Abstract
Spatial organization of the genome plays a central role in gene expression, DNA replication, and repair. But current epigenomic approaches largely map DNA regulatory elements outside of the native context of the nucleus. here we report assay of transposase-accessible chromatin with visualization (ATAC-see), a transposase-mediated imaging technology that employs direct imaging of the accessible genome in situ, cell sorting, and deep sequencing to reveal the identity of the imaged elements. ATAC-see revealed the cell-type-specific spatial organization of the accessible genome and the coordinated process of neutrophil chromatin extrusion, termed NETosis. integration of ATAC-see with flow cytometry enables automated quantitation and prospective cell isolation as a function of chromatin accessibility, and it reveals a cell-cycle dependence of chromatin accessibility that is especially dynamic in G1 phase. the integration of imaging and epigenomics provides a general and scalable approach for deciphering the spatiotemporal architecture of gene control.
Eukaryotic genomes are extensively compacted in chromatin, except for active regulatory elements, which are accessible and control gene activity. These accessible elements comprise approximately 1% of the genome in any given cell type; they include enhancers, promoters, and other regulatory sequences whose proper control is critical in development and disease1. Nuclear architecture and 3D genome organization influence gene expression, replication, and DNA repair2. Despite recent advances3–6, current epigenomic methods extract regulatory DNA from its native context in the nucleus and reconstruct regulation on an imaginary linear genome that lacks the intricate spatiotemporal organization seen in movies of living cells7,8.
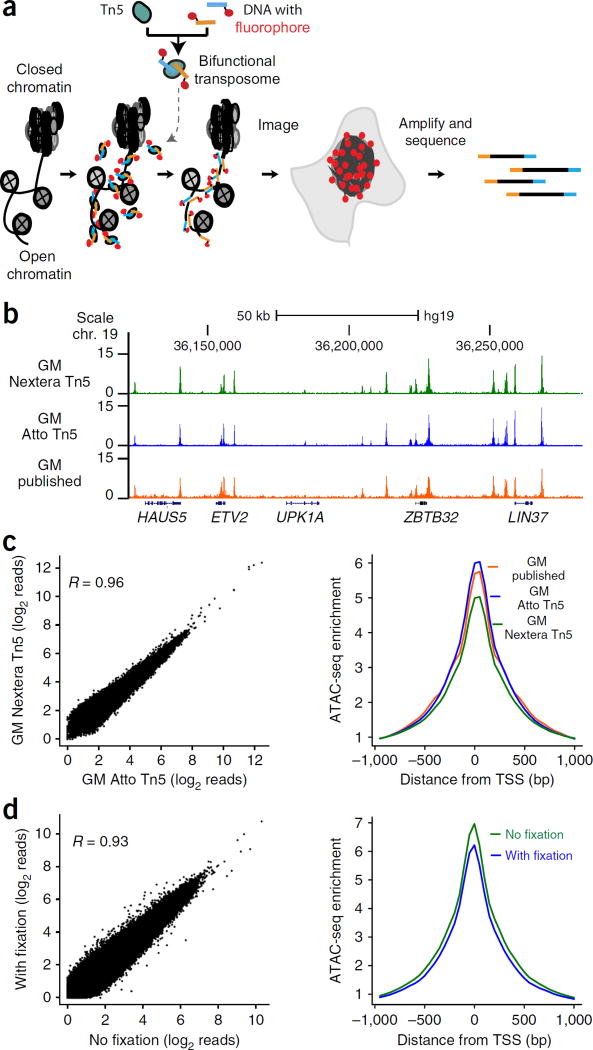
We previously reported assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq), a method in which hyperactive Tn5 transposase loaded with DNA adaptors selectively inserts the adaptors into accessible chromatin loci within living cells3. Deep sequencing of adaptor-tagged sites reveals active regulatory elements, transcription factor binding sites, and nucleosome positions3. We reasoned that if transposases could insert fluorescent DNA adaptors into accessible chromatin loci, then we could use these transposases to selectively and covalently insert fluorophores at open chromatin sites genome wide (ATAC-see, Fig. 1a). By fluorescently labeling accessible loci, ATAC-see transforms the molecular accessibility of chromatin into points of light. After imaging the spatial organization of the accessible genome in 3D, the inserted adaptors allow deep sequencing to map open chromatin sites on the same sample in a manner identical to that of ATAC-seq. Here we report the development of ATAC-see and several of its possible applications.
Figure 1.
ATAC-see visualizes the accessible genome in situ. (a) Schematic of ATAC-see. (b) Genome browser tracks of ATAC-seq libraries from GM12878 cells (GM) generated by using different Tn5 transposases: green, Nextera Tn5; blue, Atto Tn5; orange, previously published data3. A 50-kb scale bar and genome locations are indicated at the top of the tracks; gene names (HAUS5, etc.) are shown at the bottom. Chr, chromosome; hg19, human genome. (c) Left panel, genome-wide comparison of ATAC-seq reads of GM12878 libraries (GM) prepared by using either Nextera Tn5 or Atto Tn5. Right panel, TSS enrichment of GM12878 ATAC-seq libraries transposed with different Tn5. Green, Nextera Tn5; blue, Atto Tn5; orange, previously published data3. (d) Genome-wide comparison of ATAC-seq reads of HT1080 library prepared with or without fixation. Left, scatter plot of all data points; right, metagene analysis centered on transcriptional start sites (TSS).
RESULTS
ATAC-see for regulatory DNA imaging and sequencing
We designed, produced, and optimized a bifunctional Tn5 transposome with fluorescent adaptors (see Online Methods). Recombinant purified hyperactive Tn5 transposase loaded with Atto590 fluorophore-conjugated adaptors (Atto–Tn5) retained the activity and selectivity of the standard unlabeled Tn5 transposome (Nextera Tn5; Fig. 1b,c and Supplementary Fig. 1a–f). ATAC-seq of human B cells (GM12878) showed that the DNA accessibility profile generated with Atto–Tn5 is similar to that obtained using Nextera Tn5 (R = 0.96; Fig. 1c). Moreover, standard quality control metrics such as DNA insert length distribution, transcriptional start site (TSS) enrichment, and genomic distribution of ATAC-seq peaks were also similar among Atto–Tn5, Nextera Tn5, and published ATAC-seq data from the same cell line3 (Fig. 1c and Supplementary Fig. 1a–e). Comparison of Atto–Tn5 with Nextera Tn5 showed no greater variation between the two than between technical replicates of Nextera Tn5 (Supplementary Fig. 1f), suggesting that the bifunctional Tn5 transposome bearing molecular tags does not affect transposase activity. Our labeling strategy is general, and it is compatible with strategies using different fluorophores and other chemical tags (data not shown).
Because cellular fixation is a common and essential step to preserve nuclear architecture and cellular composition, we next developed methods to perform ATAC-seq in crosslinked samples. Transposition of formaldehyde-fixed cells produced biased libraries with short DNA fragments3 (Supplementary Fig. 2a,b). We optimized a reverse crosslinking method compatible with ATAC-seq (see Online Methods). ATAC-seq data from HT1080 fibrosarcoma cells fixed in this manner are comparable to data from standard ATAC-seq of living cells (R = 0.93; Fig. 1d and Supplementary Fig. 2c–f), indicating that fixation does not affect the Tn5 tagmentation efficiency in the intact nucleus. Engineering the transposome to label accessible DNA in fixed samples set the stage for sequentially imaging and sequencing the accessible genome.
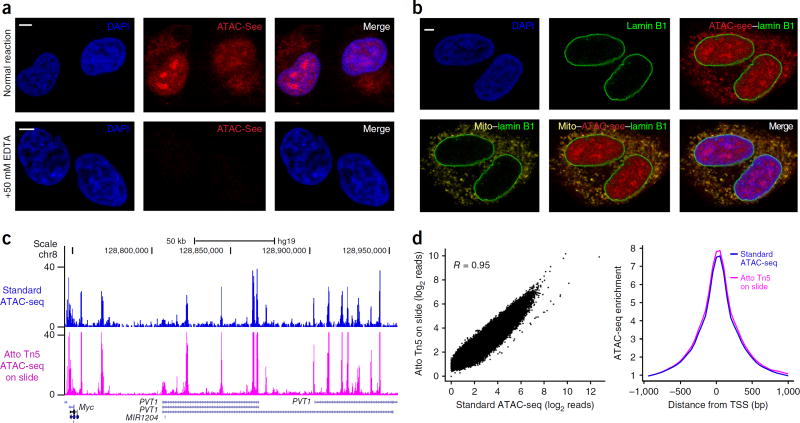
ATAC-see experiments revealed the 3D spatial organization of accessible DNA in situ. Adherent HT1080 cells were fixed by formaldehyde per standard immunofluorescence (IF) on glass slides, and they were reacted with Atto–Tn5. Samples were then stringently washed, counterstained for total DNA with 4’, 6-diamidino-2-phenylindole (DAPI), and imaged with confocal microscopy. The resulting images showed that the accessible chromatin is heterogeneously distributed throughout the nucleus and distinct from DAPI signal (which demarcates tightly packed DNA). ATAC-see signal was concentrated into several nuclear foci in some but not all cells (Fig. 2a, Supplementary Fig. 3a, and Supplementary Video 1). This result suggests that the accessible genome forms higher order structure in the intact nucleus; these foci of the accessible genome may reflect the open-chromatin-rich ‘A compartment’ detected by chromosome conformation capture techniques6. Addition of 50 mM EDTA that inactivates Tn5 activity3,9–11 greatly reduced the ATAC-see signal to 2.69% ± 0.04% of the control signal (Fig. 2a and Supplementary Fig. 3a). Thus, the ATAC-see signal shows the integration of fluorophore-labeled adaptors into accessible genomic DNA.
Figure 2.
ATAC-see enables imaging and sequencing of the accessible genome in the same cells. (a) Representative image of ATAC-see result in HT1080 cells. Very limited ATAC-see signal is observed in +EDTA negative control (quantified in supplementary Fig. 3a). Merge, merged image of DAPI and ATAC-see. Scale bars, 2 µm. (b) Multimodal imaging combining ATAC-see with immunofluorescence. The representative images employ costaining with nucleus lamin B1 and/or mitochondrial protein marker (Mito) to show ATAC-see signals overlapping with mitochondria outside of the nucleus. Scale bar, 2 µm. (c) Genomic tracks of ATAC-seq data from standard protocol and Atto-Tn5 on slide after imaging. x-axis, genomic coordinates; y-axis, normalized ATAC-seq read counts. 50-kb scale bar and genome locations are indicated at the top; gene names (Myc, etc.) are shown at the bottom. Chr, chromosome; hg19, human genome. (d) Genome-wide comparisons of standard ATAC-seq data with Atto Tn5 on-slide data after imaging. Left, scatter plot of all data points; right, metagene analysis centered on transcriptional start sites (TSS).
We developed conditions for multimodal labeling and imaging with ATAC-see. ATAC-seq is known to capture mitochondrial DNA (which is not chromatinized). Four-color imaging combining ATAC-see, DAPI, and IF of lamin B1 and a mitochondrial protein marker clearly delineated the accessible genome in the nucleus and revealed a strong overlap between mitochondrial and ATAC-see signal outside of the nucleus (Fig. 2b and Supplementary Fig. 3b). Combined ATAC-see and IF with four histone marks and two forms of RNA polymerase II (RNAPII ser-2 and ser-5 phosphorylation) showed strong colocalization of ATAC-see in nuclear territories with active marks H3K4me3, H3K27ac, and RNAPII (R ≈ 0.80) but absent or negative correlation with repressive marks H3K27me3 and H3K9me3 (R = 0 to –0.20; Supplementary Fig. 3c,d). To further confirm that ATAC-see specifically labels accessible chromatin, we combined ATAC-see with Xist RNA fluorescent in situ hybridization (FISH) (which marks the inactive X chromosome) in female cells. We found that the Xist RNA cloud lay within a ‘hole’ of reduced ATAC-see signal; the ATAC-see signal was 2.3- ± 0.14-fold lower within the Xist domain that outside of it (P < 0.005; Supplementary Fig. 3e). These results validate the specificity of ATAC-see in localizing active regulatory elements and, by exclusion, heterochromatin in the nucleus. The ability to combine ATAC-see with the extensive toolkits of IF imaging suggests that ATAC-see can be easily adopted by researchers and can impact a broad range of biomedicine.
To perform sequencing after imaging, we developed an on-slide lysis procedure compatible with ATAC-see samples (see Online Methods and Supplementary Fig. 4a). ATAC-seq data obtained from a sample after ATAC-see imaging of the same sample are highly correlated with standard ATAC-seq data from parallel cell samples (R = 0.95; Fig. 2c,d and Supplementary Fig. 4b). In addition, differential peaks analysis of ATAC-seq peaks showed that the variation between Atto–Tn5 and Nextera Tn5 was no greater than the variation between technical replicates of Nextera Tn5 (Supplementary Fig. 4c). On-slide and fixed ATAC-see with serial cell dilution showed highly reproducible mapping of DNA accessibility with input ranging from 50,000 to 500 cells (Supplementary Fig. 4d). Sequential imaging and accurate mapping of the open-chromatin landscape by sequencing suggested that ATAC-see captured a comprehensive portrait of the spatial organization of the accessible genome.
Cell-type-specific spatial organization of the accessible genome
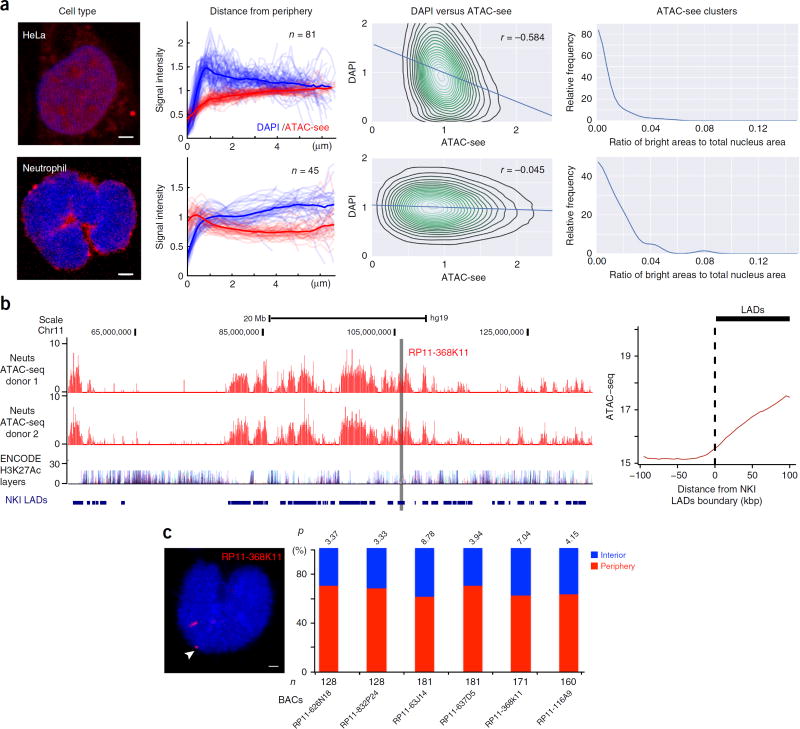
ATAC-see of five human cell types revealed that chromatin accessibility is spatially organized in a cell-type-specific fashion with overlaid single-cell variation (Fig. 3a; Supplementary Fig. 5a,b; and Supplementary Videos 1–5). Nuclear architecture is hierarchically organized into distinct compartments, topological domains, and chromosome loops1,12. Heterochromatin is believed to be physically condensed and is readily labeled by DAPI staining; and it typically resides near the nuclear periphery13. In contrast, euchromatin contains accessible regulatory elements and active genes, and it tends to be located in the nuclear interior1,10,14. We thus hypothesized that ATAC-see signal would be more prominent in the nuclear interior and anticorrelated with the DAPI signal in individual nuclei. We carefully masked out mitochondrial contribution to ATAC-see signal (Supplementary Fig. 6a). In HeLa cells, ATAC-see signal increased gradually from the nucleus periphery to the interior, and it was inversely correlated with DAPI staining (R = –0.584; Fig. 3a, Supplementary Fig. 5a, and Supplementary Video 2). However, analyses of additional cell types with the same strategy showed multiple exceptions to this simplistic picture. Primary human CD4+ T cells exhibited lower anticorrelation between ATAC-see and DAPI signal (R = –0.243), and the intranuclear distribution of the ATAC-see signal varied dramatically from cell to cell (Supplementary Fig. 5a and 7). In addition, some but not all CD4+ T cells had a cruciform pattern of clustered foci of ATAC-see signal (Supplementary Fig. 7 (right panel), Supplementary Fig. 5a, and Supplementary Video 3). Approximately 40% of CD4+ T cells had a strong ATAC-see signal in a semicircular ‘cap pattern’ at the nuclear periphery (Supplementary Fig. 6b and 7). B-lymphoblastoid GM12878 cells also exhibited single-cell ATAC-see variation, but they lacked intranuclear clusters of ATAC-see foci (Supplementary Fig. 5a and 7, Supplementary Video 4). In contrast, adherent HT1080 fibrosarcoma cells more closely resembled HeLa cells in the segregation of DAPI-rich heterochromatin to the nuclear periphery and ATAC-see to the nuclear interior, with the notable distinction that prominent nuclear clusters of ATAC-see foci were present in HT1080 cells but not in HeLa cells (Supplementary Fig. 5a and 7, Supplementary Video 1).
Figure 3.
Cell-type-specific accessible-chromatin organization in the intact nucleus. (a) Image analysis. For each cell type (organized in rows), we display four columns (i–iv, left to right): (i) a representative ATAC-see image (red, ATAC-see; blue, DAPI; scale bar, 2 µm); (ii) signal intensity of ATAC-see and DAPI as a function of distance from nuclear periphery. Each trace is one nucleus, and n = number of nuclei analyzed; (iii) correlation of ATAC-see and DAPI signal intensity. Pearson correlation (r) is indicated; (iv) ATAC-see clusters, quantified as the ratio of ATAC-see bright areas versus the total nucleus area. (b) Unique feature of ATAC-seq from human neutrophil (Neuts) after imaging. Left, genome browser track of ATAC-seq in human neutrophil, H3K27Ac layer from ENCODE 7 cell lines, and lamin-associated domains (LADs) published by the Netherlands Cancer Institute (NKI). x-axis, genomic coordinates; y-axis, ATAC-seq-normalized read counts. The gray line indicates the location of bacterial artifical chromosome (BAC) chosen for DNA FISH in c. Right, metagene plot of human neutrophil ATAC-seq signal centered on the boundary between NKI LADs and neighboring sequences. The dashed black line indicates the boundary of LADs, and the thick black line on the top of the graph presents the distance to LADs boundary. (c) Left, example of DNA FISH from indicated BAC (in b) in human neutrophil; the white arrow indicates the FISH signal (red) at the neutrophil periphery. Right, location quantification of DNA FISH signal (percentage in y-axis) from the indicated BACs (x-axis) in the human neutriophil. The nucleus periphery was defined as a distance between DNA FISH signal to DAPI staining edge <0.1 µm (see Online Methods). n, number of alleles counted in DNA FISH. P values were calculated by a binomial test.
Neutrophils are abundant immune cells that play important roles in infection control and in inflammation15; they also have distinctive multilobulated nuclei. ATAC-see of primary human neutrophils revealed a striking and unique organization of the accessible genome—the vast majority of the ATAC-see signal in neutrophils was located at the nucleus periphery and formed a rim structure (Fig. 3a; Supplementary Fig. 5a and 8a; and Supplementary Video 5). Neutrophil ATAC-see signal intensity decreases from the nuclear periphery to the interior, and there is little correlation between ATAC-see and DAPI signals (R = –0.045). The rim pattern of ATAC-see signal is not caused by mitochondria, as was evidenced by the distinct staining pattern of a mitochondrial marker in the same cells (Supplementary Fig. 8a and Supplementary Video 5). ATAC-seq of the same samples after imaging confirmed that more than 99% of ATAC-see signal arises from genomic DNA. In addition, neutrophils have ~50-fold fewer focal peaks of chromatin accessibility than do other cell types with similar sequencing depth and alignment rates16 (Supplementary Fig. 8b). Instead, neutrophils harbor large multi-kb blocks of accessible chromatin that are inversely correlated with H3K27Ac from seven ENCODE cell lines but significantly correlated with lamina-associated domain (LADs) (R = 0.31 genome wide, P < 0.0001; Fig. 3b and Supplementary Fig. 8c)17. DNA FISH validated that six of six genomic loci indicated by ATAC-see were at the nuclear periphery in neutrophils (Fig. 3c and Supplementary Fig. 8d,e).
The significant correlation between LADs in other cell types and ATAC-seq reads in neutrophils suggests the LADs in the neutrophil are now made accessible. Imaging showed that primary human neutrophils downregulated lamin B1 protein concomitantly with placement of the accessible genome at the nuclear periphery (Supplementary Fig. 8a). Our results also suggest that lamin B1 is not required for the peripheral localization of LADs, an observation that is consistent with another study18. Next we tested whether neutrophil invert their nuclear architecture as do the rod photoreceptor cells19; however, the spatial distribution of euchromatic marks and heterochromatic marks were localized in the expected pattern in neutrophil nuclei (Supplementary Fig. 9a). Thus, human neutrophils have significant transposase-accessible DNA at the nuclear periphery, but this accessible DNA does not bear the standard euchromatic marks. Collectively, these observations suggest that distinct human cell types often have a distinctive and diverse spatial organization of the accessible genome (Fig. 3a, Supplementary Fig. 5b and 7), typified here by human neutrophils.
ATAC-see uncovers the chromatin dynamics of NETosis
We next explored how the unusual spatial organization of the accessible genome in neutrophils might facilitate neutrophil functions. When mature neutrophils encounter bacteria in blood or tissue, neutrophils can release their chromatin and kill bacteria, a unique form of programmed cell death termed neutrophil extracellular traps (NETosis)20. NETosis is also believed to contribute to human inflammatory diseases, but it is not known whether the genome is randomly fragmented or processed in an organized manner for NET release. We reasoned that prepositioning of open DNA at the nuclear periphery might prepare neutrophils for initiation of NETosis. We combined molecular imaging and epigenomic sequencing by ATAC-see and ATAC-seq of the same cells and discovered two key steps in NETosis (Fig. 4).
Figure 4.
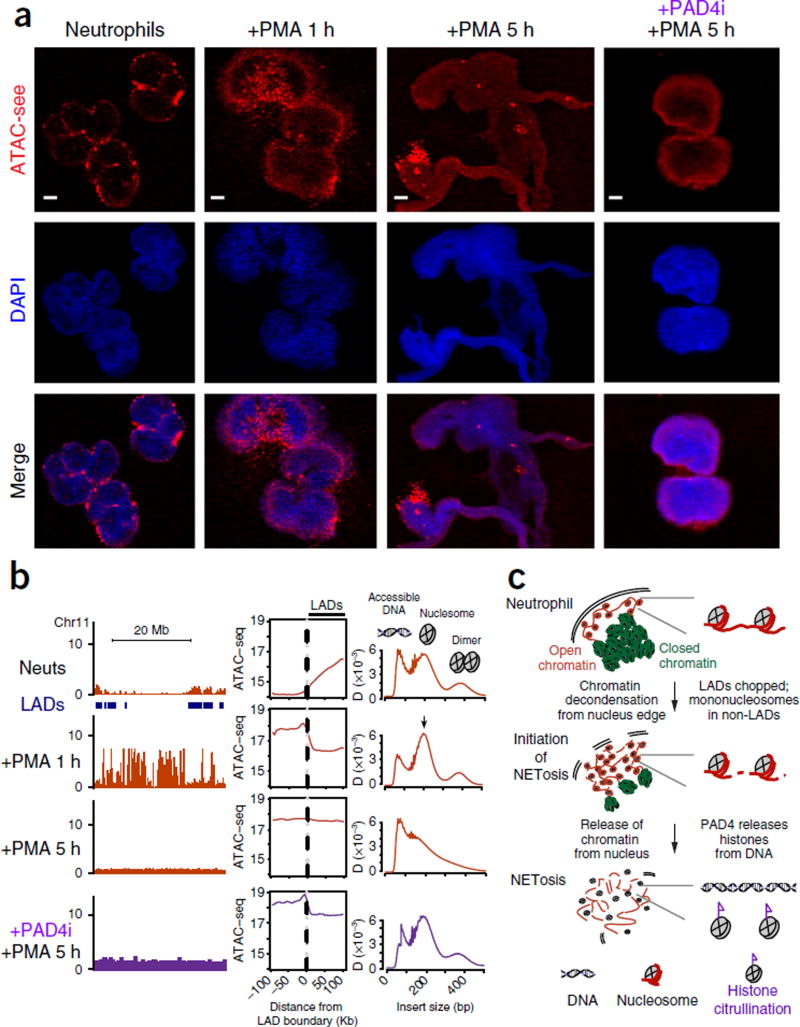
ATAC-see and ATAC-seq reveal the dynamic chromatin organization during human NETosis. DAPI, stain used to stain the nucleus; merge, merged image of DAPI and ATAC-see. (a) Representative ATAC-see images in the indicated conditions. PMA stimulation timecourse, PAD4 inhibitor treatment (PAD4i). Scale bars, 2 µm. (b) Epigenomic landscape of NETosis. Left column, genomic tracks of ATAC-seq data from the indicated conditions. Locations of NKI lamin-associated domains (LADs) are indicated. The x-axis represents genomic coordinates; the y-axis represents ATAC-seq normalized read counts. Middle column, metagene plot of ATAC-seq signal centered on the boundary between NKI LADs and neighboring sequences. The top plot is the same as the right panel in Figure 3b and is reproduced here for clarity as the baseline in this timecourse. Right column, ATAC-seq insert size distribution for the corresponding samples. Diagnostic insert sizes for accessible DNA, mononucleosome, and dinucleosome are labeled. (c) Proposed model of NETosis illustrates the coordinated dynamics of nuclear architecture, accessible genome reorganization, and genome disassembly.
First, upon neutrophil activation, the LADs located at the nuclear periphery serve as focal points for chromatin disassembly into mononucleosomes. We stimulated primary neutrophils with phorbol myristate acetate (PMA) to trigger NETosis21 for 1, 3, and 5 h. ATAC-see revealed that, after cells were stimulated for 1 h, the accessible genome was fragmented into coarse granules and extended into the nuclear interior (Fig. 4a). ATAC-seq of the same cells showed a gain of chromatin accessibility at genomic loci adjacent to LADs that were previously closed; these accessible sites were predominantly in the form of mononucleosomes (Fig. 4b). Thus, NETosis initiates with a major reorganization of chromatin accessibility around LADs and spreads from the nuclear periphery to the remainder of the genome.
Second, with sustained neutrophil activation, mononucleosomes were further disassembled into free DNA and histones, a step that requires histone citrullination. After 3 h of PMA stimulation, approximately half of the neutrophils (49.7%) had released their chromatin into the extracellular space (Supplementary Fig. 9b,c). The NETs had broadly distributed ATAC-see signal and contained multiple bright foci, consistent with the presence of decondensed chromatin (Supplementary Fig. 10a). ATAC-seq of the same samples indicated that the entire genome was accessible and that NETs were largely in the form of free DNA, essentially indistinguishable from the read-length distribution of purified genomic DNA3,10 (Supplementary Fig. 10b). This state remained after 5 h of PMA stimulation, and 59.7% of neutrophils had released NETs at this time (Fig. 4a,b and Supplementary Fig. 9b,c). Histone citrullination by the PAD4 enzyme has been reported to mediate NETosis and chromatin decondensation22,23. Concurrent treatment of neutrophils with PAD4 inhibitor Cl–amidine (PAD4i) and PMA inhibited histone citrullination and NETosis (Supplementary Fig. 9b – d). PAD4i–treated active neutrophils initiated the gain in chromatin accessibility around LADs and processed the chromatin to mononucleosomes, but they failed to release free DNA from mononucleosomes even after 5 h of neutrophil activation (Fig. 4a,b and Supplementary Fig. 9d). Morphologic and genomic analyses indicated that PAD4i-treated cells were arrested at a step between 1 and 3 h of neutrophil activation (Fig. 4a,b and Supplementary Fig. 9d). Thus, NETosis is separable into two distinct steps, and PAD4-mediated histone citrullination is only required in the second step for disassembling nucleosomes to release DNA as NETs.
These results show that NETosis is a programmed genome disassembly that is precisely spatially organized and sequence programmed (Fig. 4c). NETosis occurs outside–in, proceeding from the edge to the interior of the nucleus. Chromatin accessibility of LADs, situated at the nuclear periphery, serve as the initial trigger point to process chromatin to nucleosomes, and then histone citrullination disassembles DNA from histones. The ability to image and sequence the accessible genome of the same cells (‘see and seq’) greatly facilitated our ability to decipher the spatial choreography of this unique form of programmed cell death.
ATAC-see with flow cytometry deconvolutes cells by chromatin accessibility
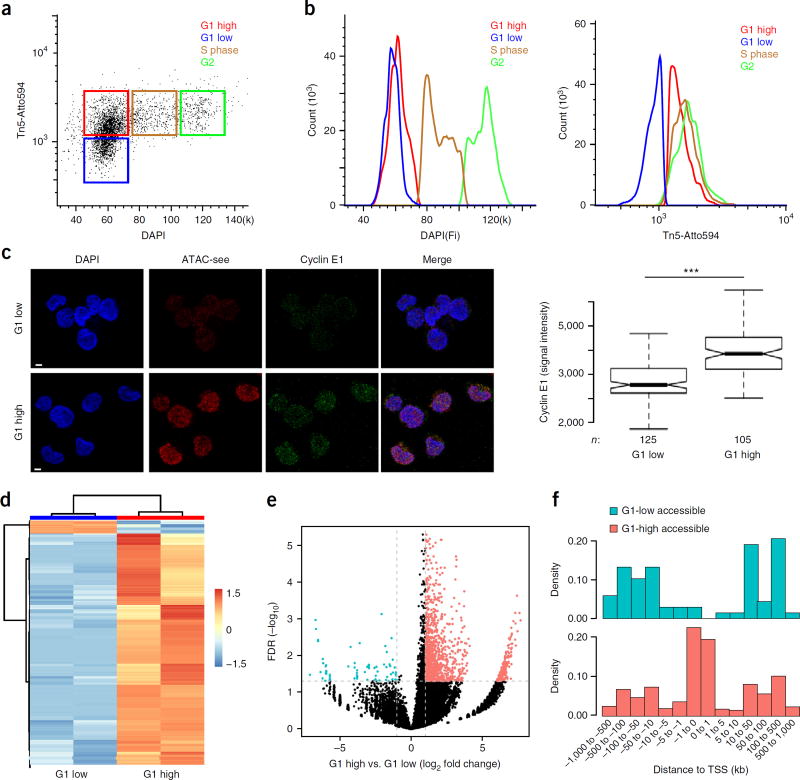
Finally, we integrated ATAC-see with flow cytometry to explore quantitative cell counting and prospective cell sorting as a function of chromatin accessibility. A central question in epigenetics is how chromatin organization is disassembled and reassembled during the cell cycle24. We employed ATAC-see and DAPI staining of DNA content for fluorescence activated cell sorting (FACS) in human B-cell line GM12878. We found four groups of cells—G1-low, G1-high, S-phase, and G2 cells (Fig. 5a,b and Supplementary Fig. 11a,b). Approximately half of the cells in G1 (2N DNA content) had a lower level of ATAC-see signal (we refer to these as ‘G1-low’), and half of the G1 cells with 2N DNA content had the same ATAC-see signal as S and G2 cells (these cells were termed ‘G1 high’). We hypothesized that the G1-low cells were in early G1, as the chromatin structure starts to decondense after maximal chromosome compaction during mitosis. Staining with cyclin E1, a late G1–S transition marker25, clearly showed that G1 cells with low amounts of accessible DNA versus G1 cells with high amounts corresponded to cells in early versus late G1 (Fig. 5c; P < 0.001).
Figure 5.
ATAC-see reveals cell-cycle-specific genome accessibility. (a) Flow cytometry with ATAC-see. Dot plot of signal intensity in dual staining for DAPI and ATAC-see of GM12878 cells; results showed four groups of cells: G1 low, G1 high, S phase, and G2. (b) Quantitation of DAPI (left) and ATAC-see (right) signals from different groups. (c) Cyclin E1 staining in ATAC-see-sorted G1-high and G1-low cells. Left panel, representative images from confocal microscopy. Scale bars, 2 µm. Right panel is a box plot depicting signal intensity measurement. n, cell number; ***, P < 0.001, Student t-test. (d) Heatmap shows cluster of different ATAC-seq accessible regions between the G1-high and G1-low cells (FD > 2, FDR < 0.05); each group has one replicate. (e) The volcano plot represents genome-wide comparisons of accessible regions in G1-high versus G1-low cells. (f) The density histograms represent the distribution of the more accessible regions in G1-high and G1-low cells across the transcription starting sites (TSS). The more accessible regions in the two groups were color coded.
ATAC-seq further showed that the two G1 populations had differential DNA accessibility profiles compared with each other and with S- and G2-phase cells (Supplementary Fig. 11c–e). Focusing on G1-low versus G1-high cells, we found that 96 loci had relatively higher accessibility (fold change (FC) > 2; FDR < 0.05) in G1-low cells; conversely, 2,067 loci had increased DNA accessibility (FC > 2; FDR < 0.05) in G1-high compared with G1-low cells (Fig. 5d,e and Supplementary Table 1). Notably, a majority (59.3%) of the more accessible loci in G1-low cells were enhancers, and the largest category (57.4%) of the more accessible loci in G1-high cells were promoters (Fig. 5f and Supplementary Fig. 12a,b). These results raised the possibility that certain enhancers become accessible first in early G1, and many promoters become accessible in mid-to-late G1. Gene ontology analysis showed that the accessible elements in G1-high cells were enriched for genes with histone acetyltransferase activity, histone modification, gene transcription, nuclear body and other biological process (Supplementary Table 2) consistent with stepwise rebuilding of chromatin and nuclear architecture from early-to-late G1 after cell division. In a separate application, we found that ATAC-see of mouse bone marrow cells could distinguish several myeloid cell populations by flow cytometry (Supplementary Fig. 13a), separating common myeloid progenitor (CMP) cells, granulocyte–monocyte progenitor (GMP) cells, and neutrophils. These FACS data were confirmed by confocal imaging (Supplementary Fig. 13b). Collectively, these experiments demonstrate the feasibility of using ATAC-see and flow cytometry to dissect cell-population heterogeneity and enhance cell-state-specific signals that may otherwise be averaged out in the bulk population.
DISCUSSION
ATAC-see unveils the spatial organization of the accessible genome in its native context for any cell type by covalently inserting fluorophore molecules in the genome. ATAC-see is compatible with multimodal imaging of landmark proteins and flow cytometry, and it enables sophisticated regulome analysis. The compatibility of ATAC-see with fixed samples makes it readily applicable to human clinical specimens. The ability to apply ATAC-see in FACS sorting should enhance the understanding of the chromatin basis of cellular heterogeneity. ATAC-see provides rich spatial and epigenomic information that together give a molecular portrait of the cell. In the future, it will be of interest to use ATAC-see to identify the unique spatial organization of the accessible genome in different human cell types and organs that comprise the human cell atlas and to reveal fine structure of accessible-chromatin organization in clinical diagnosis by using super-resolution microscopy. Finally, transposome engineering facilitated the successful combination of imaging and epigenomics; extending this approach may facilitate novel modes of genome interrogation and control.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Cell culture
GM12878 cells were grown in RPMI 1640 (cat. no. 11875-093, Gibco), 2 mM L-glutamine (cat. no. 25030-081, Gibco), 15% fetal bovine serum (FBS; cat. no. 07905, Gibco), and 1% Pen/ Strep (cat. no. 15140-122, Gibco). HT1080 cells were cultured in DMEM/F-12, GlutaMAX supplement (cat. no. 10565-018,Gibco), 10% FBS (cat. no. 07905, Gibco), and 1% Pen/Strep (cat. no. 15140-122, Gibco). HeLa cells were maintained in DMEM (cat. no. 11965-092, Gibco), 10% FBS (cat. no. 07905, Gibco), and 1% Pen/ Strep (cat. no. 15140-122, Gibco). Neural progenitor cells (NPCs) were cultured in N2B27 media (DMEM/F12, Invitrogen cat. no. 11320-033; Neurobasal, Gibco cat. no. 21103-049; NDiff Neuro-2 Medium Supplement Millipore, cat. no. SCM012; B27 Supplement, Gibco cat. no. 17504-044) supplemented with EGF (cat. no. 315-09, Peprotech) and FGF (cat. no. 100-18B, Peprotech) (10 ng/mL each). NPCs were passaged every other day with Accutase (cat. no. SCR005, Millipore) and seeded on gelatin-coated plates. All cells were kept under standard 37 °C and 5% CO2 conditions.
Study participants
This study was approved by the Stanford University Administrative Panels on Human Subjects in Medical Research, and written informed consent was obtained from all participants. All participants (three in total) were randomly and blinding selected.
Human CD4+ T cells isolation
Human CD4+ T cells were isolated from whole blood of a healthy female donor under a Stanford University IRB-approved protocol. In brief, RosetteSep Human CD4+ T Cell Enrichment Cocktail was added into the whole blood with a final concentration of 50 µL/mL, and the mixture was incubated 20 min at room temperature. After incubation, the whole blood was diluted with equal volume of PBS + 2% FBS (cat. no. 07905, Gibco). Then the diluted samples were layered on top of the density medium and centrifuged for 20 min at 1,200× g at room temperature with the brake off. The enriched cells were collected into a new tube and washed twice with PBS + 2% FBS. The residual red blood cells were lysed with buffer EL (cat. no. 79217, Qiagen) 5 min at room temperature.
Neutrophil isolation
Human neutrophils were isolated from two healthy donors’ blood, described as follows, under a Stanford University IRB-approved protocol23. In brief, 7 ml of heparinized blood was layered on 7 ml Histopaque 1119 (cat.no. 11191-100ML, Sigma Aldrich) in a 15 ml Falcon tube, and the mixture was spun 20 min at 800× g at room temperature. The interphase (lymphocytes and monocytes) was discarded and the granulocyte-rich layer of Histopaque 1119 (diffuse, red phase above red blood cell (RBC) pellet) was collected in a new tube. 5 ml of cells was washed with 10 ml of medium (HBSS+) and spun 10 min at 200× g at room temperature. The cells were resuspended in 3 ml DPBS with 0.5% human serum albumin (cat. no. 101-15–50, Lee BioSolutions), and the cell suspension was layered on 10 ml of a discontinuous Percoll gradient (85% to 65% Percoll) and spun 20 min at 800× g at room temperature. The interphase between 70% and 75% Percoll layers were collected in a new 15 ml tube. DPBS was added to the tube with 0.5% human serum albumin until the total volume reached 15 ml, and the tube was spun for 10 min at 200× g at room temperature. The cell pellet was resuspended in RPMI–HEPES (10 mM) at 1.5 × 106 cells per ml for neutrophil activation.
Human neutrophil activation
Human neutrophils were stimulated by phorbol myristate acetate (PMA) as described23. Briefly, the purified neutrophils were suspended in 3 ml RPMI 1640 with final concentration of 1.5 × 106 per ml in three 15 ml Falcon tubes with loosened caps and incubated in cell culture CO2 incubator (5% CO2, 37 °C) with final concentration of 30 nM PMA for 1 h, 3 h, and 5 h. After 2 h of simulation, protease inhibitor cocktail was added at a 1:200 dilution to both control and simulated tubes. At the respective timepoints, neutrophils were fixed with 1% formaldehyde for 10 min at room temperature, and fixed neutrophil cells spun down on the glass slide with Cytospin 1,000 r.p.m. 5 min at room temperature for ATAC-see or immunostaining.
PAD4 inhibitor treatment in human neutrophil
The PAD4 inhibition was performed as previously described25. In brief, the isolated human neutrophils were incubated with 200 µM Cl-amidine for 30 min before PMA stimulation.
Hyperactive Tn5 production
Hyperactive Tn5 was produced as previously described26. In brief, pTXB1-Tn5 plasmid (cat. no. 60240, Addgene) was introduced into T7 Express LysY/Iq Escherichia coli strain (cat. no. C3013, NEB). 10 ml of overnight-cultured E. coli was inoculated to 500 ml lysogeny broth (LB) medium. After incubation for 1.5 h at 37 °C, bacteria was incubated about 2.5 h at room temperature. When the optical density (OD) 600 = 0.9, Tn5 protein was produced by adding 0.25 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 h. E. coli pellet was resuspended in lysis buffer (20 mM HEPES–KOH pH 7.2, 0.8 M NaCl, 1mM EDTA, 10% glycerol, 0.2% triton X-100, complete proteinase inhibitor (cat. no. 11697498001, Roche)) and lysed by sonication. 10% polyethylenimine (PEI) was added to supernatant of lysate to remove bacterial genomic DNA. 10 ml chitin resin (cat. no. S6651L, NEB) was added to the supernatant and incubated with rotation for 1 h at 4 °C. The resin was washed by lysis buffer extensively. In order to cleave Tn5 protein from intein, lysis buffer containing 100 mM DTT was added to the resin and stored at 4 °C. After 48 h, protein was eluted by gravity flow and collected in 1ml fractions. 1 µl of each fraction was added to detergent-compatible Bradford assay (cat. no. 23246, ThermoFisher Scientific), and peaked fractions were pooled and dialyzed against 2× dialysis buffer (100 mM HEPE–KOH pH 7.2, 0.2 M NaCl, 0.2 mM EDTA, 2 mM DTT, 0.2% triton X-100, 20% glycerol). Dialyzed Tn5 protein was concentrated by using ultracel 30-K column (cat. no. UFC903024, Millipore), and the quantity of Tn5 was measured by Bradford assay and visualized on NuPAGE Novex 4–12% Bis-Tris gel (cat. no. NP0321, ThermoFisher Scientific) followed by Coomassie blue staining.
Adaptor sequences
The Atto-590N-labeled oligonucleotides for Tn5 transposome adaptor were synthesized at Integrated DNA Technologies (IDT), and the oligonucleotide sequences are as follows: Tn5MErev, 5’-[phos]CTGTCTCT TATACACATCT-3’; Tn5ME-A-ATTO590, 5’-/5ATTO590/ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3’;Tn5ME-B-ATTO590: 5’-/ATTO590/GTCTCGTGGGCTCGGAGATG TGTATAAGAGACAG-3’.
Tn5 transposome assembly
The assembly of Tn5 transposome was performed as described26. Briefly, oligonucleotides (oligo) (Tn5ME-A-ATTO590, Tn5ME-B-ATTO590, Tn5MErev) were resuspended in water to a final concentration of 100 µM each. Equimolar amounts of Tn5MErev/Tn5ME-A-ATTO590 and Tn5MErev/Tn5ME-B-ATTO590 were mixed in separate 200 µl PCR tubes. These two tubes of oligo mixtures were denatured on a thermocycler for 5 min at 95 °C and cooled down slowly on the thermocycler by turning off the thermocycler. The Tn5 transposome was assembled with the following components: 0.25 vol Tn5MErev/Tn5ME-A-ATTO590 + Tn5MErev/Tn5ME-B-ATTO590 (final concentration of each double-stranded oligo was 50 µM each), 0.4 vol glycerol (100% solution), 0.12 vol 2× dialysis buffer (100 mM HEPES–KOH at pH 7.2, 0.2 M NaCl, 0.2 mM EDTA, 2 mM DTT, 0.2% Triton X-100, 20% glycerol), 0.1 vol SL–Tn5 (50 µM), 0.13 vol water. The reagents were mixed thoroughly but gently, and the solution was left on the bench at room temperature for 1 h to allow annealing of oligos to Tn5.
Slide preparation and fixation
HT1080 cells and HeLa cells were grown on secureSlip cell culture system (cat. no. 0515073, Grace Bio-Labs) until 80–90% confluent and fixed with 1% formaldehyde (Sigma-Aldrich) for 10 min at room temperature. GM12878 cells were fixed with 1% formaldehyde (Sigma) for 10 min at room temperature and centrifuged on the glass slide with cytospin, 1,000 r.p.m. for 5 min, 30,000–50,000 cells per slide.
Immunostaining
The formaldehyde-fixed slide was rinsed in PBS and permeabilized in PBS 0.5% Triton-X 100 for 10 min at room temperature. Then, the slide was blocked with antibody dilution reagent (cat. no. 00–3218, ThermoFisher Scientific) for 1 h at room temperature. Primary antibodies were diluted in antibody dilution reagent 1:500: rabbit anti-LaminB1 (cat. no. ab16048, Abcam), mouse anti-mitochondria (cat. no. ab3298, Abcam), and Anti-Cyclin E1 antibody (EP435E; Alexa Fluor 488; cat. no. ab194068, Abcam), rabbit anti-H3K9me3 (cat. no. SAB4800027-100UL, Sigma), rabbit anti-H3K9me2 (cat. no. 39375, Active Motif), rabbit anti-H3K27acetylation (cat. no. 25238, BPS Bioscience), rabbit anti-H3 citrulline (cat. no. ab5103, Abcam), mouse zanti-Phospho RNA Polymerase II CTD (Ser5) mAb (cat. no. MABI0603, MBL International); and they were incubated overnight at 4 °C. After washing with PBS containing 0.05% Tween-20 three times 10 min each, slides were incubated with secondary antibodies goat anti-rabbit-ATTO488 (cat. no. 18772-1ML-F, Sigma-Aldrich) or goat anti-mouse-Atto647N (cat. no. 50185-1ML-F, Sigma-Aldrich) for 45 min at room temperature. The slides were washed with PBS containing 0.05% Tween-20 three times 10 min each, mounted using Vectashield with DAPI (cat. no. H-1200, Vector labs), and imaged with Leica SP8 or Zeiss LSM 700.
For ATAC-see costaining with epigenetic marks or RNAP II, ATAC-see was performed first, and then immunostaining was done on the same slide. All the primary antibodies was diluted at a 1:500 ratio, and the antibody information is as follows: rabbit anti-H3K9me3 (cat. no. SAB4800027-100UL, Sigma); rabbit anti-H3K4me3 (cat. no. ab8580, Abcam); rabbit anti-H3K27acetylation (cat. no. 25238, BPS Bioscience); mouse anti-Phospho RNA Polymerase II CTD (Ser5) mAb (cat. no. MABI0603, MBL International); mouse anti-H3K27me3 (cat. no. ab6147, Abcam); rabbit anti-RNA polymerase II CTD (Ser-5) antibody (cat. no. ab5095, Abcam).
ATAC-see
After fixation, the cells (either growing on slide or centrifuged on glass slide with Cytospin) were permeabilized with lysis buffer (10 mM Tris–Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.01% Igepal CA-630) for 10 min at room temperature. After permeabilization, the slides were rinsed in PBS twice and put in humid chamber box at 37 °C. The transposase mixture solution (25 µl 2× TD buffer, final concentration of 100 nM Tn5-ATTO-59ON, adding dH2O up to 50 µl) was added on the slide and incubated for 30 min at 37 °C. After the transposase reaction, slides were washed with PBS containing 0.01% SDS and 50 mM EDTA for 15 min three times at 55 °C. After washing, slides were mounted using Vectashield with DAPI (cat. no. H-1200, Vector labs) for imaging with Leica SP8 or Zeiss 700 or for additional costaining with lamin B1 or mitochondria (see the materials and methods above).
ATAC-see with Xist RNA FISH
Female mouse neural progenitor cells (NPCs) were grown on secureSlip cell culture system (cat. no. 0515073, Grace Bio-Labs) until they were 80–90% confluent, and they were then fixed with 1% formaldehyde (Sigma-Aldrich) for 10 min at room temperature. After washed with PBS three times (5 min each), the slide was permeabilized with PBS 0.1% NP-40 10 min at room temperature. ATAC-see (with Atto-480-labeled Tn5) was performed under standard protocol as stated above. After ATAC-see, the slide was dehydrated in 70% EtOH for 1 h at 4 °C and rehydrated in 2× SSC + 10% formamide for 5 min at room temperature. The Xist RNA FISH probes (Stellaris mXist) were hybridized on the slide overnight at 37 °C. The washing was conducted in the following conditions: twice for 30 min in 2× SSC + 10% formamide at 37 °C and once for 5 min in 2× SSC at 37 °C. After Xist RNA FISH washing, the slides were mounted with Vectashield (cat. no. H-1200, Vector labs).
FACS sorting and analyzing of ATAC-see
GM12878 cells were washed with PBS, then fixed with 1% formaldehyde for 10 min at room temperature. After the fixation, cells were permeabilized with lysis buffer (10 mM Tris–Cl, pH 7.4; 10 mM NaCl; 3 mM MgCl2 ;0.01% Igepal CA-630) and centrifuged at 700 g for 10 min at room temperature. 5 × 106 cells were then either stained with DAPI (for negative control) or transposed for 30 min at 37 °C using Atto-594-labeled in-house Tn5 (see materials and methods above). After the transpose reaction, cells were centrifuged and stained with DAPI, analyzed and sorted using a BD FACS-Aria II (BD Biosciences). Collected G1-high, G1-low, S-phase and G2 cells were reverse crosslinked, purified, and amplified by standard ATAC-seq PCR reaction; and the libraries were sequenced on Illumina NextSeq at the Stanford Functional Genomics Facility. The sorted ATAC-see groups were collected on glass slide using Cytospin, and images were taken under Zeiss 700. The sorted ATAC-see G1-high and G1-low cells were collected and centrifuged using Cytospin, and Anti-Cyclin E1 antibodies (EP435E; Alexa Fluor 488; cat. no. ab194068, Abcam) were stained on different groups. For bone marrow progenitor studies, cells were harvested from femur, tibia, and humerus. Bones were fragmented by mortar and pestle, and debris was removed by gradient centrifugation using Histopaque 1119 (Sigma-Aldrich). Cells were passed through a 70-µM strainer, and red blood cells were lysed with ACK lysis buffer. 5–10 × 106 cells were stained at 4 °C in FACS buffer (DPBS + 0.5% BSA + 2 mm EDTA) with the following antibodies at a 1:200 dilution: CD16/32 eFluor450 (cat. no. 93, eBioscience), CD117 FITC (cat. no. 2B8, eBioscience), CD11b–redFluor710 or APC (cat. no. M1/70, Tonbo Bioscience), B220-PE (cat. no. RA3–6B2, eBioscience), and CD105-PE (cat. no. MJ7/18, eBioscience). Cells were then washed, fixed, and transposed as described above. CMPs were identified as Lineagec kithiCD16/32−. GMPs were identified as Lineage-ckithiCD16/32+. Band neutrophils were identified as Lineage−ckit−CD11b+CD16/32+. Cells were sorted into FACS buffer, and cell purities of at least 95% were confirmed by postsorting analysis.
ATAC-seq
ATAC-seq with Illumina-Tn5-transposase- (Nextera) and Atto-590-labeled homemade Tn5 in human GM-12878 cells was performed as described3. Briefly, 50,000 cells were centrifuged 500 g 5 min at room temperature. The cell pellet was resuspended in 50 µl lysis buffer (10 mM Tris–Cl, pH 7.4; 10 mM NaCl; 3 mM MgCl2; 0.01% Igepal CA-630) and centrifuged immediately 500 g for 10 min at 4 °C. The cell pellet was resuspended in 50 µl transposase mixture (25 µl 2× TD buffer, 22.5 µl dH2O, and 2.5 µl Illumina Tn5 transposase or with final concentration of 100 nM Atto-590-labeled in-house Tn5) and incubated at 37 °C 30 min. After transposition, the mixture was purified with Qiagen Mini-purification kit and eluted in 10 µl Qiagen EB elution buffer. Sequencing libraries were prepared following the original ATAC-seq protocol3. The sequencing was performed on Illumina NextSeq at the Stanford Functional Genomics Facility.
ATAC-seq in fixed cells
Human HT1080 cells were fixed with 1% formaldehyde (Sigma, USA) for 10 min and quenched with 0.125 M glycine for 5 min at room temperature. After the fixation, cells were counted; and 50,000 cells were used per ATAC-seq reaction. The transposed reaction is the same as normal ATAC-seq except with 0.05% Igepal CA-630 in the lysis buffer. After the transposase reaction, a reverse-crosslinked solution was added (with final concentration of 50 mM Tris–Cl, 1 mM EDTA, 1% SDS, 0.2 M NaCl, 5 ng/ml proteinase K) up to 200 µl. The mixture was incubated at 65 °C at 1,000 r.p.m. with shaking in a heat block overnight, then purified with Qiagen Mini-purification kit and eluted in 10 µl Quiagen EB elution buffer. Sequencing libraries were prepared following the original ATAC-seq protocol3. The sequencing was performed on Illumina NextSeq at the Stanford Functional Genomics Facility.
ATAC-seq after imaging
80% confluent human HT1080 cells (30,000–50,000 cells) grown in eight-well-chamber slides were fixed with 1% formaldehyde (Sigma, USA) for 10 min and quenched with 0.125 M glycine for 5 min at room temperature. After fixation, slides were put in PBS at 4 °C. ATAC-see was performed under standard protocol as stated above. For the imaging and sequencing, slides were mounted using Vectorshield, and images were taken with a Zeiss LSM 700. After imaging, the coverslip was removed, and cells were lysed with 50 µl of 50 mM Tris–Cl, 1 mM EDTA, 1% SDS, and 0.2 M NaCl 5 min on slide at room temperature. The cell lysate was carefully transferred to 1.5 ml Eppendorf tube. The glass slide was washed with 50 µl of the same buffer another three times, and washing solutions were transferred to the same tube carefully. After adding final concentration of 50 µg/µl proteinase K to the cell lysate, the mixture was incubated at 65 °C at 1,000 r.p.m. with shaking in a heat block overnight, purified with Qiagen Mini-purification kit, and eluted in 10 µl Qiagen EB elution buffer. Sequencing libraries were prepared following the original ATAC-seq protocol3. The sequencing was performed on Illumina NextSeq at the Stanford Functional Genomics Facility. For ATAC-seq after imaging the human neutrophils, 50,000 cells were centrifuged on the glass slides with Cytospin. The remaining steps are the same as those for HT1080 cells, except the lysis buffer contains 0.01% Igepal CA-630.
For the systematical sensitivity assay of ATAC-seq after imaging, different amount of cells (50,000; 5,000; 500; and 50 cells) were seeded in the eight-well chamber with duplications, and cells were fixed as stated above after 6 h seeding. The following procedures are the same as those stated above.
DNA FISH probe labeling
DNA FISH probe labeling was performed as previous described27. In brief, BAC clones (ThermoFisher Scientific) RP11-626N18, RP11-832P24, RP1163J14, RP11637D5, RP11-368K11, and RP11-116A9, were cultured based on standard protocol and purified with BACMAX DNA Purification Kit (cat. no. BMAX044, Epicenter). After the purification, BACs were sheared to 300–800 bp with sonication. The sheared BAC DNA was labeled with BioPrime Array CGH Genomic Labeling System (cat. no. 18095-011, Thermo Fisher Scientific) by mixing with either Cy3-dCTP (BACs: RP11–626N18, RP1163J14) (cat. no. PA53021, GE Healthcare Life Sciences, USA), Green 496 dUTP (BACs: RP11–832P24, RP11637D5) (cat. no. ENZ-42831L-0050, Enzo Life Sciences), or Cy5-dCTP (BACs: RP11-368K11, RP11-116A9) (cat. no. PA55021, GE Healthcare Life Sciences). The labeled probes were purified with MinElute PCR Purification Kit (cat. no. 28006, Qiagen).
DNA FISH in human neutrophil
DNA FISH was done as previously described27. In brief, purified human neutrophils were fixed with 1% formaldehyde for 10 min at room temperature, centrifuged on glass slides with cytospin, and stored in 70% ethanol. The neutrophils on slides were permeabilized in PBS 0.5% Triton-X 100 for 10 min at room temperature, rinsed in PBS, denatured in 2× SSC/50% formamide for 30 min at 80 °C, and put in ice-cold 2× SSC for 5 min. At the same time, 40 µl of hybridization mixture, 100 ng BAC probes, 1× DNA FISH hybridization buffer (2× SSC, 10% dextran sulfate, 50% formamide) at a final concentration of 100 ng/µl human Cot-1 DNA (cat. no. 15279-011, Thermo Fisher Scientific), was denatured for 5 min at 95 °C and put on ice for 5 min. After adding denatured probes onto the denatured neutrophils on slide, the slide was hybridized in a humid dark box for 14 h at 37 °C. On the second day, the slide was sequentially washed twice in 2× SSC/50% formamide for 15 min at 40 °C and twice with 2× SSC for 15 min at 40 °C. After washing, the slide was mounted using Vectashield with DAPI (cat. no. H-1200, Vector labs). 3D DNA as FISH images were taken with Zeiss LSM 700. FISH at the periphery was defined as cases in which the distance between the center of DNA FISH signal and edge of DAPI staining is less than 0.1 µm, as manually measured in Volocity (PerkinElmer).
Imaging processing for ATAC-see signal
Cell nuclei were identified and segmented by Gaussian filtering raw DAPI image data and then by applying an intensity threshold to generate nuclear outlines. Nuclei were randomly selected, and over ~30 nuclei were chosen for each cell line. All nuclear outlines were verified by eye. Regions of the cell containing mitochondria were excluded from all analysis using a mitochondrial mask, which was similarly generated by Gaussian filtering Mito-Tracker image data (see Supplementary Fig. 6a), and applying an intensity threshold.
Correlations between DAPI and ATAC-see signals were measured on a per-pixel basis within the nucleus, with each channel normalized to its mean intensity per nucleus. Correlation data was fit to a linear regression on both a per-cell basis and a cell population basis. Intensity distributions of DAPI and ATAC-see signals were measured as a function of radial distance from the nuclear rim toward the nuclear center by measuring the average intensity within a series of one-pixel-wide annuli generated through iterative erosion of each nuclear outline. Each channel was again normalized to its mean intensity per nucleus.
Bright regions of ATAC-see signal within each nucleus were identified by Gaussian filtering the raw data and then thresholding to find all regions more than 50% brighter than the mean filtered intensity. The fraction of area within a bright region was calculated per cell as the total bright spot area over the total nuclear area. All analysis was performed using a custom-written Python and C++ code, drawing heavily on NumPy and SciPy.
Signal intensity measurement was performed in Volocity software (PerkinElmer), and the signal intensity correlation of ATAC-see and other marks (including H3K4me3, H3K27Ac, H3K9me3, H3K27me3, RNAPII ser-2, RNAPII ser-5) or Xist RNA FISH was calculated by Volocity software by using DAPI staining as mask and plot in custom-written R script.
ATAC-seq library data preprocessing
ATAC-seq paired-end reads were trimmed for Illumina adaptor sequences and transposase sequences using an in-house script, and they were mapped to hg19 using Bowtie2 (ref. 28) v2.1.0 with parameters –very sensitive. Over ~11 million mapped reads were generated in each sequencing library and used for downstream data mining, and there are over ~35 million mapped reads in each sequencing library from human neutrophil and NETosis. Duplicate reads were removed with Picard (http://broadinstitute.github.io/picard/) v1.79. Peak calling was performed by MACS2 (ref. 29) narrow peak mode with parameters –q 0.01 –nomodel –shift 0. Overlapping peaks from all samples were merged together to a consensus peak list, and number of unique-mapped and properly paired reads mapped to each peak for each individual samples was quantified to calculate the Pearson correlation. The insert size of fragments was estimated from the distance between the pair-ended reads and plotted against the frequency in a histogram. Genome ontology enrichment analysis was performed in GREAT software30. EdgeR was used to identify variable peaks between Atto-Tn5 versus Nextera and between technical replicates from either Atto–Tn5 or Nextera.
ATAC-seq signal intensity around TSS
A 2 kb window centered on TSS was divided into 40 equally sized bins of 50 bp. The number of unique-mapped and properly paired ATAC-seq tags overlapping each bin was counted. The average fragment count plotted in each bin was normalized to total 10 millions of reads.
ATAC-seq signal intensity around LADs
The human lamin associated domains (LADs) were downloaded from http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/. We aligned all 1,302 LADs by their left or right borders and calculated average profiles of ATAC-seq read coverage across the combined 2,604 borders. In detail, a 200 kb window centered on 2,604 LAD borders was divided into 40 equally sized bins of 50 kb. The order of the bins for the right borders of LADs was the reverse of that for the left borders. The number of unique-mapped and properly paired ATAC-seq tags was counted in each bin. The average fragment count plotted in each bin was normalized to total 10 millions of reads. To compare the ATAC-seq signal inside and outside LADs, we took 10 kb nonoverlapping sliding windows on the human genome and calculated ATAC-seq read counts in each window. We labeled the windows inside the LADs as 1 and outside the LADs as 0, and then we plotted ATAC-seq signals in two groups in a boxplot. t-test was applied to compare the mean of the two groups.
Differential analysis of ATAC-seq peaks
Differentially accessible peaks from the union peak list were identified with edgeR31 using raw counts of each sample in the overlapping peak list. edgeR was run with default settings, with a fold-change threshold of 2 and FDR < 0.05. The distribution of differential peaks on the human genome was further investigated using a functional genomic annotation based on 15 chromHMM states32 derived from ENCODE histone markers on the GM12878 cell line.
Supplementary Material
Acknowledgments
We thank S. Kim (Stanford) for FACS access. This work was supported by NIH grant P50-HG007735 (to H.Y.C. and W.J.G.), the Life Extension Foundation (to H.Y.C.), NCI Physical Sciences Oncology Center grant U54CA143836 (to J.T.L.), and National Institute of Biomedical Imaging and Bioengineering (NIBIB)/4D Nucleome Roadmap Initiative grant 1U01EB021237 (to J.T.L.).
Footnotes
Accession codes. Raw and processed data are available at the NCBI Gene Expression Omnibus, accession numbers GSE76006 and GSE79921.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
X.C., W.J.G., and H.Y.C. conceived and designed the study. X.C., J.D.B., U.L., A.T.S., A.C.C., and R.P.G. performed experiments. Y.S. and X.C. performed genomic data analysis. W.D., X.C. and J.T.L. conducted image analyses. S.W.C., A.E.-S., and J.A.D. generated reagents. X.C. and H.Y.C. wrote the manuscript with input from all authors. H.Y.C. supervised all aspects of this work.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Misteli T. Self-organization in the genome. Proc. Natl. Acad. Sci. USA. 2009;106:6885–6886. doi: 10.1073/pnas.0902010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markaki Y, et al. The potential of 3D–FISH and super-resolution structured illumination microscopy for studies of 3D nuclear architecture: 3D structured illumination microscopy of defined chromosomal structures visualized by 3D (immuno)-FISH opens new perspectives for studies of nuclear architecture. BioEssays. 2012;34:412–426. doi: 10.1002/bies.201100176. [DOI] [PubMed] [Google Scholar]
- 8.Deng W, Shi X, Tjian R, Lionnet T, Singer RH. CASFISH: CRISPR/ Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc. Natl. Acad. Sci. USA. 2015;112:11870–11875. doi: 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amini S, et al. Haplotype-resolved whole genome sequencing by contiguity preserving transposition and combinatorial indexing. Nat. Genet. 2014;46:1343–1349. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cusanovich DA, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibcus JH, Dekker J. The hierarchy of the 3D genome. MoL. Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buys CH, Anders GJ, Gouw WL, Borkent-Ypma JM, Blenkers-Platter JA. A comparison of constitutive heterochromatin staining methods in two cases of familial heterochromatin deficiencies. Hum. Genet. 1979;52:133–138. doi: 10.1007/BF00284607. [DOI] [PubMed] [Google Scholar]
- 14.Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 15.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 16.Qu K, et al. Individuality and variation of personal regulomes in primary human T cells. Cell Syst. 2015;1:51–61. doi: 10.1016/j.cels.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 18.Amendola M, van Steensel B. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep. 2015;16:610–617. doi: 10.15252/embr.201439789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solovei I, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 20.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 21.Li P, et al. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis HD, et al. Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat. Chem. Biol. 2015;11:189–191. doi: 10.1038/nchembio.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christophorou MA, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinde E, Cardarelli F, Digman MA, Gratton E. Changes in chromatin compaction during the cell cycle revealed by micrometer-scale measurement of molecular flow in the nucleus. Biophys. J. 2012;102:691–697. doi: 10.1016/j.bpj.2011.11.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 2005;7:831–836. doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- 26.Picelli S, et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24:2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, et al. Chromatin in situ proximity (ChrISP): single-cell analysis of chromatin proximities at a high resolution. Biotechniques. 2014;56:117–124. doi: 10.2144/000114145. [DOI] [PubMed] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.