Abstract
Context
In an era in which testing of patient tumor material for molecular and other ancillary studies is of increasing clinical importance for selection of therapy, the ability to test on small samplings becomes critical. Often, small samplings are rapidly depleted in the diagnostic workup or are insufficient for multiple ancillary testing approaches.
Objective
To describe technical methodologies that can be implemented to preserve and maximize tissue for molecular and other ancillary testing.
Data Sources
Retrospective analysis of a case cohort from the University of Colorado, description of techniques used at the University of Colorado, and published literature.
Conclusions
Numerous techniques can be deployed to maximize molecular and other ancillary testing, even when specimens are from small samplings. A dedicated process for molecular prioritization has a high success rate, but also increases workload, which must be factored into establishing such a process. Additionally, establishing high-fidelity communication strings is critical for success of dedicated molecular prioritization of samples. Numerous approaches can be deployed for alternative specimen types, and several technical approaches can also aid in maximizing small specimens.
Molecular testing, as well as other ancillary biomarker analyses, is rapidly becoming part of routine care in many tumor types.1–4 In many cases, such testing is used primarily for therapeutic predictive purposes, such as consideration for specific targeted therapies. In addition to predictive utility, a growing cadre of biomarkers is used for potential diagnostic utility; and as the knowledge base for a wide assortment of biomarkers grows for different tumor types, increasing utilization for these and other indications such as prognosis and disease monitoring, is expected. In this era of “precision medicine,” expectations for pathology laboratories and pathologists to effectively handle, manage, and refer specimens for ancillary testing will become paramount.5
In the context of this increasing implementation of biomarker-based analysis, key challenges include scenarios in which either more markers are being requested on the same amount of tissue or on tissue that is historically diminishing in size. Driven in large part by increased utilization of minimally invasive tissue sampling techniques—such as small-gauge needle biopsies and fine-needle aspirates (FNAs) obtained in an interventional radiology setting for deep-seated lesions, endobronchial ultrasound evaluation of thoracic lymph nodes, fine-needle aspiration of superficial lesions, and a myriad of other approaches—pathologists increasingly must not only issue diagnoses on tissues of smaller total volume, but also serve in a tissue triage role to ensure adequate distribution for ancillary testing.6,7
Other challenges exist in the ability to perform testing on small samples, such as intralesional cellular heterogeneity, in this context specifically referring to the observation that a biopsied lesion may not be entirely composed of tumor cells and commonly contains regions of inflammatory, reactive, and stromal components, and that these components may be variable depending on the area of the lesion sampled. This heterogeneity complicates tissue handling, as there is minimal ability to have upfront knowledge of the cellular composition of each tissue fragment before processing, thus making it challenging to know if specific tissue fragments (or portions thereof) are more valuable for testing than others.
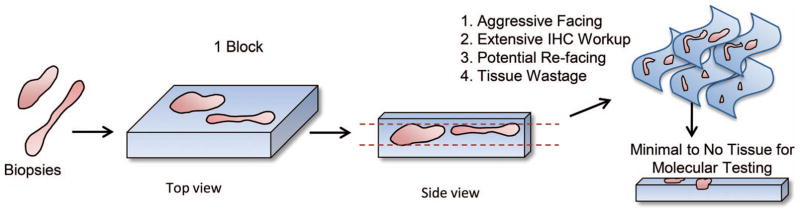
In particular, the standard approach to tissue processing in pathology is one that is optimally designed for rendering a histologic diagnosis and does not prioritize the preservation of tissue (Figure 1). Facing of blocks is most typically designed to examine at least 1 complete cross-section of each fragment of tissue. The process of obtaining a complete cross-section, by definition, results in the cutting through of some tissue that is nearly universally discarded at the microtome. The extent of discarded tissue is magnified when multiple fragments of tissue are placed in a single block, as different fragments are likely to be present at different depths in the block; therefore, obtaining a complete cross-section of all fragments can result in near depletion of some of the pieces of tissue contained within a single paraffin block. These issues are further magnified by diagnostic workups involving immunohistochemistry, which often require refacing of the block, leading to additional tissue loss.
Figure 1.
Schematic representation of the “typical” pathology process in which multiple small tissue fragments are embedded in a single paraffin block. Facing of the block to adequately examine representative cross-sections of all tissue fragments can lead to waste of tissue that could otherwise be used for molecular and ancillary studies, as visualization of 1 fragment of tissue may require substantially cutting into another fragment(s) in the same block. Abbreviation: IHC, immunohistochemistry. Reprinted with permission from the Regents of the University of Colorado. Copyright 2016.
Another factor contributing substantially to lack of tissue maximization for ancillary studies involves critical communication strings between the ordering clinician, the physician performing the procedure to obtain pathologic material, and the pathology laboratory. In an era when obtaining ancillary testing is escalating in importance, it is becoming increasingly common for molecular testing to be a major, if not the exclusive, reason for performing a tissue acquisition procedure. However, lack of effective communication can severely hamper a pathologist in the ability to maximally preserve tissue, particularly when either a prior established diagnosis, or the intent of the procedure to prioritize molecular testing, is unknown. In these situations, it may be common for pathologists to pursue a repeated diagnostic workup that may include immunohistochemical and other studies that could be avoidable if upfront knowledge and communication had been provided.
This does, however, raise the observation that even in the face of knowledge of desired molecular and ancillary testing, there are situations in which both a primary diagnostic workup and ancillary testing are performed, and pathologists can be faced with the challenging scenario of managing small specimens for multiple concurrent priorities.
Lastly, an additional complicating factor can be requests for release of residual tissue for clinical trials. Even in cases in which diagnosis remains the top priority, management of small tissues may be relevant, as diagnostic workup may deplete the material, thereby requiring the patient to undergo additional procedures in order to participate in a clinical trial.
Using strategies to maximize tissue for molecular and ancillary testing has numerous potential benefits, including minimizing patient procedures (and attendant risks and expenses), extending the ability of the tissue derived from a single procedure to fulfill multiple priorities, and reducing turnaround time to molecular and ancillary testing results. Herein we describe multiple approaches to tissue management that we have successfully used at the University of Colorado (Aurora) in order to maximize the ability to perform molecular and ancillary studies on small biopsy and cytopathology specimens.
There are increased resource requirements for all of the techniques described below. However, the benefits for overall patient care can be significant, and the potential resource and workflow implications, as well as feasibility for general adoption, are considered below.
METHODS FOR MAXIMIZING TISSUE FOR TESTING: THE MOLECULAR PRIORITY BIOPSY
Specialized Handling
A particularly challenging circumstance for patients, practitioners, and pathologists is when a biopsy requested specifically for the purpose of molecular testing is found to be insufficient for the desired testing. This leads to patient and practitioner frustration, the potential for multiple invasive procedures, and even delay or impossibility of appropriate therapy selection. To address this circumstance, our institution designed a systematic approach for the implementation of a “molecular priority” biopsy. The goal of this approach was to ensure that when molecular testing is the singular priority for a clinician requesting a biopsy, the tissue is managed and handled in a manner to maximize the probability of successful downstream testing, while continuing to issue a diagnosis and without compromising the ability to perform a complete diagnostic workup as needed.
One essential component of this approach was the attempted standardization of material acquisition standards. Based on multiple interdisciplinary conversations, it was originally determined that the baseline request was to obtain a minimum of 2 cores (or tissue fragments) plus at least 1 FNA pass per lesion, with limitations as medically appropriate for safety concerns. Subsequently, this standard was raised to 4 cores (or tissue fragments) plus at least 1 FNA pass per lesion, with the same limitations with regard to patient safety. At the discretion of the physician performing the tissue-acquiring procedure, cytopathology rapid-on-site evaluation (ROSE) may be requested. If ROSE was performed, the cytopathology section retained all material associated with the pass used for ROSE and issued a separate report (although it could be later used for molecular testing if needed), and additional pass(es) were requested for molecular prioritization. Fine-needle aspirate material for molecular testing is procured by needle rinse in formalin to generate a single cell block, and more recently, air-dried or alcohol-fixed smears have been generated specifically for molecular testing.
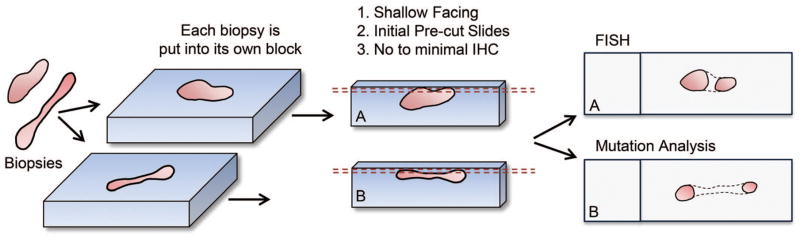
The technical elements of this approach involve modifications to routine handling of tissues for pathologic evaluation. Technical processing parameters (such as tissue processor settings) are kept identical to those otherwise used for small biopsy specimens but substantial shifts in specimen handling are used. Those include limitations placed on formalin exposure time (6–48 hours, specifically avoiding prolonged formalin exposure over weekends or holidays), separation of multiple tissue fragments into multiple blocks (when ordinarily all fragments would be embedded in a single block), generation of a cell block from FNA material without generating smeared or spun slides, shallow facing of each of the generated blocks, and avoidance of repeated diagnostic immunohistochemistry with, instead, an anatomic pathology examination focused solely on the presence or absence of tumor in comparison to the patient’s known diagnosis (Figure 2).
Figure 2.
Schematic representation of the approach used for our “molecular priority” specimen processing, in which tissue fragments are separately embedded and superficially faced. Superficial facing may result in nonvisualization of the complete tissue cross-section (represented by dotted line); however, as the goal is to evaluate for presence of tumor, superficial facing is often sufficient for this purpose. Abbreviations: FISH, fluorescence in situ hybridization; IHC, immunohistochemistry. Reprinted with permission from the Regents of the University of Colorado. Copyright 2016.
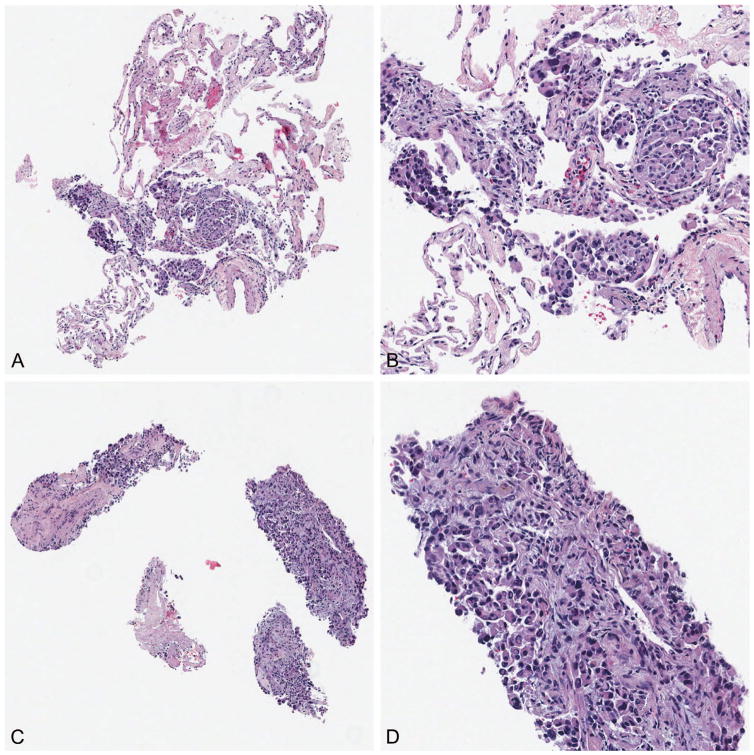
Technically, tissues are generally handled so that individual core biopsies are separately embedded (1 core per block). If an individual core biopsy is unusually long (eg, >2 cm), it may be bisected for embedding in 2 blocks. Smaller tissue fragments, such as those generated by forceps biopsy procedures, are generally embedded with 1 to 3 fragments per block, depending on size. The approach of separating tissue fragments into individual blocks and shallow facing allows for multiple advantages. First, issues of tissue wastage due to multiple fragments needing to be visualized are generally eliminated, as there is no need to cut substantially through one fragment to evaluate another fragment. Second, shallow facing allows the pathologist to assess for the presence or absence of tumor in the obtained biopsy sample without substantial tissue wastage. Third, and importantly, this approach allows each tissue fragment to be examined separately, enabling optimal distribution of tissue for specific molecular or ancillary testing. For example, a fluorescence in situ hybridization (FISH) analysis requiring ~100 intact tumor cells for successful analysis could be completed by using a selected block with the fewest excess tumor cells, particularly in cases where additional evaluated blocks on the same tumor acquisition procedure have a higher tumor burden. In this example, using a block with more than 200 tumor cells per section for a FISH analysis is an unnecessary use of valuable material, which can be directed elsewhere for additional molecular or ancillary analysis (example case in Figure 3, A through D).
Figure 3.
When separate blocks are generated for separate tissue fragments, individual fragments can be chosen by best appropriateness for individual testing. Two separate blocks from the same biopsy procedure (A and C), with (B) and (D) demonstrating high-power images. On the basis of tumor content, the block shown in (A) was directed for fluorescence in situ hybridization analysis, and the remainder preserved for immunohistochemistry, and the block shown in (C) was directed for microdissection and mutation testing (original magnifications ×40 [A and C] and ×200 [B and D]).
As this approach to specimen management is labor- and materials-intensive as compared to specimens handled by routine pathology approaches, limitations were implemented to appropriately manage the resources required for this process. Specifically, utilization of this approach was restricted to cases in which the patient had an established previous pathologic diagnosis and either (1) the diagnostic biopsy was insufficient for molecular testing or (2) the patient was receiving a biopsy for analysis of molecular features at the time of progression during therapy (in some cases associated with specific research protocols).
Communication Is the Key
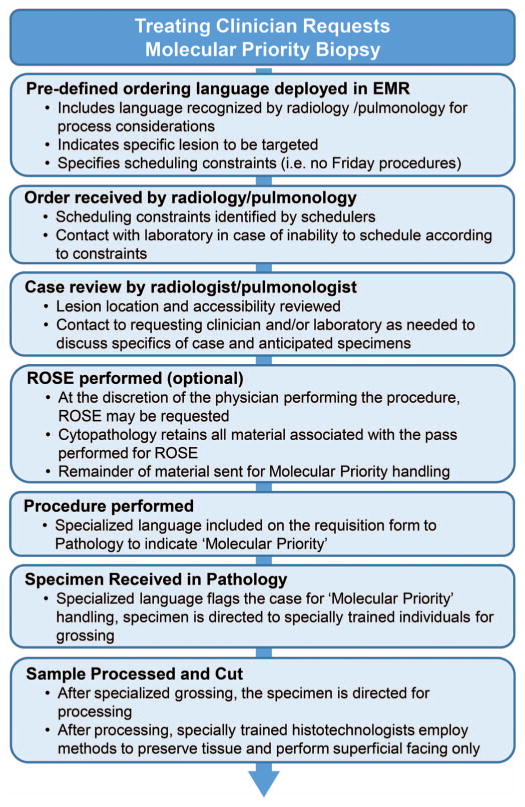
Most relevant, the institution of this process relied on substantial effort being placed into establishing well-defined routes of communication between invested parties. Establishing accurate communication processes between ordering clinician, physician performing the tissue-obtaining procedure, hospital couriers, pathology receipt, and the specialized processing laboratory was perhaps the most critical component of the success of this process (Figure 4). We held numerous prelaunch sessions establishing criteria, specific communication language to be used during ordering of tissue-obtaining procedures, and specific instructions for specimen labeling. Critical pitfalls we sought to avoid included (1) lack of knowledge on the part of the physician performing the procedure of the molecular study prioritization; (2) lack of knowledge on the part of the physician Performing the procedure as to whether a specific lesion was to be targeted for biopsy, especially in the setting of acquired resistance where growing lesions are the sites of interest; (3) lack of knowledge on the part of the evaluating pathologist of molecular study prioritization; (4) inappropriate scheduling of the patient procedure (to limit formalin exposure times, specimens are not accepted for this process on Fridays or the day before holiday weekends); (5) lack of notification that the specimen requires the specialized handling developed to prioritize specimen preservation; and (6) lack of awareness of which specific molecular or ancillary testing was an objective of the procedure. It was through establishing communication strings for all components of this process from inception to completion that we were and continue to be able to prioritize molecular testing in these specialized situations.
Figure 4.
Schematic demonstration of the workflow and multidisciplinary process involved in coordination and implementation of a “molecular priority” biopsy approach. Abbreviations: EMR, electronic medical record; ROSE, rapid on-site evaluation.
Retrospective Analysis
A retrospective analysis of cases submitted for this process between September 2011 and March 2012 was performed to examine tumor burden and molecular success rate. A total of 75 cases were submitted by using the molecular priority approach, with a distribution across sex, and indication (initial biopsy specimen insufficient for molecular testing versus analysis of progressing lesion). During this initial time frame the molecular priority process was predominantly used for patients with advanced-stage lung cancer. Therefore, these data reflect such sample acquisition and molecular ordering patterns; however, this process has now been made widely available and has been similarly applied in other disease settings.
Cases were submitted as either “primary” molecular analysis cases or for analysis of a progressing lesion. Primary molecular analysis meant the biopsy was obtained on the basis of (1) resampling due to prior insufficient tissue; (2) availability of new testing or biomarker (including for consented clinical trials); or (3) prior partial testing with clinical indication to obtain additional marker analysis. Lesions submitted for progression were characterized by previous knowledge of a targetable alteration, with rebiopsy obtained for the purpose of reevaluation of the molecular driver and/or codriver status in the setting of clinically acquired resistance, and in some cases, combined with consented clinical trial–based testing. Selected patient and specimen characteristics are summarized in Table 1.
Table 1.
Patient and Specimen Characteristics
| No. of Cases | |
|---|---|
| Sex | |
| Male | 38 |
| Female | 37 |
| Indication | |
| Initial | 41 |
| Progression | 34 |
| Biopsy location | |
| Lung | 38 |
| Liver | 12 |
| Lymph node | 11 |
| Pleura | 3 |
| Soft tissue | 2 |
| Bone | 5 |
| Body fluid | 3 |
| Brain | 1 |
| Primary tumor | |
| NSCLC | 70 |
| Melanoma | 3 |
| GIST | 1 |
| Colorectal | 1 |
Abbreviations: GIST, gastrointestinal stromal tumor; NSCLC, non–small cell lung cancer.
Although the procedural request included 2 to 4 biopsy fragments and a concomitant FNA, the actual sampling approach had variability, as indicated in Table 2. Overall, 89.3% (67 of 75) of cases had tumor present in the sampling, and of these, 89.5% (60 of 67) had successful molecular testing, defined by at least 1 molecular assay of interest performed, across a variety of platforms with analytic sensitivities ranging from 2% allelic (allele-specific real-time polymerase chain reaction) to 15% (Sanger sequencing). Given the variability of sampling approach, we evaluated how the blocks were allocated for molecular studies. Briefly, of 60 cases with successful molecular testing, the availability of multiple blocks was exploited in 20 cases (33.3%, 20 of 60), with 2 or more blocks used for testing (summarized in Table 2). Most commonly, 1 block was dedicated to microdissection and mutational analysis with tissue from a different block used for FISH or other studies. In some cases, although multiple fragments of tissue were obtained, tumor was present in only a subset of all processed tissue blocks. Thus, utilization of greater than 1 block was dictated by a combination of (1) quantity of tumor in any individual block; (2) the extent of requested testing; and (3) additional anticipated needs for the tissue (including clinical trials–based testing, often requiring release of substantial numbers of slides). Also of note, in some cases in which tumor was present in both biopsy and FNA specimen, the FNA sampling was preferentially chosen over the biopsy sampling. In this cohort, all cases in which the FNA specimen was preferentially used for testing over tissue biopsy specimen actually demonstrated some tumor present in the biopsy tissue sampling. Thus, reasons for selecting the FNA over the tissue preparation were typically related to tumor cellularity, as interfering nontumor cells can be less prominent in some FNA specimens. Furthermore, this retrospective cohort did not include the use of smeared slides, which was introduced to the laboratory after these data were collected. Our anecdotal experience is that the use of smeared slides for molecular testing can frequently salvage a case that otherwise has insufficient material through biopsy and cell block, and in some cases the smear is preferentially used owing to improved nucleic acid quality (discussed in other articles in this Special Section). This demonstrates the value of a combined approach to sampling and processing of lesions in which molecular analysis is a priority. Our current practice, with inclusion of cytopathology smears for consideration, is to simultaneously consider all available material (including biopsy, cell block, and smears when available) to identify the specimen best suited to the individual test requested.
Table 2.
Tissue Sampling, Molecular Success Rate, and Material Allocation
| N (Total) | N (Subset) | Description | |
|---|---|---|---|
| Sampling Approach | |||
| Biopsy and FNA/cytology total | 61 | … | |
| Tumor present in both | … | 40 | |
| Tumor in tissue biopsy only | … | 15 | Considering only the cell block of the FNA/cytology component |
| Tumor present in FNA/cytology only | … | 0 | |
| No tumor present in entire sampling | … | 6 | Considering only the cell block of the FNA/cytology component |
| Tumor present in any combination | … | 55 (90.2%) | Of all cases with biopsy and FNA/cytology (n = 61) |
| Biopsy only total | 12 | ||
| Tumor present | … | 11 (91.7%) | Of cases with only biopsy (n = 12) |
| FNA/cytology only total | 2 | ||
| Tumor present | … | 1 (50%) | Of cases with only FNA/cytology (n = 2) |
| Total cases | 75 | … | |
| Molecular Success | |||
| Overall tumor present | 67 (89.3%) | … | Of cases submitted (n = 75) |
| Successful molecular testing | … | 60 (89.5%) | Of cases with tumor present (n = 67) |
| Overall molecular success | 60 (80%) | … | Of all submitted cases (n = 75) |
| Unsuccessful for molecular testing | 7 (10.4%) | … | Of cases with tumor present (n = 67) |
| Tumor too scant | … | 5 (71.4%) | Of cases unsuccessful for testing (n = 7) |
| Patient died before testing (testing canceled) | … | 1 (14.3%) | Of cases unsuccessful for testing (n = 7) |
| Small cell morphology (resistance; testing canceled) | … | 1 (14.3%) | Of cases unsuccessful for testing (n = 7) |
| Material Allocation | |||
| Total successful molecular testing | 60 | … | |
| No. using >1 block | 20 (33.3%) | … | Of cases with successful molecular testing (n = 60) |
| No. using combined biopsy and FNA/cytology material | … | 3 (15%) | Of cases using >1 block (n = 20) |
| No. with FNA/cytology chosen over biopsy | 12 (30%) | … | Of cases with tumor in biopsy and FNA/cytology (n = 40) |
| No. using a single block | 28 (46.7%) | … | Of cases with successful molecular testing (n = 60) |
Abbreviation: FNA, fine-needle aspirate.
To further examine the parameters associated with success of molecular testing, we evaluated the total tumor content of each case. This was accomplished by evaluating the first cut hematoxylin-eosin (H&E)-stained slide from each block, and estimating the total number of tumor cells in each case, when considering all blocks from the case in aggregate. Cell estimates were classified as one of the following categories: no appreciable tumor (0); fewer than 10; 10 to 50; greater than 50 to 100; greater than 100 to 500; greater than 500 to 1000; and greater than 1000 tumor cells. Visual estimations of tumor quantity were performed by 2 board-certified anatomic pathologists (D.L.A. and M.D.R.). Of the 75 cases, 62 (83%) had all H&E-stained sections from each block available for analysis. Cases were excluded from analysis if all H&E sections were not available, as this could bias the evaluation of tumor content. The distribution of tumor cell estimates and proportion of successful molecular results are summarized in Table 3.
Table 3.
Success Rate of Molecular Testing Based on Cumulative Tumor Cell Count
| No. Cumulative Tumor Cellsa | No. of Cases | Molecular Success Rate,b % |
|---|---|---|
| >1000 | 11 | 100 |
| >500–1000 | 6 | 100 |
| >100–500 | 25 | 100 |
| >50–100 | 6 | 100 |
| 10–50 | 3 | 66 |
| <10 | 3 | 33 |
| NDc | 13 | 79 |
Abbreviations: H&E, hematoxylin-eosin; ND, not determined.
As determined by evaluation of 1 H&E slide per block for all blocks in the case.
Molecular success determined by successful completion of at least 1 molecular test.
All representative H&Es slides were required to be available for evaluation of tumor cell content.
The proportion of cases in which successful molecular testing was achieved began to decline as the aggregate number of tumor cells fell below 50 visualizable cells in all evaluated blocks. In cases with diminishing numbers of tumor cells, the ability to perform successful testing was aided in large part by the extensive material remaining in the block after H&E assessment (owing to initial superficial facing), combined with microdissection (further elaborated below). This evaluation has a likely bias of total tumor assessment based on examination of a single H&E level from each block, which may represent an underestimate, as the defined process included only a superficial facing for H&E staining.
Workload Considerations
The approach outlined here is substantially more labor-intensive than a traditional approach to histologic processing and evaluation. The average number of blocks processed per procedure (including FNA cell block) was 3.44, and the average number of blocks processed for biopsy fragments alone was 2.48 per case, with a range of 1 to 13 when biopsy cores were acquired. A distribution of block numbers for biopsy processing per case is demonstrated in Figure 5. The increased labor associated with this approach may be dramatically offset by reductions in repeated biopsies, creating an overall cost saving when a “big picture” view is considered. In our practice, we receive approximately 1 to 2 molecular priority biopsies per week, which represents less than 1% of our total annual volume of surgical pathology specimens, owing in part to the limitations placed on utilization. Additionally, it is highly likely that embedding tissue fragments in separate blocks reaches a point of diminishing returns with greater number of blocks generated per case. Although we have not specifically studied this issue, it seems reasonable to suggest that exceeding 4 blocks is unlikely to provide additional benefit in most cases. While there are rare exceptions in which extensive numbers of core biopsies are procured, we now generally restrict this process to a maximum of 4 to 5 blocks. If approaches such as these were to become more widely implemented across pathology laboratories, the cost considerations within a pathology department could be substantial, as typical reimbursement for biopsy samples (Current Procedural Terminology [CPT] 88305) does not account for the multiplication of materials, and technical and professional effort involved. Given the potential downstream economic and patient care advantages (namely reduction in repeated biopsies, and the resultant reduction in turnaround time to molecular results), it may ultimately be worthwhile to consider that pathology professional organizations request additional CPT codes to reflect the costs associated with mechanisms to provide specialized handling processes such as these.
Figure 5.
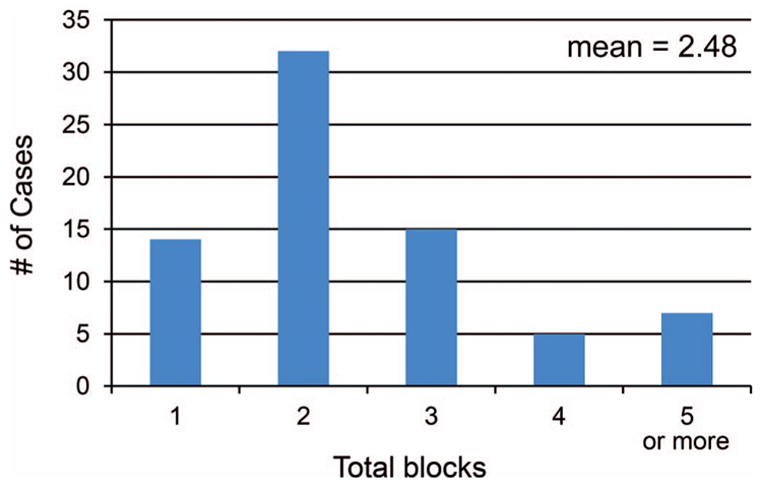
Histogram of blocks used for biopsy/tissue component in the retrospective analyses of the “molecular priority” specimens. This demonstrates that the number of blocks generated is substantially greater than if all tissue fragments had been placed into a single block, portraying the increased workload associated with this process.
Of note, specific obstacles did arise in the technical implementation of this specialized handling methodology. First, when initial (shallow) facing of all prepared paraffin blocks demonstrated no identifiable tumor, substantial additional effort was used by performing multiple successive refacings of each block with new H&E-stained sections obtained to conclusively exclude tumor in the specimen through an examination of representative cross-sections of all tissue fragments. In this situation, as the number of blocks was increased in which tissue fragments were distributed, the labor associated with excluding tumor presence was substantially increased, as compared to cases in which tumor was readily evident.
Another challenge is in regard to the writing of the pathology report associated with these specimens. As the purpose of the biopsy is to prioritize tissue for molecular analysis, every effort was made to avoid block refacing, or use of material for immunohistochemical or special stains. In many cases, pathologists responsible for these cases may issue a diagnosis of “carcinoma present” or a similarly imprecise diagnosis, as additional refinements of the diagnosis would require the use of immunohistochemical or special stains. In these situations, a disclaimer is included to indicate that the reason additional studies were not performed was based on clinical request to prioritize material for molecular and/or ancillary studies. This notwithstanding, it is not surprising that in very rare circumstances, the pathologic findings seen on shallow facing of these molecular priority biopsies are not consistent with either previously reviewed material on the same patient, or with reported history. In these circumstances, it is incumbent upon the pathologist to discuss with the referring treating physician and emphasize that it is important to prioritize a relevant change in diagnosis first and foremost. In these cases, our laboratory has used material otherwise intended for molecular study to render an unexpected diagnostic finding. This underscores the value of having tumor specimens separated across multiple blocks, as in these situations, often some portion of tumor can be used for diagnostic clarification while preserving additional material for requested molecular studies. Furthermore, this underscores the need for anatomic pathologists to be integrated into any process such as this, as processing of tissue without qualified morphologic assessment can lead to incorrect patient treatment.
An additional key challenge is in the appropriate description of limitations and caveats associated with performing molecular testing on minimal samples. While positive findings always have the potential to demonstrate value, negative findings should be interpreted with caution, and disclaimers to this effect are warranted.
The analysis of a limited cohort in our experience with the molecular priority biopsy indicates substantial potential benefit from the process of separating tissue fragments into separate blocks, combined with superficial facing and avoidance of immunohistochemical staining. A properly controlled evaluation of this process is not feasible when applied to real clinical situations, as it is not possible to handle the same set of tissues by using 2 separate conditions. However, our experience is that this process clearly expands use of limited tissues for multiple purposes. In many cases, although tumor was reasonably abundant when all blocks are considered in aggregate, the probability of rapid tissue depletion was substantial had these procedures not been followed. In numerous anecdotal situations, we have been able to provide molecular analysis services and send tissue for multiple clinical trials based on a single biopsy procedure, with high probability that this would not have been possible if “routine” approaches had been implemented. Of note, in the time frame since the analysis of this cohort, our laboratory has expanded testing on cytopathology specimens to include the ability to perform testing on non–cell block preparations such as smears. This additional capacity has led to additional successes in testing minimal samples, including samples submitted for the molecular priority biopsy specimen, particularly in cases in which the tissue biopsy is paucicellular but the smeared slides are suitable for testing.
METHODS FOR MANAGEMENT OF ALTERNATIVE SPECIMENS
Bony Lesions
Decalcification is known to be detrimental to nucleic acids, often entirely eliminating the ability to perform molecular testing on specimens subjected to this treatment. We separately use an approach to evaluation of bony specimens, whether they are referred for molecular priority biopsy handling, or routine pathology handling. In both settings, utilization of decalcification is specifically avoided on small biopsy specimens, to the greatest extent possible. In the routine pathology laboratory, samples that are received as bone biopsies are investigated for potential molecular testing needs before specimen processing. When a specimen is received for molecular-only biopsy handling (or if investigation demonstrates that a specimen submitted for routine pathology does require molecular testing), every conceivable effort is made to avoid decalcification. Bony lesions are palpated at the grossing stage and unless extensively sclerotic, are submitted for processing per routine without decalcification. In some cases, this requires manual dissection of soft tissue components of a biopsy specimen away from sclerotic components, and submission as separate blocks. However, in many cases, no decalcification is needed, as the extent of calcified bony spicules in the sample is largely diminished by the presence of metastatic disease and only scant areas of calcification remain (Figure 6, A).
Figure 6.
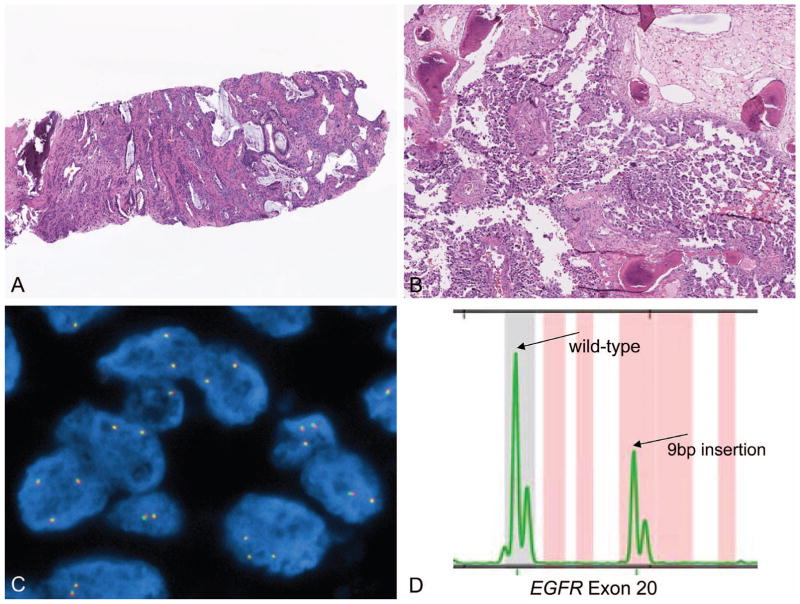
A, Specimen in which decalcification was deemed unnecessary by palpation. Only a single bony spicule is visualized, and processing and microtome cutting was feasible without decalcification. B, Sample submitted as decalcified. Histologic examination demonstrated that this sample was only partially affected by decalcification treatment, as evidenced by persistence of basophilic staining in the center of bony spicules. This sample was successful for molecular analysis, as illustrated in (C) with ALK FISH demonstrating no evidence of ALK rearrangement, but excellent FISH staining attributes, and in (D) with polymerase chain reaction–sizing assay demonstrating the presence of an EGFR exon 20 insertional mutation (hematoxylin-eosin, original magnifications ×20 [A] and ×60 [B]); original magnification ×1000 [C]). Abbreviations: bp, base pair; FISH, fluorescence in situ hybridization.
Another approach we use is encouraging paired FNA sampling for bony lesions, as these typically do not require decalcification. Although not represented in our limited review of molecular priority biopsy specimens, we have observed on multiple occasions that a paired FNA of a bony lesion can allow for molecular testing that would not have otherwise been possible. In the situation of an aspirate of a bony lesion, we also recommend to the individual performing the procedure that the needle undergo a rinse to completely expel all material.
We have also used on occasion an approach to the decalcified specimen that integrates histologic evaluation of the specimen, even in the case of decalcification treatment explicitly stated on a pathology report. The extent of decalcification can be assessed histologically, by the depth of basophilia of bone trabeculae. Complete alteration of calcified areas to homogeneously waxy, eosinophilic appearance suggests prolonged treatment with decalcifying agent, while retention of basophilic staining of calcium suggests less thorough exposure. These histologic features may correlate with degree of degradation of nucleic acid, and when decalcification appears less than complete, we will attempt molecular testing, recognizing that findings must be interpreted with caution (Figure 6, B through D).
Lastly, there is a growing recognition that different decalcification treatments may have different impacts on molecular testing. For example, ethylenediaminetetraacetic acid (EDTA) treatment may be less detrimental for subsequent molecular testing than strong acid decalcification.8,9 Despite its potential benefit for downstream testing, EDTA-based decalcification is not universally used. This may be due to requirements for longer decalcification time, potentially more frequent changes of decalcification solution, and potentially higher cost. For specimens not processed by our laboratory, we make every effort to ascertain the type of decalcification treatment used for a specimen. Particularly in the clinical scenario where the bony decalcified specimen is the only available specimen for a patient who is reluctant to undergo an additional procedure to obtain tissue, knowledge that less harsh decalcification treatments were used can justify attempts at molecular testing. As technologies continue to improve, some groups10,11 have reported successful molecular testing with decalcified specimens. This also underscores the importance of documenting the type of decalcification treatment used in anatomic pathology reports.
Utilization of Cytopathology Specimens
As minimally invasive techniques increasingly become mainstays for diagnosis, the implementation of molecular testing on fine-needle aspiration and exfoliative cytopathology specimens is increasing. Testing on cytopathology cell blocks has been fairly widely deployed12 and is separately discussed in this Special Section of Archives of Pathology & Laboratory Medicine.13 Particularly when lesional tissue is scant, when the lesion is poorly accessible, or with inability to perform additional procedures, the development and validation of testing on cytopathology material fills a key clinical need. For example, a patient with advanced-stage lung adenocarcinoma may have a diagnostic procedure consisting only of an FNA, including a cell block with a paucity of tumor cells. In the absence of testing innovation, this scenario would require an additional procedure or procedures in order to obtain material for molecular analysis, which is now considered standard of care. However, numerous approaches to testing innovation have led to the ability to test smeared slides, touch imprints, and cytospin preparations, including for multianalyte platforms such as next-generation sequencing. Our laboratory, among many others, has developed techniques for utilization of various cytopathology preparations other than cell blocks and have validated their use for molecular analysis.14–18 Some approaches to utilization of cytopathology specimens are covered in detail elsewhere in this special issue of the Archives,19,20 and thus, additional technical details and discussion are not provided here.
Use of Residual Material
In the setting of scant material, particularly material that has not been handled according to a molecular priority approach, an often overlooked source of material is that which already exists in the pathology archives. In many laboratories, biopsy processing protocols indicate a standard number of unstained slides to be cut and held. Our experience is that when affiliated laboratories forward material for molecular testing, often these precut slides are not included. When material is scant, specifically asking for such materials to be identified and included with molecular studies has proven useful.
Also of use on rare occasions are the immunohistochemically negative control slides. A negative control slide, used for the purpose of ensuring lack of cross-reactivity of secondary antibodies with the studied tissue, can be valuable for molecular testing. Utilization of this tissue type requires that the relevant slide be carefully decoverslipped, and its reuse will result in its loss from the pathology archive. Furthermore, laboratories using this approach must independently validate the specimen type. In recent years, the College of American Pathologists has eliminated the requirement for a negative control slide to be included from each evaluated tissue, thus the utility of this approach has declined as these controls are generated less often.
TECHNICAL ENHANCEMENTS
Extensive Microdissection
Tumor enrichment is a key component of molecular testing, as a minimum threshold of tumor cell percentage (tumor cellularity) is required for many assays. For example, traditional Sanger sequencing requires that material used for molecular testing be composed of a minimum of ~30% tumor cells (compared to all other nucleated cells in the tested portion of the specimen).21 As most histopathologic sections do not meet the tumor cellularity requirements without tumor enrichment, such strategies are routinely used. The most commonly used strategy is microdissection, in which regions of a specimen are identified for isolation or nonmicroscopic microdissection, meaning that H&E-stained slides are marked by microscopic evaluation and used to guide scraping of slides or procurement of tissue directly from the block (eg, tissue coring or carving; heretofore referred to as nonmicroscopic microdissection). Alternatively, microdissection can be performed under a stereoscopic microscope with the use of a tissue counter-stain, which allows for an additional level of precision in tumor enrichment. However, cases that show scant material can require extensive levels of microdissection, and either the lack of suitability of a case for nonmicroscopic microdissection or the need for extensive microdissection can lead to specimen rejection. In many circumstances, we find that a specimen may be suitable for testing only if extensive microdissection is used. In our laboratory, microdissection is performed under a stereoscopic microscope, using 10-μm cut sections on uncharged slides, with hematoxylin counterstain. While resection specimens can often be effectively microdissected with 3 or fewer consecutive cut levels, small specimens may require microdissection of an extensive number of consecutively cut sections. In our laboratory, we will microdissect up to 30 consecutive cut sections (and in exceptional circumstances, more sections will be used) to obtain sufficient material for testing, thus allowing testing in specimens that might otherwise be classified as insufficient. Representative sections of the microdissected slides after the microdissection procedure is completed are retained in the permanent record, as a form of quality assurance of the microdissection procedure, and to allow for assessment of actual tumor enrichment to be undertaken at the time of analysis of molecular data. Examples of cases requiring 30 consecutive levels of microdissection, and the paired postmicrodissection slide, are shown in Figure 7, A through F. Moreover, microdissection is used in cases in which the extent of nontumor tissue present in the specimen would preclude the use of nonmicroscopic microdissection for successful molecular testing. In these circumstances, precision micro-dissection techniques, deployed by hand, can allow for sufficient tumor enrichment for testing (Figure 7, G through I). Yields from this approach vary, although they often are sufficient for capture-based, or high-input amplicon-based next-generation sequencing. In cases with the least favorable parameters, extensive microdissection may yield only enough nucleic acid for a few selected, targeted assays. However, often the yield is substantial and can be used for assays requiring high levels of nucleic acid input. Although not used by our laboratory, other groups22–24 have also described success in using laser-capture microdissection methodologies as well.
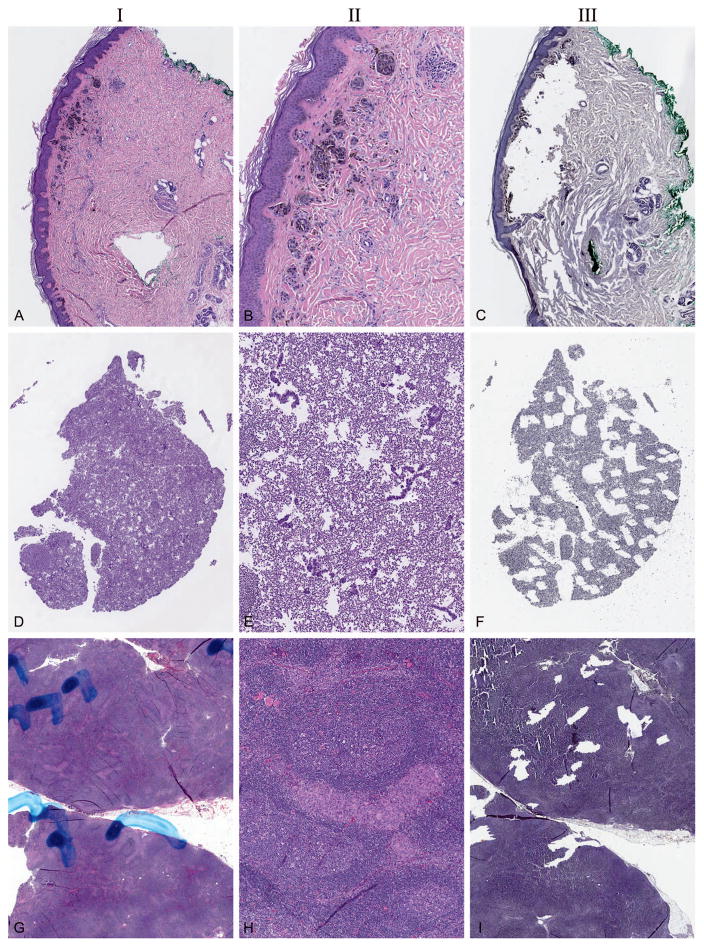
Figure 7.
Examples of extensive or high-fidelity microdissection, allowing for use of specimens that may have otherwise been inadequate or unsatisfactory for testing. In all cases, column I is low power (~×20), pre microdissection; column II is moderate power (~×60), pre microdissection; and column III is low power (~×20), post microdissection. A through C, Skin melanoma specimen, microdissection of 30 levels cut at 10 μm. D through F, Pleural fluid cell block, microdissection of 30 levels cut at 10 μm. G through I, Lymph node with metastatic melanoma in an infiltrative pattern, microdissection of 12 levels cut at 10 μm. In all cases, sufficient material was obtained for molecular testing (hematoxylin-eosin, original magnifications ~×20 [A, D, and G] and ~×60 [B, E, and H]); hematoxylin, original magnification ×20 [C, F, and I]).
Capturing Cut-ins
Small specimens are particularly prone to tissue loss during the process of facing in a paraffin block on a microtome. Obtaining an even face on the microtome can lead to many full or partial sections cut from the tissue before the specimen is amenable to capture of a “good” section on a glass slide. To maximally capitalize on the tissue in such specimens, we have initiated a process of “cut-in capture,” in which tissue generated during the facing-in process is captured, even if not amenable to producing a flat, even histologic section that might be optimal for staining or other evaluation. Multiple sections can be placed on a single glass slide, and these are floated on a heated water bath in a manner similar to other generated slides. For cases that are especially minute, this approach can particularly aid in retaining material that can be used for microdissection. However, as this process is labor-intensive, it is reserved for cases in which the material is clearly highly limited by visual inspection of the block and, when available, the corresponding H&E-stained section.
Microtome Hold Versus Precutting Slides
On rare occasions, we use a tactic in which following initial facing of a paraffin block, a single slide is generated to stain by H&E. The block is held on the microtome for the duration of the H&E staining process and immediately following H&E staining, the slide is reviewed by a pathologist to determine optimal cutting procedures for the case. For cases with extremely scant tumor, this can provide a benefit of minimizing or eliminating refacing of blocks and highlights the difficulty a molecular laboratory can face when only a block is sent for testing and a corresponding H&E slide is not included. Specifically, for scant tissues, if only a block is sent without a corresponding H&E slide, the molecular laboratory may need to face the block once to assess the specimen, and then face the block a second time to obtain the necessary cuts for preanalytic processing for desired testing. This highlights the need to identify all relevant materials to send to a molecular laboratory (further discussed below). However, this also highlights an opposite approach, which is occasionally implemented. In some cases, identification that the material is scant will result in the determination to precut multiple sections from a block either at the time of initial facing or at the time of initial histopathologic evaluation (particularly for molecular priority specimens), rather than risk the potential for multiple microtome facings. Decisions on how to precut the block at the time of intake for molecular testing depend on which specific molecular evaluations are requested. Determination of whether to use either of these tactics, and which ones to use, depends greatly on the visual inspection of the block, the specimen acquisition type, and the evaluation of descriptive language in the correlated pathology report (when received as a completed case). However, use of these approaches is minimized by proactively working to obtain the correlated H&E-stained section for evaluation before block facing (further discussed below).
FISH “Double-Cut”
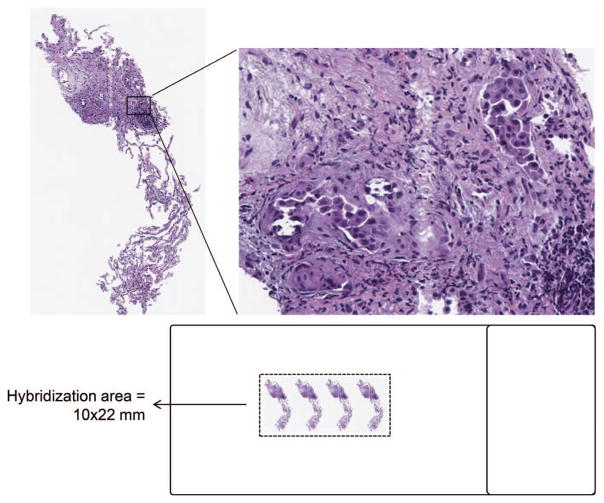
In many circumstances in which biopsy material is limited, the total number of nuclei available for FISH assays is marginal or too low for appropriate scoring. To overcome these limitations, we evaluated an approach that we termed the double-cut. In this approach, 2 (or more) sequential sections of tumor are placed close together on a single slide, falling within the area of a single hybridization area (most commonly maximally determined by the area encompassed beneath a 22-mm coverslip) (Figure 8). This technical approach can be challenging, as cutting and lifting sections with precision within this small area can itself be difficult. In our experience, tissue adherence to the slide is facilitated by trimming the excess paraffin from the tissue profile with forceps while the section is floated on a heated water bath.
Figure 8.
Schematic representation of the “double-cut” methodology, in which multiple sequential levels of a single specimen are placed onto a fluorescence in situ hybridization–compatible slide such that all tumor cells are under a single hybridization area (hematoxylin-eosin, original magnifications ~×20 [left] and ~×200 [right]).
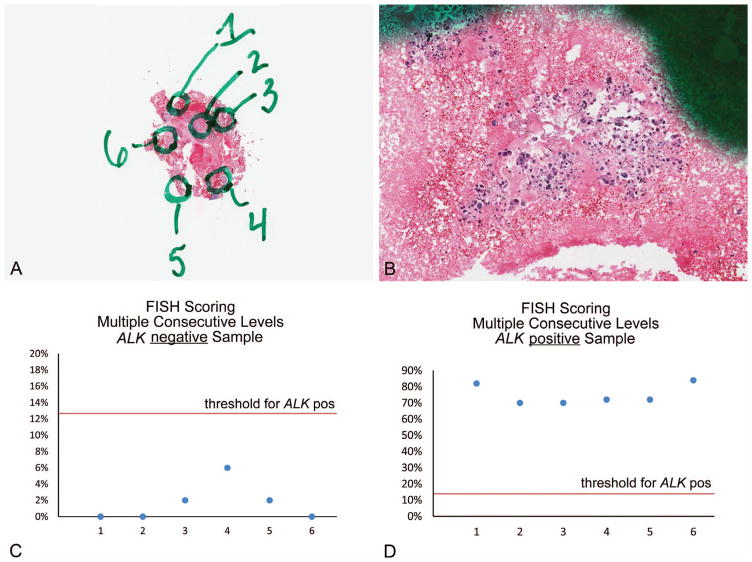
An obvious concern about this approach for FISH testing is the possibility of inaccurate results based on the potential for scoring a single nucleus more than one time. To address this concern, we undertook validation studies to demonstrate that this approach does not result in overrepresentation of truncation artifact (which could lead to false-positive ALK FISH results based on single 3′ ALK red signals) or undercounting of positive cells (which could lead to false-negative ALK FISH results). We identified numerous ALK-positive and ALK-negative samples and selected 6 subareas of each sample (example case shown in Figure 9, A and B). Sequential sections were cut, and the subareas were independently scored on each of the levels to simulate the experience of a limited sampling with multiple sequential sections evaluated. Numerous combinations of areas and levels were considered to determine percentage positive cells in various permutations of sequential level scoring. Our findings indicated that in no case was a previously characterized ALK-positive sample scored as negative, regardless of permutations of sequential levels. Similarly, no previously characterized ALK-negative sample was scored as positive (Figure 9, C and D). Thus, this approach allows for implementation of a break-apart FISH assay even when tumor cell presence is extremely sparse. The utility of this approach may become more limited with implementation of immunohistochemical screening approaches for ALK expression; however, the availability of this approach does allow for testing on samples that may otherwise be ineligible.
Figure 9.
A and B, Low- and moderate-power photomicrographs of an example case selected for validation of the “double-cut” methodology. Tumor cells were distributed in multiple foci of small clusters throughout the specimen. Six individual subareas were marked, and each area was separately scored on independently cut successive levels (5 total levels examined), to simulate a small specimen being scored with multiple successive levels. C and D, Scoring 5 consecutive levels of each marked subarea does not influence the overall result of the FISH analysis. Additional permutations of areas and levels to simulate different sized biopsy specimens and different numbers of sequentially scored levels demonstrated similar results (not shown) (hematoxylin-eosin, original magnifications ~×1 [A] and ~×100 [B]). Abbreviations: FISH, fluorescence in situ hybridization; pos, positive.
Material Referral
As indicated above, a molecular laboratory can face particular challenges with scant specimens when selected components of a case are referred for testing. Receipt of a block without an H&E-stained section can result in at least 2 facings of a block, which can create clear difficulties when the specimen is minimal at the time of molecular referral. Receipt of a set number of precut slides can result in difficulties in having sufficient material for all requested studies and having slides cut at the incorrect thickness or on the incorrect slide type for the needed testing. Recognizing that individual laboratories have policies regarding release of blocks, depletion of blocks, and release of precut slides, some considerations are likely to offer patient benefit for completing molecular testing in a timely fashion, based on available material. If a block is to be sent on a scant specimen, optimal handling would include provision of a correlated H&E-stained section from the originating institution, thereby allowing the laboratory to assess the specimen without refacing.
Another challenge in the evaluation of referred material for molecular testing is challenging specimens for which immunohistochemical studies were crucial in establishing a definitive diagnosis of malignancy. For example, a pleural fluid that demonstrates a highly cellular admixture of tumor cells, and reactive mesothelial cells in which the tumor characteristics are subtle, often requires immunohistochemical studies for confirmation of malignancy. In this circumstance, the molecular laboratory also substantially benefits from having the immunohistochemically stained slide available, as it can help to refine the assessment of tumor cellularity and options for tumor enrichment. Thus, we generally communicate to referring groups that if the immunohistochemical study was vital for the diagnosis, it will be of use in the sample assessment phase for molecular testing.
CONCLUSIONS
Mechanisms to increase the ability to maximally use specimens for ancillary testing encompass a complex series of techniques, which often must be used in sequence. Specifically, establishing communication strings to appropriately notify all invested parties that a specimen is prioritized for molecular/ancillary studies, regardless of whether special handling techniques are used, is critical in avoiding tissue waste or duplicative efforts. If special handling techniques are established, separation of tissue fragments into separate blocks with minimal facing substantially preserves tissues for ancillary testing and potential subsequent tissue release; however, it requires substantially increased resources in terms of reagents, consumables, and technical and professional time and additionally requires training of personnel in how to handle tissues in this specialized manner. In many of the listed techniques, preservation or utilization of tissue for molecular testing would not be feasible without staff who have been extensively and specifically trained in these specialized handling approaches. It is noted, however, that many of these described approaches require additional resources and that prioritizing such resource utilization in this arena is challenging. Table 4 lists many of the elaborated approaches and whether they may be viewed as readily adoptable or more likely reserved only for laboratories willing to dedicate substantial resources to this effort. Note that all efforts are predicated on the implementation of an effective communication string, as ability to undertake these additional interventions is entirely reliant on upfront knowledge that can only be fully understood with a successful approach to interdepartmental communication.
Table 4.
Highlighted Approaches and Potential for Routine Adoption
| Technical or Other Approach | Ease of Adoption | Priority for Adoptiona | Notes/Tips |
|---|---|---|---|
| Interdepartmental communication on individual Cases | Challenging | 1 |
|
| Embedding specimen fragments in multiple blocks | Reasonable and manageable to split designated specimens into 2–3 blocks | 2 |
|
| Superficial facing | Reasonable to indicate blocks for superficial facing | 1 |
|
| Capturing cut-in levels | Challenging and time-intensive | 3 |
|
| Avoidance of decalcification for bony lesions | Reasonable for widespread adoption | 1 |
|
| Extensive microdissection | Challenging | 3 |
|
| Microtome hold | Challenging | 3 |
|
| Material referral practices | Straightforward | 1 |
|
1 = highest priority, 3 = lowest priority owing to resource requirements.
Despite the obstacles, implementation of systems to maximize tissue to whatever extent is feasible at individual institutions is likely to provide patient benefit in the form of reduced requirements for repeated procedures, reduced turnaround time to results of ancillary testing, and increased availability of residual tissue for additional investigations.
Acknowledgments
The authors gratefully acknowledge the graphical and artistic support provided by Lisa Litzenberger and the logistical support provided by Nancy Hart and Keith Kinsella.
This work was supported in part by the Molecular Pathology Shared Resource and the Tissue Biobanking and Processing Shared Resource of Colorado’s NIH/NCI Cancer Center Support Grant P30CA046934. Drs Aisner and Franklin disclose partial ownership of patent number PCT/US13/38284, describing a pneumatic cell collection device that can be used for microdissection. This device was not used for any of the microdissections presented in this manuscript. Dr Merrick received honoraria for preceptorship (March 2016) from ARIAD Pharmaceuticals Inc, Cambridge, Massachusetts. The other authors have no relevant financial interest in the products or companies described in this article.
References
- 1.Berse B, Lynch JA. Molecular diagnostic testing in breast cancer. Semin Oncol Nurs. 2015;31(2):108–121. doi: 10.1016/j.soncn.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013;137(6):828–860. doi: 10.5858/arpa.2012-0720-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pillai RK, Lopategui JR, Dhall D, et al. The state of the art in colorectal cancer molecular biomarker testing. Adv Anat Pathol. 2016;23(2):92–103. doi: 10.1097/PAP.0000000000000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volmar KE, Idowu MO, Souers RJ, Nakhleh RE. Molecular testing in anatomic pathology and adherence to guidelines: a College of American Pathologists Q-Probes study of 2230 testing events reported by 26 institutions. Arch Pathol Lab Med. 2015;139(9):1115–1124. doi: 10.5858/arpa.2014-0513-CP. [DOI] [PubMed] [Google Scholar]
- 5.Bernicker E. Biomarker testing in non-small cell lung cancer: a clinician’s perspective. Arch Pathol Lab Med. 2015;139(4):448–450. doi: 10.5858/arpa.2014-0085-ED. [DOI] [PubMed] [Google Scholar]
- 6.Moreira AL, Saqi A. Diagnosing Non-Small Cell Carcinoma in Small Biopsy and Cytology. New York: Springer; 2015. [Google Scholar]
- 7.VanderLaan PA. Molecular markers: implications for cytopathology and specimen collection. Cancer Cytopathol. 2015;123(8):454–460. doi: 10.1002/cncy.21560. [DOI] [PubMed] [Google Scholar]
- 8.Choi SE, Hong SW, Yoon SO. Proposal of an appropriate decalcification method of bone marrow biopsy specimens in the era of expanding genetic molecular study. J Pathol Transl Med. 2015;49(3):236–242. doi: 10.4132/jptm.2015.03.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass BP, Engel KB, Greytak SR, Moore HM. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: how well do you know your FFPE specimen? Arch Pathol Lab Med. 2014;138(11):1520–1530. doi: 10.5858/arpa.2013-0691-RA. [DOI] [PubMed] [Google Scholar]
- 10.Goswami R, Luthra R, Singh R, et al. Identification of factors affecting the success of next-generation sequencing testing in solid tumors. Am J Clin Pathol. 2016;145(2):222–237. doi: 10.1093/ajcp/aqv023. [DOI] [PubMed] [Google Scholar]
- 11.Fu B, Wang Z, Li X, Wang SA, Zuo Z. Detection of BRAF V600E mutation in Langerhans cell histiocytosis using high-resolution melting analysis in decalcified, paraffin-embedded tissue. J Leuk (Los Angel) 2013;1:101. [Google Scholar]
- 12.Idowu MO, Dumur CI, Garrett CT. Molecular Oncology Testing for Solid Tumors: A Pragmatic Approach. Cham, Switzerland: Springer (Switzerland); 2015. [Google Scholar]
- 13.Saqi A. The state of cell blocks and ancillary testing: past, present and future. Arch Pathol Lab Med. doi: 10.5858/arpa.2016-0125-RA. published online ahead of print August 24, 2016. [DOI] [PubMed] [Google Scholar]
- 14.Oktay MH, Adler E, Hakima L, et al. The application of molecular diagnostics to stained cytology smears. J Mol Diagn. 2016;18(3):407–415. doi: 10.1016/j.jmoldx.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Harada S, Agosto-Arroyo E, Levesque JA, et al. Poor cell block adequacy rate for molecular testing improved with the addition of Diff-Quik-stained smears: need for better cell block processing. Cancer Cytopathol. 2015;123(8):480–487. doi: 10.1002/cncy.21561. [DOI] [PubMed] [Google Scholar]
- 16.Betz BL, Dixon CA, Weigelin HC, Knoepp SM, Roh MH. The use of stained cytologic direct smears for ALK gene rearrangement analysis of lung adenocarcinoma. Cancer Cytopathol. 2013;121(9):489–499. doi: 10.1002/cncy.21286. [DOI] [PubMed] [Google Scholar]
- 17.Gailey MP, Stence AA, Jensen CS, Ma D. Multiplatform comparison of molecular oncology tests performed on cytology specimens and formalin-fixed, paraffin-embedded tissue. Cancer Cytopathol. 2015;123(1):30–39. doi: 10.1002/cncy.21476. [DOI] [PubMed] [Google Scholar]
- 18.Treece AL, Montgomery ND, Patel NM, et al. FNA smears as a potential source of DNA for targeted next-generation sequencing of lung adenocarcinomas. Cancer Cytopathol. 2016;124(6):406–414. doi: 10.1002/cncy.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy-Chowdhuri S, Stewart J. Preanalytic variables in cytology: lessons learned from next-generation sequencing—The MD Anderson experience. Arch Pathol Lab Med. doi: 10.5858/arpa.2016-0117-RA. published online ahead of print June 22, 2016. [DOI] [PubMed] [Google Scholar]
- 20.Tian S, Lin O, Arcila M. More than meets the eye: the hidden potential of cytology for molecular testing. Arch Pathol Lab Med. 2016 In press. [Google Scholar]
- 21.Aisner DL, Sams SB. The role of cytology specimens in molecular testing of solid tumors: techniques, limitations, and opportunities. Diagn Cytopathol. 2012;40(6):511–524. doi: 10.1002/dc.22820. [DOI] [PubMed] [Google Scholar]
- 22.Fend F, Raffeld M. Laser capture microdissection in pathology. J Clin Pathol. 2000;53(9):666–672. doi: 10.1136/jcp.53.9.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhuri SR, Xi L, Pham TH, et al. EGFR and KRAS mutation analysis in cytologic samples of lung adenocarcinoma enabled by laser capture microdissection. Mod Pathol. 2012;25(4):548–555. doi: 10.1038/modpathol.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bongiovanni M, Molinari F, Eszlinger M, et al. Laser capture microdissection is a valuable tool in the preoperative molecular screening of follicular lesions of the thyroid: an institutional experience. Cytopathology. 2015;26(5):288–296. doi: 10.1111/cyt.12226. [DOI] [PubMed] [Google Scholar]