK1→IKK→NF-κB p65 pathway, linking this inflammatory response to EMT and self-renewal of cancer cells 21–23. The pleotropic activities of the MUC1-C subunit are dependent on a CQC motif in the cytoplasmic domain that is necessary and sufficient for the formation of MUC1-C homodimers and their import into the nucleus 20, 24, 25. Accordingly, inhibitors of MUC1-C, such as GO-203, target the CQC motif and block MUC1-C homodimerization 26. In this way, the effects of GO-203 on NSCLC cells are phenocopied by MUC1-C silencing and expression of a dominant-negative MUC1-C(CQC→AQA) mutant 26–28.
The present studies demonstrate that targeting MUC1-C in NSCLC cells is associated with downregulation of PD-L1 expression. We show that MUC1-C induces CD274 transcription by an NF-κB p65-dependent mechanism. Our results further demonstrate that activation of a MUC1-C→NF-κB→ZEB1 pathway results in repression of TLR9, IFNG, MCP-1/CCL2 and GM-CSF/CSF2 gene expression. These previously unexplored findings support a model in which MUC1-C plays a central role in integrating the induction of PD-L1 with suppression of genes encoding effectors of innate and adaptive immunity.
RESULTS
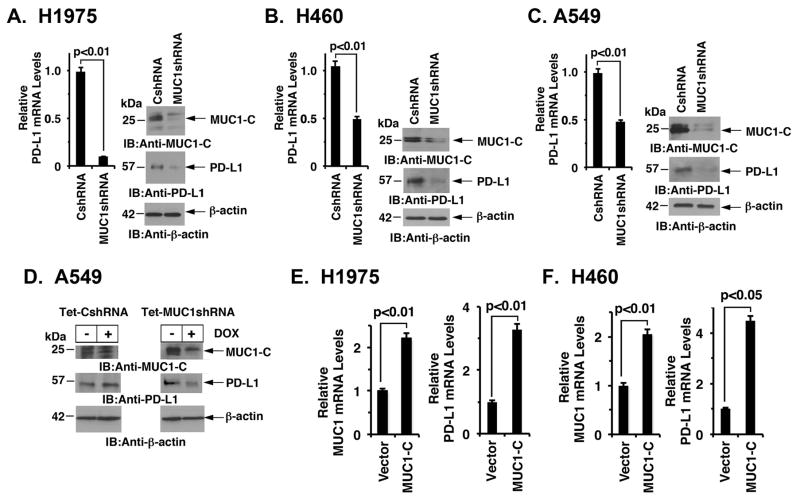
MUC1-C drives PD-L1 expression NSCLC cells. Targeting MUC1-C in H1975/EGFR(L858R/T790M) NSCLC cells is associated with suppression of EGFR activation 27. In the present work, we found that silencing MUC1-C in H1975 cells also results in downregulation of PD-L1 mRNA (Fig. 1A, left) and protein (Fig. 1A, right). Other work had shown that MUC1-C confers EMT and self-renewal capacity in A549/KRAS(G12S) and H460/KRAS(Q61H) cells 28. To extend these observations to KRAS mutant NSCLC cells, we silenced MUC1-C in H460 cells and found downregulation of PD-L1 expression (Fig. 1B, left and right). Similar results were obtained with A549 cells (Fig. 1C, left and right), indicating that stable silencing of MUC1-C in both EGFR and KRAS mutant NSCLC cells decreases PD-L1 expression. To confirm these findings, we established A549 cells stably expressing a tetracycline-inducible tet-on control shRNA (tet-CshRNA) or a MUC1 shRNA (tet-MUC1shRNA). Treatment of A549/tet-CshRNA cells with doxycycline (DOX) for 7 days had no apparent effect on MUC1-C or PD-L1 expression (Fig. 1D, left). By contrast, treatment of A549/tet-MUC1shRNA cells resulted in suppression of MUC1-C, as well as PD-L1, expression (Fig. 1D, right). In further support of a MUC1-C→PD-L1 pathway, enforced overexpression of MUC1-C in H1975 (Fig. 1E) and H460 (Fig. 1F) cells was associated with upregulation of PD-L1, indicating that MUC1-C is sufficient for this response. These results supported the premise that MUC1-C is necessary for PD-L1 expression in NSCLC cells.
Figure 1. MUC1-C drives PD-L1 expression in NSCLC cells.
A–C. H1975 (A), H460 (B) and A549 (C) NSCLC cells stably expressing a Control shRNA (CshRNA) or a MUC1 shRNA (MUC1shRNA) were analyzed for PD-L1 mRNA levels by qRT-PCR (left). The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative mRNA levels compared to that obtained with cells expressing CshRNA (assigned a value of 1). Lysates were immunoblotted with the indicated antibodies (right). D. A549 lung cancer cells were transfected to stably express an inducible control shRNA (left) or MUC1 shRNA (right). After treatment with doxycycline (DOX) for 72 h, lysates from the indicated cells were immunoblotted with antibodies against MUC1-C, PD-L1 and β-actin. E and F. H1975 (E) and H460 (F) cells were transiently transfected to express an empty vector or MUC1-C for 72 h. MUC1 (left) and PD-L1 (right) mRNA levels were determined by qRT-PCR. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative mRNA levels as compared to that obtained for cells expressing the empty vector (assigned a value of 1).
Targeting the MUC1-C cytoplasmic domain downregulates PD-L1 expression
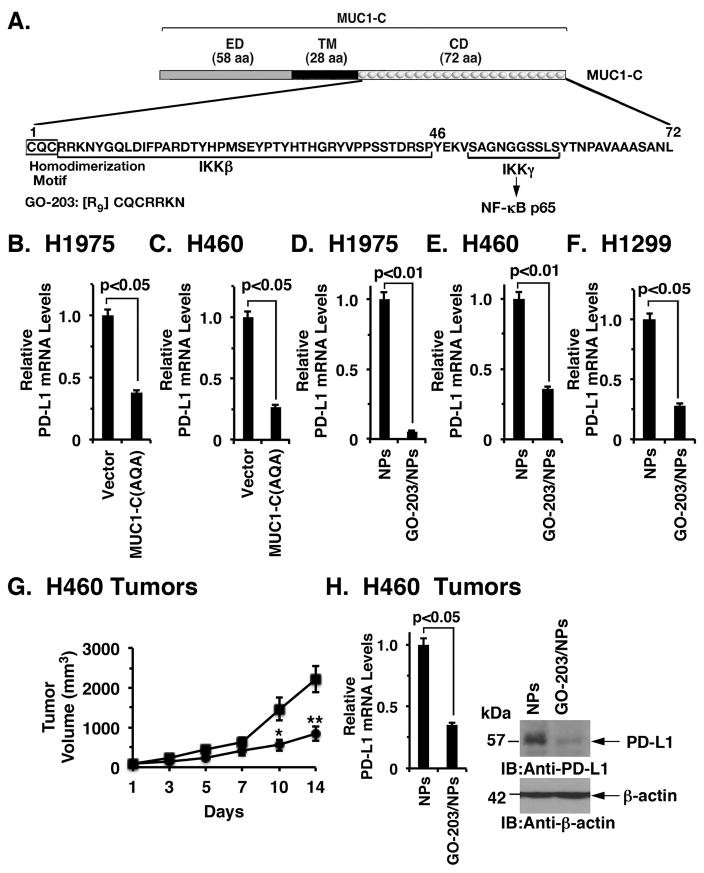
MUC1-C includes a 58 aa extracellular domain, a 28 aa transmembrane domain and a 72 aa cytoplasmic domain (Fig. 2A). The MUC1-C cytoplasmic domain contains a CQC motif that is necessary for MUC1-C homodimerization, nuclear localization and function as an oncoprotein (Fig. 2A) 18, 29. In this respect, expression of a MUC1-C(CQC→AQA) mutant in NSCLC cells blocks anchorage-independent growth and tumorigenicity, supporting a dominant-negative effect 27–29. In addition, we found that stable expression of the MUC1-C(CQC→AQA) mutant in H1975 (Fig. 2B) and H460 (Fig. 2C) cells results in suppression of PD-L1 expression, further indicating that the MUC1-C cytoplasmic domain confers this response. Targeting the MUC1-C CQC motif with the cell-penetrating peptide, GO-203, blocks MUC1-C homodimerization and signaling in NSCLC cells 26–28. GO-203 has been recently formulated in polymeric nanoparticles (NPs) for more effective intracellular delivery to cancer cells growing in mouse models 30. Treatment of H1975 and H460 cells with GO-203/NPs, but not empty NPs, was associated with downregulation of PD-L1 expression (Figs. 2D and 2E). Moreover, studies of H1299 NSCLC cells, which express wild-type EGFR and KRAS 31, demonstrated that GO-203/NP treatment decreases PD-L1 expression (Fig. 2F), indicating that the effect of targeting MUC1-C on PD-L1 repression is independent of the mutational status. GO-203/NP treatment of immunocompromised nude mice bearing H460 tumor xenografts was associated with inhibition of growth (Fig. 2G), consistent with decreases in self-renewal 28, and also suppression of PD-L1 mRNA and protein (Fig. 2H, left and right). These findings demonstrate that MUC1-C is a target for downregulating PD-L1 in NSCLC cells.
Figure 2. Targeting the MUC1-C cytoplasmic domain downregulates PD-L1 expression.
A. Schematic representation of the MUC1-C subunit with the 58 aa extracellular domain (ED), the 28 aa transmembrane domain (TM), and the sequence of the 72 aa cytoplasmic domain (CD). The MUC1-C cytoplasmic domain contains a CQC motif that is necessary and sufficient for MUC1-C homodimerization and oncogenic function. GO-203 is a cell-penetrating peptide that targets the CQC motif and blocks MUC1-C homodimerization. GO-203 has been encapsulated into nanoparticles (GO-203/NPs) for delivery in mouse tumor models. The MUC1-C cytoplasmic domain binds directly to IKKβ, IKKγ and NF-κB p65 and promotes the activation of NF-κB target genes. B and C. H1975 (B) and H460 (C) cells were infected with lentiviral vectors to stably express an empty vector or MUC1-C(AQA). The indicated cells were analyzed for PD-L1 mRNA levels by qRT-PCR. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative PD-L1 mRNA levels as compared to that obtained for the vector cells (assigned a value of 1). D-F. H1975 (D), H460 (E) and H1299 (F) cells were treated with empty NPs or 2.5 μM GO-203/NPs for 0 and 72 h, and then harvested at 144 h. PD-L1 mRNA levels were determined by qRT-PCR. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative PD-L1 mRNA levels as compared to that obtained for the empty NP-treated cells (assigned a value of 1). G. Mice bearing established H460 tumor xenografts (~150 mm3) were treated weekly with intraperitoneal injections of empty NPs (squares) or 15 mg/kg GO-203/NPs (circles). The results are expressed as tumor volume (mean±SEM, 6 mice per group). * denotes p<0.05. ** denotes p<0.01. H. Tumors obtained on day 14 were analyzed for PD-L1 mRNA levels by qRT-PCR (left). The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative PD-L1 mRNA levels as compared to that obtained for the tumors obtained in control mice (assigned a value of 1). Tumor lysates from empty NP- and GO-203/NP-treated mice (day 14) were immunoblotted with the indicated antibodies (right).
MUC1-C drives CD274/PD-L1 transcription by an NF-κB p65-dependent mechanism
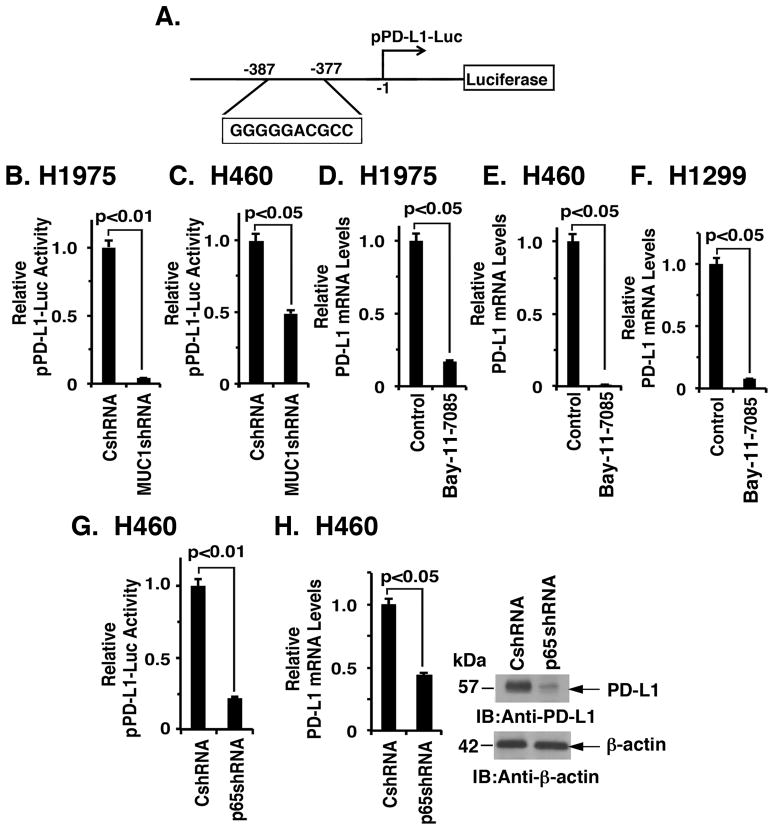
To define the mechanism by which MUC1-C drives PD-L1 expression, we first transfected H1975/CshRNA and H1975/MUC1shRNA cells with a CD274 promoter-luciferase reporter (pPD-L1-Luc) (Fig. 3A). The experiment revealed that silencing MUC1-C decreases pPD-L1-Luc activity (Fig. 3B). Similar results were obtained in H460 (Fig. 3C) and A549 (Supplemental Fig. S1A) cells, indicating that MUC1-C induces CD274 transcription. Along these lines, expression of the MUC1-C(CQC→AQA) mutant in H1975 (Supplemental Fig. S1B) and H460 (Supplemental Fig. S1C) cells also suppressed CD274 promoter activity. MUC1-C activates the inflammatory TAK1→IKK→NF-κB p65 pathway 21–23. Accordingly, we asked if treatment of NSCLC cells with Bay-11-7085, an irreversible inhibitor of IκBα phosphorylation, affects PD-L1 expression. Using this approach, we found that inhibiting the NF-κB pathway in H1975 (Fig. 3D), H460 (Fig. 3E) and H1299 (Fig. 3F) cells suppresses PD-L1 expression. In concert with these findings, silencing NF-κB p65 in H460 cells was associated with downregulation of CD274 transcription (Fig. 3G) and PD-L1 mRNA and protein (Fig. 3H, left and right). We also found that treatment of H1975/MUC1-C and H460/MUC1-C cells with Bay-11-7085 decreases MUC1-C-induced upregulation of PD-L1 expression (Figs. 1E and 1F; Supplemental Figs. S2A and S2B). Moreover, our results show that the effects of silencing MUC1-C on PD-L1 expression (Figs. 1A and 1B) are potentiated by adding Bay-11-7085 (Supplemental Figs. S2C and S2D), further supporting the notion that the MUC1-C→NF-κB p65 pathway drives PD-L1 expression.
Figure 3. MUC1-C drives CD274/PD-L1 transcription by an NF-κB p65-dependent mechanism.
A. Schema of the pPD-L1-Luc reporter with positioning of the putative the NF-κB binding site at −377 to −387 upstream to the transcription start site. B and C. The indicated H1975 (B) and H460 (C) cells were transfected with the pPD-L1-Luc reporter for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SEM of three biological replicates each performed in triplicate) compared with that obtained from cells expressing the CshRNA (assigned a value of 1). D–F. H1975 (D), H460 (E) and H1299 (F) cells were treated with 5 μM Bay-11-7085 or DMSO as the vehicle control for 18 h. PD-L1 mRNA levels were determined by qRT-PCR. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative PD-L1 mRNA levels as compared to that obtained for the control cells (assigned a value of 1). G. H460 cells stably expressing a CshRNA or an NF-κB p65 shRNA (NF-κBshRNA) were transfected with the pPD-L1-Luc reporter for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SEM of three biological replicates each performed in triplicate) compared with that obtained from cells expressing the CshRNA (assigned a value of 1). H. The indicated H460 cells were analyzed for PD-L1 mRNA levels by qRT-PCR (left). The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative PD-L1 mRNA levels as compared to that obtained for cells expressing the CshRNA (assigned a value of 1). Lysates were immunoblotted with the indicated antibodies (right).
MUC1-C/NF-κB p65 complexes occupy the CD274 promoter
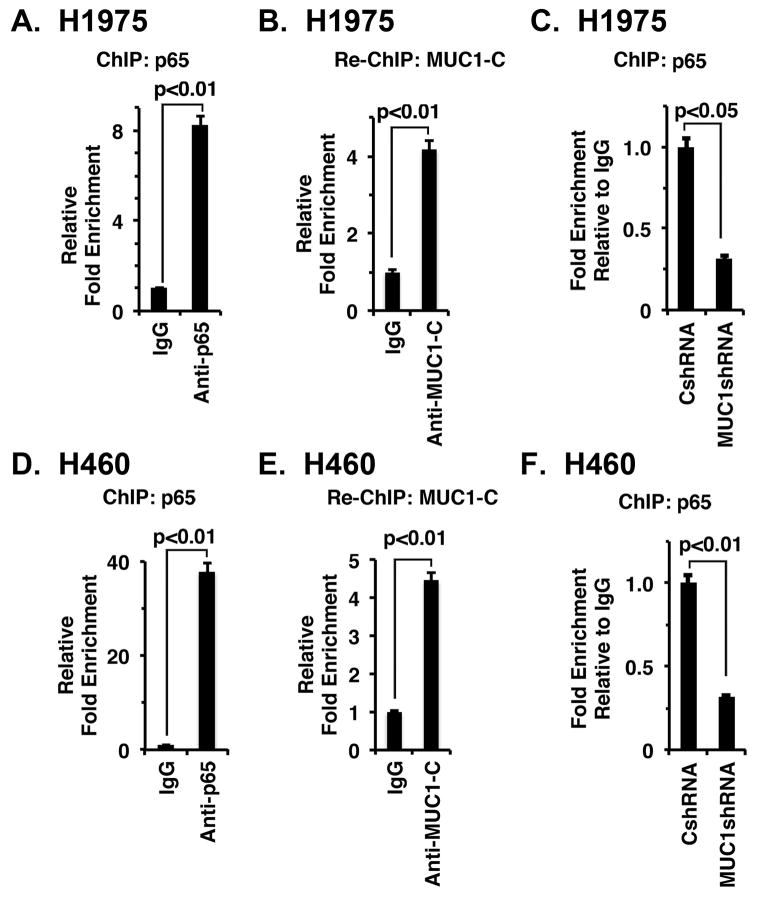
A potential NF-κB binding site (GGGGGACGCC) is located in the CD274 promoter at position −377 to −387 upstream to the transcription start site (Fig. 3A). However, to our knowledge there is no available evidence showing that NF-κB occupies the CD274 promoter. ChIP analysis of H1975 cell chromatin demonstrated occupancy of the CD274 promoter by NF-κB p65 (Fig. 4A). Moreover and in concert with the finding that MUC1-C binds directly to NF-κB p65 22, re-ChIP studies demonstrated that MUC1-C forms a complex with NF-κB p65 on the CD274 promoter (Fig. 4B). We also found that silencing MUC1-C decreases NF-κB p65 occupancy (Fig. 4C), consistent with previous studies on promoters of other NF-κB target genes, including MUC1 itself 22. Similar results were obtained in experiments with H460 cells; that is, (i) occupancy of the CD274 promoter by NF-κB p65/MUC1-C complexes (Figs. 4D and 4E), and (ii) silencing MUC1-C decreases occupancy by NF-κB p65 (Fig. 4F).
Figure 4. MUC1-C/NF-κB p65 complexes occupy the CD274/PD-L1 promoter.
A. Soluble chromatin from H1975 cells was precipitated with anti-NF-κB or a control IgG. B. In the re-ChIP experiments, NF-κB precipitates were released and then reimmunoprecipitated with an anti-MUC1-C. The final DNA samples were amplified by qPCR with primers for the CD274promoter NF-κB binding region or GAPDH as a control. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1). C. Soluble chromatin from H1975/CshRNA and H1975/MUC1shRNA cells was precipitated with anti-NF-κB or a control IgG. The final DNA samples were amplified by qPCR with primers for the PD-L1 promoter NF-κB binding region or GAPDH as a control. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as the relative fold enrichment compared to that obtained for H1975/CshRNA cell chromatin (assigned a value of 1). D. Soluble chromatin from H460 cells was precipitated with anti-NF-κB or a control IgG. E. In re-ChIP experiments, NF-κB precipitates were released and then reimmunoprecipitated with an anti-MUC1-C. The final DNA samples were amplified by qPCR with primers for the PD-L1 promoter NF-κB binding region or as a control GAPDH. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as the relative fold enrichment compared with that obtained with the IgG control (assigned a value of 1). F. Soluble chromatin from H460/CshRNA and H460/MUC1shRNA cells was precipitated with anti-NF-κB or a control IgG. The final DNA samples were amplified by qPCR with primers for the PD-L1 promoter NF-κB binding region or GAPDH as a control. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as the relative fold enrichment compared to that obtained for H460/CshRNA cell chromatin (assigned a value of 1).
Targeting MUC1-C induces TLR7 expression by an NF-κB p65-mediated mechanism
Our findings that MUC1-C induces PD-L1 expression through NF-κB p65 invoked the possibility that the MUC1-C→NF-κB p65 pathway might regulate other genes involved in immune responses. However, we found that targeting MUC1-C in H1975 (Supplemental Fig. S3A) and H460 (Supplemental Fig. S3B) cells had no significant effect on PD-1, PD-L2, CTLA-4, TIM-3 and LAG-3 expression. We also investigated the potential role of MUC1-C in regulation of the toll-like receptor 7 (TLR7) gene, which is constitutively expressed in NSCLC cells, activates the NF-κB pathway and confers chemoresistance and poor survival 32–34. In concert with the demonstration that the TLR7 promoter is activated by NF-κB p65 (Fig. S4A) 35, we found that targeting MUC1-C results in marked downregulation of TLR7 mRNA levels (Supplemental Fig. S4B). Moreover targeting the NF-κB pathway with Bay-11-7085 (Supplemental Fig. S4C) or silencing NF-κB p65 (Supplemental Fig. S4D) was associated with decreases in TLR7 mRNA levels, indicating that, like PD-L1, the MUC1-C→NF-κB p65 pathway induces TLR7 in NSCLC cells.
MUC1-C represses immune-related genes by a ZEB1-mediated mechanism
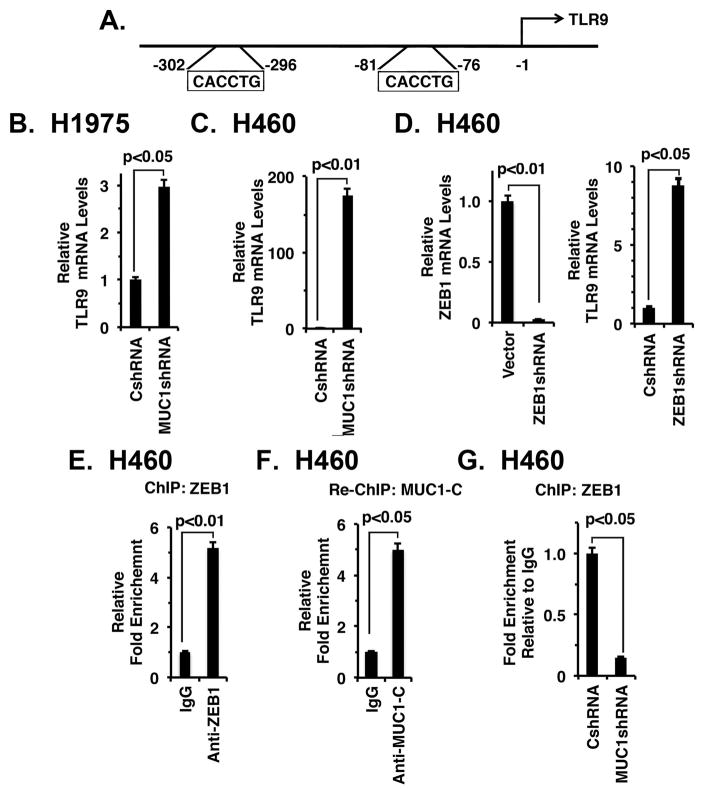
The MUC1-C→NF-κB p65 pathway activates the ZEB1 gene, which encodes the EMT-inducing transcription factor 36. In turn, MUC1-C interacts with ZEB1, represses miR-200c and induces EMT 36. Based on these findings, we reasoned that the MUC1-C→NF-κB→ZEB1 pathway might link EMT with the suppression of certain immune-related genes. As one candidate, we studied the TLR9 gene, which encodes the innate TLR9 receptor, is downregulated by NF-κB signaling 37 and has two GC-rich E-boxes as potential binding sites for MUC1-C/ZEB1 complexes (Fig. 5A). Notably, targeting MUC1-C (Figs. 5B and 5C) was associated with marked upregulation of TLR9 mRNA levels. Moreover, silencing ZEB1 (Fig. 5D, left) resulted in induction of TLR9 expression (Fig. 5D, right). ChIP and re-ChIP studies further demonstrated that ZEB1 occupies the TLR9 promoter in a complex with MUC1-C (Figs. 5E and 5F). Silencing MUC1-C also decreased ZEB1 occupancy on the TLR9 promoter (Fig. 5G). Consistent with these results, analysis of TCGA and RNA-seq datasets demonstrated that MUC1 correlates negatively with TLR9 expression in NSCLCs (Supplemental Fig. S5A, left and right).
Figure 5. Targeting MUC1-C derepresses TLR9 expression.
A. Schema of the TLR9 promoter with positioning of the E-boxes upstream to the transcription start site. B and C. The indicated H1975 (B) and H460 (C) cells were analyzed for TLR9 mRNA levels by qRT-PCR. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative mRNA levels as compared to that obtained for the CshRNA cells (assigned a value of 1). D. H460 cells stably express a control CshRNA or a ZEB1shRNA were analyzed for TLR9 mRNA levels by qRT-PCR. The results (mean±SEM of two biological replicates each performed in triplicate) are expressed as relative mRNA levels as compared to that obtained for the CshRNA cells (assigned a value of 1). E. Soluble chromatin from H460 cells was precipitated with anti-ZEB1 or a control IgG. F. In re-ChIP experiments, ZEB1 precipitates were released and then reimmunoprecipitated with anti-MUC1-C. The final DNA samples were amplified by qPCR with primers for the TLR9 promoter ZEB1 binding region or as a control GAPDH. The results (mean±SEM of two biological replicates each performed in triplicate) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1). G. Soluble chromatin from H460/CshRNA and H460/MUC1shRNA cells was precipitated with anti-ZEB1 or a control IgG. The final DNA samples were amplified by qPCR with primers for the TLR9 promoter ZEB1 binding region or as a control GAPDH. The results (mean±SEM of two biological replicates each performed in triplicate) are expressed as the relative fold enrichment compared to that obtained with H460/CshRNA cell chromatin (assigned a value of 1).
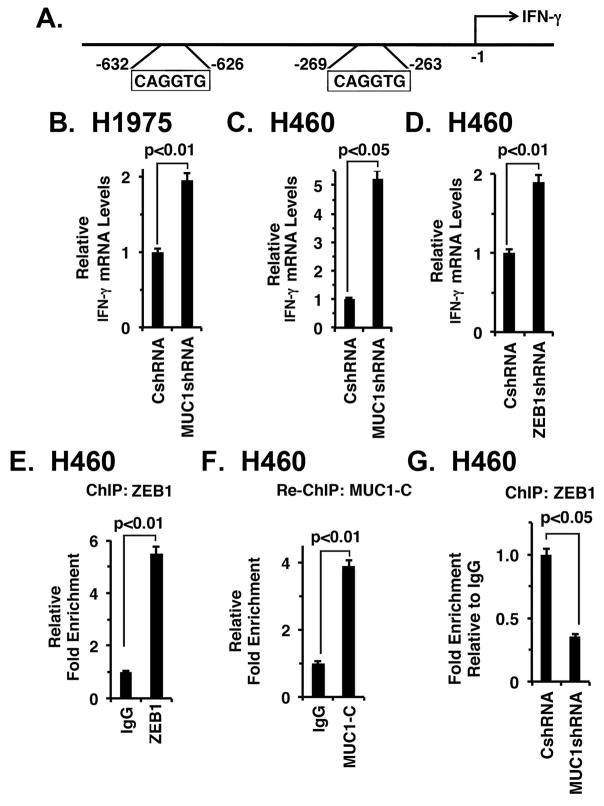
We also studied effects of targeting MUC1-C on the IFNG gene, which encodes IFN-γ and contains E-boxes in its promoter, but is not known to be regulated by ZEB1 (Fig. 6A). As shown for TLR9, silencing MUC1-C in H1975 and H460 cells induced IFN-γ expression (Figs. 6B and 6C). In addition, silencing ZEB1 was associated with increases in IFN-γ mRNA levels (Fig. 6D). ChIP experiments further demonstrated that (i) ZEB1 occupies the IFNG promoter in a complex with MUC1-C (Figs. 6E and 6F) and (ii) MUC1-C promotes ZEB1 occupancy (Fig. 6G), supporting the notion that MUC1-C suppresses IFNG activation by a ZEB1-mediated mechanism. In this respect, MUC1 correlated negatively with IFN-γ expression in NSCLCs (Supplemental Fig. S5B, left and right). Interestingly, IFN-γ induces CD274 transcription by a mechanism involving IRF1 38, 39; however, our findings that MUC1-C downregulates IFN-γ and activates the CD274 promoter by occupancy with MUC1-C/NF-κB p65 complexes support a distinct pathway.
Figure 6. Targeting MUC1-C activates IFN-γ expression.
A. Schema of the IFNG promoter with positioning of the E-boxes upstream to the transcription start site. B and C. The indicated H1975 (B) and H460 (C) cells were analyzed for IFN-γ mRNA levels by qRT-PCR. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative mRNA levels as compared to that obtained for the CshRNA cells (assigned a value of 1). D. H460 cells stably express a control CshRNA or a ZEB1shRNA were analyzed for IFN-γ mRNA levels by qRT-PCR. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative mRNA levels as compared to that obtained for the CshRNA cells (assigned a value of 1). E. Soluble chromatin from H460 cells was precipitated with anti-ZEB1 or a control IgG. F. In the re-ChIP experiments, ZEB1 precipitates were released and then reimmunoprecipitated with anti-MUC1-C. The final DNA samples were amplified by qPCR with primers for the IFNG promoter ZEB1 binding region or as a control GAPDH. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1). G. Soluble chromatin from H460/CshRNA (left) and H460/MUC1shRNA was precipitated with anti-ZEB1 or a control IgG (right). The final DNA samples were amplified by qPCR with primers for the IFNG promoter ZEB1 binding region or as a control GAPDH. The results (mean±SD of three biological replicates each performed in triplicate) are expressed as the relative fold enrichment compared to that obtained with the CshRNA cells.
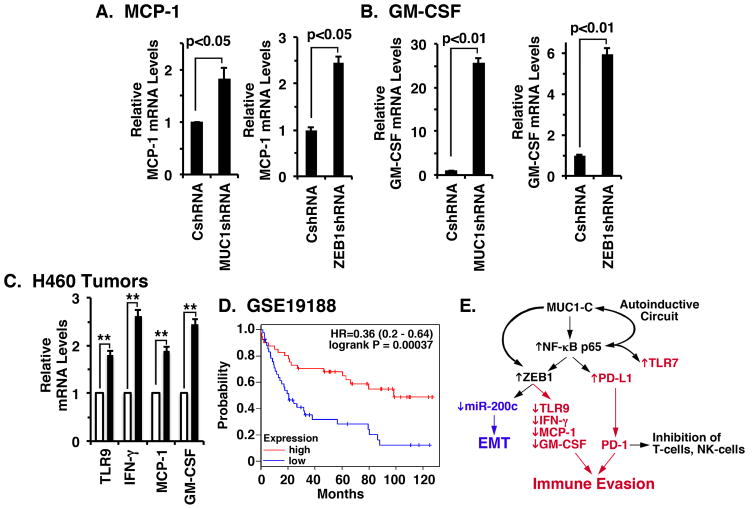
Along these same lines, we found that targeting MUC1-C and ZEB1 was associated with upregulation of (i) MCP-1/CCL2, a key chemokine that regulates the migration and infiltration of monocytes/macrophages 40(Fig. 7A), and (ii) GM-CSF, a key hematopoietic growth factor and immune modulator 41 (Fig. 7B), supporting the previously unexplored notion that MUC1-C/ZEB1 complexes contribute to repression of the MCP-1 and GM-CSF genes. In concert with these findings, (i) the MCP-1 gene intron 1 region has 4 putative ZEB1 binding sites (CAGCTG) at +294 to +300, +328 to +334, +400 to +406 and +616 to +622 and (ii) the GM-CSF promoter contains a potential E-box (CACGTG) at −1097 to −1103 relative to their transcription start sites. Additionally, like TLR9 and IFN-γ, we found that MUC1 negatively correlates with MCP-1 in NSCLCs (Supplemental Fig. S5C, left and right). In contrast, a negative correlation between MUC1 and GM-CSF was not statistically significant. Nonetheless and in concert with these results, treatment of H460 tumors with GO-203/NPs was associated with increases in TLR9, IFN-γ, MCP-1 and GM-CSF expression (Fig. 7C), indicating that targeting MUC1-C in vivo reverses this program of immune evasion. We also found that low levels of TLR9, IFN-γ, MCP-1 and GM-CSF in the major NSCLC pathologic subtypes (GSE19188) 42 are associated with significant decreases in overall survival (Fig. 7D). Additionally, similar results were obtained from analysis of an independent dataset of NSCLCs of the adenocarcinoma subtype (GSE29013)43 (Supplemental Fig. S6), supporting the notion that suppression of these immune-related genes in NSCLCs is associated with a poor clinical outcome.
Figure 7. Targeting MUC1-C induces MCP-1 and GM-CSF expression by ZEB1-mediated mechanisms.
A and B. The indicated H460 cells were analyzed for MCP-1 (A) and GM-CSF (B) mRNA levels by qRT-PCR. The results (mean±SEM of two biological replicates each performed in triplicate) are expressed as relative mRNA levels as compared to that obtained for the CshRNA cells (assigned a value of 1). C. H460 tumors obtained on day 14 (see Fig. 2F) of treatment with empty NPs (open bars) or GO-203/NPs (solid bars) were analyzed for TLR9, IFN-γ, MCP-1 and GM-CSF mRNA levels by qRT-PCR. The results (mean±SEM of three biological replicates each performed in triplicate) are expressed as relative mRNA levels as compared to that obtained for the tumors obtained in empty NP-treated mice (assigned a value of 1). ** denotes p value <0.05. D. Kaplan-Meier plot comparing the overall survival of patients with NSCLC in the GSE19188 dataset. Patients were stratified with the high (red) or low (blue) expression of TLR9, IFN-γ, MCP-1 and GM-CSF against the median average. The survival curves were compared using Log-rank (Mantel-Cox) test. HR: Hazard Ratio. E. Proposed schema of a MUC1-C-induced inflammatory pathway linking EMT (blue) and regulation of immune-related genes (red) in NSCLC cells. MUC1-C activates the inflammatory NF-κB p65 pathway 21–23 and thereby PD-L1 expression. MUC1-C also upregulates TLR7, which contributes to NF-κB p65 activation, survival and chemoresistance of NSCLC cells 32. MUC1-C forms a complex with NF-κB p65 and induces the activation of NF-κB target genes, including MUC1 itself, in an autoinductive circuit 22. MUC1-C also promotes occupancy of NF-κB p65 on the ZEB1 36 and CD274/PD-L1 promoters and contributes to activation of these genes. The upregulation of ZEB1 and the formation of MUC1-C/ZEB1 complexes suppresses miR-200c and thereby induces EMT 36. Of note, PD-L1 is also a target of miR-200 66, invoking the possibility that the MUC1-C→NF-κB p65→ZEB1 pathway could increase PD-L1 expression by both transcriptional and post-transcriptional mechanisms. Our results further support a role for MUC1-C/ZEB1 complexes in suppression of TLR9, IFNG, MCP-1 and GM-CSF, linking these responses with EMT. Thus, targeting MUC1-C suppresses PD-L1 and induces TLR9, IFN-γ, MCP-1 and GM-CSF expression in NSCLC tumors, supporting the notion that MUC1-C is of importance for integrating the regulation of these genes.
DISCUSSION
Blockade of the PD-1/PD-L1 immune checkpoint is broadly effective in the treatment of NSCLCs and can extend survival in patients with tumors not responsive to targeted therapy. Nonetheless, PD-1/PD-L1 blockade is associated with a response rate of about 20% in NSCLC and these responses can be of short duration 3. Those findings have supported the premise that the signaling pathways responsible for induction of CD274 in NSCLCs may be integrated with the regulation of other immune-related genes. Our results demonstrate that targeting MUC1-C in NSCLC cells is effective in decreasing PD-L1 mRNA and protein. MUC1-C activates the proinflammatory TAK1→IKK→NF-κB p65 pathway 21–23. A critical aspect of this autoinductive circuit is that MUC1-C binds directly to NF-κB p65 and increases occupancy of NF-κB p65 on the promoters of its target genes, including MUC1 itself 22, 36 (Fig. 7E). The present results extend this circuit by demonstrating that MUC1-C forms a complex with NF-κB p65 on the CD274 promoter, increases NF-κB p65 occupancy and, in turn, activates CD274 transcription (Fig. 7E). Interestingly, activation of monocytes with lipopolysaccharide is also associated with NF-κB occupancy of the CD274 promoter, consistent with involvement of this pathway in the inflammatory response 44. However, NF-κB has not been previously shown to occupy the CD274 promoter in cancer cells or to constitutively activate the CD274 gene 45, supporting the importance of MUC1-C in driving the inflammatory TAK1→IKK→NF-κB p65 pathway and thereby PD-L1 expression. Notably, our findings do not exclude the possibility that MUC1-C contributes to the regulation of PD-L1 by additional mechanisms. For instance, recent work has demonstrated that MUC1-C drives MYC in KRAS mutant NSCLC cells 46 and, by extension, MYC activates CD274 transcription 47. Thus, in addition to the present findings, MUC1-C could regulate PD-L1 expression by alternative mechanisms that are dependent on context of the cancer cell. Indeed, the complexity of PD-L1 regulation may reflect the lack of a significant direct relationship between MUC1 and PD-L1 expression in NSCLCs.
MUC1-C associates with NF-κB p65 on the ZEB1 promoter and drives ZEB1 transcription (Fig. 7E) 36. In turn, MUC1-C binds to ZEB1 and MUC1-C/ZEB1 complexes contribute to the transcriptional suppression of miR-200c and the induction of EMT (Fig. 7E) 36. In this way, our results demonstrate that MUC1-C represses TLR9 transcription by a ZEB1-dependent mechanism (Fig. 7E). TLR9 is an innate immune receptor that recognizes unmethylated double-stranded DNA CpG motifs present in the genomes of bacteria and certain viruses 48, 49. TLR9 is expressed in plasmacytoid DCs and B-cells 48–50. Certain carcinomas also express TLR9, a finding that has linked low TLR9 levels with poor outcomes in renal and triple-negative breast cancers 51, 52. Little is known about the regulation of TLR9 in NSCLC cells and, to our knowledge, ZEB1 is not a known suppressor of TLR9 expression. Our results support a model in which MUC1-C promotes ZEB1 occupancy of E-boxes in the TLR9 promoter and thereby suppresses TLR9 transcription (Fig. 7E). The prognostic importance of TLR9 in NSCLC is not known; however, the TLR9 immune response induces the production of type I IFNs and proinflammatory cytokines, linking innate to adaptive immunity 53. In contrast to TLR9, other studies have demonstrated that TLR7 is overexpressed in NSCLC cells, and confers chemoresistance and poor survival 32–34. In addition, TLR7 activates the NF-κB pathway in a positive-regulatory loop (Fig. 7E) 32, 33, 35. The present results demonstrate that MUC1-C also induces TLR7 expression by an NF-κB p65-dependent mechanism, invoking the possibility that TLR7 may contribute to the MUC1-C→NF-κB p65 autoinductive circuit (Fig. 7E). The findings with TLR9 and TLR7 further indicate that MUC1-C differentially regulates expression of the TLR family members.
Based on the finding that MUC1-C represses TLR9 expression, we identified other immune-related genes that are upregulated by silencing MUC1-C and have potential ZEB1 binding motifs in their promoters. Using this approach, we found that the IFNG promoter contains E-boxes and that MUC1-C/ZEB1 complexes occupy this promoter in NSCLC cells. Additionally, targeting MUC1-C decreased ZEB1 occupancy and induced IFN-γ expression. IFN-γ stimulates the JAK/STAT pathway and induces pleotropic immune responses that activate both innate and acquired immunity 54. In this way, IFN-γ upregulates MHC Class I molecules with the enhancement of antigen priming and presentation 55. IFN-γ also induces multiple aspects of Th1-mediated immune responses by regulating the activation of T-cells 56. Moreover, IFN-γ activates macrophages and the production of chemokines that recruit effectors to inflammatory sites 57. By extension, we found that targeting MUC1-C and ZEB1 induces (i) MCP-1, which promotes the migration and infiltration of monocytes/macrophages 40 and (ii) GM-CSF, an immune modulatory cytokine that expands granulocyte and macrophage populations, induces dendritic cell recruitment and maturation, and contributes to T-cell activation 41. Importantly and like TLR9, we are unaware of previous findings associating ZEB1 with the repression of the IFNG, MCP-1 and GM-CSF genes.
The finding that MUC1-C activates the NF-κB p65 pathway has linked inflammation with diverse oncogenic signals that are conveyed by MUC1-C at the cell membrane and in the nucleus 23. The interaction between MUC1-C and NF-κB p65 induces EMT through the ZEB1/miR-200c regulatory loop (Fig. 7E) 28, 36. The mesenchymal phenotype has been associated with expression of PD-L1 in NSCLCs and other carcinomas 58, 59. The present results thus provide support for a model in which MUC1-C drives EMT and PD-L1 expression by an NF-κB p65-dependent mechanism, indicating that MUC1-C could play a pivotal role in coordinating both of these responses (Fig. 7E). In turn, our results extend this model by demonstrating that MUC1-C drives ZEB1-mediated suppression of key innate and adaptive immune-related genes and that their downregulation confers a poor prognosis for patients with NSCLC. Importantly, similar findings were obtained by targeting MUC1-C with GO-203/NPs in a NSCLC tumor xenograft model in immunocompromised mice. Based on these results, we will be studying the effects of GO-203/NPs in immune-competent GEMMs to assess targeting of the MUC1-C→NF-κB p65→ZEB1 pathway and thereby immune recognition and destruction. In this respect, GO-203 has been evaluated in a Phase I trial for patients with advanced malignancies and, based on a highly favorable toxicity profile, the GO-203/NP formulation is being advanced for the treatment of patients with NSCLC and potentially in combination with immunotherapeutic agents.
MATERIALS AND METHODS
Cell culture
Human A549/KRAS(G12S), H460/KRAS(Q61H) and H1975/EGFR(L858R/T790M) NSCLC cells (ATCC, Manassas, VA, USA) were grown in RPMI1640 medium supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS), 100 μg/ml streptomycin, 100 units/ml penicillin and 2 mM L-glutamine. Authentication of the cells was performed by short tandem repeat (STR) analysis. Cells were monitored monthly for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza, Rockland, MA, USA). Cells were transfected with lentiviral vectors to stably express a scrambled control shRNA (CshRNA; Sigma, St. Louis, MO, USA), a MUC1 shRNA (MUC1shRNA; Sigma), a NF-κB p65 shRNA (Sigma), MUC1-C or MUC1-C(AQA)27, 28, 46. Puromycin and hygromycin concentrations used for optimal selection of transfected cells were determined by generating survival curves for the different cell lines. For stable transfectants, single cell clones were identified by limiting dilution and expanded in the presence of 2 μg/ml puromycin. Cells were treated with (i) doxycycline (DOX; Sigma) and (ii) the IκB inhibitor Bay-11-7085 (Sigma) or DMSO as the vehicle control. Cells were also treated with empty nanoparticles (NPs) or GO-203/NPs 30.
Quantitative real-time, reverse transcriptase PCR (qRT-PCR)
Whole cell RNA was isolated using the RNeasy mini kit (Qiagen, Germantown, MD, USA). The High Capacity cDNA Reverse Transcription kit (Life Technologies, Carlsbad, CA, USA) was used to synthesize cDNAs from 2 μg RNA. The GAPDH gene was used as an internal control. The SYBR green qPCR assay kit and the ABI Prism Sequence Detector (Applied Biosystems, Foster City, CA, USA) were used to amplify the cDNAs. Relative mRNA expression levels were calculated using the ΔΔCt method 60. Primers used are listed in Supplemental Table S1.
Immunoblot analysis
Whole cell lysates were prepared in NP-40 lysis buffer and immunoblotted with (i) anti-MUC1-C (ThermoFisher Scientific, Waltham, MA, USA; Cat. #HM-1630-P1, 1:50 dilution) and an anti-Armenian hamster secondary antibody (Abcam, Cambridge, MA, USA; Cat. #ab5745, 1:300 dilution), (ii) anti-PD-L1 (Cell Signaling Technology, Danvers, MA, USA; Cat. #13684S, 1:1000 dilution) and an anti-rabbit secondary antibody (GE Healthcare Life Sciences, Marlborough, MA, USA; Cat. #NA934V, 1:3000 dilution) and anti-β-actin (Sigma; Cat. A5316, 1:1000 dilution) and an anti-mouse secondary antibody (GE Healthcare Life Sciences; Cat. #NA931, 1:3000 dilution). Horseradish peroxidase secondary antibodies and enhanced chemiluminescence (GE Healthcare Life Sciences) were used for the detection of immune complexes. Immunoblot results were each confirmed with a second analysis.
Promoter-reporter assays
Cells were transfected with 1.5 μg of PD-L1 promoter-luciferase reporter (pPD-L1-Luc) or control vector (Active Motif, Carlsbad, CA, USA; Cat. S701930) and SV-40-Renilla-Luc (100:1 ratio) with Superfect according to the manufacturer’s recommendations (Qiagen). After 48 h, the cells were lysed in passive lysis buffer. Lysates were analyzed using the Dual-Luciferase assay kit (Promega, Madison, WI, USA).
Chromatin immunoprecipitation (ChIP) assays
Soluble chromatin was isolated from 3 x 106 cells and immunoprecipitated with anti-NF-κB p65 (Santa Cruz Biotechnology, Dallas, TX, USA) or a control IgG as described 36. In re-ChIP experiments, NF-κB complexes obtained were reimmunoprecipitated with anti-MUC1-C (ThermoFisher Scientific) or a control IgG. qPCR analyses were performed using the SYBR green kit and the ABI Prism 7000 Sequence Detector (Applied Biosystems, Foster City, CA, USA). Primers used for the CD274, TLR9 and IFNG promoters and GAPDH as a control are listed in Supplemental Table S2. Relative fold enrichment was calculated as described 36.
NSCLC tumor xenograft studies
H460 cells (5 x 106) were injected subcutaneously in the flank of six-week old female NCR nu/nu mice. After reaching a tumor size of ~150 mm3, mice were pair-matched in two groups and treated with empty NPs or 15 mg/kg GO-203/NPs. The formula V=L x W2/2, where L and W are the larger and smaller diameters, respectively, was used to calculate tumor volumes in an unblinded fashion. These studies were performed under Protocol #03-029 approved by the Dana-Farber Institutional Animal Care and Use Committee.
Data analysis
Statistical significance was determined using the Student’s t-test (GraphPad Software, LaJolla, CA, USA). The Shapiro-Wilk test was used to confirm normal distribution of the data (GraphPad Software).
Bioinformatic analyses
NSCLC clinical datasets were obtained from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) under the accession number (GSE72094) 23, 46, 61. GSE72084 microarray gene expression data were normalized with IRON as described 62. TCGA data were obtained from Firehose by using RTCGAToolbox 63. Log2 expression values of MUC1, IFN-γ, TLR9 and MCP-1/CCL2 from both datasets were assessed for correlation using Spearman’s coefficient. The prognostic value of TLR9, IFN-γ, MCP-1/CCL2 and CSF2/GMCSF expression in NSCLC datasets GSE19188 42 and GSE29013 43 was performed as previously described 64, 65. Expression values of TLR9, IFN-γ, MCP-1/CCL2 and CSF2/GMCSF were averaged and NSCLC patients were divided by the median expression. The Kaplan-Meier survival probability plot with the hazard ratio (95% confidence interval) and long rank p value was calculated and plotted in R.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers CA97098 and CA166480, and by the Lung Cancer Research Foundation.
ABBREVIATIONS
- NSCLC
non-small cell lung cancer
- EGFR
epidermal growth factor receptor
- PD-1
programmed death 1
- PD-L1
programmed death ligand 1
- MUC1
mucin 1
- MUC1-C
MUC1 C-terminal subunit
- CD274
gene encoding PD-L1
- GEMMs
genetically engineered mouse models
- DOX
doxycycline
- ChIP
chromatin immunoprecipitation
- TLR7
toll-like receptor 7
- TLR9
toll-like receptor 9
- IFN-γ
interferon-gamma
- MCP-1
monocyte chemoattractant protein-1
- GM-CSF
granulocyte-monocyte colony-stimulating factor
Footnotes
CONFLICT OF INTEREST
The authors declare competing financial interests; D.K. holds equity in Genus Oncology and is a consultant to the company. The other authors disclosed no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
AB, HR, KKW, SK and DK designed the research
AB, HR, CJ, MA, MH, TM, XH and DA performed the research and data analysis
MS and AT performed bioinformatics analysis
AB, KKW and DK wrote the manuscript
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005;7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Carrizosa DR, Gold KA. New strategies in immunotherapy for non-small cell lung cancer. Transl Lung Cancer Res. 2015;4:553–559. doi: 10.3978/j.issn.2218-6751.2015.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reck M, Paz-Ares L. Immunologic checkpoint blockade in lung cancer. Semin Oncol. 2015;42:402–417. doi: 10.1053/j.seminoncol.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Mayor M, Yang N, Sterman D, Jones DR, Adusumilli PS. Immunotherapy for non-small cell lung cancer: current concepts and clinical trials. Eur J Cardiothorac Surg. 2015;49:1324–1333. doi: 10.1093/ejcts/ezv371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T cell activity in the lung tumor microenvironment. Cancer Res. 2016;76:999–1008. doi: 10.1158/0008-5472.CAN-15-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Situ D, Wang J, Ma Y, Zhu Z, Hu Y, Long H, et al. Expression and prognostic relevance of MUC1 in stage IB non-small cell lung cancer. Med Oncol. 2010;28:596–604. doi: 10.1007/s12032-010-9752-4. [DOI] [PubMed] [Google Scholar]
- 9.Jarrard JA, Linnoila RI, Lee H, Steinberg SM, Witschi H, Szabo E. MUC1 is a novel marker for the type II pneumocyte lineage during lung carcinogenesis. Cancer Res. 1998;58:5582–5589. [PubMed] [Google Scholar]
- 10.Awaya H, Takeshima Y, Yamasaki M, Inai K. Expression of MUC1, MUC2, MUC5AC, and MUC6 in atypical adenomatous hyperplasia, bronchioloalveolar carcinoma, adenocarcinoma with mixed subtypes, and mucinous bronchioloalveolar carcinoma of the lung. Am J Clin Pathol. 2004;121:644–653. doi: 10.1309/U4WG-E9EB-FJN6-CM8R. [DOI] [PubMed] [Google Scholar]
- 11.Khodarev N, Pitroda S, Beckett M, MacDermed D, Huang L, Kufe D, et al. MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res. 2009;69:2833–2837. doi: 10.1158/0008-5472.CAN-08-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacDermed DM, Khodarev NN, Pitroda SP, Edwards DC, Pelizzari CA, Huang L, et al. MUC1-associated proliferation signature predicts outcomes in lung adenocarcinoma patients. BMC Medical Genomics. 2010;3:16. doi: 10.1186/1755-8794-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu F, Liu F, Zhao H, An G, Feng G. Prognostic significance of mucin antigen MUC1 in various human epithelial cancers: a meta-analysis. Medicine (Baltimore) 2015;94:e2286. doi: 10.1097/MD.0000000000002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kufe D. Mucins in cancer: function, prognosis and therapy. Nature Reviews Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kufe D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raina D, Agarwal P, Lee J, Bharti A, McKnight C, Sharma P, et al. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One. 2015;10:e0135156. doi: 10.1371/journal.pone.0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, et al. MUC1 oncoprotein activates the IκB kinase β complex and constitutive NF-κB signaling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad R, Raina D, Joshi MD, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the NF-κB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi H, Jin C, Rajabi H, Pitroda S, Alam M, Ahmad R, et al. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene. 2015;34:5187–5197. doi: 10.1038/onc.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, et al. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 25.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol. 2012;40:1643–1649. doi: 10.3892/ijo.2011.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Ther. 2011;10:806–816. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kharbanda A, Rajabi H, Jin C, Tchaicha J, Kikuchi E, Wong K, et al. Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin Cancer Res. 2014;20:5423–5434. doi: 10.1158/1078-0432.CCR-13-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kharbanda A, Rajabi H, Jin C, Alam M, Wong K, Kufe D. MUC1-C confers EMT and KRAS independence in mutant KRAS lung cancer cells. Oncotarget. 2014;5:8893–8905. doi: 10.18632/oncotarget.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kufe D. Functional targeting of the MUC1 oncogene in human cancers. Cancer biology & therapy. 2009;8:1201–1207. doi: 10.4161/cbt.8.13.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasegawa M, Sinha RK, Kumar M, Alam M, Yin L, Raina D, et al. Intracellular targeting of the oncogenic MUC1-C protein with a novel GO-203 nanoparticle formulation. Clin Cancer Res. 2015;21:2338–2347. doi: 10.1158/1078-0432.CCR-14-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunaga N, Imai H, Shimizu K, Shames DS, Kakegawa S, Girard L, et al. Oncogenic KRAS-induced interleukin-8 overexpression promotes cell growth and migration and contributes to aggressive phenotypes of non-small cell lung cancer. Int J Cancer. 2012;130:1733–1744. doi: 10.1002/ijc.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherfils-Vicini J, Platonova S, Gillard M, Laurans L, Validire P, Caliandro R, et al. Triggering of TLR7 and TLR8 expressed by human lung cancer cells induces cell survival and chemoresistance. J Clin Invest. 2010;120:1285–1297. doi: 10.1172/JCI36551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee S, Crozet L, Damotte D, Iribarren K, Schramm C, Alifano M, et al. TLR7 promotes tumor progression, chemotherapy resistance, and poor clinical outcomes in non-small cell lung cancer. Cancer Res. 2014;74:5008–5018. doi: 10.1158/0008-5472.CAN-13-2698. [DOI] [PubMed] [Google Scholar]
- 34.Dajon M, Iribarren K, Cremer I. Dual roles of TLR7 in the lung cancer microenvironment. Oncoimmunology. 2015;4:e991615. doi: 10.4161/2162402X.2014.991615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Hayashi M, Lo JF, Fearns C, Chu WM, Luo Y, et al. Nuclear factor kappaB (NF-kappaB) activation primes cells to a pro-inflammatory polarized response to a Toll-like receptor 7 (TLR7) agonist. Biochem J. 2009;421:301–310. doi: 10.1042/BJ20090013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajabi H, Alam M, Takahashi H, Kharbanda A, Guha M, Ahmad R, et al. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2014;33:1680–1689. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeshita F, Suzuki K, Sasaki S, Ishii N, Klinman DM, Ishii KJ. Transcriptional regulation of the human TLR9 gene. J Immunol. 2004;173:2552–2561. doi: 10.4049/jimmunol.173.4.2552. [DOI] [PubMed] [Google Scholar]
- 38.Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, Park YM, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274) FEBS Lett. 2006;580:755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 39.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, et al. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 40.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metcalf D. The colony-stimulating factors and cancer. Nat Rev Cancer. 2010;10:425–434. doi: 10.1038/nrc2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Y, Xiao G, Coombes KR, Behrens C, Solis LM, Raso G, et al. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res. 2011;17:5705–5714. doi: 10.1158/1078-0432.CCR-11-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang G, Wen Q, Zhao Y, Gao Q, Bai Y. NF-kappaB plays a key role in inducing CD274 expression in human monocytes after lipopolysaccharide treatment. PLoS One. 2013;8:e61602. doi: 10.1371/journal.pone.0061602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gowrishankar K, Gunatilake D, Gallagher SJ, Tiffen J, Rizos H, Hersey P. Inducible but not constitutive expression of PD-L1 in human melanoma cells is dependent on activation of NF-kappaB. PLoS One. 2015;10:e0123410. doi: 10.1371/journal.pone.0123410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouillez A, Rajabi H, Pitroda S, Jin C, Alam M, Kharbanda A, et al. Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Res. 2016;76:1538–1548. doi: 10.1158/0008-5472.CAN-15-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 49.Takeshita F, Leifer CA, Gursel I, Ishii KJ, Takeshita S, Gursel M, et al. Cutting edge: Role of Toll-like receptor 9 in CpG DNA-induced activation of human cells. J Immunol. 2001;167:3555–3558. doi: 10.4049/jimmunol.167.7.3555. [DOI] [PubMed] [Google Scholar]
- 50.Pivarcsi A, Muller A, Hippe A, Rieker J, van Lierop A, Steinhoff M, et al. Tumor immune escape by the loss of homeostatic chemokine expression. Proc Natl Acad Sci USA. 2007;104:19055–19060. doi: 10.1073/pnas.0705673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronkainen H, Hirvikoski P, Kauppila S, Vuopala KS, Paavonen TK, Selander KS, et al. Absent Toll-like receptor-9 expression predicts poor prognosis in renal cell carcinoma. J Exp Clin Cancer Res. 2011;30:84. doi: 10.1186/1756-9966-30-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandholm J, Selander KS. Toll-like receptor 9 in breast cancer. Frontiers in immunology. 2014;5:330. doi: 10.3389/fimmu.2014.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 54.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 55.Seliger B, Ruiz-Cabello F, Garrido F. IFN inducibility of major histocompatibility antigens in tumors. Adv Cancer Res. 2008;101:249–276. doi: 10.1016/S0065-230X(08)00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agnello D, Lankford CS, Bream J, Morinobu A, Gadina M, O’Shea JJ, et al. Cytokines and transcription factors that regulate T helper cell differentiation: new players and new insights. J Clin Immunol. 2003;23:147–161. doi: 10.1023/a:1023381027062. [DOI] [PubMed] [Google Scholar]
- 57.Hu X, Chakravarty SD, Ivashkiv LB. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol Rev. 2008;226:41–56. doi: 10.1111/j.1600-065X.2008.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lou Y, Diao L, Parra Cuentas ER, Denning WL, Chen L, Fan YH, et al. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-1434. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gnemmi V, Bouillez A, Gaudelot K, Hemon B, Ringot B, Pottier N, et al. MUC1 drives epithelial-mesenchymal transition in renal carcinoma through Wnt/beta-catenin pathway and interaction with SNAIL promoter. Cancer Lett. 2014;346:225–236. doi: 10.1016/j.canlet.2013.12.029. [DOI] [PubMed] [Google Scholar]
- 61.Tagde A, Rajabi H, Bouillez A, Alam M, Gali R, Bailey S, et al. MUC1-C drives MYC in multiple myeloma. Blood. 2016;127:2587–2597. doi: 10.1182/blood-2015-07-659151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Welsh EA, Eschrich SA, Berglund AE, Fenstermacher DA. Iterative rank-order normalization of gene expression microarray data. BMC Bioinformatics. 2013;14:153. doi: 10.1186/1471-2105-14-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samur MK. RTCGAToolbox: a new tool for exporting TCGA Firehose data. PLoS One. 2014;9:e106397. doi: 10.1371/journal.pone.0106397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goodwin JM, Svensson RU, Lou HJ, Winslow MM, Turk BE, Shaw RJ. An AMPK-independent signaling pathway downstream of the LKB1 tumor suppressor controls Snail1 and metastatic potential. Mol Cell. 2014;55:436–450. doi: 10.1016/j.molcel.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nature communications. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.