Abstract
The Critical Assessment of Genome Interpretation (CAGI) experiment is the first attempt to evaluate the state-of-the-art in genetic data interpretation. Among the proposed challenges, Crohn disease (CD) risk prediction has become the most classic problem spanning three editions. The scientific question is very hard: can anybody assess the risk to develop CD given the exome data alone? This is one of the ultimate goals of genetic analysis, which motivated most CAGI participants to look for powerful new methods.
In the 2016 CD challenge we implemented all the best methods proposed in the past editions. This resulted in 10 algorithms, which were evaluated fairly by CAGI organizers. We also used all the data available from CAGI 11 and 13 to maximize the amount of training samples. The most effective algorithms used known genes associated with CD from the literature. No method could evaluate effectively the importance of unannotated variants by using heuristics. As a downside, all CD datasets were strongly affected by sample stratification. This affected the performance reported by assessors. Therefore, we expect that future datasets will be normalized in order to remove population effects. This will improve methods comparison and promote algorithms focused on causal variants discovery.
Keywords: Disease Risk Prediction, Crohn Disease, Variants prioritization, SNV evaluation, Machine Learning, Linear Models, Methods Comparison, Genetic Analysis, Next-Generation Sequencing, Exome Data
Introduction
One of the main applications of next-generation sequencing is related to human health diagnostics. Although there are already solid demonstrations that disease causal variants could be identified (MacArthur et al., 2014), only a few studies have tried to build predictive models for disease risk and phenotype prediction (Weedon et al., 2006)(Morrison et al., 2007)(Giollo et al., 2015). The Critical Assessment of Genome Interpretation (CAGI) is the first effort aimed at objectively assessing the state of the art for genome interpretation. Introduced in 2010, in its four editions it proposed dozens of datasets where researchers could try to predict the effects of genetic variants. During each challenge, the CAGI organizers release unpublished data and formulate a specific question related to it. During a prediction season, participants can analyze the data and try to answer the challenge question. After this season, the CAGI assessors evaluate the quality of the submissions, and a conference is organized to discuss prediction performance and emerging ideas.
In the literature there are a vast number of bioinformatics tools available to perform predictions and risk assessments, mostly based on statistical methods and machine learning (Giollo et al., 2014). It is possible to build a disease risk estimation tool using the same principles, but the curse of dimensionality (Friedman) and limited sample size together represent a huge challenge in CAGI. The former issue is due to the high number of variants that can be observed in each sample, on the order of several thousands. Just a few of them are likely to be important for human health, but in most situations the key variants for a disease are unknown. Ideally, one should first perform feature selection (Saeys et al., 2007) in CAGI, with the aim to discard irrelevant variants for disease onset. This step is the main result of Genome-Wide Association Studies (GWAS) (WTCC Consortium, 2007) and linkage analysis (Easton et al., 1993), but there is still a huge number of variants that need to be annotated. Tools for pathogenicity prediction like SIFT (Sim et al., 2012) and PolyPhen2 (Adzhubei et al., 2010) can mitigate the problem just partially. In fact, these tools (1) only work on Single-Nucleotide Polymorphisms (SNPs), (2) have a limited accuracy (Thusberg et al., 2011) and (3) predict protein loss-of-function, which is not the same as predicting disease risk. The interaction among different variants (GAP & WTCC Consortium, 2010) and environmental relationships (Molodecky and Kaplan, 2010) are even harder to assess for a proper disease risk prediction. These problems must be considered in CAGI, but their solutions require a large sample size which is not available.
In this paper, we decided to focus on the CAGI Crohn disease (CD) challenges. Given the exomes of a few cases and controls, participants should predict their disease risk. CD is a multifactorial disease which has received vast attention in the last decade due to its burden on human health (Barrett et al., 2008). As a result, a considerable amount of prior knowledge can be found in the literature. CAGI proposed CD datasets in 2011, 2013 and 2016. This is a unique challenge that enables the study over time of the performance in this disease prediction problem. We participated successfully in all CD challenges, and during CAGI 2016 we tested all the best methods ever proposed for disease risk prediction. Here, we report the state-of-the-art in this challenge, and emphasize the key features of the most effective methods. We also highlight some issues related with all datasets and the proper evaluation of algorithm performance.
Materials and Methods
Datasets
CAGI published three different CD datasets over the last three editions. For each of them, the task was always the same. Prediction of a disease risk indicator based on exome data. Genotype sequences were collected from German patients, and part of them lead to the association of PRDM1 and NDP52 variants to CD (Ellinghaus et al., 2013). It is therefore clear that careful study can extract valuable knowledge from CAGI exomes. From the experimental point of view, Illumina instruments were used for sequencing. In 2011, reads were aligned with respect to the human genome build 18 (hg18), and base calling was obtained by a combination of BWA (Li and Durbin, 2009), Picard and SAMtools (Li et al., 2009). The main differences in the 2013 and 2016 editions were the introduction of GATK (DePristo et al., 2011) and the use of hg19. Finally, data was provided to CAGI participants as a VCF formatted file (see Table 1 for a summary).
Table 1. Summary of the CD challenge data.
| Cases | Controls | Ref. Genome | |
|---|---|---|---|
| CAGI 11 | 42 | 14 | hg18 |
| CAGI 13 | 51 | 15 | hg19 |
| CAGI 16 | 64 | 47 | hg19 |
Over time, the number of samples increased significantly, with special attention for controls in the latest edition. All exomes data is provided by Andre Franke and Britt-Sabina Petersen.
An interesting peculiarity of CAGI 13 was the presence of exomes from 28 pedigrees, which included a pair of monozygous discordant twins. The number of cases and controls was declared during the prediction season. In addition, during the last two editions data from the previous challenges could be used for training. This idea will be explored heavily over the next sections.
Algorithms
This section explains the ten algorithms used to prioritize exome variants and predict disease risk. These were implemented because they proved to be among the most effective methods in CAGI 11 and 13. The goal in CAGI 16 was to validate them on a new dataset. All implementations were written in R, with the intention of obtaining a fully automatic prediction. At first, coding variants with Minor Allele Frequency (MAF) < 0.04 were selected using information from dbSNP (Sherry et al., 2001) and a collection of BioConductor packages (Gentleman et al., 2004) (Obenchain et al., 2014) (Durinck et al., 2009). Let S ∈ {0, 1, 2, NA}n×m be the resulting matrix of exome variants, where n is the sample size and m represents the observed Single-Nucleotide Variants (SNVs). By construction, sij represents the number of variants at genomic position j for sample si. In other words, 0 and 2 are equivalent to observing twice the nucleotide with the major and minor allele frequency, respectively. A heterozygous variant is encoded with 1. Finally, NA is used to denote unobserved nucleotides in a sample, e.g. due to technical issues or different experimental setup.
Algorithms developed for CD risk prediction exploit either a weighting scheme or machine learning (ML). A weighting scheme w is used as a linear model. Positive weights correspond to pathogenic mutations, whereas negative coefficients are protective variants. By computing the dot product between a genotype si and w a disease risk can be computed. Machine Learning instead assumes that one can identify patterns to predict a disease risk from a training set, i.e. CAGI 11 and 13 CD data. Based on these two ideas, the following methods were tested.
Key variants weighting
this is the simplest form of weighting, which looks at the presence of a predetermined set of important SNVs to predict disease risk. These variants are given a weight of 1, while all other variants are set to 0. This is the typical model used for Mendelian diseases, when a single SNV is evaluated.
Odds Ratio weighting
GWAS estimated odds ratios for variants related to a disease (Beck et al., 2014). This is a risk measure of developing a disease based on direct associations on real data. Given this information, let define the weight wg as follows:
where ORg is the set of CD associated variants with known odds ratios in gene g. In other words, all variants sij within the same gene g will share the same average weight wg.
Publication weighting
genes related to CD are reported in the literature. Independent studies also corroborate some associations, providing additional belief in previous findings. As an example, 623 genes were linked CD (Yu et al., 2010) so far. 67% of them were reported just once, while NOD2 alone appears in 356 publications. Phenopedia (Yu et al., 2010) is a public database that stores the number of times cg that a gene g was linked to a disease in a scientific publication. In this case, the weighting scheme wg is defined as follows:
Once again, variants within the same gene share the same weight.
NA weighting
DNA sequencing techniques can measure a wide range of information, e.g. single SNVs, exomes and full genomes. Based on the experimental setup, some DNA regions are accessible or not detectable. On top of this, sequencing errors and computational limitations might lead to the impossibility of observing the nucleotides in some DNA regions. These are called Not Available (NA) variables, and pose big issues in data analysis. In this method, disease risk r for sample si is defined as
where NA(si) is the set of not observable variants in si.
Overrepresented weighting
more than 50% of the samples in CD challenges are expected to be cases. This proportion is much larger than the disease prevalence in Europe (Shivananda et al., 1996). Thus, variants that are overrepresented with respect to these in a reference populations (e.g. 1000 genomes (1000GP Consortium, 2012)) might be the causal CD mutations. In this method, dbSNP (Sherry et al., 2001) was used to obtain the minor allele frequency of variants. The overrepresented ones were identified using a binomial test. Bonferroni correction was applied to correct for the standard p-value threshold of 0.05. The variants selected in this way were given a weight of 1, and 0 for the others.
Bi-clustering
this is a type of unsupervised learning techniques that can divide automatically samples in two groups by looking at their reciprocal similarity. During CAGI, we used both k-means (with k = 2) and Hierarchical Clustering in order to solve this problem (Jain, 2010). We assumed that the smaller cluster is the one with healthy samples. Other insights from additional data sources were used to confirm this last hypothesis.
Transductive clustering
this method is based on the Transductive Learning principle (Chapelle et al., 2009). Similarity among samples was estimated using the cophenetic distance (Jain, 2010). Small clusters (n ≤ 5) of highly similar samples were identified. Within such groups, the average disease onset age of CD was estimated using knowledge from past CD editions (where available) and transferred to the CAGI 16 samples. Healthy samples were assumed to have a disease onset age of 1000, just to allow the computation of a trend within the group.
Manual prediction
by using the evidence gained from the previous methods, a manual assessment of each sample was performed. Clearly, this is not an algorithm.
Transductive SVM
to construct a suitable feature set for classification, an initial assessment of each variant sequence element was carried out against the CAGI 13 training data. The Fisher exact test was used to select variants associated with disease onset. Overall, 43 variants had a p-value lower than 5×10-6 and were used to build a machine learning classifier. CAGI 13 labels were attached to CAGI 15 samples using transductive learning (Chapelle et al., 2009). The fully labeled dataset was used to train an SVM classifier with RBF kernel.
Logistic Regression
Variant relevance was estimated using log-odds ratios on the CAGI 13 dataset to build a classifier. Only protective SNVs were selected by looking at negative log-odds scores and logistic regression algorithm was used train a classifier.
Ensemble
the disease risk was estimated by combining the previous techniques with bootstrap (Efron, 1992). In this case, the disease indicator of a given sample was equal to the proportion of methods that ranked it higher than the 67th percentile of all samples.
Performance Measures
The main task in CAGI was the definition of a method that could estimate a disease risk probability given the exome data. The CAGI assessors used Receiver Operating Characteristic (ROC) curves to identify the best submissions (Bradley, 1997). ROC represents the relationship between True Positive Rate (TPR) and False Positive Rate (FPR):
where TP and FP are the number of True Positives and False Positives, while P and N are the total number of Positive and Negative examples in the test set. The ROC curve integral provides the well-known Area Under the Curve (AUC) metric (Bradley, 1997). Accuracy (ACC) and Pearson correlations (COR) are also used to indicate the association between variables:
where TN are True Negatives, while o′ is the standard deviation of a population.
Results
Each CD challenge proposed during CAGI has peculiarities that should be addressed properly to maximize performance. In this section a description for all three CD datasets is presented, with considerations about the best method to use in each case.
CAGI 11
CAGI 11 proposed a disease risk challenge based on exome data for the first time. It represents an important event in establishing best practice that should be taken into account in similar tasks. Unfortunately, there was a critical issue in this challenge, which hampered a proper evaluation of submissions. Cases and controls in the dataset were collected using different experimental setups, which produced incomparable sequencing results. Cases appeared to be sequenced in a limited amount of exome regions, whereas controls were covered in a much wider range. As a result, one could observe a very large variation in the number of genetic variants reported depending on the two groups, which influenced most prediction algorithms and the submissions. Interestingly, the NA weighting strategy would pick-up this signal, and achieves a 95% classification accuracy. Given this huge bias, it is clear that bi-clustering can achieve the same results. An implementation of the publication weighting strategy by Yana Bromberg proved to be the most effective CAGI11 submission among all participants, leading to a nearly perfect ranking. However, it is very likely that this submission converged implicitly to NA weighting due to the dataset bias.
In this edition, only a manual strategy was implemented by our group, which was based on ANNOVAR (Wang et al., 2010) variants selection of rare SNVs (MAF < 0.04) for genes related to CD according to PheGenI (Ramos et al., 2014) and String (Szklarczyk et al., 2011). Samples with fewer variants were assumed to be controls (see Table 2). The five submissions were very similar in terms of reciprocal correlation, mainly because they were based on the same variant set. However, rs76982592 was the one that dominated all submissions. By just looking at its presence, according to the key variants weighting, one would achieve 89% accuracy.
Table 2.
Performance on the CAGI 11 dataset.
| Method | Ranking Details | AUC |
|---|---|---|
| Uniform weight | Count variants in CD genes. | 0.66 |
| SNV co-occurrence | Count twice variants that occurs frequently paired with others, according to association rules. | 0.678 |
| Bi-clustering | K-means on the variants. Rank according to the number of variants in CD genes. | 0.666 |
| Ensemble | Average of the previous | 0.626 |
| Manual | A manual evaluation of variants | 0.678 |
The five methods rely on 9 variants identified in PTPN11, DSPP, TDG, NCOA3, RBMX, PRKRA, ZFHX3, RUNX2 and CELA1 genes. Methods have a very high correlation, which ranged between 0.76 and 0.96.
CAGI 13
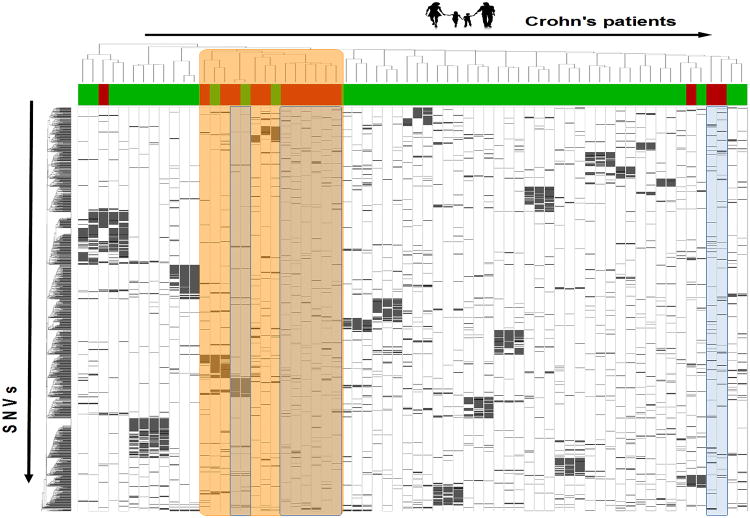
Given the experience from CAGI11, one might try to use the same idea on the CAGI 13 challenge. However, this was quite a unique dataset, with knowledge about (1) of the number of controls (Table 1), (2) 28 clear pedigrees, and (3) a pair of discordant twins. The CAGI 11 data was available for training as well. Given this specific setting, any CAGI11 strategy should be refined to achieve better performance. In fact, the plain publication weighting strategy was not as effective as in the previous challenge, probably due to the complexity of this structured dataset. Clustering is a key technique to highlight the 28 pedigrees and the twins (see Figure 1). The overall best submissions used bi-clustering and CAGI 11 as training data. Using this, 94% accuracy could be achieved, with 13 out of 15 controls detected. Overrepresented weighting implemented by Rita Casadio's group also proved to be very effective, even though this technique would be strongly biased by the presence of pedigrees.
Figure 1.
Heatmap of CAGI13 data. Columns represent the 66 dataset samples (red: controls, green: cases), grouped by genetic similarity using hierarchical clustering. Rows contain mutations clustered by sample similarity. The orange columns are outliers forming a sub-cluster with most of the controls. Prior knowledge is available for samples blue due to control-group membership in CAGI11.
In this challenge, we implemented the most effective strategy. Transductive clustering managed to classify effectively 8 samples (blue samples in Figure 1), as they proved to be very similar to CAGI 11 controls. The twins had mostly an identical set of variants, except for a variant in the MOC2 gene. This was assumed to be the causal mutation for CD in all submissions. After this initial screening, 9 out of 15 controls were detected. Interestingly, the majority of healthy samples are part of an outlier group, marked orange in Figure 1. In the best submission (Bi-clustering in Table 3), it was assumed that the blue and orange groups contained all controls. Within each group, disease risk was proportional to the number of variants – the same principle used in CAGI 11. The bi-clustering submission was somehow related to the same result seen in the CAGI11 dataset. Healthy samples were very dissimilar with respect to the CD ones, probably due to a different experimental setup. Transductive clustering confirmed that this group was largely composed of controls, boosting significantly belief in this submission. It is still unclear if the controls were actually reused in the two challenges. However, CAGI 13 confirmed once again the need for better controls in a proper challenge setup.
Table 3.
Performance on the CAGI 13 dataset.
| Method | Ranking Details | AUC |
|---|---|---|
| Uniform weight 1 | Count SNVs in CD genes. | 0.743 |
| Uniform weight 2 | Count SNVs in CD genes and their STRINGdb interactors. | 0.736 |
| Mixed pedigree 1 | Max one control per family. Count SNVs in CD genes. | 0.844 |
| Mixed pedigree 2 | Max one control per family. Count pathogenic SNVs (according to SIFT) in CD genes. | 0.688 |
| Mixed pedigree 3 | Max one control per family. Look for families with large difference in SNVs count in CD genes. | 0.798 |
| Bi-clustering | Bi-clustering on the dataset. Rank according to the number of SNVs in CD genes. | 0.866 |
The submissions have a high correlation, ranging between 0.67 and 0.89. This is mainly due to (1) a similar set of variants selected for all methods and (2) the use of transductive clustering from CAGI 11. The use of clustering is critical to maximize performance.
CAGI 16
This last challenge is probably the hardest ever proposed in CAGI. Samples are apparently quite uniform and there are no obvious issues with experimental settings, like the one described for CAGI 11 or any prior information available like in CAGI 13. The dataset size is also much larger compared to previous editions (see Table 1). Transductive clustering was not very effective, since it could match just a single sample as a control. In addition, just two pedigrees could be detected in the entire dataset.
CAGI16 is hence composed of independent cases and controls, with basically no relationship to past challenges. This is a good dataset to validate the methods that proved to be most effective in previous challenges. The simplest approach to look for data bias is NA weighting. As can be seen in Table 4, it would be probably one of the best methods, with an AUC of 0.7. Methods dealing explicitly with missing data, like variable imputation, are likely to exploit the same source of information. The three methods using published SNVs associated to CD are significantly effective, with an AUC ranging between 0.59 and 0.61. Key variants weighting worked well with rs2066844, a SNP known to increase significantly CD risk (Ningappa et al., 2011). On the other hand, Odds Ratio weighting proved that published GWAS variants are informative. Publication weighting and Overrepresented weighting results were not statistically significant, suggesting that ad-hoc weighting strategies are not effective. Similar findings were highlighted by Marco Carraro work, which showed that key variants weighting could achieve good performance when it combines multiple CD variants reported in clinical studies. However, his submissions based on manual weighting of a larger set of SNVs lead to a decrease in discriminative power (down to 0.59).
Table 4.
Results of the methods on CAGI 16 dataset.
| Method | AUC | Pearson Correlation |
|---|---|---|
| NA weighting | 0.7 * | 0.36 * |
| Publication weighting | 0.56 | 0.05 |
| Key variants weighting (rs2066844) | 0.59 * | 0.23 * |
| Overrepresented weighting | 0.47 | -0.06 |
| Transductive clustering | 0.52 | 0.06 |
| Odds Ratio weighting | 0.59 * | 0.14 |
| Manual prediction | 0.63 * | 0.2 * |
| Transductive SVM | 0.6 * | 0.2 * |
| Logistic regression | 0.57 | 0.16 |
| Ensemble | 0.66 * | 0.29 * |
| Key variants weighting (clinical studies SNVs) | 0.61 * | 0.21 * |
Performances marked with a star are statistically significant.
Important, tested methods rely on very different assumptions. Key variants weighting and Odds Ratio weighting are the most similar by design, with a correlation of 0.37. Nevertheless, this value is much lower than any method tested in the previous challenges. All other methods have a correlation between -0.1 and 0.26, proving that these strategies explore more divergent hypotheses than in the former CD challenges. Finally, NA weighting is apparently strongly related to bi-clustering on the dataset with a 0.74 correlation between both.
David Jones tested machine learning effectiveness on this dataset. Transductive SVM and Logistic regression were used, with statistically significant results for the former. This is probably due to the clustering assumption for this specific type of algorithm. An ensemble prediction was realized with bootstrap, where the disease risk of a given sample was estimated as the amount of methods in agreement for a high risk.
Discussion
Present over the last three editions, the Crohn disease challenge represents well the idea behind CAGI: developing novel tools for genetic interpretation. During these years, the organizers proposed ever larger datasets where methods could be tested more accurately. The prior knowledge provided to participants and the sequencing pipelines varied slightly over time, but it is still obvious that the CD challenge is the only one enabling an analysis over time in this context. There are a few lessons that are clear from the reported results.
Our goal in this CAGI 16 edition was to test thoroughly all top performing approaches previously proposed in CAGI, implementing from scratch 10 methods. Many of these were previously proposed by other participants, so we could implement the core ideas based on our understanding.
From the results, weighting schemes managed to assign proper importance to exome variants only when numerical coefficients were based on solid studies like GWAS. SNP rs2066844 is a clear example, as it is well known to be associated to CD and it was indeed a powerful discriminative variable in CAGI 16. No heuristic could address effectively this problem in a similar way. From a perspective of machine learning methods, use of CAGI 11 data was critically important in CAGI 13 for the best submissions. During CAGI 16, past training sets were not as useful as in CAGI 13, probably due to the high dissimilarity of new samples. It is clear that ML is very effective in simple scenarios where samples share high similarity. However, new methods for improved sample comparison are needed to boost the performance of ML algorithms. Overall, both weighting schemes and machine learning performance are limited in the same way, due to the curse of dimensionality. To deal with this issue it is critical to use training sets with a large number of samples. This would help with the selection of key variants and proper estimation of their effect on disease onset. This idea was already explored using data collected during the International IBD Genetics Consortium's Immunochip project (Wei et al., 2013), where a simple regularized logistic model achieved an AUC of 0.86 on a very large CD test set. Interestingly, models based on Support Vector Machines and Gradient Boosted Trees decreased the predictive performance, suggesting that more complex algorithms are overfitting the data. Most of CAGI methods do not use any training set at all, mainly due to the efforts needed to request such controlled datasets to Data Access Committees. As a result, a significant number of CAGI submissions were no better than random. We believe that the main improvements in CD prediction will be enabled by just using regularized additive data-driven models We therefore hope to see a simplified access to large datasets (Wei et al., 2013) for all CAGI participants in the future.
The huge number of variables in all datasets is the main motivation for the use variant selection tools like ANNOVAR. This tool was heavily used in CAGI 11 as a black box, where its filtering process lead to the variants reported in Table 2. Variant rs76982592 was the one with the highest impact given our weighting strategy. Just at the time of the first CAGI conference, it became clear that the variant was strongly associated with the different experimental setup. This is surely an important contribution of CAGI highlighting a bi-clustering pattern in the data. ANNOVAR was also used in CAGI 13 for the annotation step. In that edition the tool was used carefully, as its blind usage might filter away important disease variants. Bi-clustering was once again an effective method. ANNOVAR filtering was finally replaced completely by a collection of R packages in CAGI 16. Having an in-house tool for variant selection helped to move from manual predictions, like in CAGI 11, to a fully automated pipeline where human decisions are limited. This is a key step for reproducibility, which is achievable with a full control and understanding of the tools at hand. In this implementation, it was much simpler to identify the relationship between NAs and bi-clustering, ANNOVAR did not emphasize at all the unobserved variables. BioConductor packages allowed controlling in detail the entire filtering step, and learning more about the samples.
Over these three editions, best method performances kept decreasing from an AUC of ∼0.9 to an AUC of ∼0.7. This negative trend is probably unexpected, but we believe that this is the result of an improper performance evaluation, especially in the first CAGI editions. We believe that all submission results reported so far are not truly representative of our current ability to predict disease risk, but they are inflated due to the strong bias in the datasets. Bi-clustering appeared to be an effective method for predicting disease risk in CAGI datasets, but it is unlikely that this approach would work well in a real-world context. Case-control separation is in fact induced largely by NA variants and experimental issues. In well-designed genetic studies, a data normalization step typically adjusts the dataset for population stratification issues. NA variants are also addressed in this step, as they may lead to spurious associations. By evaluating submissions through a crude ranking of samples with AUC, CD submissions that exploit patient stratification (intentionally or not) will be the most effective. Therefore, it is important to improve CD challenge either by normalizing the dataset provided to participants or by asking for a set of causal SNVs in the submission. The former step is probably easier to implement, so we hope to see this improvement in future CAGI editions. By doing so, we believe that submission performance will reduce even further, leading to a truly effective validation of our current ability to predict disease risk. A well-structured dataset would also be a step forward to promote automated methods for disease risk prediction, as this should remove the manual analysis used to detect irrelevant facts like twins or pedigrees.
Overall, CAGI was successful in increasing the attention toward genome interpretation. In CAGI 11, many participants tested a number of approaches. Many submissions were much worse than random (AUC < 0.5), suggesting that there was a substantial lack of expertise in the field. Over time, the assessor and participant talks shed light on the common pitfalls and shared the most effective ideas. This has increased remarkably our understanding of this field, and raised the interest of many researchers. Even though submission results are currently inflated, we believe that CAGI organizers are also in the process of learning how to evaluate properly disease risk predictions, and there are positive signals of improvement in this direction. Therefore, we believe that with the next editions, and with bigger and well normalized datasets, CAGI will play a role in evaluating and testing innovative methods and provide novel ideas for disease risk evaluation.
Acknowledgments
The data providers Prof. Dr. Andre Franke & Dr Britt-Sabina Petersen, Institute of Clinical Molecular Biology, Christian-Albrechts-University Kiel, Germany.
The CAGI experiment coordination is supported by NIH U41 HG007446 and the CAGI conference by NIH R13 HG006650.
The project was partially supported by the Italian Ministry of Health grants GR-2011-02347754 (to EL and ST) and GR-2011-02346845 (to ST).
References
- 1000G P Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, Hastings RK, Gollapudi S, Free RC, Brookes AJ. GWAS Central: a comprehensive resource for the comparison and interrogation of genome-wide association studies. Eur J Hum Genet. 2014;22:949–952. doi: 10.1038/ejhg.2013.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997;30:1145–1159. [Google Scholar]
- Chapelle O, Scholkopf B, Eds AZ. Semi-Supervised Learning. In: Chapelle O, et al., editors. IEEE Trans Neural Networks. Vol. 20. 2009. pp. 542–542. 2006 [Book reviews] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- Efron B. Bootstrap Methods: Another Look at the Jackknife. In: Kotz S, Johnson NL, editors. Breakthroughs in Statistics. Springer; New York: 1992. pp. 569–593. [Google Scholar]
- Ellinghaus D, Zhang H, Zeissig S, Lipinski S, Till A, Jiang T, Stade B, Bromberg Y, Ellinghaus E, Keller A, et al. Association between variants of PRDM1 and NDP52 and Crohn's disease, based on exome sequencing and functional studies. Gastroenterology. 2013;145:339–347. doi: 10.1053/j.gastro.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JH. On Bias, Variance, 0/1—Loss, and the Curse-of-Dimensionality. Data Min Knowl Discov. 1:55–77. [Google Scholar]
- GAP & WTCC Consortium. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giollo M, Martin AJ, Walsh I, Ferrari C, Tosatto SC. NeEMO: a method using residue interaction networks to improve prediction of protein stability upon mutation. BMC Genomics. 2014;15:1–11. doi: 10.1186/1471-2164-15-S4-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giollo M, Minervini G, Scalzotto M, Leonardi E, Ferrari C, Tosatto SCE. BOOGIE: Predicting Blood Groups from High Throughput Sequencing Data. PLoS ONE. 2015;10:e0124579. doi: 10.1371/journal.pone.0124579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK. Data clustering: 50 years beyond K-means. Pattern Recognit Lett. 2010;31:651–666. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodecky NA, Kaplan GG. Environmental Risk Factors for Inflammatory Bowel Disease. Gastroenterol Hepatol. 2010;6:339–346. [PMC free article] [PubMed] [Google Scholar]
- Morrison AC, Bare LA, Chambless LE, Ellis SG, Malloy M, Kane JP, Pankow JS, Devlin JJ, Willerson JT, Boerwinkle E. Prediction of Coronary Heart Disease Risk using a Genetic Risk Score: The Atherosclerosis Risk in Communities Study. Am J Epidemiol. 2007;166:28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- Ningappa M, Higgs BW, Weeks DE, Ashokkumar C, Duerr RH, Sun Q, Soltys KA, Bond GJ, Abu-Elmagd K, Mazariegos GV, et al. NOD2 Gene Polymorphism rs2066844 Associates With Need for Combined Liver–Intestine Transplantation in Children With Short-Gut Syndrome. Am J Gastroenterol. 2011;106:157–165. doi: 10.1038/ajg.2010.322. [DOI] [PubMed] [Google Scholar]
- Obenchain V, Lawrence M, Carey V, Gogarten S, Shannon P, Morgan M. VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics. 2014;30:2076–2078. doi: 10.1093/bioinformatics/btu168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos EM, Hoffman D, Junkins HA, Maglott D, Phan L, Sherry ST, Feolo M, Hindorff LA. Phenotype–Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur J Hum Genet. 2014;22:144–147. doi: 10.1038/ejhg.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivananda S, Lennard-Jones J, Logan R, Fear N, Price A, Carpenter L, van Blankenstein M. Incidence of inflammatory bowel disease across Europe: is there a difference between north and south? Results of the European Collaborative Study on Inflammatory Bowel Disease (EC-IBD) Gut. 1996;39:690–697. doi: 10.1136/gut.39.5.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, McCarthy MI, Hitman G, Walker M, Groves CJ, Zeggini E, Rayner NW, Shields B, Owen KR, Hattersley AT, et al. Combining Information from Common Type 2 Diabetes Risk Polymorphisms Improves Disease Prediction. PLOS Med. 2006;3:e374. doi: 10.1371/journal.pmed.0030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Wang W, Bradfield J, Li J, Cardinale C, Frackelton E, Kim C, Mentch F, Van Steen K, Visscher PM, et al. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Hum Genet. 2013;92:1008–1012. doi: 10.1016/j.ajhg.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WTCC Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Clyne M, Khoury MJ, Gwinn M. Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26:145–146. doi: 10.1093/bioinformatics/btp618. [DOI] [PMC free article] [PubMed] [Google Scholar]