Abstract
The performance of a classifier is largely dependent on the size and representativeness of data used for its training. In circumstances where accumulation and/or labeling of training samples is difficult or expensive, such as medical applications, data augmentation can potentially be used to alleviate the limitations of small datasets. We have previously developed an image blending tool that allows users to modify or supplement an existing CT or mammography dataset by seamlessly inserting a lesion extracted from a source image into a target image. This tool also provides the option to apply various types of transformations to different properties of the lesion prior to its insertion into a new location. In this study, we used this tool to create synthetic samples that appear realistic in chest CT. We then augmented different size training sets with these artificial samples, and investigated the effect of the augmentation on training various classifiers for the detection of lung nodules. Our results indicate that the proposed lesion insertion method can improve classifier performance for small training datasets, and thereby help reduce the need to acquire and label actual patient data.
Index Terms: Data augmentation, computer aided diagnosis, Poisson editing, pulmonary nodules
I. Introduction
Classifier performance is known to be largely dependent on the size and representativeness of the data used in its training [1], [2], [3], [4]. In natural image classification and speech recognition, state-of-the-art systems rely on millions of labeled samples to train complex models consisting of hundreds of thousands to millions of parameters [5], [6]. In most applications, collection and storage of large amounts of data are no longer the primary obstacles in assembling large datasets due to falling storage costs. Interpretation and labeling of samples are thus the major remaining bottlenecks in creation of large repositories for classifier training. In some fields such as computer vision for natural image analysis, it is possible to use a crowd-sourcing mechanism such as Amazon’s Mechanical Turk to obtain annotations and labels in a relatively cheap and timely manner. Unlike natural images or speech data, however, interpretation and labeling of medical images requires a higher level of expertise. It is therefore generally more difficult to obtain reliable ground truth or annotations through crowd-sourcing, although some recent efforts have been able to use a combination of expert and crowd-sourced truthing in their systems [7]. Additionally, some diseases, abnormalities, and lesion types have a very low rate of occurrence in the general population, thereby making it even more difficult and expensive to collect large datasets of abnormal cases. Combined with the fact that data sharing in the medical imaging community is still not as commonplace as some other fields, creation of large medical imaging repositories remains a very difficult and expensive task, and such datasets are not only currently much smaller than their counterparts in natural images but may never reach the size of those databases.
Augmentation of small and/or imbalanced training sets through addition of synthetically generated samples has had a long successful history in alleviating the issues associated with small datasets across many classification tasks. The introduction of the synthetic samples into the training set can provide the classifier with a better representation of data for the classes with a small number of samples, and thereby help a classifier avoid overtraining and improve its generalization performance. The mechanisms for producing the artificial samples typically fall under three categories. In the first one, new samples are created in the feature space by adding noise (also called jittering) or applying other modifications to feature vectors of the existing samples that belong to the under-represented class in the training data [8], [9], [10], [11], [12]. It may be difficult to arrive at a physical interpretation of what the new samples represent given that they are directly created in the feature space. Alternatively, new samples can be created by applying simple transformations to existing samples [13], [5], [14]. This technique is particularly popular in deep learning for image classification where flipped, rotated, and translated copies of each image are added to the training set in order to reduce the sensitivity of the classifier to trivial variations of samples belonging to a particular class. Lastly, new samples can be created based on a generative model that mimics the variations of data observed in the general population of real samples [15], [16]. For instance, the generative model may have parameters that determine size, shape, deformations, and level of random noise such that each combination of settings results in a new sample that appears very similar to real samples, but whose properties may not have been represented in the existing small training set. When the parameters are properly selected, samples generated by a well-crafted generative model may even be used to create training sets consisting entirely of artificial samples.
In medical imaging, many techniques for creating new artificial samples and altering the distribution of lesion properties in an existing dataset have been developed. In one approach, new lesions are first mathematically simulated based on various deformation or cluster growth models. These lesions are subsequently inserted into the raw projection data or reconstructed clinical images. A few examples of this approach can be found in [17], [18], [19] for lung nodules, in [20], [21] for mammography, and in [22] for digital breast tomosynthesis (DBT). An important factor when using a simulated lesion generated based on mathematical models (rather than biologically inspired models) is to assure the realism of the characteristics of the artificial samples. In a second approach, a real lesion is first extracted from a clinical image and then inserted into a new location on the same or different image using image processing techniques [23], [24], [25], [26], [27]. Unlike the first approach, the realism of the nodule characteristics is guaranteed in the latter group of methods since the lesions are real by design. So far none of the techniques cited here have actually been used or evaluated for training a computer-aided diagnosis (CAD) system.
In this study we use our previously developed image blending tool [26], [27], [28] to create additional training samples for a CAD system designed for lung nodule detection. This tool provides a drag and drop capability such that a user can simply draw a rectangular region of interest (ROI) around a nodule, and then select the center of the desired insertion area in the target slice. This produces a seamless insertion of the nodule in a new location, while preserving the background texture and salient features in the target image. In addition, this tool provides the capability to change different properties of a nodule prior to its insertion into a new location. The combination of these factors allows us to more effectively augment a given dataset by altering its distribution of nodule shapes, contrasts, and locations, as well as creating new lesion-background combinations. We use these synthetic samples to increase the number of nodules in different size training sets, and demonstrate that the performance of various classifiers can be improved by this augmentation approach instead of relying on costly incremental collection of additional real samples from patients. A preliminary version of this work was published in a conference proceedings [29] where inserted nodules were not changed in any way prior to insertion, fewer features were extracted from each volume of interest (VOI), and only a single type of classifier was considered. The augmentation and training procedure in this paper is most similar in its concept to the method in [30] which uses a 3D shape model to generate synthetic persons based on a few real subjects. The artificial persons are subsequently superimposed onto real pictures and a people detection system is trained on the resulting pictures.
The main contributions of this paper are that to our knowledge, this is the first time the effectiveness of lesion insertion as a data augmentation method in CAD training is investigated. Moreover, we explore the effect of using each real nodule as a basis to generate multiple new nodules with different properties on CAD training, and compare the performance of classifiers trained using this augmentation method with those using the minority over-sampling technique in [8].
The remainder of this paper is organized as follows. In Section II we describe briefly the background of our image blending system, as well as our CAD system. Results of the training procedure and the effect of data augmentation for small training sets on various classifiers are subsequently presented in Section III before concluding in Section IV.
II. Methods
A. Lesion Blending
In this section we provide an overview of our lesion insertion tool. We refer the interested reader to [26], [27], [28] for full details of the individual procedures that constitute the overall system and their computationally efficient implementation. Our tool allows a user to select a nodule of interest and insert it into a new location on a different image, and is mainly based on Poisson image editing [31], which is a well-known and versatile seamless image composition algorithm. Using the Poisson equation with Dirichlet boundary conditions, this technique can perform an inward interpolation of the boundary values of the target image’s paste area to seamlessly recreate the source object of interest in a new location within the target image. The interpolation is guided by a guidance field, which in its simplest form is set equal to the gradient field of the source object. Using the notation D for the domain under ℝ2 over which the target image is defined, Ω for a closed subset of D with boundary ∂Ω, fs and ft for the source and target images, respectively, and f for the desired blend result over the interior of Ω, we can find f by solving the following minimization problem:
| (1) |
where is the gradient operator.
Equivalently, the unique solution to Eq. (1) can be obtained using the Poisson equation with Dirichlet boundary conditions:
| (2) |
where is the Laplacian operator and denotes the divergence operator. Eq. (2) above can be discretized to obtain a sparse system of linear equations which can be efficiently solved using sparse solvers.
The blending of a nodule into a new location is intended to increase the diversity of samples in a training set. It is therefore essential to maintain the texture and salient features in the background of the target image so that the blended image emulates the presence of a nodule within the selected background texture and surrounding tissue. However, the image composite f obtained by solving Eq. (2) completely replaces the original contents of the target image within Ω with the contents of fs. Hence we modify the guidance field by replacing the gradient field of the source with the gradient field of the target image at spatial locations where the magnitude of the gradient in the target image is higher than that of the source image. The combination of the gradient fields of source and target images in this fashion will then allow the salient elements of both images to coexist alongside each other in the composite f:
| (3) |
Another feature of the blending tool is its capability to remove extraneous objects located within the source ROI such as undesirable anatomical structures or vascular attachments prior to the insertion of a nodule into a new location. This is made possible using two procedures called masking and gradient field shaping, which modify the guidance field in different ways to achieve the desired outcome. These options make the selection of the source ROI easier, as the user can identify the source nodule by simply drawing a casual ROI around it even when other salient features are present in the ROI. Finding an appropriate insertion area in the target image is similarly simplified as the user does not need to find an exact match in the target image for vascular attachments that may exist in the source image. Finally, the insertion process is further simplified using an additional procedure that finds the optimal boundary conditions given an initial source ROI and the center of the desired insertion area. Poisson editing is essentially a diffusion process, and is prone to halos and bleeding artifacts when the differences between source and target images along the insertion boundary are not uniform. Manually finding and tracing a boundary that satisfies this condition is nearly impossible. A graph based iterative approach is therefore used to automatically find the optimal boundary for Poisson reconstruction [32] given the source and target images, and a segmentation of the nodule. When the nodule segmentation is not available from an automated algorithm, a rough segmentation (drawn manually or using simple intensity thresholding) is sufficient to guide the calculation of the optimal boundary.
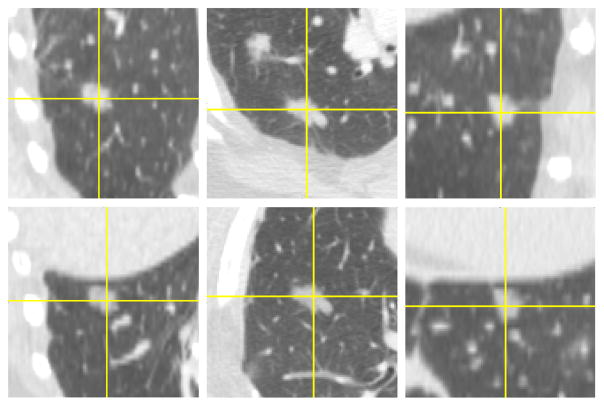
Fig. 1 shows an example of blending consecutive slices of a nodule onto a new location. User involvement was limited to selecting the initial ROI encompassing the nodule, and the center of the insertion area on a single slice of the source and target images, respectively. The algorithm subsequently followed the steps outlined in this section to seamlessly blend the whole volume of the nodule in a new location, while preserving the texture and salient features in the target background slices. Figs. 2 and 3 shows the orthogonal views of the same nodule and its inserted counterpart. It can be seen that the appearance of the inserted nodule is consistent with its appearance in its original location in every view. However, changing the position of the nodule, e.g. by placing it into a different lung or lobe, from the back of the patient to his/her front, or from one patient onto a different patient, introduces more diversity in the dataset in terms of interactions of the nodule with background tissue or texture.
Fig. 1.
Inserting consecutive slices of a nodule into a new location. In each row from left to right: source image, target image, blend results using optimal boundary with both boundaries superimposed, blend results using optimal boundary without boundaries superimposed. Orange and red boxes show the initial boundary drawn by the user, and the optimal boundary is shown in yellow.
Fig. 2.
Orthogonal views of the nodule in Fig. 1. Original (first row) and its inserted counterpart (second row).
Fig. 3.
Zoomed in orthogonal views of the same nodule as in Figs. 1 and 2. Original (first row) and its inserted counterpart (second row)
B. Nodule deformation model
The procedure described in Sec. II-A to create new samples only requires a pair of source and target images. Since nodule-free target ROIs are plentiful, this procedure can be used to obtain multiple nodule-background combinations from a single nodule. However, repeatedly inserting one nodule to multiple locations may produce features that are quite similar to each other, since the diversity introduced by the insertion is limited to differences in the local background while the lesion remains unchanged. We therefore use a deformation model to create additional synthetic nodules with different properties compared to the original ones using different parameterizations of the model. Using proper model parameters, and given the fact that the synthetic nodules are created based on real nodules, we can obtain nodule properties that should better approximate that which might be seen in the general population of real nodules. The synthetic nodules can then be inserted into the same target location as the one selected for the non-modified nodule, and thereby amplify the number of samples for classifier training in a single pass after initial target locations are specified.
We apply one or more affine transformations of rotation, shear transform, and size scaling to each real nodule in the image space. The contrast of the nodule can additionally be changed by weighting the gradient field of an original nodule. The transformations can be combined, such that multiple properties are altered simultaneously. Each transformation can take different parameters to generate many new synthetic nodules that are each different from one another and the original nodule. Other transformations such as warping are also possible, but we considered them more extreme in the sense that they have the potential to induce dramatic and biologically implausible effects on nodule properties.
Fig. 4 shows examples of nodules that have undergone single transformations across a range of parameters. Aside from rotation, the choice of parameters for the other transformations is important so that changes to the properties of a nodule remain reasonable. Fig. 5 shows examples of changing several nodule properties simultaneously by applying a combination of all four transformations to a nodule. From these examples, it can be seen that characteristics of a nodule can be modified across a wide range, from those that are reasonable to ones that are extreme. Depending on the properties of samples in the original training set, parameters of the deformation model can be adjusted such that the synthetic samples exhibit minor to moderate variations of the original nodules in order to maintain their biological plausibility. The parameters can also be selected to more specifically supplement the training data for the under-represented sample properties. For instance, if most or all of the existing nodules in a dataset are smaller than 5mm in diameter or have high contrast, the transformation parameters can be tuned to generate a library of new synthetic samples that are larger or have lower contrast, respectively. As with the real nodules, the diversity of the training set is further increased by inserting the synthetic samples into locations with different backgrounds, textures, and surrounding tissue.
Fig. 4.
Examples of nodule insertion after a single transformation of the original nodule. In each row from left to right: rotation (−135, −90, −45, 45, 90, 135), size scaling (0.7, 0.8, 0.9, 1.1, 1.2, 1.3), horizontal shear (−0.3, −0.2, −0.1, 0.1, 0.2, 0.3), and contrast scaling (0.55, 0.7, 0.85, 1.15, 1.30, 1.45). Only a single slice of the nodule is shown here.
Fig. 5.
Insertion of a nodule after a combination transformation. Parameters used in each column correspond to parameters of the single transformations in same column of examples in Fig. 4. Only a single slice of the nodule is shown here.
C. VOI selection and feature extraction
The CAD system in our study uses some of the components of a previously developed CAD system [33], [34], [35], [36], [37]. This system consisted of three main modules: pre-screening, feature extraction, and classification. In this work we kept the first two modules unchanged, and modified the classification module and the way it was trained. We will therefore provide only an overview of the earlier components here. We will describe the classifier module and its training in detail in Sec II-D.
The pre-screening stage consists of three separate steps. First, an adaptive scheme is used to detect and segment the left and right lungs in each CT slice. A low threshold is initially used to separate the thorax and mediastinum from the air surrounding the patient. A weighted k-means clustering technique is then applied to the pixel gray values of these regions to segment the lung regions, followed by partitioning each lung into central and peripheral regions. The last step of pre-screening generates binary nodule candidate objects within the lungs using a weighted k-means technique with a variable threshold (adjusted based on the location of a slice within the patient) that controls the relative number of voxels assigned to the background and object clusters.
The purpose of the pre-screening stage is to filter out obviously irrelevant areas of the lung. Any structure that is excluded in this stage cannot be recovered in subsequent processing steps. The sensitivity of the pre-screening stage is therefore very high, while the specificity is very low. Most false-positives from the pre-screening stage occur in high CT-value areas within the lung, which are typically located along the vessels. Most non-juxtapleural nodules are nearly spherical or ellipsoid shaped (except spiculated ones). Vessels on the other hand are elongated tube like structures. In order to further analyze the structures found in pre-screening and to reduce the number of false positives, two types of segmentations are obtained for each detected object. The first segmentation, referred to as nodule-oriented segmentation, uses a 3D region growing algorithm and aims at segmenting objects in the VOI that are expected to be more compact in shape. The second segmentation, referred to as vessel-oriented segmentation, is obtained by applying expectation maximization to Hessian-enhanced CT volumes and aims at segmenting objects that are elongated and connected to other densities in the VOI being examined [36], [37].
A total of 82 features are extracted from VOIs enclosing the segmentations for each detected object. The features fall into four general categories: (a) 12 features describing the shape and general properties of the candidate structure based on 3D ellipsoid shape fitting, (b) 38 features showing the statistical distribution of voxel intensities, (c) 24 features describing the distribution of the 3D gradient field within the VOI, and (d) 8 features related to eigenvalues of the Hessian matrix.
Features in group (a) are defined by assessing the sphericity of a candidate object based on properties of a 3D ellipsoid fitted to the segmented object. For spherical objects, the ratios of lengths of any two out of the three major axes will be close to 1. For each of the objects’ two segmentations, six features are extracted based on 3D ellipsoid shape fitting that consist of the lengths of the three major axes, and their ratios (12 total) [34]. Another set of 19 features describing the shape and distribution of gray-scale values within the detected object are extracted from both segmentations of the object (total of 38) for feature group (b), and include features such as object volume, surface area, average and standard deviation of the gray-scale values, compactness, skewness, and kurtosis of the gray-level histogram, etc [33].
Features in group (c) were defined by analyzing the 3D gradient field around the object. As the object may not encompass the entire nodule (or other non-nodule structure), the VOI being considered is expanded by a fixed amount in all dimensions. The expansion of the VOI also allows the capture of the background information surrounding the candidate object. A 3D isotropic interpolation is then used to create voxels with equal side-lengths so that different processing steps and filters can be applied to the object more easily. Three concentric spherical shells are subsequently centered at the object’s centroid, and 26 uniformly distributed points are selected on each shell. For each point on a shell, the orientation of the 3D gradient at that point relative to the radial vector connecting the centroid to that point and its magnitude are computed. A total of 24 features describing the statistics and distribution of magnitudes and relative orientations of the gradient field across the points on the three shells are then extracted from the nodule-oriented segmentation of each object [34]. The features in group (d) are related to eigenvalues of the Hessian matrix. For each voxel within the VOI, line and blob responses are derived based on the three eigenvalues. Statistics representing the average, standard deviation, etc. of the responses within the VOI were extracted as 8 additional Hessian features [35].
Based on the aforementioned description of the four categories of features, it is expected that the introduction of new foreground-background combinations through lesion insertion will largely impact features in group (c) as they are extracted from an expanded VOI containing the detected object and its immediate background, while the insertion of deformed lesions will affect all feature groups (a–d).
D. Classifiers, training, & experimental setup
In order to evaluate the effect of nodule insertion on training a CAD system with a limited dataset, we developed an experimental setup that emulated having an initially small number of training cases, and the incremental accrual of additional patients to increase the sample size. We then compared the effect of training multiple classifiers based on the original datasets and ones that were augmented using our method. Each classifier was tested against an independent dataset containing only real (i.e. non-inserted) nodules and the comparison of performance was based on the area under the ROC curve (AUC).
We employed a range of classifiers with different levels of complexity in our evaluations: nearest neighbors (k = 15), SVM with radial basis function (RBF) kernel (c = 1, γ = 0.01), random forest (RF) with 100 decision trees, AdaBoost with a maximum of 50 decision trees, Gaussian Naive Bayes (NB), and linear discriminant analysis (LDA). The parameters of each classifier were selected experimentally according to a small validation set (independent from the test set) for performance of TR1 in the situation where 20 patients were included in training. Each classifier’s parameters were then fixed across all training scenarios and number of patients used in training.
In each CT exam used in training, a single original nodule was extracted from its original location and inserted into a different location (i.e. different slice, lobe, lung, and/or background texture) within the same patient’s CT exam with or without additional transformations to its properties. As both the source and target images belonged to the same patient, they had the same pixel size, slice thickness, and overall noise properties. It was therefore not necessary to make any adjustments to the nodule prior to blending it into a new location (in [26], we describe a noise transfer procedure when the noise properties of source and target images do not match). The insertion of nodules into new locations was done by the first author who does not have medical training by using commonsense rules (e.g. a nodule was not inserted outside of the lung, or such that it would significantly overlap a major structure in the CT image on any slice), but not necessarily based on anatomically informed rules. For instance, nodules (especially larger ones) often have visible vascular connections, but in our insertions some of the nodules were placed along or at the end of vessels to produce the appearance of a vascular connection to the nodule (e.g. see Fig. 6 or second slice of inserted nodule in Fig. 1), while others were not.
Fig. 6.
Inserting a nodule along a vessel. From left to right: source image, target image, blend results using optimal boundary with both boundaries superimposed, blend results using optimal boundary without boundaries superimposed.
The total insertion time for a nodule includes the time needed to find a proper insertion area and the processing time for the blending algorithm. This time depends on the size of the nodule, but for a typical nodule enclosed by a 30 × 30 pixel ROI and spanning across 5 slices the average time to locate an insertion area was three minutes, while the time for the blending procedure was under 5s.
The pre-screening procedure was applied to each CT exam to obtain candidate VOIs for feature extraction. If an exam contained multiple nodules, only a single one was randomly selected for insertion and the other ones were excluded from the training data to control the experiments and eliminate other confounding factors during training of the classifier. Juxta-pleural nodules often exhibit directional growth towards the inside of the lung due to the fact that their growth is inhibited from the side attached to the pleural boundary. This reduces the utility and usability of pleural nodules for data augmentation in terms of locations they can be inserted into as well as transformations that can be applied to them. We therefore also excluded pleural nodules from both training and testing the classifiers.
The classifiers were trained in several different ways to assess the impact of data augmentation compared to using the original samples only. In each experiment, the training set consisting only of natural nodules plus the true negatives is referred to as TR1. The pre-screening stage finds a large number of true negatives per case, resulting in an unbalanced set of samples that include a large number of negatives. A common way to avert issues associated with class imbalance is to prune the majority negative class samples such that a subset of hard negatives (i.e. those that are more difficult to classify) are kept and the rest are discarded [38], [39], [40]. We under-sample the negative samples according to one of the Hessian features from Section II-C and keep a subset of negative samples belonging to objects that appear most spherical per case. A second training set, TR2, is obtained by augmenting TR1 with feature vectors corresponding to the inserted counterparts of the positive samples in TR1, and is used to evaluate the effect of our image blending technique to change the location of nodules but not any of their other properties. This new training set has the same number of negatives, but twice as many positives as the original training set TR1. A third training set, TR3, is constructed by augmenting TR2 with feature vectors corresponding to nodules that underwent a transformation prior to being inserted into a new location (here we chose the same insertion location for the deformed nodules as their non-modified counterparts in TR2 in each case). TR3 is used to evaluate the effect of increasing the number of positives in the training data by modifying different properties of the nodules, as well their location, on classifier performance.
E. Training and test datasets
The source of data in our experiments is the publicly available Lung Image Database Consortium (LIDC) dataset [41]. Each case in LIDC was reviewed by four experienced thoracic radiologists who marked the location of ≥ 3mm nodules by delineating their boundaries on every slice in which the nodule was detected. Not all nodules were detected by all four radiologists. We therefore unified the decisions in assigning the ground truth using the following rule: a marked location was designated as a nodule if at least two radiologists had identified that nodule, and if there was at least a 20% overlap in the nodule volume (percent overlap was defined as the intersection of volumes according to delineations from the individual radiologists who marked that nodule divided by the union of those volumes). We took a random subset of 60 cases from the LIDC database that contained at least one > 3mm nodule that satisfied the aforementioned conditions, and followed the procedure outlined in Sec. II-D to perform the data augmentation. Of the 60 total original nodules, 24 (40% of total) were inserted in a different location within the same lung, out of which only 12 (20% of total) were inserted into the same lobe as the original nodule. Table I shows additional information regarding the change in location of the centroid of the inserted nodules relative to that of their original counterparts in terms of in-plane distance (i.e. only considering change in x and y coordinates) and Z distance (i.e. only considering change in slice number).
TABLE I.
Change in location of centroid of inserted nodules relative to that of their original counterparts
| Min | Max | Mean ± Std | |
|---|---|---|---|
|
| |||
| Z Distance (slices) | 0 | 61 | 13±11 |
| In-Plane Distance (pixels) | 6 | 325 | 161±84 |
Another random subset of 100 cases fitting the same criteria for nodules as above was used as the independent test set for our study, containing 138 and 492 positive and negative samples (i.e. detected objects), respectively.
III. Performance Evaluation
Fig. 7 shows the results of the experiments described in Section. II-D. Starting from a small subset of 20 patients, each classifier was trained using the original samples, as well as using training sets obtained from several different augmentation methods. Additional patient data were incrementally added until all 60 patients were included, and the classifiers were retrained each time from scratch and tested against the test set. The distribution of nodule sizes in TR1 across different number of patients and in the test set are shown in Table III.
Fig. 7.
Performance of various classifiers across different number of patient cases used in training. The classifiers were trained using the original training sets, as well as using different augmentation methods. The independent test set contains only real patient cases from LIDC (i.e. no inserted nodules were included in the test set). The vertical axis is extended for Naive Bayes and LDA due to their lower performance compared to other classifiers. Error bars are only shown for TR3 to avoid clutter in the plots.
TABLE III.
Principal axis length (in mm) of nodules in different datasets
| Dataset | Length (mean±standard deviation) | min | Max |
|---|---|---|---|
|
| |||
| TR1 @ 20 | 14.84±5.95 | 9.17 | 33.12 |
| TR1 @ 30 | 14.65±6.06 | 7.04 | 33.12 |
| TR1 @ 40 | 14.14±5.90 | 6.36 | 33.12 |
| TR1 @ 50 | 14.21±5.67 | 6.31 | 33.12 |
| TR1 @ 60 | 13.89±5.44 | 5.21 | 33.12 |
|
| |||
| Test set | 12.28±7.54 | 5.21 | 43.93 |
We used the four transformations listed in Table II to create TR3, thereby resulting in a training set that had 6 times the number of positives as TR1 (including the original nodule and the insertion without any transformation) but the same number of negatives. Each row in this table shows the combination of transformations applied to the nodule prior to its insertion into a new location. The baseline performance in each experiment is the AUC obtained from training using TR1, and the augmented training sets should perform at least as well as TR1 in order to be useful. It can be seen that in nearly every experiment, TR2 performs at least as well as TR1, with TR3 outperforming both TR1 and TR2. We also compared the AUC values from TR3 to those of TR1 for each of the classifiers and number of cases included in training using the non-parametric method by DeLong et al. [42] as implemented in the iqmodelo software package [43]. Table IV shows the 95% confidence interval for the AUC difference between TR3 and TR1 (TR3-TR1) for the various classifiers and sample sizes in Fig. 7. Although the lower limit of the confidence interval is above 0 for most combinations, this cannot be interpreted as a significant result for our experiments because we did not prospectively define a hypothesis to be tested and we had multiple comparisons.
TABLE II.
List of transformations used in TR3
| Rotation (degrees) | Scale | Horizontal Shear | Contrast |
|---|---|---|---|
|
| |||
| −60 | 0.75 | −0.15 | 0.92 |
| 60 | 1.25 | 0.15 | 1.08 |
| −120 | 0.8 | −0.2 | 0.88 |
| 120 | 1.2 | 0.2 | 1.12 |
TABLE IV.
95% Confidence interval of difference of AUC of TR3 relative to that of TR1 for different classifiers and numbers of patients
| Classifier | @20 | @30 | @40 | @50 | @60 |
|---|---|---|---|---|---|
|
| |||||
| Nearest Neighbors | (0.004, 0.036) | (−0.001, 0.020) | (−0.011, 0.016) | (−0.012, 0.014) | (−0.014, 0.017) |
| RBF SVM | (0.019, 0.073) | (0.009, 0.047) | (0.013, 0.046) | (0.007, 0.035) | (0.005, 0.031) |
| Random Forest | (−0.001, 0.026) | (0.003, 0.021) | (0.003, 0.027) | (0.001, 0.027) | (−0.006, 0.019) |
| AdaBoost | (−0.012, 0.054) | (0.008, 0.062) | (0.011, 0.056) | (−0.013, 0.021) | (−0.027, 0.016) |
| Naive Bayes | (0.020, 0.105) | (0.071, 0.153) | (0.056, 0.135) | (0.026, 0.090) | (0.009, 0.058) |
| LDA | (0.037, 0.165) | (−0.010, 0.041) | (0.002, 0.054) | (0.010, 0.062) | (−0.002, 0.036) |
Another interesting observation based on the AUC values is that for all classifiers but LDA, TR3 has a comparable or better performance than TR1 using only half the number of patients. For easier comparison, we extracted this information from Fig. 7 and show it in Tables V and VI. This demonstrates the effectiveness of the deformation model for creating realistic synthetic nodules in our augmentation technique.
TABLE V.
Comparison of AUC of TR3 at 20 patients with TR1 at 40 patients (point estimate ± standard deviation)
| TR3 @20 | TR1 @40 | |
|---|---|---|
|
| ||
| Nearest Neighbors | 0.925 ± 0.013 | 0.940 ± 0.014 |
| RBF SVM | 0.923 ± 0.013 | 0.928 ± 0.013 |
| Random Forest | 0.951 ± 0.009 | 0.944 ± 0.009 |
| AdaBoost | 0.898 ± 0.016 | 0.902 ± 0.017 |
| Naive Bayes | 0.854 ± 0.020 | 0.810 ± 0.024 |
| LDA | 0.814 ± 0.024 | 0.907 ± 0.018 |
TABLE VI.
Comparison of AUC of TR3 at 30 patients with TR1 at 60 patients (point estimate ± standard deviation)
| TR3 @30 | TR1 @60 | |
|---|---|---|
|
| ||
| Nearest Neighbors | 0.941 ± 0.012 | 0.946 ± 0.013 |
| RBF SVM | 0.948 ± 0.009 | 0.940 ± 0.011 |
| Random Forest | 0.952 ± 0.009 | 0.955 ± 0.008 |
| AdaBoost | 0.935 ± 0.010 | 0.934 ± 0.014 |
| Naive Bayes | 0.909 ± 0.017 | 0.882 ± 0.020 |
| LDA | 0.878 ± 0.019 | 0.926 ± 0.016 |
In order to summarize the performance of classifiers at a fixed sensitivity, the false positive rates (FPR) for TR1-3 for 40 patients at 90% sensitivity were extracted from their corresponding ROC curves and are shown in Table VII. The numbers in parentheses show the percent improvement in FPR for TR2 and TR3 over TR1. It should be noted that the relative ranking of classifiers in this table according to FPR at fixed sensitivity should not be confused with the rankings observed in Fig. 7 since the ROC curves may cross each other, whereas the AUC is the average sensitivity across all values of FPR. In order to examine whether the improvement in performance for the augmented datasets TR2 and TR3 is only due to noise injection, or if the new background combinations and nodule transformations are contributing to improved learning and better generalization, we also used the popular synthetic minority over-sampling technique (SMOTE) [8] to boost the number of positive samples in each experiment. Rather than over-sampling with replacement, this data augmentation method creates new synthetic samples directly in the feature space. The artificial sample is generated on a random point along the line connecting each minority sample and its nearest neighbor as follows: for each minority sample, take the difference between the feature vector of that sample and its nearest neighbor, multiply the difference by a random number between 0 and 1, and add to the feature vector of the original minority sample. The new feature vector is then added to the training set. This technique has two parameters: N for desired percent over-sampling, and k that denotes the number of nearest neighbors to consider and select from. The number of synthetic samples generated by SMOTE with N = 100% is equal to that with TR2, and SMOTE with N = 500% is equal to that in TR3 (these two will be referred to as SMOTE100 and SMOTE500 from here on). We ran our experiments with k ranging between 2 and 5 (first nearest neighbor is considered to be the sample itself), and for each k repeated the experiments 10 times and computed the average AUC. For a fair comparison, we show the maximum average AUC across different values of k for SMOTE100 and SMOTE500 in Fig. 7. The performance of SMOTE is found to be equivalent or inferior to that of TR2 in most experiments, and in some even inferior to that of TR1. In almost all other ones, the performance of SMOTE lies somewhere between the baseline performance of TR1 and the upper performance level obtained from training the classifier on TR3. The difference in performance between SMOTE and our method can be explained by the fact that samples generated by SMOTE are essentially points that are interpolated between existing samples in the dataset. As a result, these samples may result in a smoother decision surface and therefore less over-training, but cannot extend the training set to include unobserved areas of the minority sample space. Our augmentation procedure on the other hand allows for more direct control of the properties of the synthetic samples, and thereby better generalization performance for a classifier. For instance, if we consider the largest diameter of the nodule as a feature, and if all nodules in the limited training dataset are between 5mm to 8mm in diameter, the samples generated by SMOTE will also have a diameter that lies between 5mm to 8mm, whereas our augmentation procedure allows for generation of samples that are less than 5mm or larger than 8mm in diameter. A similar observation can be made about other features such as nodule contrast, various foreground-background interactions that affect the gradient features, etc. This can also be seen in Fig. 8 by visualizing TR3 and SMOTE500 for 60 patients in two dimensions using the t-SNE toolbox [44]. This technique performs dimensionality reduction on high-dimensional datasets so that they can be visualized in a low dimensional (2D or 3D) space, and is capable of preserving both the local and global structure of high-dimensional data very well. In Fig. 8, the synthetic samples are clustered tightly around the original positive samples for SMOTE500. For TR3 on the other hand, the synthetic samples are more dispersed in space, and without substantial intermixing with the negative samples which would erode the boundary between the two classes. It should be noted that t-SNE is a data driven method that learns a non-parametric mapping for the points, and not an explicit mapping function that projects the high dimensional points to the lower dimensional space. This is the reason why the locations of the real points in the two scatter plots are different. However, here we are only concerned with the distribution of the synthetic positive points around the real positive points as discussed earlier.
TABLE VII.
Improvement in False Positive Rate for 40 Patients at 90% Sensitivity
| Classifier | FPRTR1 | FPRTR2 | FPRTR3 |
|---|---|---|---|
|
| |||
| Nearest Neighbors | 0.152 | 0.125(17.8%) | 0.148(2.6%) |
| RBF SVM | 0.214 | 0.214(0.0%) | 0.145(32.2%) |
| Random Forest | 0.171 | 0.141(17.5%) | 0.129(24.6%) |
| AdaBoost | 0.276 | 0.417(−51.1%) | 0.170(38.4%) |
| Naive Bayes | 0.715 | 0.420(41.3%) | 0.287(59.9%) |
| LDA | 0.347 | 0.191(45.0%) | 0.151(56.5%) |
Fig. 8.
2D visualization of SMOTE500 (left) and TR3 (right) for 60 patients using t-SNE.
We also examined the effect of including an increasing number of combination transformations for each nodule in TR3, as shown in Fig. 9. We built a table of parameters similar to the one shown in Table II, where the values for the various transformations were selected from −150 degrees to 150 degrees for rotation, 0.7 to 1.3 for size scaling, −0.3 to 0.3 for shear, and 0.75 to 1.25 for contrast scaling. The transformations were added in symmetric pairs, such that one transformation used the higher value of the parameters listed above and the other used the corresponding lower value (e.g. the rotation component in a pair of transformations was 30 degrees in one and −30 degrees in the other). The results of these experiments, indicate that for most classifiers the performance level of TR3 is fairly stable for the range of transformation parameters selected here, and the number of combination transformations. As a result, selection of parameters in TR3, namely the combination transformation parameters and number of transformations per nodule, requires very little experimentation as long as the parameters are selected such that they fall within a reasonable range of values for each transformation type. Another way to interpret the relative stability of performance of TR3 across an increasing number of deformed nodules is that pulmonary nodules exhibit a high level of heterogeneity in terms of their various properties as well as their interactions with surrounding tissue. The deformations used in our model and the insertion process can approximate some of these variations, but not all of them, when only a limited pool of nodules and cases are available. As a result, the performance of a classifier trained on a dataset augmented using our method improves but reaches a plateau that can only be overcome if additional real nodules containing the unmodeled properties are included in the training set.
Fig. 9.
Performance of various classifiers across different numbers of combination transformations included in TR3 for 40 patients in the original dataset. The baseline AUC of each classifier trained on TR1 for 40 patients is taken from Fig. 7 and provided in parentheses after the classifier’s name in the legend for easy comparison with the performance results using TR3.
IV. Conclusion
We have demonstrated the effectiveness of our technique for seamless insertion of pulmonary nodules in augmenting limited datasets for training classifiers with varying degrees of complexity. This technique can be used to increase the count and variability of the positive training samples, which can be particularly important for lesions with rare shapes, locations, or background interactions. By minimizing user involvement, this augmentation process requires far less manual labor and time compared to other existing image augmentation methods. Each nodule in the original dataset can also be used as a basis to create new synthetic nodules with properties that might be unobserved in the original dataset. The combination of nodule transformations and nodule insertion is shown to result in a larger improvement in performance for multiple classifiers compared to only insertion into a new background. The increase in the number and variability of lesions and the resulting improvement in performance of the classifier comes at a small cost of the time required to perform the insertions to the CAD developer, and can reduce the need for accumulation of actual patient exams when data is scarce or expensive to obtain.
In this study, we used lung nodule detection in CT images as a surrogate example to demonstrate the potential of lesion insertion to augment data for training CAD systems for combinations of focal diseases and imaging modalities where annotated public datasets may not exist, or may be small. In principle, Poisson editing and our technique are applicable to other imaging modalities and organs, with the realism of the blending result depending on the compatibility and context of the source and target images. In a previous publication, we have already shown that our lesion insertion technique can be directly applied to the insertion of masses in mammography [27]. We have made the source code of our algorithm publicly available [28] so that it can be tailored to the specific needs of the disease or imaging modality of other studies.
Work is currently under way to develop a procedure for automatic selection of suitable insertion areas so that each nodule can be replicated in multiple new locations without manual user involvement. Combined with our deformation model for applying various transformations to the nodule characteristics, such a procedure could efficiently create a large number of new nodules in different backgrounds based on the original nodules from a dataset. The deformation model could also easily be extended to 3D transformations on the volume containing a lesion, instead of our current method where the lesion is processed on each slice using a series of 2D transformations. This can further extend the capabilities of this technique in terms of generation of synthetic samples with properties similar to real but unobserved lesions. Another interesting area of future work would be to assess whether instead of common-sense rules, a more medically informed set of rules could be devised to guide the manual or automatic placement of nodules into new locations.
Acknowledgments
The authors acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study. This work was supported in part through an Office of Women’s Health grant from the U.S. Food and Drug Administration, and by an appointment to the Research Participation Program at the Center for Devices and Radiological Health administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
Contributor Information
Aria Pezeshk, Division of Imaging, Diagnostics, and Software Reliability (DIDSR), Office of Science and Engineering Laboratories (OSEL), Center for Devices and Radiological Health (CDRH), U.S. Food and Drug Administration (FDA), Silver Spring, MD 20993 USA.
Nicholas Petrick, DIDSR/OSEL/CDRH/FDA, Silver Spring, MD 20993 USA.
Weijie Chen, DIDSR/OSEL/CDRH/FDA, Silver Spring, MD 20993 USA.
Berkman Sahiner, DIDSR/OSEL/CDRH/FDA, Silver Spring, MD 20993 USA.
References
- 1.Raudys S, Jain A. Small sample size effects in statistical pattern recognition: recommendations for practitioners. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1991;13(3):252–264. [Google Scholar]
- 2.Chan HP, Sahiner B, Wagner RF, Petrick N. Classifier design for computer-aided diagnosis: Effects of finite sample size on the mean performance of classical and neural network classifiers. Medical Physics. 1999;26(12):2654–2668. doi: 10.1118/1.598805. [DOI] [PubMed] [Google Scholar]
- 3.Sahiner B, Chan HP, Petrick N, Wagner RF, Hadjiiski LM. Feature selection and classifier performance in computer-aided diagnosis: The effect of finite sample size. Medical Physics. 2000;27(7):1509–1522. doi: 10.1118/1.599017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanczar B, Hua J, Sima C, Weinstein J, Bittner M, Dougherty ER. Small-sample precision of ROC-related estimates. Bioinformatics. 2010 Mar;26(6):822–830. doi: 10.1093/bioinformatics/btq037. [DOI] [PubMed] [Google Scholar]
- 5.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Bartlett P, Pereira F, Burges C, Bottou L, Weinberger K, editors. Advances in Neural Information Processing Systems (NIPS) Red Hook, NY: Curran Associates, Inc; 2012. pp. 1106–1114. [Google Scholar]
- 6.Hannun AY, Case C, Casper J, Catanzaro BC, Diamos G, Elsen E, Prenger R, Satheesh S, Sengupta S, Coates A, Ng AY. Deep speech: Scaling up end-to-end speech recognition. CoRR. 2014;abs/1412.5567 [Online]. Available: http://arxiv.org/abs/1412.5567. [Google Scholar]
- 7.Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N. Aggnet: Deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans Med Imaging. 2016;35(5):1313–1321. doi: 10.1109/TMI.2016.2528120. [DOI] [PubMed] [Google Scholar]
- 8.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: Synthetic minority over-sampling technique. Journal of Artificial Intelligence Research. 2002 Jun;16(1):321–357. [Google Scholar]
- 9.Melville P, Mooney R. Constructing diverse classifier ensembles using artificial training examples. Proceedings of the 18th International Joint Conference on Artificial Intelligence; Acapulco, Mexico. 2003; pp. 505–510. [Google Scholar]
- 10.Bishop CM. Training with noise is equivalent to Tikhonov regularization. Neural Comput. 1995 Jan;7(1):108–116. [Google Scholar]
- 11.Raviv Y, Intrator N. Bootstrapping with noise: An effective regularization technique. Connection Science. 1996;8:355–372. [Google Scholar]
- 12.van der Maaten L, Chen M, Tyree S, Weinberger KQ. Learning with marginalized corrupted features. Proc ICML. 2013;28:410–418. [Google Scholar]
- 13.Papageorgiou C, Poggio T. A trainable system for object detection. Int J Comput Vision. 2000 Jun;38(1):15–33. [Google Scholar]
- 14.Laptev I. Improving object detection with boosted histograms. Image Vision Comput. 2009 Apr;27(5):535–544. [Google Scholar]
- 15.Ho T, Baird H. Large-scale simulation studies in image pattern recognition. IEEE Trans on PAMI. 1997;19:1067–1079. [Google Scholar]
- 16.Pezeshk A, Tutwiler RL. Automatic feature extraction and text recognition from scanned topographic maps. IEEE Trans Geoscience and Remote Sensing. 2011;49(12):5047–5063. [Google Scholar]
- 17.Zhang X, Olcott E, Raffy P, Yu N, Chui H. Simulating solid lung nodules in MDCT images for CAD evaluation: modeling, validation, and applications. Proc. SPIE Medical Imaging: Computer-Aided Diagnosis; Mar, 2007. pp. 65 140Z0–65 140Z8. [Google Scholar]
- 18.Li X, Samei E, Delong DM, Jones RP, Gaca AM, Hollingsworth CL, Maxfield CM, Carrico CWT, Frush DP. Three-dimensional simulation of lung nodules for paediatric multidetector array CT. British Journal of Radiology. 2009;82(977):401–411. doi: 10.1259/bjr/51749983. [DOI] [PubMed] [Google Scholar]
- 19.Shin HO, Blietz M, Frericks B, Baus S, Savellano D, Galanski M. Insertion of virtual pulmonary nodules in CT data of the chest: development of a software tool. European Radiology. 2006;16(11):2567–2574. doi: 10.1007/s00330-006-0254-x. [DOI] [PubMed] [Google Scholar]
- 20.Saunders R, Samei E, Baker J, Delong D. Simulation of mammographic lesions. Academic Radiology. 2006;13(7):860–870. doi: 10.1016/j.acra.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Rashidnasab A, Elangovan P, Yip M, Diaz O, Dance DR, Young KC, Wells K. Simulation and assessment of realistic breast lesions using fractal growth models. Physics in Medicine and Biology. 2013;58(16):5613–5627. doi: 10.1088/0031-9155/58/16/5613. [DOI] [PubMed] [Google Scholar]
- 22.Vaz MS, Besnehard Q, Marchessoux C. 3D lesion insertion in digital breast tomosynthesis images. Proc SPIE Medical Imaging: Physics of Medical Imaging. 2011;7961:79 615Z1–79 615Z10. [Google Scholar]
- 23.Ambrosini RD, O’Dell WG. Realistic simulated lung nodule dataset for testing CAD detection and sizing. Proc SPIE Medical Imaging: Computer-Aided Diagnosis. 2010 Mar;7624:76 242X1–76 242X8. [Google Scholar]
- 24.Peskin AP, Dima AA. Modeling clinical tumors to create reference data for tumor volume measurement. In: Bebis G, Boyle R, Parvin B, Koracin D, Chung R, Hammound R, Hussain M, KarHan T, Crawfis R, Thalmann D, Kao D, Avila L, editors. Advances in Visual Computing, ser. Lecture Notes in Computer Science. Vol. 6454. Springer; Berlin Heidelberg: 2010. pp. 736–746. [Google Scholar]
- 25.Madsen MT, Berbaum KS, Schartz KM, Caldwell RT. Improved implementation of the abnormality manipulation software tools. Proc SPIE Medical Imaging: Image Perception, Observer Performance, and Technology Assessment. 2011 Mar;7966:7 966 121–7 966 127. [Google Scholar]
- 26.Pezeshk A, Sahiner B, Zeng R, Wunderlich A, Chen W, Petrick N. Seamless insertion of pulmonary nodules in chest CT images. IEEE Trans Biomedical Engineering. 2015;62(12):2812–2827. doi: 10.1109/TBME.2015.2445054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pezeshk A, Sahiner B, Petrick N. Seamless lesion insertion in digital mammography: Methodology and reader study; Proc SPIE Medical Imaging: Computer-Aided Diagnosis; 2016. pp. 97 850J-97 850J–6. [Google Scholar]
- 28.Pezeshk A. Lesionblender. 2015 https://github.com/DIDSR/LesionBlender.
- 29.Pezeshk A, Sahiner B, Chen W, Petrick N. Improving CAD performance by seamless insertion of pulmonary nodules in chest CT exams. Proc SPIE Medical Imaging: Computer-Aided Diagnosis. 2015:94 140A–94 140A–6. [Google Scholar]
- 30.Pishchulin L, Jain A, Wojek C, Andriluka M, Thormhlen T, Schiele B. Learning people detection models from few training samples. Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR’11); 2011; pp. 1473–1480. [Google Scholar]
- 31.Perez P, Gangnet M, Blake A. Poisson image editing. Proc ACM SIGGRAPH. 2003:313–318. [Google Scholar]
- 32.Jia J, Sun J, Tang C-K, Shum H-Y. Drag and drop pasting. Proc ACM SIGGRAPH. 2006:631–636. [Google Scholar]
- 33.Gurcan MN, Sahiner B, Petrick N, Chan HP, Kazerooni EA, Cascade PN, Hadjiiski LM. Lung nodule detection on thoracic computed tomography images: Preliminary evaluation of a computer-aided diagnosis system. Medical Physics. 2002;29(11):2552–2258. doi: 10.1118/1.1515762. [DOI] [PubMed] [Google Scholar]
- 34.Ge Z, Sahiner B, Chan HP, Hadjiiski LM, Cascade PN, Bogot N, Kazerooni EA, Wei J, Zhou C. Computer-aided detection of lung nodules: False positive reduction using a 3D gradient field method and 3D ellipsoid fitting. Medical Physics. 2005;32(8):2443–2454. doi: 10.1118/1.1944667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahiner B, Ge Z, Chan H-P, Hadjiiski LM, Bogot P, Kazerooni E. False positive reduction using Hessian features in computer-aided detection of pulmonary nodules on thoracic CT images. Proc SPIE Medical Imaging: Image Processing. 2005:790–795. [Google Scholar]
- 36.Sahiner B, Hadjiiski LM, Chan H-P, Zhou C, Wei J. Computerized lung nodule detection on screening CT scans: performance on juxta-pleural and internal nodules. Proc SPIE Medical Imaging: Image Processing. 2006:61 445S–61 445S–6. [Google Scholar]
- 37.Zhou C, Chan HP, Sahiner B, Hadjiiski L, Chughtai A, Patel S, Wei J, Ge J, Cascade P, Kazerooni EA. Automatic multiscale enhancement and hierarchical segmentation of pulmonary vessels in CT pulmonary angiography (CTPA) images for CAD applications. Medical Physics. 2007;34(12):4567–4577. doi: 10.1118/1.2804558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Japkowicz N. The class imbalance problem: Significance and strategies. Proceedings of the 2000 International Conference on Artificial Intelligence (ICAI); 2000; pp. 111–117. [Google Scholar]
- 39.Canevet O, Lefakis L, Fleuret F. Sample distillation for object” detection and image classification. Proceedings of the 6th Asian Conference on Machine Learning (ACML), ser. JMLR: Workshop and Conference Proceedings; Nov, 2014. [Google Scholar]
- 40.Liu XY, Wu J, Zhou ZH. Exploratory undersampling for class-imbalance learning. IEEE Trans Sys Man Cyber Part B. 2009 Apr;39(2):539–550. doi: 10.1109/TSMCB.2008.2007853. [DOI] [PubMed] [Google Scholar]
- 41.Armato SG, III, et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): A completed reference database of lung nodules on CT scans. Medical Physics. 2011;38(2):915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeLong ER, DeLong DM, Clarkse-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 43.Wunderlich A. iqmodelo: Statistical software for image quality assessment. https://github.com/DIDSR/IQmodelo.
- 44.van der Maaten L, Hinton GE. Visualizing high-dimensional data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]