Abstract
Human genetic studies have been the driving force in bringing to light the underlying biology of psychiatric conditions. As these studies fill in the gaps in our knowledge of the mechanisms at play, we will be better equipped to design therapies in rational and targeted ways, or repurpose existing therapies in previously unanticipated ways. This review is intended for those unfamiliar with psychiatric genetics as a field and provides a primer on different modes of genetic variation, the technologies currently used to probe them, and concepts that provide context for interpreting the gene–phenotype relationship. Like other subfields in human genetics, psychiatric genetics is moving from microarray technology to sequencing-based approaches as barriers of cost and expertise are removed, and the ramifications of this transition are discussed here. A summary is then given of recent genetic discoveries in a number of neuropsychiatric conditions, with particular emphasis on neurodevelopmental conditions. The general impact of genetics on drug development has been to underscore the extensive etiological heterogeneity in seemingly cohesive diagnostic categories. Consequently, the path forward is not in therapies hoping to reach large swaths of patients sharing a clinically defined diagnosis, but rather in targeting patients belonging to specific “biotypes” defined through a combination of objective, quantifiable data, including genotype.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-017-0551-x) contains supplementary material, which is available to authorized users.
Key Words: Psychiatric genetics, copy number variant, sequencing, microarrays, GWAS
Introduction
The familiality of psychiatric illness has long been appreciated, but only in the past decade or so have the tools and resources become available to probe its genetic determinants directly. This new era of genetics is particularly meaningful in psychiatry, where, unlike other medical fields, disorders have often been diagnosed and treated subjectively and in the absence of a clear biological framework. Furthermore, the historically unappreciated link between mental illness and biology has led to centuries of stigma for those suffering. By using genetics to frame mental illness as a biomedical phenomenon, there is hope that stigma can be alleviated.
Because many aspects of psychiatric illness are uniquely human and have no adequate animal or cellular model, human genetic research studies are particularly crucial for illuminating the underlying biology of these conditions. As these studies fill in the gaps in our knowledge, we will be better equipped to design therapies in a rational and targeted way.
This review is intended for outsiders to the field of psychiatric genetics and perhaps even genetics in general. I begin by laying a foundation of basic aspects of genetic variation so that the most salient genetic findings from a number of major neurodevelopmental and neuropsychiatric conditions can be appreciated in their context in the latter half of the review. First, an overview of different kinds of genetic variation is given, followed by technologies currently used to probe them. General concepts that routinely come up in these genetic studies are then reviewed, and finally a brief synopsis of current trajectories of discovery in the major neurodevelopmental and neuropsychiatric conditions is given.
Types of Genetic Variation
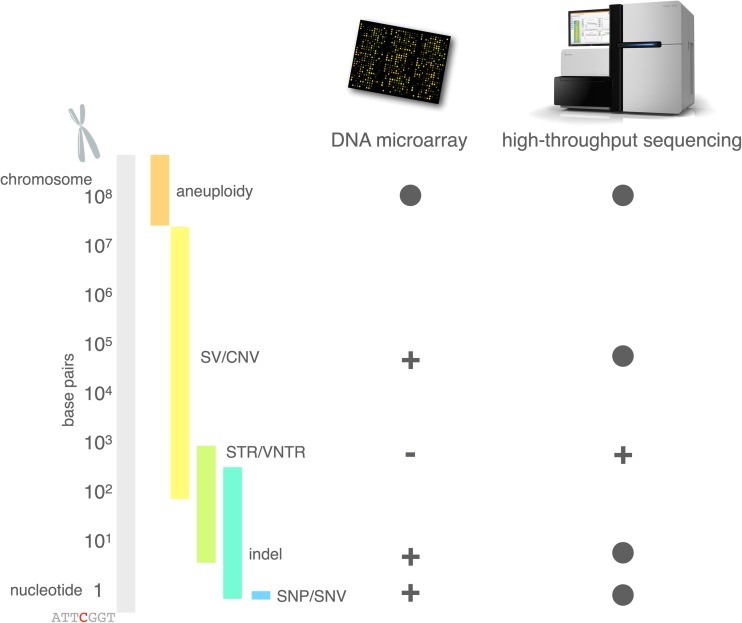
Aneuploidy, the largest of the genetic variations (Fig. 1), is an abnormal chromosome count resulting from errors in chromosome segregation during cell division. Aneuploidy increases in the maternal germline with advancing maternal age [1]. Most aneuploidies that occur in the germline are embryonic lethal, but some result in viable individuals with developmental syndromes, the most well-studied examples being Down syndrome (trisomy 21) and Klinefelter syndrome (47, XXY), as well as Turner syndrome (monosomy X), Edward syndrome (trisomy 18), and Patau syndrome (trisomy 13). Exciting approaches that re-engineer X-inactivation mechanisms for potential treatment of some aneuploidies are currently under investigation [2, 3].
Fig. 1.
Size distributions of classes of genetic variation, and the ability of microarray and sequencing technology to detect them. Solid circle = routinely detectable; (+) = detectable depending on platform, or with special protocols, or with limitations on size or coverage; (–) = not detectable; SNP = single-nucleotide polymorphism; SNV = single nucleotide variant; indel = insertion/deletion; STR = short tandem repeat; VNTR = variable number tandem repeat; SV = structural variant; CNV = copy number variant
Genomic structural variants (SVs) occur on a subchromosomal scale (Fig. 1) and include inversions and translocations (where there is no net loss of genetic material, such as the balanced translocation involving DISC1 in schizophrenia (SZ) [4]), as well as deletions and duplications (where there is a net loss or gain, respectively, of genetic material). Deletions and duplications comprise a subclass of SVs called copy number variants (CNVs). SVs are sometimes arbitrarily defined as larger than 1 kb [5, 6]; however, in actuality, their size distribution is continuous, with smaller variants being more numerous within an individual genome and larger variants being less numerous. Ascertainment of CNVs has played a crucial role in psychiatric genetics over the past decade, as it has helped to turn the spotlight from the common variants popularized by genome-wide association studies (GWAS) to rare variants of large effect [7]. CNVs are a normal form of human genetic variation [8, 9], but neuropsychiatric populations show enrichment for de novo and rare, large CNVs [7, 10–16]. Genes that support the development and function of the brain tend to be larger than other genes, and so are more frequently affected by structural variation [17]. Furthermore, segmental duplications, which can act as a catalyst for SVs, are expanding in the primate and specifically the human lineage [18], with brain genes particularly affected [19]. Taken together, structural variation is an important mode of genetic variation in human evolution and disease in general, and in neuropsychiatric conditions in particular.
Short tandem repeats (STRs) and variable number tandem repeats (VNTRs), are smaller genetic variations (typically a few to hundreds of base pairs, and up to thousands of base pairs for VNTRs; see Fig. 1), consisting of a number of repeats of some core polynucleotide sequences (2–6 for STRs and 7–100 for VNTRs) . STRs arise chiefly as a result of slippage of DNA replication machinery and have high mutation rates compared with other forms of genetic variation [20]. Well-studied conditions with STRs as the causal factor include Huntington disease (HD) and Fragile X syndrome. In huntingtin (HTT), the gene that underlies HD, a CAG trinucleotide repeat encodes a polyglutamine tract in the corresponding huntingtin protein. If this STR expands past 35 repeats, the polyglutamine tract of the corresponding protein causes protein aggregation, which is pathogenic, and results in progressive neurodegeneration, chorea, psychiatric systems, and early death [21]. The CAG repeat is unstable and expansion between generations can lead to earlier onset of the disease, a phenomenon known as anticipation. In contrast to HD, the STR that underlies Fragile X syndrome lies in a noncoding regulatory sequence, upstream of FMR1. If this CGG repeat expands past ~200 units, the promoter becomes hypermethylated and gene expression of FMR1 is shut down [22]. FMR1 encodes a vital regulatory protein that binds mRNAs, in particular those related to synaptic function, and loss of FMR1 results in mass dysregulation of protein expression and autism-like neurodevelopmental features. Genotyping STRs at a genome-wide scale using high-throughput sequencing technology is not routinely done because read lengths are not currently long enough to read through many STRs while still having enough flanking sequence to reliably map them. Consequently, while specific markers are used for forensics or diagnosing well-known diseases, relatively little is known about the role of STRs in disease at the genomic scale.
Indels (insertion/deletions) are polynucleotide expansions (insertion) or contractions (deletion) relative to the reference genome. Like SVs, the size distribution of indels is continuous, but generally variants ranging from 1 to 50 base pairs are considered indels (with larger indels becoming indistinguishable from small SVs; see Fig. 1) [23]. In protein-coding sequence, indels that are not multiples of 3 are under selective pressure because they would induce a frame shift in the subsequent transcript and protein, either leading to nonsense-mediated decay or an entirely different structure downstream of the indel.
Single nucleotide variants (SNVs) are the smallest type of genetic variant (Fig. 1), and are by far the most thoroughly used in genetic association studies. In protein coding sequence, SNVs can change codons to alter the eventual sequence of amino acids in the protein (missense variant), introduce a premature STOP codon (nonsense variant), or alter splicing properties (splice variants). SNVs may also change the codon to another codon for the same amino acid (synonymous variants). SNVs in a noncoding sequence may exert an effect by altering binding affinities for DNA-binding proteins, such as transcription factors, or by affecting epigenetic properties of the chromatin, such as DNA methylation. These single-letter changes to the genetic code are, from a technical standpoint, the easiest class of genetic variation to identify in a massively parallel way, which led to them being the basis for the first generation of GWAS. SNVs that are common in the population are called single-nucleotide polymorphisms (SNPs), and these are the variants that are probed by the microarrays used in massive studies like those of the Psychiatric Genomics Consortium (PGC; http://pgc.unc.edu). Because SNPs are common in the population, they have generally withstood a reasonable amount of selective pressure, and, consequently, associations between a SNP and a deleterious condition such as a disease will generally have a small effect size. Such small effects require large studies to provide the power necessary to declare a statistically significant association. However, rare SNVs (or any kind of rare genetic variant) have generally not undergone the same level of purifying selection, and so disease associations can have larger effect sizes. However, owing to the rarity of these variants, they are often pooled within a gene or even a gene set or pathway to provide the necessary power to demonstrate a statistically significant association. A number of properties of SNPs plagued early GWAS [24], including confounding effects with population structure, as well as underpowered designs (as common SNP effects are small and require large samples to achieve significance). Furthermore common SNPs often merely tag causal variants by virtue of their “hitchhiking” together through recombination events (contributing to a phenomenon called linkage disequilibrium). This tagging effect can make locating and interpreting the causal variant a challenging exercise.
Technologies
Two basic technologies dominate current genetic studies of psychiatric conditions: DNA microarrays and high-throughput sequencing (Fig. 1). Microarrays use DNA oligonucleotide probes, which give a sparse picture of the genome, to genotype SNPs and CNVs. The low cost of these arrays (typically around $100–200) enabled the massive GWAS projects of the past decade, to the point that “microarray” and “GWAS” have become nearly synonymous. While the future is clearly in sequencing technologies (see below), microarray platforms, particularly those with custom probe content (such as the Illumina PsychArray, which probes an assortment of variants nominated by researchers in psychiatric genetics) still have their place in certain study designs. Specifically, in studies where assessment of common genetic risk is prioritized over gene discovery, microarrays can be a cost-effective tool.
As DNA sequencing prices have fallen dramatically over the past decade, investigators have more options to consider when designing genetic studies [25]. Whole-exome sequencing (WES) was the first form of high-throughput sequencing to be widely adopted in psychiatric genetics studies, with early studies in autism demonstrating enrichment of deleterious protein-coding mutations [26–29]. In WES, a preliminary capture step is performed that limits input DNA to protein-coding exons, thus reducing the DNA to be sequenced by more than an order of magnitude. While cost was the major consideration that birthed WES as a stopgap technology, investigators found that in contrast to GWAS, where top hits were often in functionally ambiguous noncoding regions, data from WES were comparatively straightforward to interpret: changes to coding DNA sequence affect splicing and amino-acid sequence of the resulting protein in a relatively straightforward way.
As sequencing throughput continues to increase with new technology, the price differential between WES and whole-genome sequencing (WGS) is shrinking. A whole genome sequence currently costs around 2 to 3 times the cost of an exome, yet yields ~30 times more data. Furthermore, the protein-coding fraction of the genome is covered more uniformly in WGS [30], and SVs can be called far more reliably and comprehensively in WGS. However, compared with WES, all these additional data require a substantial investment in storage and computational infrastructure, as well as the expertise to interpret noncoding variation. By far the most widely used technology for WGS is Illumina’s sequencing by synthesis, which sequences about 150 base pairs (75–300 base pairs, depending on the instrument and chemistry) of the ends of DNA fragments. While Illumina’s technology has proven popular and cost-effective, short-read (SR) approaches to sequencing have limitations. SR cannot read through long STRs, and consequently cannot always genotype them. SR can only be used for short-range phasing of variants; it struggles to resolve complex structural variation and low-complexity sequences. Competitors have focused on developing long-read technologies in the hope of luring customers who have run into Illumina’s limitations. PacBio uses a single-molecule real-time approach to achieve reads averaging > 10 kb [31]. This technology was demonstrated to great effect in the sequencing of a hydatidiform mole genome, resulting in a far more comprehensive view into SVs than can be provided by Illumina’s technology [32]. This work also enabled the closure of many interstitial assembly gaps in the human reference genome. While PacBio’s technology delivers impressive results and throughput is improving, its high cost pushes it out of the reach of most laboratories and makes it a poor solution for sequencing large cohorts. A somewhat more cost-effective approach to long-read sequencing has been developed by 10X Genomics [33], called linked read sequencing. This approach uses microfluidics to partition high-molecular-weight DNA molecules into droplets where barcodes are added to amplified fragments, thus introducing the means to “link” the resulting sequencing reads back to a single DNA molecule. The library is sequenced on standard Illumina instrumentation, and a custom alignment and variant-calling pipeline is used to identify and phase SNVs, indels, and SVs. On average, 97% of SNVs were phased into phase blocks ranging from 0.9 to 2.8 Mb in length [33].
While WGS is a powerful technology for comprehensive discovery of genetic variations, targeted sequencing offers a more economical approach for replication studies or other hypothesis-driven designs that target a smaller number of genes and loci. Technologies such as Agilent’s SureSelect use RNA probes to capture targeted genomic regions prior to sequencing library preparation. Another cost-effective approach to targeted sequencing is molecular inversion probes [34]. These are linear DNA oligos whose ends target sequences that flank regions of interest of about 200 base pairs. A gap filling and ligation reaction fills in the targeted sequence and the probe is circularized, the residual linear DNA digested, and the circular capture products amplified through PCR. The pool of PCR products is then sequenced with standard Illumina chemistry. Although the up-front investment in the synthesis of the oligo probes can be substantial, the number of individuals that can be sequenced with the resulting probe pools is, for all practical purposes, limitless. Cost analyses suggest that a panel of 1000 targeted regions can be sequenced for < $10/per individual [35].
Concepts
Here I introduce some concepts that come up routinely in psychiatric genetics. While some of these concepts are illustrated with examples from the field, the major findings are presented in more detail in the “Current Trajectories” section.
GWAS data have seen a renaissance in recent years with an increasing appreciation of the role of common polygenic risk [36–39]. Focus has shifted from array-based GWAS data as the primary means of disease gene discovery to microarrays as a tool to calculate the aggregate genetic risk of individuals for various diseases. Predictive models are trained using large discovery cohorts, such as from the PGC, and then a linear combination of risk alleles is computed in the cohort of interest for each individual, essentially producing a single numerical value for each individual that represents their polygenic risk for the disease in question. These polygenic risk scores are then correlated with other variables of interest to draw inferences about the role of common polygenic risk in the phenotypes of interest. Such analyses, using either a polygenic risk score or an alternative approach called LD score regression [40], are often referred to as studies of genetic correlations [41, 42]. Notable examples of the application of polygenic risk scores in the general population include the finding that polygenic risk for autism is positively correlated with cognitive ability [37] and that polygenic risk for SZ is predictive of creativity [38]. Such examples may serve to explain, at least in part, the evolutionary double-edged sword of polygenic psychiatric risk. Furthermore, it has been shown through genetic correlations that major depressive disorder, bipolar disorder, and SZ share genetic risk factors; and that anorexia and SZ share genetic risk factors [41]. A further permutation of the concept of polygenic risk is the use of predictive models to infer gene expression (rather than disease risk) based on large-scale genotyping data [43, 44]. In this way, the collection of imputed gene expression values may be used as an intermediate phenotype to link trait or disease state to genotype.
As parallel efforts to discover the genetic basis for different common psychiatric conditions have progressed, it has become clear that a substantial number of risk genes confer risk for multiple conditions [42], a phenomenon called pleiotropy [45]. Consequently, the notion that there are “schizophrenia genes” and “autism genes”, and so on, has evolved into the idea that there are simply “brain genes” that can be perturbed in ways and combinations so as to predispose individuals to either SZ or autism or any other neuropsychiatric condition [46]. Certain classes of genes may be more likely to be involved in one condition than another [e.g., chromatin and transcriptional regulators in autism spectrum disorder (ASD)], but the field is young enough that it is difficult to determine whether or not these perceptions of thematic segregation by condition are merely a product of ascertainment bias.
A very specific kind of pleiotropy has been observed in connection with some well-studied CNVs: reciprocal (or mirror) phenotypes. When a deletion and its reciprocal duplication (i.e., of the same genetic material) results in phenotypes that are correspondingly mirrored at opposite ends of a spectrum, that locus is said to be involved in a reciprocal phenotype. The implication is that phenotypes associated with these loci vary in a semi-quantitative way with gene dosage. Notable examples include variation of head circumference (i.e., trends toward microcephaly or macrocephaly) and body mass index with comorbid neuropsychiatric features observed in 16p11.2 [47, 48], head circumference in 1q21.1 [49], and head circumference, stature, and bone maturation rates at 5q35 [50]. Reciprocal CNVs at a number of genomic loci that confer reciprocal risk for autism or SZ (1q21.1, 16p11.2, 22q11.21, and 22q13.3) have led to a hypothesis that, in some respects, these 2 conditions might be considered reciprocal phenotypes [51].
As sequencing prices have continued to fall, it has become practical to sequence large numbers of trios with the aim of identifying putatively causal de novo mutations (DNMs). These are new mutations that do not affect the parents but that occur sporadically in either the sperm or egg haploid genomes, thus propagating to all cells in the offspring. This approach to gene discovery is particularly fruitful when family history is strongly suggestive of the condition being sporadic: the proband’s genome is compared with the parents’ genomes, and genetic variants that are absent in both parents are candidate DNMs (sequencing errors lead to many false-positives and best practices require confirmation of putative DNMs with an additional genotyping technology). Early exome studies of autism rapidly expanded the list of autism risk genes by identifying genes that were hit by de novo and presumed damaging mutations in multiple individuals [26–29]. Further, these studies showed that individuals with autism do not have a greater burden of exonic DNMs, although their DNMs are more likely to be damaging. WGS studies of autism and other samples showed that a disproportionate number of DNMs (~75%) are transmitted from the father, that DNM burden is positively correlated with paternal age (about 1 DNM per year of paternal age), that DNMs cluster together in a nonrandom fashion, that humans harbor about 50 to 100 single-nucleotide DNMs each, and that single-nucleotide mutation rate varies substantially across the genome [52, 53]. As DNMs have undergone only a single round of selective pressure, they are attractive candidates for potentially causal variants of large effect.
Depending on the comprehensiveness of the genetic study, there may be many thousands of variants of interest, and a variant annotation scheme must be used to prioritize candidates for further investigation. For protein-coding variants, indicators of the functional consequence (amino-acid change, premature STOP codon, splice-site disrupting, etc.) can be assigned in a straightforward manner. Variants are also annotated according to the frequency of the minor allele in a population sample, and this is used to classify the variant as common or rare (with thresholds varying, but usually either < 0.05 or < 0.01 qualifies as rare). Measures of selective constraint, such as PolyPhen [54], Genomic Evolutionary Rate Profiling (GERP) [55], or Combined Annotation Dependent Depletion (CADD) [56] are often used as an indicator of how deleterious the variant is. In addition, noncoding variants are sometimes annotated according to whether they intersect known regulatory elements or epigenetic marks, which may give further clues as to their regulatory consequences.
The impact of a genetic variant does not occur in a vacuum; it is often modulated by other genetic factors or the environment. This modulatory effect can result in incomplete penetrance (when some carriers of a damaging variant do not show the corresponding phenotype) or variable expressivity (when the associated phenotype manifests itself differently in individuals). When the effect of a variant depends upon the genetic background of an individual or the genotype at another locus, the effect is said to be epistatic. Epistatic effects are often called gene–gene interactions, and epistasis has been used in model organisms as a means to flesh out functional gene networks [57], though in humans the phenomenon has not been observed as extensively. Nevertheless, clear examples of epistasis have been observed in human neuropsychiatric and developmental conditions [58–64].
In a way that is comparable to epistasis, the sex of the variant carrier can have a decisive effect on the phenotypic expression of the variant. Perhaps the most striking example of this is the well-known male bias in ASD [65]. This work has led to a broader idea of the “female protective effect” in neurodevelopment in general [66–68], where females can carry higher levels of genetic risk than males, while remaining largely asymptomatic. The idea of a female protective effect is attractive, in part, because of its therapeutic implications: if the biological pathways that make females more resilient in the face of genetic insult can be better understood, then perhaps they can be exploited to therapeutic effect [66, 68, 69]. The level of sexual dimorphism in neuropsychiatric conditions varies along a spectrum ranging from extreme male bias in autism and other neurodevelopmental conditions to extreme female bias in eating disorders and anxiety [70]. Because of the crucial role that sex plays in the phenotypic expression of genetic conditions, it should be a core factor in experimental designs, and not ignored for convenience sake.
As with sex, the environment can play a crucial modulatory role in the expression of genetic disease, including autism [71, 72], major depression [73], SZ [74, 75], and others. The prototypical example of the gene–environment interaction in neuropsychiatry is post-traumatic stress disorder (PTSD) [76, 77], where not all individuals who suffer a traumatic event develop PTSD. It is thought that there is a latent genetic risk that, when combined with a traumatic event, manifests as PTSD.
Current Trajectories of Genetic Discovery
In the wake of the completion of the Human Genome Project, psychiatric geneticists have exploited the above technologies and concepts to learn more about the biological nature of neuropsychiatric conditions. Below I provide a brief synopsis of the recent trajectory of genetic discovery for some of the major neurodevelopmental and neuropsychiatric conditions. Emphasis is given to neurodevelopmental conditions, and these are not intended to be comprehensive; indeed, the breadth of the field makes a comprehensive review impossible. Rather, the goal is to illustrate patterns of inquiry currently in use across conditions. Table 1 summarizes the various consortia and working groups (with websites, where available) undertaking research in each condition. While not exhaustive, Table 1 focuses mostly on consortia that have demonstrated their productivity through multiple peer-reviewed publications over the last 5 years.
Table 1.
List of active consortia, initiatives, and working groups by condition
| Condition | Group | URL |
|---|---|---|
| ID | Simons Foundation Variation in Individuals (VIP) Project | https://simonsvipconnect.org/ |
| Care4Rare Canada Consortium | http://care4rare.ca/ | |
| FORGE Canada Consortium | https://www.genomebc.ca/research-programs/projects/health/finding-of-rare-disease-genes-in-canada-forge-canada/ | |
| Cardiff University Experiences of Children With Copy Number Variants (ECHO) Study | http://www.cardiff.ac.uk/mrc-centre-neuropsychiatric-genetics-genomics/research/themes/developmental-disorders/echo-study-cnv-research | |
| 16p11.2 European Consortium | ||
| EuroEPINOMICS RES Consortium | http://archives.esf.org/coordinating-research/eurocores/programmes/euroepinomics.html | |
| Array Referral Consortium | ||
| Childhood Overgrowth Consortium | ||
| Autism Sequencing Consortium | https://genome.emory.edu/ASC/ | |
| Urea Cycle Disorders Consortium | https://www.rarediseasesnetwork.org/cms/ucdc | |
| Japanese Mental Retardation Consortium | ||
| International Molecular Genetic Study of Autism Consortium (IMGSAC) | ||
| South-East Netherlands NIPT Consortium | http://niptconsortium.nl/ | |
| Unique Rare Chromosome Disorder Support Group | http://www.rarechromo.org/html/home.asp | |
| Pediatric Craniofacial Malformation (PECRAM) Study Group | ||
| Danish Fetal Medicine Research Group | ||
| Danish Clinical Genetics Study Group | ||
| Tuberous Sclerosis 2000 Study Group | ||
| Deciphering Developmental Disorders (DDD) Study | https://www.ddduk.org/ | |
| ASD | Simons Foundation Autism Research Initiative (SFARI) | https://sfari.org |
| Simons Foundation Variation in Individuals (VIP) Project | https://simonsvipconnect.org/ | |
| Autism Sequencing Consortium | https://genome.emory.edu/ASC/ | |
| Autism Genome Project (AGP) Consortium | http://nationalautismnetwork.com/research/research-initiatives/autism-genome-project.html | |
| 16p11.2 European Consortium | ||
| Psychiatric Genomics Consortium (Autism Spectrum Disorders Working Group) | https://www.med.unc.edu/pgc/pgc-workgroups | |
| iPSYCH | http://ipsych.au.dk/ | |
| ADHD | Psychiatric Genomics Consortium (ADHD Working Group) | https://www.med.unc.edu/pgc/pgc-workgroups |
| IMAGEN Consortium | https://imagen-europe.com | |
| IMpACT | http://www.impactadhdgenomics.com/ | |
| Comorbid Conditions of Attention deficit / hyperactivity disorder (CoCa) | ||
| iPSYCH | http://ipsych.au.dk/ | |
| DLD | SLI Consortium (not active) | |
| TS | European Multicentre Tics in Children Study (EMTICS) | http://www.emtics.eu |
| TS-EUROTRAIN | http://ts-eurotrain.eu | |
| Tourette International Collaborative Genetics (TIC Genetics) | https://tic-genetics.org | |
| SZ | Psychiatric Genomics Consortium (Schizophrenia Working Group) | https://www.med.unc.edu/pgc/pgc-workgroups |
| International Schizophrenia Consortium | https://www.ncbi.nlm.nih.gov/pubmed/19571811 | |
| Molecular Genetics of Schizophrenia Consortium | ||
| MooDS Consortium | ||
| Genetic Risk Outcome of Psychosis (GROUP) Consortium | ||
| Psychosis Endophenotypes International Consortium | https://www.ucl.ac.uk/psychiatry/biomarkers-for-psychosis/peic | |
| Wellcome Trust Case Control Consortium | https://www.wtccc.org.uk | |
| iPSYCH | http://ipsych.au.dk/ | |
| BPD | Psychiatric Genomics Consortium (Bipolar Disorder Working Group) | https://www.med.unc.edu/pgc/pgc-workgroups |
| MooDs Consortium | ||
| Bipolar Genome Study (BiGS) Consortium | ||
| Wellcome Trust Case Control Consortium | https://www.wtccc.org.uk | |
| NIMH Genetics Initiative Bipolar Disorder Consortium | https://www.nimhgenetics.org/available_data/bipolar_disorder/ | |
| Bipolar Sequencing Consortium | http://metamoodics.org/bsc/consortium/ | |
| iPSYCH | http://ipsych.au.dk/ | |
| MDD | Psychiatric Genomics Consortium (Major Depressive Disorder Working Group) | https://www.med.unc.edu/pgc/pgc-workgroups |
| CONVERGE Consortium | ||
| MooDS Consortium | ||
| GenRED Consortium | ||
| iPSYCH | http://ipsych.au.dk/ |
ID = intellectual disability; ASD = autism spectrum disorders; ADHD = attention deficit/hyperactivity disorder; DLD = developmental language disorder; TS = Tourette syndrome; BPD = bipolar disorder; MDD = major depressive disorder
Intellectual Disability
Intellectual disability (ID) is a genetically heterogeneous neurodevelopmental disorder characterized by impaired adaptive functioning and low IQ. Prevalence of ID is estimated to be between 0.05% and 1.55% [78]. ID is often a comorbidity in other neuropsychiatric conditions [79, 80] and is especially prevalent in SZ and ASD, as well as attention deficit/hyperactivity disorder (ADHD) [81]. Because of its profound effect on fecundity, severe ID is presumed to be largely monogenic and not familial [82], though the total number of risk genes has yet to be enumerated. At the same time, ID has varying levels of severity and it has been shown that some less severe forms of ID are the result of complex polygenic inheritance [82], essentially representing the lower range of intelligence, which itself has been shown to have a heritability of 0.4 to 0.8 [83].
ID has classically been linked to large-scale structural variations in the genome, and these have been extensively reviewed [84]. Recent WGS and WES studies, which can resolve deleterious genetic variants at a much finer resolution, have shored up known ID genes, discovered new ones, and demonstrated emergent patterns. One recent study [85] performed WGS on 50 individuals diagnosed with severe ID and their unaffected parents and found a clear genetic cause in 42% of the subjects (this was estimated to generalize to a rate of 62% in a new sample where microarray and WES analyses had not already been performed). This study, like others, found an excess of protein-coding de novo mutations, as well as enrichment of de novo hits among a list of 528 known ID genes. The use of WGS was vindicated by the discovery of several de novo SVs that escaped detection by microarray technology. As expected, slightly lower diagnostic yields are obtained with WES [86]. More recently, a meta-analysis of > 2000 trios with ID found statistical associations of rare and de novo variants in 10 newly identified candidates: DLG4, PPM1D, RAC1, SMAD6, SON, SOX5, SYNCRIP, TCF20, TLK2, and TRIP12 [87]. These genes were shown to be intolerant to functional nonsynonymous variants.
As a complement to the unbiased, genome-wide studies, many recent studies have followed the “genotype-first” approach [88], where a sample of individuals carrying damaging mutations in the same gene is assembled, and then characterized extensively from a phenotypic standpoint. Examples include POGZ [89], DYRK1A [90], TBCK [91], EBF3 [92], CHAMP1 [93], and IARS [94].
Inborn errors of metabolism represent a well-known cause of ID [95], and are attractive from a research perspective because of the potential for addressing or even preventing negative outcomes through dietary supplementation or restriction [95, 96]. Well-studied examples include phenylketonuria [97], defects in creatine transport [98], branched-chain amino-acid metabolism [99, 100], glycosylation [101], and lysosomal storage disorders [102]. A recent network analysis of ID risk genes underscored the central role of metabolic pathways, and highlighted other areas of convergence in ID, including genes involved in nervous system development, RNA metabolism, transcription, hedgehog signaling, glutamate signaling, peroxisomes, glycosylation, and cilia [103].
A number of pharmacological strategies targeting ID are under development [104]. Risperidone was shown in early studies to significantly ameliorate problematic behaviors [105, 106], though significant side effects temper enthusiasm about its use [107]. Furthermore, because it does not directly target pathways known to be dysregulated in ID, it cannot be considered a specific therapy for ID. Other approaches justified by the known molecular pathologies have been investigated including mGluR5 antagonists [108–111], ampakines [112–114], and γ-aminobutryic acid B agonists [115]. The mTOR pathway has also been a target of drug development in the context of ID [116], where rapamycin and other inhibitors of mTOR function show promise [117].
ASD
ASD is a phenotypically and genetically heterogeneous collection of neurodevelopmental conditions that entail impairments in social communication and restrictive and repetitive behaviors. Heritability of ASD ranges widely depending on study design and confounds, but recent estimates put it at 0.5 to 0.54 [36, 118]. Current estimates of prevalence are at 1 in 68 [119], and ASD encompasses enormous phenotypic diversity, from profoundly affected nonverbal individuals with comorbid ID to highly intelligent but socially impaired individuals. Correspondingly, there is no single genetic architecture for ASD. Some well-known monogenic syndromes show ASD as a common feature, including Rett syndrome (MECP2), Fragile X syndrome (FMR1), and Angelman syndrome (UBE3A). However, recent work has shown that most risk for ASD lies in the cumulative effect of thousands of common risk variants [36]. It is estimated that there are likely hundreds of ASD risk genes [26], and recent coordinated and individual efforts have been aimed at enumerating them. No other neurodevelopmental condition has been investigated with sequencing technology as intensively as ASD, and many early analytical and methodological advances were made to enable the analysis of ASD sequencing data [120–122].
In contrast to sequencing studies, ASD GWAS projects have yet to suggest robustly associated loci, and data from these studies have shown more traction in developing polygenic risk scores for ASD [37, 123] than for gene discovery. Array-based CNV studies have been more productive from a locus discovery perspective, through their focus on detecting rare variation [14, 124]. In 2012, a series of coordinated WES studies of ASD were published that used de novo mutation as a means for gene discovery [26–29]. Shortly thereafter came the first WGS studies of autism [52, 125]. These studies, especially the WES studies with their larger numbers, quickly expanded the list of candidate ASD genes, and multiple follow-up studies confirmed the association of some of these genes with ASD, notably CHD8, DYRK1A, GRIN2B, TBR1, PTEN, and TBL1XR1 [126], POGZ [89], and ADNP [127]. One emerging theme is that ASD risk genes are likely to be regulatory targets of fragile X mental retardation protein (FMRP), the protein encoded by the Fragile X gene, FMR1 [27]. Other risk genes are involved in synaptic structure and integrity, such as neuroligins/neurexins [128, 129], SHANK proteins [130], and contactin proteins [131]. Still other ASD risk genes are involved in broad regulation of chromatin structure and transcriptional regulation, such as CHD8 [28, 29, 126, 132, 133], TBR1 [126, 134], and TCF4 [132, 135].
Effective treatment options for ASD are likely to be highly individualized, owing to the extensive heterogeneity of the condition. Indeed, much of the most promising treatment development work is done in genetically defined forms of ASD, such as Fragile X [136], Rett syndrome [137–139], and others [100, 140–143].
Several consortia and foundations have been and continue to be driving forces in ASD genetics. The Simons Foundation has built extensive infrastructure and data sets for ASD researchers to use, including SFARI gene, which offers a curated list of ASD candidate genes. In addition, sequencing data and biospecimens from the Simons Simplex Collection are available to qualified investigators. The Simons Foundation also sponsors the Variation in Individuals Project, which focuses on genetically defined forms of ASD such as 16p11.2 and 1q21.1. Other consortia and projects actively contributing to research in the genetics of ASD include the ARRA Autism Sequencing Collaboration, the Autism Genome Project Consortium, the 16p11.2 European Consortium, and the Psychiatric Genomics Consortium.
ADHD
ADHD is a highly heritable neurodevelopmental condition that presents with impairments in sustaining attention and an inability to control impulses and activity level. In addition to the previously mentioned comorbidity with ID, ADHD shows high comorbidity with ASD within individuals and family members [144]. Prevalence of ADHD is estimated at 5% to 7% in children [145, 146] and 3% to 5% in adults [147, 148]. Twin studies estimate high heritability (70–80% [149, 150]), and ADHD is genetically heterogeneous and a host of candidate genes have been suggested [151–153]. Candidate gene studies (i.e., where only one or a handful of polymorphisms are studied) suffer from ascertainment bias, and many of the positive associations in candidate gene studies of neurotransmitter pathway genes are not represented in genome-wide studies, where a higher burden of proof exists owing to multiple hypothesis testing. However, some integrative analyses have indicated that pooling of common variants within these neurotransmission pathways may lead to statistical significance [154, 155].
Perhaps the most compelling theme to emerge from the genome-scale studies of ADHD is glutamatergic neurotransmission. A large study of CNVs implicated metabotropic glutamate receptors, as well as functionally related genes, in the etiology of ADHD [156]. This work led to the repositioning of the drug NFC-1 (fasoracetam monohydrate), which stimulates metabotropic glutamate receptors, for treatment of ADHD in individuals with confirmed mutations in mGluR genes (clinical trial ID NCT02777931). A subsequent study of CNVs in ADHD also found a link to metabotropic glutamate receptors [157]. Imaging studies have suggested abnormal glutamate levels in cortical and subcortical brain regions [158]. Glutamatergic signaling has gradually emerged as a point of overlap between ADHD and ASD [159, 160]. Methylphenidate, a common medication for ADHD that acts as a dopamine–norepinephrine reuptake inhibitor, was found to modulate the number of surface glutamate receptor subunits in a dose-dependent, bidirectional fashion [161]. Furthermore, genetic variation in GRM7 was associated with response to methylphenidate [162].
Synaptic adhesion molecule LPHN3, which regulates synaptic density and development [163], was found to be associated with ADHD by a linkage study [164] and has been robustly replicated since then [165–175]. LPHN3 variants have been shown to be predictive of methylphenidate response in ADHD [173]. FLRT3 encodes a ligand for LPHN3 [176–178], and has itself shown suggestive genetic associations to ADHD [179–182].
Other studies of rare variation in ADHD have implicated signal transduction genes NT5DC1, PSD, SEC23IP, and ZCCHC4 [183], and a small-scale exome sequencing study found a significant excess of rare variation in 51 preselected ADHD candidate genes [184].
Developmental Language Disorder
Developmental Language Disorder (DLD), also known as specific language impairment, language impairment, or language disorder, is a neurodevelopmental condition that impairs expressive and receptive language ability that is not attributable to hearing loss or severe ID. Prevalence is estimated at 7% [185] and heritability is moderate to high [186–189]. It is highly comorbid with ADHD, with ~40% of individuals with DLD also having an ADHD diagnosis [190].
Early genetic studies of language ability were driven by pedigrees with very pronounced phenotypes, and these led to seminal discoveries such as disruptive variation in FOXP2 being associated with impaired language abilities [191–194]. However, subsequent studies of common variation have not been able to provide strong support for these genes playing a major role in DLD [195, 196]. Genome-wide studies of common variation are generally woefully underpowered and have not produced findings that withstand multiple testing correction [197, 198].
A recent study that combined GWAS and exome sequencing in an isolated population implicated SETBP1 as a language-associated gene, and this association was replicated in an admixed sample [199]. Furthermore, through exome sequencing of the most severely affected individuals in the sample, a number of putatively disrupting variants were found and, together with GWAS candidates, these genes were enriched as transcriptional targets of MEF2A. WES was used in another study that implicated NFXL1, and then confirmed the association with language ability in 2 other samples [200]. These studies illustrate that movement toward sequencing technologies will bear fruit because of its ability to combine analysis of common and rare variation together.
Tourette Syndrome
Tourette syndrome (TS) is a neurodevelopmental condition that presents with disruptive motor and vocal tics [201]. Although early work suggested a monogenic, autosomal dominant mode of inheritance [202], subsequent findings demonstrated substantial genetic and phenotypic heterogeneity [203–205]. TS shows strong comorbidities with obsessive–compulsive disorder and ADHD [206], with other mood, anxiety, and disruptive behavior disorders occurring in about 30% of probands. While many of the specific genes underlying TS remain elusive, over the last decade, histidine decarboxylase, a key enzyme in the biosynthesis of histamine, has emerged from human genetic studies as an important player in the etiology of the disease [207–209], with animal models displaying TS-like phenomenology [210] and implicating an interaction between dopaminergic and histaminergic systems in the basal ganglia. Administration of haloperidol and histamine were shown to rescue TS-like behavioral and molecular characteristics in this model.
A number of common variant studies of TS have been carried out, some at the genome-wide scale, with less conclusive results. The first GWAS of TS failed to reach significance for any SNP [211]. A follow-up study [212] that included 42 of the top candidates from the initial GWAS found a significant association at rs2060546, near the gene that encodes the axon guidance protein netrin 4 (NTN4), which shows strong expression in the striatum. A recent attempt at replicating this association failed [213]; however, a meta-analysis of 3 cohorts showed significance and consistent direction of effect.
Several consortia are currently undertaking large-scale genetic studies of TS. The European Multicentre Tics in Children Study (EMTICS) seeks to elucidate gene–environment interactions, including the involvement of infection and immune mechanisms in TS etiology. Two patient cohorts form the basis of EMTICS: the ONSET study involves follow-up of 375 high-risk children aged 3 to 10 years who have an immediate family member with a diagnosis of TS and at study entry have no tics. COURSE is a longitudinal study that is following 700 children and adolescents (aged 3–16 years) with a known chronic tic disorder or TS. The study began in March 2013 and the study will conclude in 2017.
TS-EUROTRAIN is a training network that acts as a platform to unify large-scale TS studies and educate the next generation of experts. TS-EUROTRAIN is notable for completing the first epigenome-wide association study for tics, analyzing data from the Netherlands Twin Register [214]. This study interrogated 411,469 autosomal methylation sites in 1678 individuals. Although no site reached genome-wide significance, the top hits include several genes and regions previously associated with neurological disorders and warrant further investigation.
The Tourette International Collaborative Genetics (TIC Genetics) study [215, 216] is currently finishing analysis of WES data from 325 simplex TS trios, with the main focus being the detection of de novo SNVs and indels.
Adult-Onset Disorders
Schizophrenia
Schizophrenia (SZ) is a complex, highly heritable, and heterogeneous psychiatric disorder that presents with positive (psychosis, hallucinations) and negative (apathy, blunted affect, social withdrawal, poverty of speech, anhedonia) symptoms, with associated cognitive deficits. Its prevalence is estimated at 1% [217] and it is highly heritable (65–81% [218, 219]). Significant comorbidities include substance abuse (47%) and depression (50%), as well as anxiety disorders (29% PTSD, 15% panic disorder, and 23% obsessive–compulsive disorder) [220]. Despite the wide appreciation of the genetic roots of SZ, its etiology and origins are still not fully understood [221], though some recent exciting progress has been made that is beginning to chip away at the complex physiology that underlies SZ.
It is clear that SZ is polygenic [222, 223], and, indeed, the most compelling study of the last several years identified 108 loci as SZ-risk loci [224]. One of these hits resides in the major histocompatibility complex, a notoriously difficult region to resolve from a genotype standpoint. Despite this challenge, recent work that focused on elucidating the underlying source of the association signal in this region found that the complement component gene C4 drives the underlying association, and that risk alleles result in higher expression of C4A [225, 226]. C4 protein is expressed at the synapse and plays a role in synaptic pruning, thus implicating overactive synaptic pruning as a mechanism underlying schizophrenia risk.
A recent large-scale WES study of SZ [222] implicated voltage-gated calcium ion channels and proteins comprising the ARC postsynaptic protein complex as harboring an overabundance of putatively functional rare variation. As has been noted for genes affected by damaging variation in ASD [27], this study showed an enrichment of FMRP targets affected by damaging variation in SZ probands. These and other genetic findings implicate synaptic genes as a major theme in the genetic etiology of SZ [227], suggesting that development of therapeutics should involve the targeting of synaptic proteins and processes.
Bipolar Disorder
Bipolar disorder (BPD) is a heritable psychiatric condition marked by alternating episodes of mania and depression. Prevalence is 2% to 3% [228], and heritability may be as high as 80% [229]. Individuals with BPD are at an 8- to 10-fold increased risk for suicide [230]. BPD is a complex and genetically heterogeneous condition; however, compared with other conditions such as SZ, discovery of genetic risk factors robustly and specifically connected to the condition has been slow. The largest GWAS in BPD to date [231] showed associations in 4 known (MAD1L1, 6q16.1, DDN, and TRANK1) and 2 novel loci (intergenic 9p21.3 and intronic variants in ERBB2). Exome sequencing in a large cohort of familial BPD showed an enrichment of predicted damaging variation in genes previously associated with ASD and SZ, as well as targets of the fragile X protein, FMRP [232], suggesting overlapping genetic risk with these disorders. The trickle of genetic findings in BPD, in contrast to its high heritability, speak to the extensive complexity and heterogeneity of the disorder, and support the need for larger studies of this condition. In the near term, perhaps one of the most promising approaches is in the genetics of lithium response, reviewed in detail in this issue.
Major Depressive Disorder
Major depressive disorder (MDD) is characterized by prolonged depressed mood or loss of interest or pleasure in nearly all activities, together with other disturbances in areas such as sleep, appetite, and psychomotor activity. Its prevalence varies by geographic location but is mostly 8% to 12% [233, 234], and women show more susceptibility than men. Significant comorbidities include dysthymia (20%) and anxiety disorders (21%) [235] . Heritability is estimated at about 37% [236, 237]; however, like BPD, genetic findings that provide compelling biological insights into the disorder have been elusive [238]. Nevertheless, recent studies show promise. The largest MDD GWAS to date, which used data from direct-to-consumer genetics company 23andMe, uncovered 15 loci associated with MDD [239]. Another study used low-coverage WGS and identified SIRT1 and LHPP as potential MDD risk genes [240].
While genetic studies of MDD have not yet yielded enough robust results for pathway analyses to gain traction, a recent gene expression study of individuals with MDD implicated inflammation/immune pathways (specifically interleukin-6 and natural killer signaling pathways) as a marker of MDD [241]. DVL3, a gene that regulates cell proliferation and previously implicated by PGC results [242], was also found to be differentially expressed in this analysis. These findings raise the possibility of targeting inflammation pathways as a means to treat depression [243].
Conclusion
Human genetic studies have been the driving force in bringing to light the underlying biology of psychiatric conditions. The complex nature and genetic heterogeneity of these conditions requires vast sample sizes to power statistically robust associations, and such studies can only be accomplished through continued intra- and international collaboration and an increase in data sharing. While these massive collaborative efforts are necessary for gene discovery, individual laboratories and investigators still play a vital role in contextualizing and deepening our understanding of these genetic associations through focused and hypothesis-driven investigation.
As a complement to these gene-discovery efforts, it is clear that much of the future of psychiatric genetics lies in the “genotype-first” approach to studying genetically defined neuropsychiatric conditions. These studies, which are perhaps the greatest near-term boon that genetics can bestow on therapeutic research, can inform the design of clinical trials and improve the odds of their success while providing crucial insight into the penetrance and variable expressivity displayed among carriers of functionally comparable genetic variations. The Simons Variation in Individuals Project project provides a useful model for considering the promise and the challenges of clustering patients by genetic etiology. Furthermore, emerging online patient networks such as patientslikeme.com and the Interactive Autism Network, though not explicitly genetically defined, also show potential in the way they remove geographic boundaries and allow self-clustering of patients.
From a technological standpoint, microarray technology is waning, though for certain applications it remains an economical option. Sequencing approaches, and, in particular, WGS, allow for comprehensive discovery of all modes and frequencies of genetic variation, not just those designed into an array. Cost and analytical expertise are the prevailing barriers for wider adoption, but these are rapidly diminishing.
Common polygenic risk is becoming a useful tool for comparing the shared genetic basis of disparate psychiatric conditions, for stratifying population samples according to their polygenic risk as part of the study design, as well as for studying genetic correlates in the general population while drawing conclusions about the link between psychiatric risk and traits that may be under positive selection (such as creativity or cognitive ability).
Finally, as genetic and other molecular studies of psychiatric conditions increase our understanding of the basic biology of these disorders, we may find that drugs (or supplements) already on the market may be repurposed to treat underlying causes that manifest as mental illness [100, 243].
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 1224 kb)
Acknowledgments
I thank Brooke McKenna, Dabney Hofammann, Kevin Vervier, Iman Dehzangi, Ethan Bahl, and Tanner Koomar for assisting in the review of the literature, as well as the anonymous reviewers for their constructive feedback. JJM received support from National Institutes of Health grants MH105527 and DC014489, the Simons Foundation (SPARK Clinical Recruitment Site at the University of Iowa), and the Brain and Behavior Research Foundation (NARSAD Young Investigator Award # 22967).
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-017-0551-x) contains supplementary material, which is available to authorized users.
References
- 1.Kurahashi H, Tsutsumi M, Nishiyama S, Kogo H, Inagaki H, Ohye T. Molecular basis of maternal age-related increase in oocyte aneuploidy. Congenit Anom (Kyoto) 2012;52(1):8–15. doi: 10.1111/j.1741-4520.2011.00350.x. [DOI] [PubMed] [Google Scholar]
- 2.Jiang J, Jing Y, Cost GJ, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500(7462):296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonarakis SE. Down syndrome and the complexity of genome dosage imbalance. Nat Rev Genet. 2017;18(3):147–163. doi: 10.1038/nrg.2016.154. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69(2):428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12(5):363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genom Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148(6):1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 9.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 10.Itsara A, Wu H, Smith JD, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20(11):1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy D, Ronemus M, Yamrom B, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70(5):886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Sanders SJ, Ercan-Sencicek AG, Hus V, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25(12):528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 17.Raychaudhuri S, Korn JM, McCarroll SA, et al. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLOS Genet. 2010;6(9):e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7(7):552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 19.Stankiewicz P, Shaw CJ, Withers M, Inoue K, Lupski JR. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res. 2004;14(11):2209–2220. doi: 10.1101/gr.2746604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan H, Chu JY. A brief review of short tandem repeat mutation. Genomics Proteomics Bioinforma. 2007;5(1):7–14. doi: 10.1016/S1672-0229(07)60009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labbadia J, Morimoto RI. Huntington's disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci. 2013;38(8):378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin P, Warren ST. Understanding the molecular basis of fragile X syndrome. Hum Mol Genet. 2000;9(6):901–908. doi: 10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- 23.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teo YY. Common statistical issues in genome-wide association studies: a review on power, data quality control, genotype calling and population structure. Curr Opin Lipidol. 2008;19(2):133–143. doi: 10.1097/MOL.0b013e3282f5dd77. [DOI] [PubMed] [Google Scholar]
- 25.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iossifov I, Ronemus M, Levy D, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neale BM, Kou Y, Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner TN, Hormozdiari F, Duyzend MH, et al. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am J Hum Genet. 2016;98(1):58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhoads A, Au KF. PacBio sequencing and its applications. Genomics Proteomics Bioinforma. 2015;13(5):278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaisson MJ, Huddleston J, Dennis MY, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517(7536):608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng GX, Lau BT, Schnall-Levin M, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34(3):303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantsilieris S, Stessman HA, Shendure J, Eichler EE. Targeted capture and high-throughput sequencing using molecular inversion probes (MIPs) Meth Mol Biol. 2017;1492:95–106. doi: 10.1007/978-1-4939-6442-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niedzicka M, Fijarczyk A, Dudek K, Stuglik M, Babik W. Molecular Inversion Probes for targeted resequencing in non-model organisms. Sci Rep. 2016;6:24051. doi: 10.1038/srep24051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaugler T, Klei L, Sanders SJ, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46(8):881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clarke TK, Lupton MK, Fernandez-Pujals AM, et al. Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Mol Psychiatry. 2016;21(3):419–425. doi: 10.1038/mp.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Power RA, Steinberg S, Bjornsdottir G, et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18(7):953–955. doi: 10.1038/nn.4040. [DOI] [PubMed] [Google Scholar]
- 39.Kong SW, Lee IH, Leshchiner I, et al. Summarizing polygenic risks for complex diseases in a clinical whole-genome report. Genet Med. 2015;17:536–544. doi: 10.1038/gim.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cross-Disorder Group of the Psychiatric Genomics C. Lee SH, Ripke S, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gamazon ER, Wheeler HE, Shah KP, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vervier K, Michaelson JJ. SLINGER: large-scale learning for predicting gene expression. Sci Rep. 2016;6:39360. doi: 10.1038/srep39360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14(7):483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu X, Need AC, Petrovski S, Goldstein DB. One gene, many neuropsychiatric disorders: lessons from Mendelian diseases. Nat Neurosci. 2014;17(6):773–781. doi: 10.1038/nn.3713. [DOI] [PubMed] [Google Scholar]
- 47.Golzio C, Willer J, Talkowski ME, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485(7398):363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacquemont S, Reymond A, Zufferey F, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478(7367):97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brunetti-Pierri N, Berg JS, Scaglia F, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40(12):1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franco LM, de Ravel T, Graham BH, et al. A syndrome of short stature, microcephaly and speech delay is associated with duplications reciprocal to the common Sotos syndrome deletion. Eur J Hum Genet. 2010;18(2):258–261. doi: 10.1038/ejhg.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crespi B, Stead P, Elliot M. Evolution in health and medicine Sackler colloquium: comparative genomics of autism and schizophrenia. Proc Natl Acad Sci U S A. 2010;107(Suppl; 1):1736–1741. doi: 10.1073/pnas.0906080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michaelson JJ, Shi Y, Gujral M, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151(7):1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong A, Frigge ML, Masson G, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7:Unit7.20. [DOI] [PMC free article] [PubMed]
- 55.Cooper GM, Stone EA, Asimenos G, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costanzo M, VanderSluis B, Koch EN, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitra I, Lavillaureix A, Yeh E, et al. Reverse pathway genetic approach identifies epistasis in autism spectrum disorders. PLOS Genet. 2017;13(1):e1006516. doi: 10.1371/journal.pgen.1006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fatjo-Vilas M, Prats C, Pomarol-Clotet E, et al. Involvement of NRN1 gene in schizophrenia-spectrum and bipolar disorders and its impact on age at onset and cognitive functioning. World J Biol Psychiatry. 2016;17(2):129–139. doi: 10.3109/15622975.2015.1093658. [DOI] [PubMed] [Google Scholar]
- 60.Schott BH, Assmann A, Schmierer P, et al. Epistatic interaction of genetic depression risk variants in the human subgenual cingulate cortex during memory encoding. Transl Psychiatry. 2014;4:e372. doi: 10.1038/tp.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szczepankiewicz A, Leszczynska-Rodziewicz A, Pawlak J, et al. Epistatic interaction between CRHR1 and AVPR1b variants as a predictor of major depressive disorder. Psychiatr Genet. 2013;23(6):239–246. doi: 10.1097/YPG.0000000000000007. [DOI] [PubMed] [Google Scholar]
- 62.Maciukiewicz M, Dmitrzak-Weglarz M, Pawlak J, et al. Analysis of genetic association and epistasis interactions between circadian clock genes and symptom dimensions of bipolar affective disorder. Chronobiol Int. 2014;31(6):770–778. doi: 10.3109/07420528.2014.899244. [DOI] [PubMed] [Google Scholar]
- 63.Nicodemus KK, Hargreaves A, Morris D, et al. Variability in working memory performance explained by epistasis vs polygenic scores in the ZNF804A pathway. JAMA Psychiat. 2014;71(7):778–785. doi: 10.1001/jamapsychiatry.2014.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chiesa A, Lia L, Lia C, et al. Investigation of possible epistatic interactions between GRIA2 and GRIA4 variants on clinical outcomes in patients with major depressive disorder. J Int Med Res. 2013;41(3):809–815. doi: 10.1177/0300060513477295. [DOI] [PubMed] [Google Scholar]
- 65.Werling DM, Geschwind DH. Sex differences in autism spectrum disorders. Curr Opin Neurol. 2013;26(2):146–153. doi: 10.1097/WCO.0b013e32835ee548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Werling DM, Geschwind DH. Recurrence rates provide evidence for sex-differential, familial genetic liability for autism spectrum disorders in multiplex families and twins. Mol Autism. 2015;6:27. doi: 10.1186/s13229-015-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacquemont S, Coe BP, Hersch M, et al. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gockley J, Willsey AJ, Dong S, Dougherty JD, Constantino JN, Sanders SJ. The female protective effect in autism spectrum disorder is not mediated by a single genetic locus. Mol Autism. 2015;6:25. doi: 10.1186/s13229-015-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoffman EJ, Turner KJ, Fernandez JM, et al. Estrogens suppress a behavioral phenotype in zebrafish mutants of the autism risk gene, CNTNAP2. Neuron. 2016;89(4):725–733. doi: 10.1016/j.neuron.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bao AM, Swaab DF. Sexual differentiation of the human brain: relation to gender identity, sexual orientation and neuropsychiatric disorders. Front Neuroendocrinol. 2011;32:214–226. doi: 10.1016/j.yfrne.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 71.Mazina V, Gerdts J, Trinh S, et al. Epigenetics of autism-related impairment: copy number variation and maternal infection. J Dev Behav Pediatr. 2015;36(2):61–67. doi: 10.1097/DBP.0000000000000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mandy W, Lai MC. Annual Research Review: The role of the environment in the developmental psychopathology of autism spectrum condition. J Child Psychol Psychiatry. 2016;57(3):271–292. doi: 10.1111/jcpp.12501. [DOI] [PubMed] [Google Scholar]
- 73.Klengel T, Binder EB. Gene-environment interactions in major depressive disorder. Can J Psychiatry. 2013;58(2):76–83. doi: 10.1177/070674371305800203. [DOI] [PubMed] [Google Scholar]
- 74.Ayhan Y, McFarland R, Pletnikov MV. Animal models of gene-environment interaction in schizophrenia: a dimensional perspective. Prog Neurobiol. 2016;136:1–27. doi: 10.1016/j.pneurobio.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.European Network of National Networks studying Gene–Environment Interactions in Schizophrenia (EU-GEI) van Os J, Rutten BP, et al. Identifying gene-environment interactions in schizophrenia: contemporary challenges for integrated, large-scale investigations. Schizophr Bull. 2014;40(4):729–736. doi: 10.1093/schbul/sbu069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Voisey J, Young RM, Lawford BR, Morris CP. Progress towards understanding the genetics of posttraumatic stress disorder. J Anxiety Disord. 2014;28(8):873–883. doi: 10.1016/j.janxdis.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 77.Smoller JW. The genetics of stress-related disorders: PTSD, depression, and anxiety disorders. Neuropsychopharmacology. 2016;41(1):297–319. doi: 10.1038/npp.2015.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McKenzie K, Milton M, Smith G, Ouellette-Kuntz H. Systematic review of the prevalence and incidence of intellectual disabilities: current trends and issues. Curr Dev Disord Rep. 2016;3(2):104–115. doi: 10.1007/s40474-016-0085-7. [DOI] [Google Scholar]
- 79.Einfeld SL, Ellis LA, Emerson E. Comorbidity of intellectual disability and mental disorder in children and adolescents: a systematic review. J Intellect Develop Disabil. 2011;36(2):137–143. doi: 10.1080/13668250.2011.572548. [DOI] [PubMed] [Google Scholar]
- 80.Morgan VA, Leonard H, Bourke J, Jablensky A. Intellectual disability co-occurring with schizophrenia and other psychiatric illness: population-based study. Br J Psychiatry. 2008;193(5):364–372. doi: 10.1192/bjp.bp.107.044461. [DOI] [PubMed] [Google Scholar]
- 81.Faraone SV, Ghirardi L, Kuja-Halkola R, Lichtenstein P, Larsson H. The familial co-aggregation of attention-deficit/hyperactivity disorder and intellectual disability: a register-based family study. J Am Acad Child Adolesc Psychiatry. 2017;56(2):167–174. doi: 10.1016/j.jaac.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 82.Reichenberg A, Cederlof M, McMillan A, et al. Discontinuity in the genetic and environmental causes of the intellectual disability spectrum. Proc Natl Acad Sci U S A. 2016;113(4):1098–1103. doi: 10.1073/pnas.1508093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Plomin R. Genetics and general cognitive ability. Nature. 1999;402(6761 Suppl.):C25–C29. doi: 10.1038/35011520. [DOI] [PubMed] [Google Scholar]
- 84.Nevado J, Mergener R, Palomares-Bralo M, et al. New microdeletion and microduplication syndromes: a comprehensive review. Genet Mol Biol. 2014;37(1 Suppl.):210–219. doi: 10.1590/S1415-47572014000200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511(7509):344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 86.Reuter MS, Tawamie H, Buchert R, et al. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiatry. 2017;74:293–299. doi: 10.1001/jamapsychiatry.2016.3798. [DOI] [PubMed] [Google Scholar]
- 87.Lelieveld SH, Reijnders MR, Pfundt R, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19(9):1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 88.Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014;156(5):872–877. doi: 10.1016/j.cell.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stessman HA, Willemsen MH, Fenckova M, et al. Disruption of POGZ is associated with intellectual disability and autism spectrum disorders. Am J Hum Genet. 2016;98:541–552. doi: 10.1016/j.ajhg.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bronicki LM, Redin C, Drunat S, et al. Ten new cases further delineate the syndromic intellectual disability phenotype caused by mutations in DYRK1A. Eur J Hum Genet. 2015;23(11):1482–1487. doi: 10.1038/ejhg.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bhoj EJ, Li D, Harr M, et al. Mutations in TBCK, encoding TBC1-domain-containing kinase, lead to a recognizable syndrome of intellectual disability and hypotonia. Am J Hum Genet. 2016;98(4):782–788. doi: 10.1016/j.ajhg.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harms FL, Girisha KM, Hardigan AA, et al. Mutations in EBF3 disturb transcriptional profiles and cause intellectual disability, ataxia, and facial dysmorphism. Am J Hum Genet. 2017;100(1):117–127. doi: 10.1016/j.ajhg.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hempel M, Cremer K, Ockeloen CW, et al. De novo mutations in CHAMP1 cause intellectual disability with severe speech impairment. Am J Hum Genet. 2015;97(3):493–500. doi: 10.1016/j.ajhg.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kopajtich R, Murayama K, Janecke AR, et al. Biallelic IARS mutations cause growth retardation with prenatal onset, intellectual disability, muscular hypotonia, and infantile hepatopathy. Am J Hum Genet. 2016;99(2):414–422. doi: 10.1016/j.ajhg.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Karnebeek CD, Stockler S. Treatable inborn errors of metabolism causing intellectual disability: a systematic literature review. Mol Genet Metab. 2012;105(3):368–381. doi: 10.1016/j.ymgme.2011.11.191. [DOI] [PubMed] [Google Scholar]
- 96.Picker JD, Walsh CA. New innovations: therapeutic opportunities for intellectual disabilities. Ann Neurol. 2013;74(3):382–390. doi: 10.1002/ana.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Al Hafid N, Christodoulou J. Phenylketonuria: a review of current and future treatments. Transl Pediatr. 2015;4(4):304–317. doi: 10.3978/j.issn.2224-4336.2015.10.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van de Kamp JM, Mancini GM, Salomons GS. X-linked creatine transporter deficiency: clinical aspects and pathophysiology. J Inherit Metab Dis. 2014;37(5):715–733. doi: 10.1007/s10545-014-9713-8. [DOI] [PubMed] [Google Scholar]
- 99.Burrage LC, Nagamani SC, Campeau PM, Lee BH. Branched-chain amino acid metabolism: from rare Mendelian diseases to more common disorders. Hum Mol Genet. 2014;23(R1):R1–R8. doi: 10.1093/hmg/ddu123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Novarino G, El-Fishawy P, Kayserili H, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338(6105):394–397. doi: 10.1126/science.1224631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scott K, Gadomski T, Kozicz T, Morava E. Congenital disorders of glycosylation: new defects and still counting. J Inherit Metab Dis. 2014;37(4):609–617. doi: 10.1007/s10545-014-9720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Platt FM, Boland B, van der Spoel AC. The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J Cell Biol. 2012;199(5):723–734. doi: 10.1083/jcb.201208152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kochinke K, Zweier C, Nijhof B, et al. Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet. 2016;98(1):149–164. doi: 10.1016/j.ajhg.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Verpelli C, Galimberti I, Gomez-Mancilla B, Sala C. Molecular basis for prospective pharmacological treatment strategies in intellectual disability syndromes. Dev Neurobiol. 2014;74(2):197–206. doi: 10.1002/dneu.22093. [DOI] [PubMed] [Google Scholar]
- 105.Matson JL, Bamburg JW, Mayville EA, et al. Psychopharmacology and mental retardation: a 10 year review (1990–1999) Res Dev Disabil. 2000;21:263–296. doi: 10.1016/S0891-4222(00)00042-1. [DOI] [PubMed] [Google Scholar]
- 106.Matson JL, Bielecki J, Mayville SB, Matson ML. Psychopharmacology research for individuals with mental retardation: methodological issues and suggestions. Res Dev Disabil. 2003;24(3):149–157. doi: 10.1016/S0891-4222(03)00030-1. [DOI] [PubMed] [Google Scholar]
- 107.Matson JL, Mahan S. Antipsychotic drug side effects for persons with intellectual disability. Res Dev Disabil. 2010;31(6):1570–1576. doi: 10.1016/j.ridd.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 108.Jacquemont S, Curie A, des Portes V, et al. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci Transl Med. 2011;3(64):64ra1. doi: 10.1126/scitranslmed.3001708. [DOI] [PubMed] [Google Scholar]
- 109.Berry-Kravis E, Des Portes V, Hagerman R, et al. Mavoglurant in fragile X syndrome: Results of two randomized, double-blind, placebo-controlled trials. Sci Transl Med. 2016;8(321):321ra5. doi: 10.1126/scitranslmed.aab4109. [DOI] [PubMed] [Google Scholar]
- 110.Gantois I, Pop AS, de Esch CE, et al. Chronic administration of AFQ056/Mavoglurant restores social behaviour in Fmr1 knockout mice. Behav Brain Res. 2013;239:72–79. doi: 10.1016/j.bbr.2012.10.059. [DOI] [PubMed] [Google Scholar]
- 111.Pop AS, Levenga J, de Esch CE, et al. Rescue of dendritic spine phenotype in Fmr1 KO mice with the mGluR5 antagonist AFQ056/Mavoglurant. Psychopharmacology. 2014;231:1227–1235. doi: 10.1007/s00213-012-2947-y. [DOI] [PubMed] [Google Scholar]
- 112.Seese RR, Maske AR, Lynch G, Gall CM. Long-term memory deficits are associated with elevated synaptic ERK1/2 activation and reversed by mGluR5 antagonism in an animal model of autism. Neuropsychopharmacology. 2014;39(7):1664–1673. doi: 10.1038/npp.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Berry-Kravis E, Krause SE, Block SS, et al. Effect of CX516, an AMPA-modulating compound, on cognition and behavior in fragile X syndrome: a controlled trial. J Child Adolesc Psychopharmacol. 2006;16(5):525–540. doi: 10.1089/cap.2006.16.525. [DOI] [PubMed] [Google Scholar]
- 114.Goff DC, Lamberti JS, Leon AC, et al. A placebo-controlled add-on trial of the Ampakine, CX516, for cognitive deficits in schizophrenia. Neuropsychopharmacology. 2008;33(3):465–472. doi: 10.1038/sj.npp.1301444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berry-Kravis EM, Hessl D, Rathmell B, et al. Effects of STX209 (arbaclofen) on neurobehavioral function in children and adults with fragile X syndrome: a randomized, controlled, phase 2 trial. Sci Transl Med. 2012;4(152):152ra27. doi: 10.1126/scitranslmed.3004214. [DOI] [PubMed] [Google Scholar]
- 116.Troca-Marin JA, Alves-Sampaio A, Montesinos ML. Deregulated mTOR-mediated translation in intellectual disability. Prog Neurobiol. 2012;96(2):268–282. doi: 10.1016/j.pneurobio.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 117.Troca-Marin JA, Casanas JJ, Benito I, Montesinos ML. The Akt-mTOR pathway in Down's syndrome: the potential use of rapamycin/rapalogs for treating cognitive deficits. CNS Neurol Disord Drug Targets. 2014;13(1):34–40. doi: 10.2174/18715273113126660184. [DOI] [PubMed] [Google Scholar]
- 118.Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A. The familial risk of autism. JAMA. 2014;311(17):1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Christensen DL, Bilder DA, Zahorodny W, et al. Prevalence and characteristics of autism spectrum disorder among 4-year-old children in the autism and developmental disabilities monitoring network. J Dev Behav Pediatr. 2016;37(1):1–8. doi: 10.1097/DBP.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 120.He X, Sanders SJ, Liu L, et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLOS Genet. 2013;9(8):e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu L, Lei J, Sanders SJ, et al. DAWN: a framework to identify autism genes and subnetworks using gene expression and genetics. Mol Autism. 2014;5(1):22. doi: 10.1186/2040-2392-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Michaelson JJ, Sebat J. forestSV: structural variant discovery through statistical learning. Nat Methods. 2012;9(8):819–821. doi: 10.1038/nmeth.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Robinson EB, St Pourcain B, Anttila V, et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet. 2016;48(5):552–555. doi: 10.1038/ng.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 125.Jiang YH, Yuen RK, Jin X, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93(2):249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O'Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338(6114):1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Helsmoortel C, Vulto-van Silfhout AT, Coe BP, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Reissner C, Runkel F, Missler M. Neurexins. Genome Biol. 2013;14(9):213. doi: 10.1186/gb-2013-14-9-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Krueger DD, Tuffy LP, Papadopoulos T, Brose N. The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr Opin Neurobiol. 2012;22(3):412–422. doi: 10.1016/j.conb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 130.Guilmatre A, Huguet G, Delorme R, Bourgeron T. The emerging role of SHANK genes in neuropsychiatric disorders. Develop Neurobiol. 2014;74(2):113–122. doi: 10.1002/dneu.22128. [DOI] [PubMed] [Google Scholar]
- 131.Zuko A, Kleijer KT, Oguro-Ando A, et al. Contactins in the neurobiology of autism. Eur J Pharmacol. 2013;719(1-3):63–74. doi: 10.1016/j.ejphar.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 132.Blumenthal I, Ragavendran A, Erdin S, et al. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am J Hum Genet. 2014;94(6):870–883. doi: 10.1016/j.ajhg.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Krumm N, Turner TN, Baker C, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47(6):582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Deriziotis P, O'Roak BJ, Graham SA, et al. De novo TBR1 mutations in sporadic autism disrupt protein functions. Nat Commun. 2014;5:4954. doi: 10.1038/ncomms5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sweatt JD. Pitt-Hopkins Syndrome: intellectual disability due to loss of TCF4-regulated gene transcription. Exp Mol Med. 2013;45:e21. doi: 10.1038/emm.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Michalon A, Sidorov M, Ballard TM, et al. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74(1):49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chapleau CA, Lane J, Pozzo-Miller L, Percy AK. Evaluation of current pharmacological treatment options in the management of Rett syndrome: from the present to future therapeutic alternatives. Curr Clin Pharmacol. 2013;8(4):358–369. doi: 10.2174/15748847113086660069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Buchovecky CM, Turley SD, Brown HM, et al. A suppressor screen in Mecp2 mutant mice implicates cholesterol metabolism in Rett syndrome. Nat Genet. 2013;45(9):1013–1020. doi: 10.1038/ng.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sztainberg Y, Chen HM, Swann JW, et al. Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature. 2015;528(7580):123–126. doi: 10.1038/nature16159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shcheglovitov A, Shcheglovitova O, Yazawa M, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503(7475):267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Han S, Tai C, Westenbroek RE, et al. Autistic-like behaviour in Scn1a+/– mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489(7416):385–390. doi: 10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tsai PT, Hull C, Chu Y, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488(7413):647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tian D, Stoppel LJ, Heynen AJ, et al. Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nat Neurosci. 2015;18(2):182–184. doi: 10.1038/nn.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ghirardi L, Brikell I, Kuja-Halkola R, et al. The familial co-aggregation of ASD and ADHD: a register-based cohort study. Mol Psychiatry 2017 Feb 28. [DOI] [PMC free article] [PubMed]
- 145.Polanczyk G, de Lima MS, Horta BL, Biederman J, Rohde LA. The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am J Psychiatry. 2007;164(6):942–948. doi: 10.1176/ajp.2007.164.6.942. [DOI] [PubMed] [Google Scholar]
- 146.Willcutt EG. The prevalence of DSM-IV attention-deficit/hyperactivity disorder: a meta-analytic review. Neurotherapeutics. 2012;9(3):490–499. doi: 10.1007/s13311-012-0135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Simon V, Czobor P, Balint S, Meszaros A, Bitter I. Prevalence and correlates of adult attention-deficit hyperactivity disorder: meta-analysis. Br J Psychiatry. 2009;194(3):204–211. doi: 10.1192/bjp.bp.107.048827. [DOI] [PubMed] [Google Scholar]
- 148.Franke B, Faraone SV, Asherson P, et al. The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol Psychiatry. 2012;17(10):960–987. doi: 10.1038/mp.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Burt SA. Rethinking environmental contributions to child and adolescent psychopathology: a meta-analysis of shared environmental influences. Psychol Bull. 2009;135:608–637. doi: 10.1037/a0015702. [DOI] [PubMed] [Google Scholar]
- 150.Faraone SV, Perlis RH, Doyle AE, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 151.Li Z, Chang SH, Zhang LY, Gao L, Wang J. Molecular genetic studies of ADHD and its candidate genes: a review. Psychiatry Res. 2014;219(1):10–24. doi: 10.1016/j.psychres.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 152.Akutagava-Martins GC, Rohde LA, Hutz MH. Genetics of attention-deficit/hyperactivity disorder: an update. Exp Rev Neurother. 2016;16:145–156. doi: 10.1586/14737175.2016.1130626. [DOI] [PubMed] [Google Scholar]
- 153.Hawi Z, Cummins TD, Tong J, et al. The molecular genetic architecture of attention deficit hyperactivity disorder. Mol Psychiatry. 2015;20(3):289–297. doi: 10.1038/mp.2014.183. [DOI] [PubMed] [Google Scholar]
- 154.Bralten J, Franke B, Waldman I, et al. Candidate genetic pathways for attention-deficit/hyperactivity disorder (ADHD) show association to hyperactive/impulsive symptoms in children with ADHD. J Am Acad Child Adolesc Psychiatry. 2013;52(11):1204–1212 e1. doi: 10.1016/j.jaac.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 155.Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated genome-wide association study findings: identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am J Psychiatry. 2011;168(4):365–377. doi: 10.1176/appi.ajp.2010.10070948. [DOI] [PubMed] [Google Scholar]
- 156.Elia J, Glessner JT, Wang K, et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet. 2011;44:78–84. doi: 10.1038/ng.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Akutagava-Martins GC, Salatino-Oliveira A, Genro JP, et al. Glutamatergic copy number variants and their role in attention-deficit/hyperactivity disorder. Am J Med Genet Part B Neuropsychiatr Genet. 2014;165B(6):502–509. doi: 10.1002/ajmg.b.32253. [DOI] [PubMed] [Google Scholar]
- 158.Maltezos S, Horder J, Coghlan S, et al. Glutamate/glutamine and neuronal integrity in adults with ADHD: a proton MRS study. Transl Psychiatry. 2014;4:e373. doi: 10.1038/tp.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hadley D, Wu ZL, Kao C, et al. The impact of the metabotropic glutamate receptor and other gene family interaction networks on autism. Nat Commun. 2014;5:4074. doi: 10.1038/ncomms5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Naaijen J, Bralten J, Poelmans G, et al. Glutamatergic and GABAergic gene sets in attention-deficit/hyperactivity disorder: association to overlapping traits in ADHD and autism. Transl Psychiatry. 2017;7(1):e999. doi: 10.1038/tp.2016.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cheng J, Xiong Z, Duffney LJ, et al. Methylphenidate exerts dose-dependent effects on glutamate receptors and behaviors. Biol Psychiatry. 2014;76(12):953–962. doi: 10.1016/j.biopsych.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Park S, Kim BN, Cho SC, et al. The metabotropic glutamate receptor subtype 7 rs3792452 polymorphism is associated with the response to methylphenidate in children with attention-deficit/hyperactivity disorder. J Child Adolesc Psychopharmacol. 2014;24(4):223–227. doi: 10.1089/cap.2013.0079. [DOI] [PubMed] [Google Scholar]
- 163.O'Sullivan ML, Martini F, von Daake S, Comoletti D, Ghosh A. LPHN3, a presynaptic adhesion-GPCR implicated in ADHD, regulates the strength of neocortical layer 2/3 synaptic input to layer 5. Neural Dev. 2014;9:7. doi: 10.1186/1749-8104-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Arcos-Burgos M, Jain M, Acosta MT, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry. 2010;15(11):1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- 165.Arcos-Burgos M, Velez JI, Solomon BD, Muenke M. A common genetic network underlies substance use disorders and disruptive or externalizing disorders. Hum Genet. 2012;131(6):917–929. doi: 10.1007/s00439-012-1164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ribases M, Ramos-Quiroga JA, Sanchez-Mora C, et al. Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes Brain Behav. 2011;10(2):149–157. doi: 10.1111/j.1601-183X.2010.00649.x. [DOI] [PubMed] [Google Scholar]
- 167.Fallgatter AJ, Ehlis AC, Dresler T, et al. Influence of a latrophilin 3 (LPHN3) risk haplotype on event-related potential measures of cognitive response control in attention-deficit hyperactivity disorder (ADHD) Eur Neuropsychopharmacol. 2013;23(6):458–468. doi: 10.1016/j.euroneuro.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Acosta MT, Velez JI, Bustamante ML, Balog JZ, Arcos-Burgos M, Muenke M. A two-locus genetic interaction between LPHN3 and 11q predicts ADHD severity and long-term outcome. Transl Psychiatry. 2011;1:e17. doi: 10.1038/tp.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jain M, Velez JI, Acosta MT, et al. A cooperative interaction between LPHN3 and 11q doubles the risk for ADHD. Mol Psychiatry. 2012;17(7):741–747. doi: 10.1038/mp.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Choudhry Z, Sengupta SM, Grizenko N, et al. LPHN3 and attention-deficit/hyperactivity disorder: interaction with maternal stress during pregnancy. J Child Psychol Psychiatry. 2012;53(8):892–902. doi: 10.1111/j.1469-7610.2012.02551.x. [DOI] [PubMed] [Google Scholar]
- 171.Labbe A, Liu A, Atherton J, et al. Refining psychiatric phenotypes for response to treatment: contribution of LPHN3 in ADHD. Am J Med Genet Part B Neuropsychiatr Genet. 2012;159B(7):776–785. doi: 10.1002/ajmg.b.32083. [DOI] [PubMed] [Google Scholar]
- 172.Hwang IW, Lim MH, Kwon HJ, Jin HJ. Association of LPHN3 rs6551665 A/G polymorphism with attention deficit and hyperactivity disorder in Korean children. Gene. 2015;566(1):68–73. doi: 10.1016/j.gene.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 173.Bruxel EM, Salatino-Oliveira A, Akutagava-Martins GC, et al. LPHN3 and attention-deficit/hyperactivity disorder: a susceptibility and pharmacogenetic study. Genes Brain Behav. 2015;14(5):419–427. doi: 10.1111/gbb.12224. [DOI] [PubMed] [Google Scholar]
- 174.Song J, Kim SW, Hong HJ, et al. Association of SNAP-25, SLC6A2, and LPHN3 with OROS methylphenidate treatment response in attention-deficit/hyperactivity disorder. Clin Neuropharmacol. 2014;37(5):136–141. doi: 10.1097/WNF.0000000000000045. [DOI] [PubMed] [Google Scholar]
- 175.Martinez AF, Abe Y, Hong S, et al. An ultraconserved brain-specific enhancer within ADGRL3 (LPHN3) underpins attention-deficit/hyperactivity disorder susceptibility. Biol Psychiatry. 2016;80(12):943–954. doi: 10.1016/j.biopsych.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.O'Sullivan ML, de Wit J, Savas JN, et al. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73(5):903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lu YC, Nazarko OV, Sando R, 3rd, Salzman GS, Sudhof TC, Arac D. Structural basis of latrophilin-FLRT-UNC5 interaction in cell adhesion. Structure. 2015;23:1678–1691. doi: 10.1016/j.str.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lesch KP, Merker S, Reif A, Novak M. Dances with black widow spiders: dysregulation of glutamate signalling enters centre stage in ADHD. Eur Neuropsychopharmacol. 2013;23(6):479–491. doi: 10.1016/j.euroneuro.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 179.Elia J, Gai X, Xie HM, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15(6):637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lionel AC, Crosbie J, Barbosa N, et al. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med. 2011;3(95):95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- 181.Williams NM, Zaharieva I, Martin A, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376(9750):1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lesch KP, Timmesfeld N, Renner TJ, et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm (Vienna) 2008;115(11):1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- 183.Zayats T, Jacobsen KK, Kleppe R, et al. Exome chip analyses in adult attention deficit hyperactivity disorder. Transl Psychiatry. 2016;6(10):e923. doi: 10.1038/tp.2016.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Demontis D, Lescai F, Borglum A, et al. Whole-exome sequencing reveals increased burden of rare functional and disruptive variants in candidate risk genes in individuals with persistent attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2016;55(6):521–523. doi: 10.1016/j.jaac.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 185.Tomblin JB, Records NL, Buckwalter P, Zhang X, Smith E, O'Brien M. Prevalence of specific language impairment in kindergarten children. J Speech Lang Hear Res. 1997;40:1245–1260. doi: 10.1044/jslhr.4006.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lewis BA, Thompson LA. A study of developmental speech and language disorders in twins. J Speech Hear Res. 1992;35(5):1086–1094. doi: 10.1044/jshr.3505.1086. [DOI] [PubMed] [Google Scholar]
- 187.Bishop DV, North T, Donlan C. Genetic basis of specific language impairment: evidence from a twin study. Develop Med Child Neurol. 1995;37(1):56–71. doi: 10.1111/j.1469-8749.1995.tb11932.x. [DOI] [PubMed] [Google Scholar]
- 188.Tomblin JB, Buckwalter PR. Heritability of poor language achievement among twins. J Speech Lang Hear Res. 1998;41(1):188–199. doi: 10.1044/jslhr.4101.188. [DOI] [PubMed] [Google Scholar]
- 189.Dale PS, Simonoff E, Bishop DV, et al. Genetic influence on language delay in two-year-old children. Nat Neurosci. 1998;1(4):324–328. doi: 10.1038/1142. [DOI] [PubMed] [Google Scholar]
- 190.Tomblin JB, Mueller KL. How Can the comorbidity with ADHD aid understanding of language and speech disorders? Top Lang Disord. 2012;32(3):198–206. doi: 10.1097/TLD.0b013e318261c264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413(6855):519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 192.MacDermot KD, Bonora E, Sykes N, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76(6):1074–1080. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Shriberg LD, Ballard KJ, Tomblin JB, Duffy JR, Odell KH, Williams CA. Speech, prosody, and voice characteristics of a mother and daughter with a 7;13 translocation affecting FOXP2. J Speech Lang Hear Res. 2006;49(3):500–525. doi: 10.1044/1092-4388(2006/038). [DOI] [PubMed] [Google Scholar]
- 194.Feuk L, Kalervo A, Lipsanen-Nyman M, et al. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79(5):965–972. doi: 10.1086/508902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mueller KL, Murray JC, Michaelson JJ, Christiansen MH, Reilly S, Tomblin JB. Common genetic variants in FOXP2 are not associated with individual differences in language development. PLOS ONE. 2016;11(4):e0152576. doi: 10.1371/journal.pone.0152576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Newbury DF, Bonora E, Lamb JA, et al. FOXP2 is not a major susceptibility gene for autism or specific language impairment. Am J Hum Genet. 2002;70(5):1318–1327. doi: 10.1086/339931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gialluisi A, Newbury DF, Wilcutt EG, et al. Genome-wide screening for DNA variants associated with reading and language traits. Genes Brain Behav. 2014;13(7):686–701. doi: 10.1111/gbb.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Simpson NH, Ceroni F, Reader RH, et al. Genome-wide analysis identifies a role for common copy number variants in specific language impairment. Eur J Hum Genet. 2015;23(10):1370–1377. doi: 10.1038/ejhg.2014.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kornilov SA, Rakhlin N, Koposov R, et al. Genome-wide association and exome sequencing study of language disorder in an isolated population. Pediatrics. 2016;137(4):e20152469. doi: 10.1542/peds.2015-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Villanueva P, Nudel R, Hoischen A, et al. Exome sequencing in an admixed isolated population indicates NFXL1 variants confer a risk for specific language impairment. PLOS Genet. 2015;11(3):e1004925. doi: 10.1371/journal.pgen.1004925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Paschou P, Fernandez TV, Sharp F, Heiman GA, Hoekstra PJ. Genetic susceptibility and neurotransmitters in Tourette syndrome. Int Rev Neurobiol. 2013;112:155–177. doi: 10.1016/B978-0-12-411546-0.00006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Comings DE, Comings BG, Devor EJ, Cloninger CR. Detection of major gene for Gilles de la Tourette syndrome. Am J Hum Genet. 1984;36(3):586–600. [PMC free article] [PubMed] [Google Scholar]
- 203.Pauls DL, Leckman JF. The inheritance of Gilles de la Tourette's syndrome and associated behaviors. Evidence for autosomal dominant transmission. N Engl J Med. 1986;315(16):993–997. doi: 10.1056/NEJM198610163151604. [DOI] [PubMed] [Google Scholar]
- 204.Walkup JT, LaBuda MC, Singer HS, Brown J, Riddle MA, Hurko O. Family study and segregation analysis of Tourette syndrome: evidence for a mixed model of inheritance. Am J Hum Genet. 1996;59(3):684–693. [PMC free article] [PubMed] [Google Scholar]
- 205.State MW. The genetics of Tourette disorder. Curr Opin Genet Dev. 2011;21:302–309. doi: 10.1016/j.gde.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hirschtritt ME, Lee PC, Pauls DL, et al. Lifetime prevalence, age of risk, and genetic relationships of comorbid psychiatric disorders in Tourette syndrome. JAMA Psychiat. 2015;72(4):325–333. doi: 10.1001/jamapsychiatry.2014.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ercan-Sencicek AG, Stillman AA, Ghosh AK, et al. L-histidine decarboxylase and Tourette's syndrome. N Engl J Med. 2010;362(20):1901–1908. doi: 10.1056/NEJMoa0907006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fernandez TV, Sanders SJ, Yurkiewicz IR, et al. Rare copy number variants in tourette syndrome disrupt genes in histaminergic pathways and overlap with autism. Biol Psychiatry. 2012;71(5):392–402. doi: 10.1016/j.biopsych.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Karagiannidis I, Dehning S, Sandor P, et al. Support of the histaminergic hypothesis in Tourette syndrome: association of the histamine decarboxylase gene in a large sample of families. J Med Genet. 2013;50(11):760–764. doi: 10.1136/jmedgenet-2013-101637. [DOI] [PubMed] [Google Scholar]
- 210.Castellan Baldan L, Williams KA, Gallezot JD, et al. Histidine decarboxylase deficiency causes tourette syndrome: parallel findings in humans and mice. Neuron. 2014;81(1):77–90. doi: 10.1016/j.neuron.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Scharf JM, Yu D, Mathews CA, et al. Genome-wide association study of Tourette's syndrome. Mol Psychiatry. 2013;18(6):721–728. doi: 10.1038/mp.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Paschou P, Yu D, Gerber G, et al. Genetic association signal near NTN4 in Tourette syndrome. Ann Neurol. 2014;76(2):310–315. doi: 10.1002/ana.24215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Padmanabhuni SS, Houssari R, Esserlind AL, et al. Investigation of SNP rs2060546 Immediately upstream to NTN4 in a Danish Gilles de la Tourette syndrome cohort. Front Neurosci. 2016;10:531. doi: 10.3389/fnins.2016.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zilhao NR, Padmanabhuni SS, Pagliaroli L, et al. Epigenome-wide association study of tic disorders. Twin Res Hum Genet. 2015;18(6):699–709. doi: 10.1017/thg.2015.72. [DOI] [PubMed] [Google Scholar]
- 215.Abdulkadir M, Tischfield JA, King RA, et al. Pre- and perinatal complications in relation to Tourette syndrome and co-occurring obsessive-compulsive disorder and attention-deficit/hyperactivity disorder. J Psychiatr Res. 2016;82:126–135. doi: 10.1016/j.jpsychires.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dietrich A, Fernandez TV, King RA, et al. The Tourette International Collaborative Genetics (TIC Genetics) study, finding the genes causing Tourette syndrome: objectives and methods. Eur Child Adolesc Psychiatry. 2015;24(2):141–151. doi: 10.1007/s00787-014-0543-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Regier DA, Narrow WE, Rae DS, Manderscheid RW, Locke BZ, Goodwin FK. The de facto US mental and addictive disorders service system. Epidemiologic catchment area prospective 1-year prevalence rates of disorders and services. Arch Gen Psychiatry. 1993;50:85–94. doi: 10.1001/archpsyc.1993.01820140007001. [DOI] [PubMed] [Google Scholar]
- 218.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 219.Lichtenstein P, Yip BH, Bjork C, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373(9659):234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Buckley PF, Miller BJ, Lehrer DS, Castle DJ. Psychiatric comorbidities and schizophrenia. Schizophr Bull. 2009;35(2):383–402. doi: 10.1093/schbul/sbn135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rees E, O’Donovan MC, Owen MJ. Genetics of schizophrenia. Curr Opin Behav Sci. 2015;2:8–14. doi: 10.1016/j.cobeha.2014.07.001. [DOI] [Google Scholar]
- 222.Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Neale BM, Sklar P. Genetic analysis of schizophrenia and bipolar disorder reveals polygenicity but also suggests new directions for molecular interrogation. Curr Opin Neurobiol. 2015;30:131–138. doi: 10.1016/j.conb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 224.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sekar A, Bialas AR, de Rivera H, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530(7589):177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dhindsa RS, Goldstein DB. Schizophrenia: from genetics to physiology at last. Nature. 2016;530(7589):162–163. doi: 10.1038/nature16874. [DOI] [PubMed] [Google Scholar]
- 227.Hall J, Trent S, Thomas KL, O'Donovan MC, Owen MJ. Genetic risk for schizophrenia: convergence on synaptic pathways involved in plasticity. Biol Psychiatry. 2015;77(1):52–58. doi: 10.1016/j.biopsych.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 228.Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62(6):617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Barnett JH, Smoller JW. The genetics of bipolar disorder. Neuroscience. 2009;164(1):331–343. doi: 10.1016/j.neuroscience.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Crump C, Sundquist K, Winkleby MA, Sundquist J. Comorbidities and mortality in bipolar disorder: a Swedish national cohort study. JAMA Psychiat. 2013;70(9):931–939. doi: 10.1001/jamapsychiatry.2013.1394. [DOI] [PubMed] [Google Scholar]
- 231.Hou L, Bergen SE, Akula N, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet. 2016;25:3383–3394. doi: 10.1093/hmg/ddw181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Goes FS, Pirooznia M, Parla JS, et al. Exome sequencing of familial bipolar disorder. JAMA Psychiat. 2016;73(6):590–597. doi: 10.1001/jamapsychiatry.2016.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Demyttenaere K, Bruffaerts R, Posada-Villa J, et al. Prevalence, severity, and unmet need for treatment of mental disorders in the World Health Organization World Mental Health Surveys. JAMA. 2004;291(21):2581–2590. doi: 10.1001/jama.291.21.2581. [DOI] [PubMed] [Google Scholar]
- 234.Kessler RC, Berglund P, Demler O, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289(23):3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 235.Thaipisuttikul P, Ittasakul P, Waleeprakhon P, Wisajun P, Jullagate S. Psychiatric comorbidities in patients with major depressive disorder. Neuropsychiatr Dis Treat. 2014;10:2097–2103. doi: 10.2147/NDT.S72026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kendler KS, Gatz M, Gardner CO, Pedersen NL. A Swedish national twin study of lifetime major depression. Am J Psychiatry. 2006;163(1):109–114. doi: 10.1176/appi.ajp.163.1.109. [DOI] [PubMed] [Google Scholar]
- 237.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157(10):1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 238.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81(3):484–503. doi: 10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hyde CL, Nagle MW, Tian C, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48(9):1031–1036. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.CONVERGE Consortium Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523(7562):588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jansen R, Penninx BW, Madar V, et al. Gene expression in major depressive disorder. Mol Psychiatry. 2016;21(3):339–347. doi: 10.1038/mp.2015.57. [DOI] [PubMed] [Google Scholar]
- 242.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. Ripke S, Wray NR, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18(4):497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kappelmann N, Lewis G, Dantzer R, Jones PB, Khandaker GM. Antidepressant activity of anti-cytokine treatment: a systematic review and meta-analysis of clinical trials of chronic inflammatory conditions. Mol Psychiatry 2016 Oct 18 . [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1224 kb)