Abstract
Pigs have been recognised as a reservoir of livestock associated methicillin-resistant Staphylococcus aureus (LA-MRSA) in Europe, Asia and North America. However, little is known about the presence and distribution of MRSA in the Australian pig population and pig industry. This study describes the presence, distribution and molecular characteristics of the human adapted Australian CA-MRSA ST93 isolated from pigs, people, and the environment within a piggery. Isolates were subjected to antibiotic susceptibility testing, DNA microarray, whole genome sequencing, multi locus sequence typing, virulence and resistance gene characterization and phylogenetic analysis. MRSA were isolated from 60% (n = 52) of farm workers where 84% of isolates returned ST93 and the rest ST398. Of the thirty-one pig isolates tested further, an equal number of ST398 and ST93 (15 each) and one as ST30-V were identified. Four of six environmental isolates were identified as ST93 and two as ST398. This study has identified for the first time in Australia the occurrence of CA-MRSA ST93 and LA-MRSA ST398 amongst farm workers, pigs, and the farm environment. Comparative genome analysis indicates that ST398 is likely to have been introduced into Australia from Europe or North America. This study also reports the first linezolid resistant MRSA isolated in Australia.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA), in addition to being resistant to most beta-lactams, are typically resistant to multiple classes of antibiotics. First reported in a human hospitalised patient in the United Kingdom1, MRSA are now frequently isolated worldwide. Although MRSA infections were initially exclusively associated with the hospital setting, a change in epidemiology occurred in the 1990s when infections began to emerge amongst people who had no prior hospital association2. These isolates were categorised as community-associated MRSA (CA-MRSA), and based on their accessory genome, staphylococcal cassette chromosome mec (SCCmec) and epidemiology were easily distinguishable from hospital-associated MRSA (HA-MRSA)3, 4. Clinically, CA-MRSA infections are typically skin and soft tissue-related and occur in community members who do not have traditional health care risk factors. In contrast, HA-MRSA are more frequently associated with surgical and systemic infections in the hospital setting5. CA-MRSA typically harbors a SCCmec IV or V element and additional virulence factors such as Panton-Valentine leukocidin (PVL). Although resistant to beta-lactams, CA-MRSA are generally susceptible to most antimicrobial classes6. In contrast, HA-MRSA usually harbour a SCCmec I, II, or III element, are PVL negative and are normally resistant to two or more non- beta-lactam antimicrobial classes7, 8. However, the distinction between CA-MRSA and HA-MRSA has become blurred with HA- and CA-MRSA crossing environmental boundaries, and in some cases exchanging genetic material9. CA-MRSA ST93-IV is the most common CA-MRSA in Australia and it is the second most common strain to cause infection in people, following HA-MRSA ST22-IV10.
A third category of MRSA, known as livestock-associated MRSA (LA-MRSA), was reported in the last decade11. LA-MRSA emerged independently from CA-MRSA and HA-MRSA with the most frequently isolated LA-MRSA belonging to clonal complex 398 (CC398). CC398 MRSA, first identified in pigs and pig farmers in 200512, has been reported worldwide in a variety of food-animal species13 and is the predominant lineage of LA-MRSA in Europe and North America. Although CC398 has been isolated in Asia, LA-MRSA ST9 predominates in pigs in the Asian region14. CC398 MRSA has been isolated from up to 50% of pigs in some European countries15 and in up to 86% of pig workers15–17. While CC398 is frequently reported to be associated with nasal carriage or colonization, infections resulting from this strain in humans, although rare, may also occur18. Consequently, CC398 MRSA is recognised as an occupational hazard for people working in the pig industry, including farm workers, veterinarians and abattoir workers19, 20.
LA-MRSA transmission and the extent of its ability to persist in pigs and humans are not fully understood. However, it is known that direct animal contact plays an important role in human carriage19. A study by Voss and colleagues demonstrated a 760-fold higher MRSA carriage rate among pig farmers compared to the general Dutch population21. Duration and intensity of animal contact and the number of MRSA positive animals on a farm have been linked with human CC398 colonization and infection18, 22. Furthermore, as CC398 has the ability to survive in the environment, environmental contamination may contribute toward further dissemination. Although occupational exposure to pigs is a risk factor for carriage, compared to CA-MRSA and HA-MRSA, CC398 is not easily transmissible from human to human, and has few virulence-associated genes23–25. Furthermore, the prevalence of CC398 has been shown to rapidly decrease during the absence of animal contact22, 24, 25. While colonization with CC398 rarely results in human infection26, various forms of skin and soft tissue infections, usually minor or localized, have been reported18.
In contrast to the knowledge about HA- and CA-MRSA in Australia27, little is known about the presence and distribution of MRSA in the Australian pig population and in the Australian pig industry. One cross-sectional study in 2011 examined nasal carriage of MRSA among veterinarians28. A single ST398 MRSA29 from a pig veterinarian was identified. A second Australian study in 2014 isolated CC398 from less than 1% of 324 pigs sampled from five different commercial pig farms and one feral herd located across Australia30.
Despite the limited documentation of LA-MRSA in pigs and pig professionals in Australia, an Australian pig farm was recently identified as being the focus of ongoing MRSA infections amongst its piggery employees over a three-year period. This study aims to describe the presence, distribution and molecular characteristics of MRSA isolated from the people, pigs and environment of the pig production facility where an ongoing outbreak in humans was occurring.
Methods and Methodology
Approval for the recruitment of human participants into this study was granted by the Charles Sturt University Human Research Ethics Committee (Protocol number 2015/016). Approval for the sample collection from pigs was obtained from the Charles Sturt University Animal Care and Ethics Committee (Protocol number 14/096). All methods were performed in accordance with the relevant guidelines and regulations.
Sampling
A cross-sectional study was performed at a commercial pig enterprise in Australia to detect the presence or absence of MRSA in people, pigs and the environment. The facility was identified by the farm’s jurisdictional health department due to the recurrent detection of MRSA amongst farm workers with clinical staphylococcal disease. The farm consisted of two sites (designated Farm-A and Farm-B) geographically separated by approximately 40 km. Sampling at each site occurred over a two-day period, with Farm-A visited in May and Farm-B in August 2015.
All farm workers (n = 52) were approached to participate in the study. Participation was voluntary, and signed informed consent was obtained from each participant. A sterile cotton applicator swab (Liquid Amies Elution Swab, Copan ESwab™) was provided and each participant was instructed to rotate the swab in both nostrils for five seconds, before placing the swab in a covered sterile transport tube.
On both farms, pigs were housed in separate sheds based on their age group and class (i.e. dry sows, farrowing, weaners, growers, and finishers). On Farm-A, to optimise sensitivity of the testing procedure, swabs from pigs were collected from the skin caudal and adjacent to the pinnae and from the external nares31. With the aim of determining if MRSA was present in the pigs on Farm-A, a previously described pooled sampling method was used: to detect MRSA carriage at a prevalence of at least 2% and assuming 90% test sensitivity with 95% confidence the minimum number of swabs per pool and the minimum number of pools required was calculated to be 10 and 17 respectively, using Epitools online32 http://epitools.ausvet.com.au. In addition, in order to obtain estimates of prevalence specific for each shed, six pools (60 swabs) were collected from each shed33, 34. From the seven sheds on Farm-A, 420 swabs were collected, providing 42 pools. Animals were randomly selected by farm workers and animals were marked after sampling so re-sampling of the same animals could not occur.
As a large number of MRSA positive pools were found on Farm-A, the sampling procedure was refined for Farm-B. Swabs were collected from the external nares only and samples were processed individually, rather than being pooled. As the prevalence was unknown (and therefore set at 50%) and considering a precision of 5% and 95% confidence, a minimum sample size of 385 swabs was required to determine the prevalence of MRSA in the pigs on this farm. Subsequently, nasal swabs were collected from 408 pigs of different stages of production housed in 13 sheds.
Using established protocols for dust sampling, environmental samples were collected and pooled (five swabs in each pool) from all sheds and effluent collection ponds on both farms35. Samples were collected from inside each shed and included walk ways, pens floors, feeders, fences, and walls. Swabs were also collected from the human environment (offices, showers, toilets, and kitchens/sitting rooms).
Laboratory analysis
All swabs were transported to the microbiology laboratory on ice within 12 hours of collection and immediately refrigerated upon arrival. Swabs were processed within a week of arriving in the laboratory using the isolation method recommended by the European Union Reference Laboratory for AMR (EURL-AMR) for the testing of MRSA in food-producing animals and food samples36, 37.
MRSA isolation
Swabs were pre-enriched in Mueller-Hinton broth (BD™ Difco™) containing 6.5% sodium chloride for 24 hours at 37 °C. Post incubation selective enrichment was performed by transferring one ml of pre-enriched broth into Tryptone Soy Broth (CM0129, Oxoid™) containing 3.5 mg/L cefoxitin and 75 mg/L aztreonam and incubating overnight at 37 °C. Following incubation, a loopful of the selective enriched cultured was inoculated onto chromogenic MRSA agar (CHROMagar™ MRSA) and incubated for 24 hours. As per the manufacturer’s instructions, rose to mauve coloured round colonies were preliminarily identified as MRSA. A single presumed MRSA colony from chromogenic agar was plated onto 5% sheep blood agar and incubated for 24 hours at 37 °C. Cultures were subsequently identified as S. aureus based on the Gram stain, catalase production and the S. aureus Protein-A latex agglutination test (staphylase, Oxoid™). S. aureus ATCC 29213 was used as the control strain.
Antimicrobial Susceptibility Testing
Antimicrobial disc diffusion susceptibility testing was performed on all 400 S. aureus identified. Clinical Laboratory Standards Institute (CLSI) disc concentrations and interpretive criteria were used38. Antimicrobials tested were: cefoxitin, tetracycline, erythromycin, gentamicin, penicillin, neomycin, ciprofloxacin, chloramphenicol, linezolid, trimethoprim/sulfamethoxazole, vancomycin, teicoplanin, clindamycin, quinupristin-dalfopristin, rifampin, and mupirocin.
Linezolid non-susceptibility was confirmed by minimum inhibitory concentration (MIC) testing using Etest® strips according to the manufacturer’s recommendations (bio Mérieux).
Molecular characterisation
Molecular testing was performed on 68 MRSA isolates which included the 31 MRSA isolated from humans, and a subset of the pig (n = 31) and environmental (n = 6) MRSA isolates. Pig and environmental MRSA isolates were selected based on their phenotypic antimicrobial susceptibility profiles to include as much phenotypic diversity as possible. The pig isolates were selected to include at least two isolates from each age group on both farms.
nuc and mecA characterization
S. aureus species and methicillin resistance were confirmed by the detection of nuc (thermostable extracellular nuclease) and mecA (methicillin resistance) genes respectively using multiplex PCR39.
DNA microarray
Isolates were initially characterised using a S. aureus DNA microarray (Alere Technologies, Jena Germany). Arrays and reagents were obtained from Alere Technologies, Jena Germany. The principle of the assay, related procedures, and a list of targets have been described previously40. The microarray was used to detect the presence of virulence and antimicrobial resistance genes. Probes for mecA, ugpQ, xylR, and two probes for mecR were used for SCCmec typing. The last two probes allowed detection and discrimination for untruncated mecR and mecR, respectively. Probes for the recombinase genes ccrA1, ccrB1, ccrA2, ccrB2, ccrA3, ccrB3, ccrA4, ccrB4 and ccrC1; the fusidic acid resistance marker Q6GD50; and the J region proteins, dcs, plsSCC and the kdp-operon also were included. Ambiguous array results were considered negative.
Whole genome sequencing (WGS)
Whole genome sequencing was performed using Illumina MiSeq. Genomic DNA libraries were prepared using the Nextera XT kit (Illumina) and sequenced on the 300-bp pair-ended chemistry. Raw sequences reads were denovo assembled using CLC genomics workbench (8.5.1).
Multilocus sequence typing (MLST)
DNA sequences collated at http://saureus.mlst.net/ belonging to 3,044 sequence types (ST) were downloaded in FASTA format and used as the database in the SRST2 pipeline to match the corresponding MLST profiles to the Illumina sequence data41.
In-silico virulence and resistance gene characterization
The presence of virulence and resistance genes were confirmed using VirulenceFinder, ResFinder tools from Centre for Genomic Epidemiology (CGE) website (www.genomicepidemiology.org) from the FASTA files generated using Illumina sequencing. Additional virulence factors, resistance gene determinants, beyond those detected by VirulenceFinder and ResFinder were determined using CLC Genomics Workbench.
Comparative Genomics
The MRSA identified in this study were compared to previously published MRSA strains. For ST93 the reference genome JKD6159 (NC_017338) was used as a template and aligned against all contigs to determine SNP locations42. The comparative genomic analysis was performed using a comprehensive Australian collection of MRSA ST93 using the 519 SNPs previously reported43. The comparative genome analysis of MRSA ST398 was performed by aligning the ST398 genomes sequenced in this study against the reference genome HO 5096 0412 (HE681097)44. The reference genome was modified to exclude all phage and intergenic regions. An international collection of MRSA ST398 from a previous study was utilised to perform comparative genomics23. SNPs were obtained using Panseq45, Maximum parsimony phylogenetic trees, based on the SNPs, were generated for both strains using MEGA (V6.06)46.
Statistical Analysis
All analyses were performed using the R statistical package47. Data on inhibition zone size for each drug were converted to dichotomous classification of resistant or susceptible based on inhibition zone diameters recommended in the CLSI documents M100-S2438 and VET01-S248. All intermediate resistance isolates were considered as susceptible. Confidence intervals (CI) were calculated at the 95% level of significance.
Results
Isolation of MRSA from farm workers, pigs and the environment
MRSA were isolated from 31 (60%) of the 52 farm workers who participated in the study: 10/19 (53%) on Farm-A and 21/33 (64%) on Farm-B.
Forty of the 42 pools of pig samples from Farm-A were MRSA positive. On Farm-B, the prevalence of MRSA-positive pigs was estimated to be 74% (302/408; 95% CI 70–78%).
Twenty five of the 40 pooled environmental samples were MRSA positive (63%; 95% CI 47–76%). MRSA was isolated from 18 of the 20 sheds (90%; 95% CI 70–97%). All seven sheds on Farm-A were positive while 11 were positive on Farm-B. Apart from sheds, the farms’ toilets (2/5), showers (2/4), and kitchens/sitting areas (2/3) were also positive for MRSA. None of the effluent collection ponds or farm-offices grew MRSA.
Molecular characterization of MRSA
The 68 MRSA selected for molecular characterization were mecA and nuc gene positive. Initial characterization of the isolates by DNA microarray identified three MRSA strains: ST93 (n = 45), ST398 (n = 22) and ST30 (n = 1). Forty-four of the 45 ST93 isolates carried a SCCmec IV element. A SCCmec V element was identified in one ST93 and in the ST30 and ST398 isolates.
The 31 human MRSA were identified as either ST93-IV (26 isolates) or ST398-V (5 isolates). Of the 31 pig isolates typed further, 15 were identified as ST398-V, 15 as ST93 (14 harbouring SCCmec-type IV and one with SCCmec-type V), and one as ST30-V. ST93-IV (4 isolates) and ST398-V (2 isolates) were isolated in the environment.
On Farm-A, all ten MRSA (100%) isolated from the human participants were characterised as ST93. The pig isolates were characterised as ST93 (n = 5) and ST398 (n = 3). ST93 and ST398 were also isolated in the environment.
On Farm-B, 16 of 21 MRSA (76%) isolated from the human participants were characterised as ST93-IV. The remaining five isolates were characterised as ST398-V (24%). Amongst the pig isolates four different MRSAs were characterised: 12 ST398-V, nine ST93-IV, and single isolates of ST93-V and ST30-V. MRSA ST398-V and ST93-IV were also identified in the environment.
Antimicrobial resistance phenotypes and genotypes
The antimicrobial resistance phenotype and clonal association are described in Table 1. In addition to the β-lactams, in ST93 and ST398 chloramphenicol, clindamycin, erythromycin, neomycin, and tetracycline, resistance was detected. Quinupristin-dalfopristin and linezolid resistance were also detected in ST398. The ST30 isolate was only resistant to the β-lactams and tetracycline.
Table 1.
Number and proportion of each methicillin-resistant Staphylococcus aureus strain resistant to non-β-lactam antimicrobials.
| Strain | CHL | CIP | CLI | ERY | GEN | LNZ | MUP | NEO | QDA | RIF | SXT | TEC | TET | VAN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ST398 | 10 (45.45) | 0 (0) | 19 (86.36) | 18 (81.82) | 0 (0) | 1 (4.54) | 0 (0) | 1 (4.54) | 8 (36.36) | 0 (0) | 0 (0) | 0 (0) | 22 (100) | 0 (0) |
| ST93 | 45 (100) | 0 (0) | 37 (82.22) | 35 (77.78) | 0 (0) | 0 (0) | 0 (0) | 2 (4.44) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 12 (26.67) | 0 (0) |
| ST30 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
CHL (chloramphenicol), CIP (ciprofloxacin), CLI (clindamycin), ERY (erythromycin), GEN (gentamicin), LNZ (linezolid), MUP (mupirocin), NEO (neomycin), QDA (quinupristin-dalfopristin), RIF (rifampin), SXT (trimethoprim/sulfamethoxazole), TEC (teicoplanin), TET (tetracycline), VAN (vancomycin).
All ST93 MRSA carried the chloramphenicol fexA resistant determinant (Table 2). Erythromycin (n = 35) and clindamycin (n = 37) resistance was conferred by ermC. For the 12 tetracycline resistant isolates 10 carried either tetL (9 isolates) or tetK (1 isolate).
Table 2.
Molecular characteristics and phenotypic antimicrobial resistance profile of MRSA isolated from farmworkers, pigs, and environment.
| No | Resistance genes | enterotoxin genes | IEC | PVL | Phenotypic antimicrobial Resistance |
|---|---|---|---|---|---|
| Human Isolates (31 isolates) | |||||
| ST398-V (5 isolates) | |||||
| HT19 | blaZ, ermC, tetK, tetM, norA | − | − | − | BLA, TET, ERY, CLI |
| H20 | dfrG, blaZ, ermC, vgaA, tetK, tetM, fexA, norA, lnuB, aadE | − | − | − | BLA, TET, ERY, CHL, CLI, QDA |
| H21 | blaZ, ermC, tetK, tetM, norA | − | − | − | BLA, TET, ERY, CLI |
| H23 | ermC, tetK, tetM, norA | − | − | − | BLA, TET, ERY, CLI |
| HW-31 | aadD, blaZ, norA, vgaA, lnuB, cfr, fexA, tetK, tetM, dfrG | − | − | − | BLA, TET, CHL, LNZ, CLI |
| ST93-IV (26 isolates) | |||||
| HT3 | blaZ, fexA, norA | ORFCM14 | + | − | BLA, CHL |
| H2 | blaZ, ermC, fexA, norA | ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| H3 | blaZ, ermC, fexA, norA | ORFCM14 | + | + | BLA, ERY, CHL, CLI |
| H4 | tetL, blaZ, ermC, aadD, fexA, norA | ORFCM14 | − | + | BLA, TET, ERY, NEO, CHL, CLI |
| HT13 | blaZ, ermC, aadD, fexA, norA | ORFCM14 | + | − | BLA, TET, ERY, NEO, CHL, CLI |
| H6 | blaZ, fexA, norA | ORFCM14 | − | − | BLA, ERY, CHL, CLI |
| H7 | blaZ, ermC, fexA, norA | ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| H8 | blaZ, ermC, fexA, norA | ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| H9 | blaZ, ermC, fexA, norA | ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| H11 | blaZ, ermC, fexA, norA | ORFCM14 | − | − | BLA, ERY, CHL, CLI |
| HW-2 | blaZ, ermC, fexA, norA | ORFCM14 | + | + | BLA, ERY, CHL, CLI |
| H13 | blaZ, ermC, tetL, fexA, norA | ORFCM14 | − | + | BLA, TET, ERY, CHL, CLI |
| H14 | blaZ, ermC, tetK, fexA, norA | ORFCM14 | − | + | BLA, TET, ERY, CHL, CLI |
| H15 | blaZ, ermC, fexA, norA, tetL | ORFCM14 | + | + | BLA, TET, ERY, CHL, CLI |
| H16 | blaZ, ermC, fexA, norA | ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| H17 | blaZ, ermC, fexA, norA | seg, egc ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| H18 | blaZ, ermC, fexA, norA | ORFCM14 | − | − | BLA, ERY, CHL, CLI |
| H19 | blaZ, ermC, fexA, norA | ORFCM14 | − | − | BLA, CHL |
| HW-15 | blaZ, ermC, fexA, norA | ORFCM14 | + | − | BLA, ERY, CHL, CLI |
| H24 | blaZ, ermC, fexA, fosB, norA, tetL | seg, egc ORFCM14 | + | + | BLA, TET, ERY, CHL, CLI |
| H25 | blaZ, ermC, fexA, norA | ORFCM14 | − | − | BLA, ERY, CHL, CLI |
| H26 | blaZ, ermC, fexA, norA | ORFCM14 | + | − | BLA, ERY, CHL, CLI |
| H27 | blaZ, ermC, fexA, norA | ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| H29 | ermC, fexA, norA | ORFCM14 | + | + | BLA, ERY, CHL, CLI |
| HW-24 | blaZ, fexA, norA | ORFCM14 | + | + | BLA, CHL |
| H31 | blaZ, fexA, norA | ORFCM14 | + | − | BLA, ERY, CHL, CLI |
| Pig Isolates (31 isolates) | |||||
| ST30-V(1 isolate) | |||||
| W1Bb-25 | blaZ, norA, vgaA, tetL, tetK | seg, sei, sem, sen, seo, seu, egc | − | − | BLA, TET |
| ST398-V (15 isolates) | |||||
| PTDrAP2 | aadD, blaZ, norA, vgaA, ermC, lnuB, cfr, fexA, tetK, tetM, dfrG | − | − | − | BLA, TET, ERY, CHL, CLI, QDA |
| PTWeP5 | aadE, blaZ, norA, ermC, tetM, tetK, tetL | sed | − | − | BLA, TET, ERY, NEO, CLI |
| PTGrBP1 | aadE, norA, lnuB, cfr, vgaA, ermC, fexA, tetM, tetK, dfrG | − | − | − | BLA, TET, ERY, CHL, CLI, QDA |
| W1FPa-4 | blaZ, norA, ermC, tetK, tetM | − | − | − | BLA, TET |
| W1FSb-2 | blaZ, norA, tetK, tetM | sed | − | − | BLA, TET |
| W1FPB-20 | aadE, blaZ, norA, ermC, lnuB, vgaA, fexA, tetK, tetM, dfrG | − | − | − | BLA, TET, ERY, CHL, CLI, QDA |
| W1Gr-12 | blaZ, norA, ermC, tetK, tetM | sed | − | − | BLA, TET, ERY, CLI |
| WweU-9 | aadE, blaZ, norA, ermC, lnuB, vgaA, fexA, tetK, tetM, dfrG | − | − | − | BLA, TET, CHL |
| P216 | ermC, vgaA, tetK, tetM, fexA, norA, aadE, dfrG, lnuB | − | − | − | BLA, TET, ERY, CHL, CLI, QDA |
| P221 | blaZ, ermC, tetK, tetM, norA | − | − | − | BLA, TET, ERY, CLI |
| P223 | blaZ, ermC, tetK, tetM, norA | sea | − | − | BLA, TET, ERY, CLI |
| P236 | tetK, tetM, norA | sea | − | − | BLA, TET, ERY, CLI |
| P244 | blaZ, ermC, vgaA, tetK, tetM, fexA, norA, aadE, dfrG, lnuB | − | − | − | BLA, TET, ERY, CHL, CLI, QDA |
| P257 | blaZ, ermC, vgaA, tetK, tetM, fexA, norA, aadE, dfrG, lnuB | tst1 | − | − | BLA, TET, ERY, CHL, CLI, QDA |
| P330 | blaZ, ermC, tetK, tetM, norA | − | − | − | BLA, TET, ERY, CLI |
| ST93-IV (14 isolates) | |||||
| PTDrAP4 | blaZ, ermC, fexA, norA | ORFCM14 | − | − | BLA, ERY, CHL, CLI |
| PTDrBP4 | blaZ, ermC, fexA, norA | ORFCM14 | − | + | BLA, CHL |
| PTPgP1 | blaZ, norA, fexA | sed, seg ORFCM14 | − | + | BLA, CHL, CLI |
| PTGrAP5 | blaZ, norA, ermC, fexA | ORFCM14 | − | + | BLA, CHL |
| PTF1P3 | blaZ, ermC, norA, fexA | sed, seh, ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| W1Bb-4 | blaZ, norA, ermC, fexA | sed, seg, ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| W1FPb-22 | blaZ, norA, fexA | ORFCM14 | − | − | BLA, CHL |
| W1Gr-24 | blaZ, norA, ermC, fexA, tetL | sed, ORFCM14 | + | + | BLA, TET, CHL, CLI |
| W1Fi-5 | blaZ, ermC, fexA, norA | ORFCM14 | − | + | BLA, TET, ERY, CHL, CLI |
| W1Fi-9 | blaZ, norA, fexA, tetL | sed, ORFCM14 | − | + | BLA, TET, CHL |
| WWeU-6 | ermC, fexA, tetL | ORFCM14 | + | + | BLA, TET, ERY, CHL, CLI |
| P271 | blaZ, ermC, fexA, norA | tst1, ORFCM14 | + | − | BLA, ERY, CHL, CLI |
| P306 | blaZ, ermC, tetL, fexA, norA | ORFCM14 | − | + | BLA, TET, ERY, CHL, CLI |
| P325 | blaZ, ermC, fexA, norA | seg, egc ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| ST93-V (1 isolate) | |||||
| W1Dr-3 | blaZ, ermC, fexA, norA | seg, ORFCM14 | − | + | BLA, ERY, CHL, CLI |
| Environmental Isolates (6 isolates) | |||||
| ST93-IV (4 isolates) | |||||
| ETDrA | blaZ, ermC, fexA | sed, ORF CM14 | − | − | BLA, ERY, CHL, CLI |
| E008 | blaZ, ermC, fexA, norA | ORF CM14 | − | + | BLA, ERY, CHL, CLI |
| E017 | blaZ, fexA, norA | ORF CM14 | − | − | BLA, CHL, |
| E021 | blaZ, ermC, fexA, tetL, norA | seg, egc ORF CM14 | − | + | BLA, TET, ERY, CHL, CLI |
| ST398-V (2 isolates) | |||||
| E026 | aadE, ermC, vga(A), tet(M), fexA, norA | − | − | BLA, TET, ERY, CHL, CLI, QDA | |
| ETWeP | blaZ, ermC, tetK, tetM, norA | sed | − | − | BLA, TET, ERY, CLI |
BLA (β-lactam), CHL (chloramphenicol), CLI (clindamycin), ERY (erythromycin), LNZ (linezolid), NEO (neomycin), QDA (quinupristin-dalfopristin), TET (tetracycline).
A large proportion of ST398 were co-resistant to tetracycline, erythromycin, and clindamycin (Table 2). Resistance to erythromycin (n = 18) and clindamycin (n = 19) was conferred by ermC (19/22). Tetracycline resistance (n = 22) was conferred by the carriage of the tetM, tetK, and tetL genes. Ten isolates carried the vgaA gene which encodes resistance to lincosamides, pleuromutilins and streptogramin A. The lincosamides resistant isolates also carried the lnuB gene. Chloramphenicol resistance (10 isolates) was conferred by the fexA gene and aminoglycoside resistance (11 isolates) by the aadD (2 isolates) and aadE (9 isolates) genes. Three ST398 MRSA harboured the cfr gene cassette; two from the pig samples and one from a human participant. Only the human isolate was phenotypically linezolid resistant (MIC 32 mg/L). Although the dfrG gene was identified in nine isolates, none were phenotypically co-trimoxazole (trimethoprim/sulfamethoxazole) resistant.
The ST30-V isolate was found to carry tetK, tetL and vgaA antimicrobial resistance genes.
Virulence gene data
The carriage of virulence genes was influenced by the MRSA type (Table 2).
Thirty of the 45 ST93 isolates carried the luk-F-PV/lukS-PV PVL associated genes: Sixteen of the 26 ST93 human isolates; 12 of the 15 pig isolates; and two of the four environmental isolates. Fourteen ST93 (11 from the human participants and three from pigs) also carried the type B (sak + chp + scn) human immune evasion gene cluster (IEC) genes. All ST93 isolates carried enterotoxin-like ORF CM14 gene. Enterotoxins sed (n = 6), seg (n = 7), seh (n = 1) and the enterotoxin gene cluster [egc] (n = 4) was also detected.
None of the ST398-V isolates carried lukf-PV/lukS-PV or IEC genes. Enterotoxins sea (n = 2), sed (n = 4), and toxic shock syndrome toxin-1 tst1 (n = 1) genes were detected.
The ST30-V isolate harboured the egc complex. The isolate was negative for the PVL and IEC associated genes.
ST93 phylogeny
Between the ST93 MRSA isolates very few SNP mutations were detected. The isolates were closely related to the reference genome JKD6159 (NC_017338) and shared nine common SNP markers. Based on the SNPs the isolates could be classified into three clusters (Fig. 1). There was no relation between the source of the isolates and the cluster.
Figure 1.
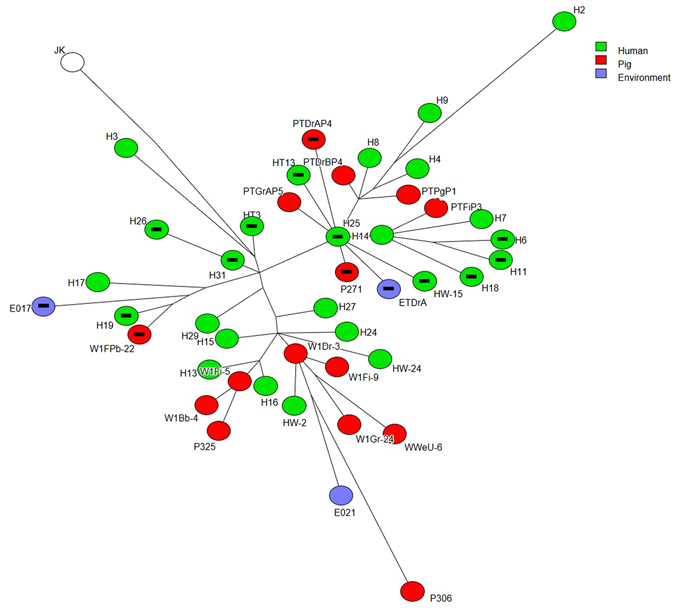
Phylogenetic tree constructed by core genome SNPs of MRSA ST93 isolated from humans, pigs and piggery environment. Each circle represents individual MRSAs. Isolates from humans, pigs, and environment are identified by green, red and purple colours respectively. Isolates that did not carry pvl gene are identified by a black bar in the centre.
ST398 phylogeny
The maximum parsimony tree constructed from ST398 isolates showed three clusters suggesting either multiple introductions of ST398 or an ongoing evolution of the strain. When compared to the international collection of ST398 MRSA genomes, the Australian ST398 isolates from the farmworkers and pigs were similar to the ST398 isolates from Europe and North America (Fig. 2).
Figure 2.
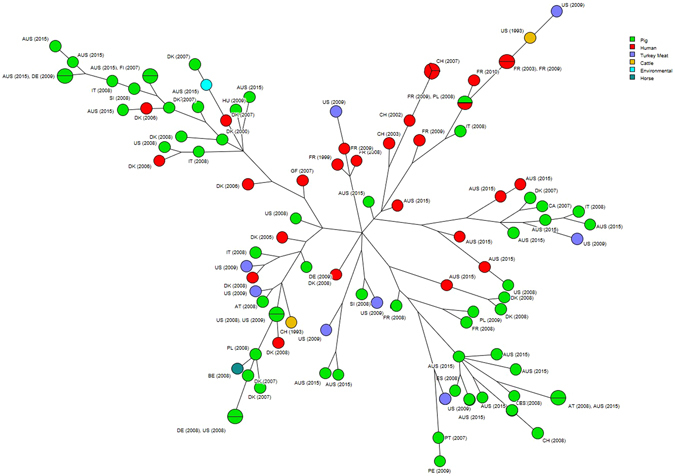
Phylogenetic tree constructed by core genome SNPs of MRSA ST398 isolated from humans, pigs, and piggery environment from this study compared to isolates from different parts of the world. Each circle represents individual MRSAs. Isolates from pigs, humans, turkey meat, cattle, environment and horses identified by green, red, purple, orange, blue and dark green colours respectively. Isolates are coded by the country of origin and year in which they were isolated. Isolates from this study is coded as AUS (2015).
Discussion
This study has investigated the carriage and the molecular characteristics of MRSA isolated from an Australian pig farm spanning two sites (Farm A and B) that had on-going clinical MRSA infections amongst its farm workers.
The study has shown concurrent carriage of MRSA in humans and pigs on both farms with two predominant strains; the Australian CA-MRSA ST93-IV and the international LA-MRSA ST398-V. Furthermore, whole genome sequencing suggests not only has anthropozoonotic and zoonotic transmission occurred, the movement of ST93-IV from human to pig and then back into humans in the presence of the multi-resistant ST398-V has resulted in the acquisition of multiple antimicrobial resistance determinants, such as those encoding for resistance to tetracycline, clindamycin and chloramphenicol, that are not normally found in ST93-IV.
The three major findings arise from this study are as follow: First, we have identified for the first time in Australia the dissemination of CA-MRSA ST93-IV and LA-MRSA ST398-V amongst farm workers, pigs and the farm environment; Second, we have found, using comparative genome analysis, that ST398-V was likely to have been introduced into Australia from Europe or North America and the ST93-IV pig MRSA isolates came from humans with subsequent pig adaptation; and third, to the best of our knowledge we report the first linezolid resistant MRSA isolated in Australia.
ST93-IV, colloquially known as “Queensland CA-MRSA” was first described in the early 2000s in a population of Caucasian humans in Queensland49. Typically, ST93-IV has been associated with a range of skin and soft tissue infections in humans only. A singleton by MLST eBURST analysis, ST93-IV has been shown to be highly virulent compared to other well-characterised Australian MRSA, including ST1-IV [2B], ST30-IV [2B] and ST239-III [3B], and the epidemic North American strain, USA30050. Since 2014 ST93-IV has become the dominant CA-MRSA across Australia27, accounting for approximately 40% of CA-MRSA, 25% of MRSA and 5% of S. aureus community-onset infections51. Consequently, the acquisition of ST93-IV in pigs is likely to have been anthropozoonotic (i.e. human to animal). Although comparative genome analysis showed the porcine and human ST93-IV isolates in our study were closely related to ST93-IV previously characterised in the region (Fig. 1 Supplementary data), the isolates had a lower than expected carriage of the bacteriophage encoded PVL-associated luk-F PV and lukS-PV genes. In previous studies upto 100% of ST93-IV have been identified as PVL positive10, 52. In contrast, one-third of ST93-IV isolates in our study were PVL negative; 39% of farmworker isolates and 20% of pig isolates. Similarly, a lower than expected carriage of the фSa3 prophage-mediated type B human invasion gene cluster (IEC; sak/scn/chp) was observed. Unlike previous studies, which have shown 100% of ST93-IV harbor a type B IEC, the majority of the pig (80%) and the farmworker (58%) ST93-IV isolates lacked an IEC. The loss of the human associated virulence lukF-PV and lukS-PV and sak/scn/chp genes in a large proportion of the ST93-IV isolates suggests the strain has successfully adapted to the porcine host. A similar adaption for S. aureus moving from the human to porcine host has been reported in CC39823.
Contrary to the loss of virulence factors, the ST93-IV identified in our study were phenotypically multi-resistant (including chloramphenicol, clindamycin, erythromycin, neomycin and tetracycline) and harboured multiple resistance genes. Typically, ST93-IV is only beta-lactam resistant with only up to 20% and 2% of isolates resistant to erythromycin and ciprofloxacin respectively52–54. It is possible that the observed elevated frequency of resistance to non-beta lactam antimicrobials among the ST93-IV isolates in our study is due to selection within the pig environment, linked to antimicrobial use on the pig farm. The anthropozoonotic transmission of the highly successful CA-MRSA ST93-IV in pigs and the acquisition of additional antimicrobial resistance genes and loss of human associated virulence genes indicate the strain has successfully evolved in the porcine host. This successful evolution may lead to the co-selection, long term maintenance, and further adaptation of ST93-IV in pigs and in the piggery environment55.
Prior to our study, ST398-V has only been reported in Australia on two occasions29, 30. In our study, 22 isolates were identified as ST398-V (representing 23% of MRSA isolates from farmworkers and also isolated from pigs and the environment). Previous studies have distinguished the livestock-associated S. aureus ST398 clade from the human clade by the presence of the tetM gene and the absence of IEC56. As the ST398-V isolated in our study lacked an IEC and a high proportion carried the tetM gene it can be assumed the isolates belong to the livestock clade56. Comparative phylogenetic analysis suggests the ST398-V was introduced into Australia from Europe or North America56. The identification of three ST398-V clusters suggests either multiple introductions into Australia have occurred or the strain has diverged within the farm environment. However, as one of the clusters (cluster 3 versus clusters 1 and 2) differed by more than 34 SNPs over a short period it is more likely that both events have occurred (Fig. 2 Supplementary data).
The importation of CC398 into a country previously free of CC398 and the subsequent transmission into the country’s pig population followed by zoonotic spread is not unique57. In Europe, the spread of LA-MRSA between countries is often mediated by animal trading of piglets sold by specialised producers58. However, in Australia, the importation of live pigs ceased over 40 years ago. Consequently, it is possible that the introduction of ST398-V into Australia has occurred from pig farmers or pig veterinarians who have had contact with European or North American pig farms, or through the entry of other animal species, such as horses, where the same movement restrictions are not present.
Although S. aureus does not cause much illness in pigs, CC398 MRSA does cause a wide spectrum of infections in humans, ranging from relatively minor or localised infections to more serious or invasive infections59. Furthermore, nosocomial infections and outbreaks have occurred associated with this MRSA. However, similar to other S. aureus, transmission of CC398 MRSA is primarily mediated by physical contact and overall relatively few cases of CC398 MRSA have occurred in people who are not directly involved in livestock production. The low prevalence of CC398 MRSA in people not associated with pig farming is probably due to the low transmissibility of the organism60. Consequently, the clinical significance of CC398 MRSA within the community is thought to be low. However, the emergence of ST398 in the Australian pig industry is a public health concern for those working within the industry. Although ST398 MRSA is less virulent and less transmissible compared to other S. aureus, the organism exhibits co-resistance to many non β-lactam antimicrobials used in medical and veterinary practice including macrolides, tetracycline, trimethoprim, gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole, lincosamides and streptogramin B57, 59. Although ST398 remains susceptible to the glycopeptides, daptomycin, tigecyclines, fusidic acid and rifampicin, isolated cases of linezolid resistance have been reported57. In ST398 MRSA, linezolid resistance is due to the acquisition of a plasmid harbouring the cfr gene which in addition to linezolid resistance mediates resistance to lincosamides, fenicols and pleuromutilins61. The selective pressure in favour of the spread of the cfr containing plasmid may be influenced by the use of linezolid in human medicine and florfenicol and tiamulin in veterinary medicine. The linezolid resistance identified in a ST398 MRSA isolated from a human in this study is the first instance of linezolid resistance in MRSA to be reported in Australia.
Although multidrug resistance may compromise the treatment of ST398 MRSA infections, the reservoir of resistance genes harboured by ST398 MRSA is more concerning from a clinical standpoint. This concern was recently highlighted by Brennan et al. following the introduction of CC398 into the Republic of Ireland’s pig industry who suggested that the reservoir of resistance genes harboured by the less virulent and less transmissible CC398 MRSA could potentially spread to other animal and human strains62. Unfortunately, this may have occurred in Australia. It appears from our study the movement of the highly virulent and highly transmissible ST93-IV CA-MRSA from humans to pigs and the importation of ST398-V LA-MRSA into the Australian pig farms has resulted in the emergence of a multi-resistant PVL-positive ST93-IV strain which continues to harbour a type B IEC. However, a comprehensive molecular comparison of the antimicrobial resistance genes and the associated mobile genetic elements found in the ST93-IV and ST398-V is required. Furthermore, the fitness cost to ST93-IV following the acquisition of the additional genetic material will need to be determined in order to assess the human health risk, if any, to those animals and humans not exposed to the particular pig herds investigated in this study.
Conclusion
In conclusion, our study has demonstrated for the first time the widespread occurrence of the highly virulent Australian ST93-IV CA-MRSA and the international ST398-V LA-MRSA strains in an Australian piggery. Furthermore, the evidence suggests that anthropozoonotic and zoonotic transmission may have occurred. The presence of multiple antimicrobial resistance determinants that are not usually found in ST93-IV could be a consequence of the transmission of ST93-IV from humans to pigs and then back into humans in the presence of the multi-resistant ST398-V. The outcomes of this study indicate that in Australia, and probably elsewhere, greater scrutiny of staphylococcal infections in humans and animals is warranted in the form of coordinated investigation of outbreaks to identify the factors predisposing each host species to infection and emergence of virulent pathogens with novel resistance traits.
Electronic supplementary material
Acknowledgements
This study was funded by Australian Pork Limited (Project number 2015/013). The authors would like to acknowledge Mrs Michelle Ayton, Senior Technical Officer at Charles Sturt University and other laboratory members for their tremendous help in microbiological analysis. We are indebted to the producer who allow us to collect swabs from their farm. The authors would like to thank the workers who participated in this study. The authors would also like to acknowledge Ms Sharon Nielsen and Dr David Luckett for their assistance in R. We are grateful to Dr Muhammad Shoaib Tufail for his help in sampling.
Author Contributions
S.S. designed the study and conceived the experiments, analysed the data and results. J.H., M.H., and D.J., designed and supervised the whole study. S.A., S.P., and G.C. conducted the molecular work. S.S., S.A., G.C., and J.H. drafted the manuscript. All authors reviewed and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04789-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jevons MP. Celbenin-resistant Staphylococci. Brit. Med. J. 1961;1:124–124. doi: 10.1136/bmj.1.5219.124-a. [DOI] [Google Scholar]
- 2.Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J. Hosp. Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-E. [DOI] [PubMed] [Google Scholar]
- 3.Enright M, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc. Natl. Acad. Sci. USA. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandenesch F, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uhlemann A, Otto M, Lowy FD, DeLeo FR. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect., Genet. Evol. 2014;21:563–574. doi: 10.1016/j.meegid.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsay JA. Hospital-associated MRSA and antibiotic resistance—What have we learned from genomics? Int. J. Med. Microbiol. 2013;303:318–323. doi: 10.1016/j.ijmm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Hiramatsu, K., Cui, L., Kuroda, M. & Ito, T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9, 486–493, doi:10.1016/S0966-842X(01)02175-8 (2001). [DOI] [PubMed]
- 9.Boakes E, et al. Molecular diversity within clonal complex 22 methicillin-resistant Staphylococcus aureus encoding Panton–Valentine leukocidin in England and Wales. Clin. Microbiol. Infect. 2011;17:140–145. doi: 10.1111/j.1469-0691.2010.03199.x. [DOI] [PubMed] [Google Scholar]
- 10.Coombs GW, et al. Australian Group on Antimicrobial Resistance Australian Staphylococcus aureus Sepsis Outcome Programme annual report, 2014. Communicable diseases intelligence quarterly report. 2016;40:E244–254. doi: 10.33321/cdi.2016.40.20. [DOI] [PubMed] [Google Scholar]
- 11.Huijsdens XW, et al. Community-acquired MRSA and pig-farming. Ann. Clin. Microbiol. Antimicrob. 2006;5:1–4. doi: 10.1186/1476-0711-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armand-Lefevre L, Ruimy R, Andremont A. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg Infect Dis. 2005;11:711–714. doi: 10.3201/eid1105.040866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuny C, et al. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int. J. Med. Microbiol. 2010;300:109–117. doi: 10.1016/j.ijmm.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Larsen J, et al. Methicillin-resistant Staphylococcus aureus ST9 in pigs in Thailand. PLoS ONE. 2012;7:e31245. doi: 10.1371/journal.pone.0031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.EFSA. Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008, Part B: factors associated with MRSA contamination of holdings; on request from the European Commission. The EFSA Journal8, 1–67, doi:10.2903/j.efsa.2010.1597 (2010).
- 16.Cuny C, et al. Nasal colonization of humans with methicillin-resistant Staphylococcus aureus (MRSA) CC398 with and without exposure to pigs. PLoS ONE. 2009;4:e6800. doi: 10.1371/journal.pone.0006800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Cleef BAGL, et al. Dynamics of methicillin-resistant Staphylococcus aureus and methicillin-susceptible Staphylococcus aureus carriage in pig farmers: a prospective cohort study. Clin. Microbiol. Infect. 2014;20:764–771. doi: 10.1111/1469-0691.12582. [DOI] [PubMed] [Google Scholar]
- 18.Köck R, et al. Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS One. 2013;8:e55040. doi: 10.1371/journal.pone.0055040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wardyn SE, et al. Swine farming is a risk factor for infection with and high prevalence of carriage of multidrug-resistant Staphylococcus aureus. Clin. Infect. Dis. 2015;61:59–66. doi: 10.1093/cid/civ234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadimpalli M, et al. Persistence of livestock-associated antibiotic-resistant Staphylococcus aureus among industrial hog operation workers in North Carolina over 14 days. Occup. Environ. Med. 2015;72:90–99. doi: 10.1136/oemed-2014-102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005;11:1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graveland H, Wagenaar JA, Bergs K, Heesterbeek H, Heederik D. Persistence of livestock associated MRSA CC398 in humans is dependent on intensity of animal contact. PLoS One. 2011;6:e16830. doi: 10.1371/journal.pone.0016830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price LB, et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio. 2012;3:e305–311. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamrozy DM, Fielder MD, Butaye P, Coldham NG. Comparative genotypic and phenotypic characterisation of methicillin-resistant Staphylococcus aureus ST398 isolated from animals and humans. PLoS One. 2012;7:e40458. doi: 10.1371/journal.pone.0040458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Cleef BAGL, et al. Persistence of livestock-associated MRSA after short term occupational exposure to pigs and veal calves. J. Clin. Microbiol. 2011;49:1030–1033. doi: 10.1128/JCM.00493-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monaco M, et al. Livestock-associated methicillin-resistant Staphylococcus aureus responsible for human colonization and infection in an area of Italy with high density of pig farming. BMC Infect. Dis. 2013;13:1–6. doi: 10.1186/1471-2334-13-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coombs GW, et al. Community-onset Staphylococcus aureus surveillance programme annual report, 2012. Commun. Dis. Intell. 2014;38:E26–S36. doi: 10.33321/cdi.2014.38.12. [DOI] [PubMed] [Google Scholar]
- 28.Jordan D, et al. Carriage of methicillin-resistant Staphylococcus aureus by veterinarians in Australia. Aust. Vet. J. 2011;89:152–159. doi: 10.1111/j.1751-0813.2011.00710.x. [DOI] [PubMed] [Google Scholar]
- 29.Groves MD, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolated from Australian veterinarians. PloS ONE. 2016;11:e0146034. doi: 10.1371/journal.pone.0146034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groves MD, et al. Staphylococcus aureus ST398 detected in pigs in Australia. J. Antimicrob. Chemother. 2014;69:1426–1428. doi: 10.1093/jac/dkt526. [DOI] [PubMed] [Google Scholar]
- 31.Pletinckx LJ, et al. Evaluation of salt concentrations, chromogenic media and anatomical sampling sites for detection of methicillin-resistant Staphylococcus aureus in pigs. Vet. Microbiol. 2012;154:363–368. doi: 10.1016/j.vetmic.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 32.Sergeant, E. S. G. Epitools epidemiological calculators, http://epitools.ausvet.com.au/ (2015).
- 33.Broens EM, Graat EAM, Van Der Wolf PJ, Van De Giessen AW, De Jong MCM. Prevalence and risk factor analysis of livestock associated MRSA-positive pig herds in the Netherlands. Prev. Vet. Med. 2011;102:41–49. doi: 10.1016/j.prevetmed.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 34.EFSA. Report of the task force on zoonoses data collection on a proposal for technical specifications for a baseline survey on the prevalence of methicillin resistant Staphylococcus aureus (MRSA) in breeding pigs The EFSA Journal129, 1–14, doi:10.2903/j.efsa.2007.129r (2007).
- 35.EFSA. Scientific opinion of the panel on biological hazards on a request from the European commission on assessment of the public health significance of methicillin resistant Staphylococcus aureus (MRSA) in animals and foods. The EFSA Journal993, 1–73 (2009).
- 36.EU Reference Laboratory - Antimicrobial Resistance. Protocol for screening for MRSA: isolation of MRSA from dust samples, http://www.eurl-ar.eu/data/images/tc_april-2009/4-protocol%20for%20screening%20mrsa.pdf (2009).
- 37.EFSA. Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin-resistant Staphylococcus aureus in food-producing animals and food. The EFSA Journal10, 1-56, doi:10.2903/j.efsa.2012.2897 (2012).
- 38.CLSI. Performance standards for antimicrobial susceptibility testing: Twenty-fourth informational supplement. Vol. 34 (Clinical and Laboratory Standards Institute, 2014).
- 39.Costa A, Kay I, Palladino S. Rapid detection of mecA and nuc genes in staphylococci by real-time multiplex polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 2005;51:13–17. doi: 10.1016/j.diagmicrobio.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Monecke S, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011;6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inouye M, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:1–16. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chua K, et al. Complete genome sequence of Staphylococcus aureus strain JKD6159, a unique Australian clone of ST93-IV community methicillin-resistant Staphylococcus aureus. J. Bacteriol. 2010;192:5556–5557. doi: 10.1128/JB.00878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stinear TP, et al. Adaptive change inferred from genomic population analysis of the ST93 epidemic clone of community-associated methicillin-resistant Staphylococcus aureus. Genome Biology and Evolution. 2014;6:366–378. doi: 10.1093/gbe/evu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holden MTG, et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013;23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laing C, et al. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics. 2010;11:1–14. doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R: A language and environment for statistical computing [Computer software, version 3.2.2] (R foundation for statistical computing, Vienna, Austria, 2015).
- 48.CLSI. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; second informational supplement. Document VET01-S2. Vol. 28 (Clinical and Laboratory Standards Institute, 2013).
- 49.Munckhof WJ, Schooneveldt J, Coombs GW, Hoare J, Nimmo GR. Emergence of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) infection in Queensland, Australia. Int. J. Infect. Dis. 2003;7:259–267. doi: 10.1016/S1201-9712(03)90104-4. [DOI] [PubMed] [Google Scholar]
- 50.Chua K, Laurent F, Coombs G, Grayson ML, Howden BP. Not community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA)! A clinician’s guide to community MRSA - Its evolving antimicrobial resistance and implications for therapy. Clin. Infect. Dis. 2011;52:99–114. doi: 10.1093/cid/ciq067. [DOI] [PubMed] [Google Scholar]
- 51.Coombs GW, et al. The molecular epidemiology of the highly virulent ST93 Australian community Staphylococcus aureus strain. PLoS ONE. 2012;7:e43037. doi: 10.1371/journal.pone.0043037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coombs GW, et al. Australian Group on Antimicrobial Resistance hospital-onset Staphylococcus aureus surveillance programme annual report, 2011. Commun. Dis. Intell. 2013;37:E210–218. doi: 10.33321/cdi.2013.37.32. [DOI] [PubMed] [Google Scholar]
- 53.Coombs GW, et al. Prevalence of MRSA strains among Staphylococcus aureus isolated from outpatients, 2006. Report from the Australian Group for Antimicrobial Resistance. Commun. Dis. Intell. 2009;33:10–20. doi: 10.33321/cdi.2009.33.2. [DOI] [PubMed] [Google Scholar]
- 54.Coombs GW, et al. Methicillin-resistant Staphylococcus aureus clones, Western Australia. Emerg Infect Dis. 2006;12:241–247. doi: 10.3201/eid1202.050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahms C, Hübner N, Cuny C, Kramer A. Occurrence of methicillin-resistant Staphylococcus aureus in farm workers and the livestock environment in Mecklenburg-Western Pomerania, Germany. Acta Vet. Scand. 2014;56:1–8. doi: 10.1186/s13028-014-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stegger M, et al. Rapid differentiation between livestock-associated and livestock-independent Staphylococcus aureus CC398 clades. PLoS ONE. 2013;8:e79645. doi: 10.1371/journal.pone.0079645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cuny C, Wieler LH, Witte W. Livestock-associated MRSA: the impact on humans. Antibiotics. 2015;4:521–543. doi: 10.3390/antibiotics4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Broens EM, et al. MRSA CC398 in the pig production chain. Prev. Vet. Med. 2011;98:182–189. doi: 10.1016/j.prevetmed.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 59.Verkade E, Kluytmans J. Livestock-associated Staphylococcus aureus CC398: Animal reservoirs and human infections. Infect., Genet. Evol. 2014;21:523–530. doi: 10.1016/j.meegid.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 60.Verkade E, et al. Transmission of methicillin-resistant Staphylococcus aureus CC398 from livestock veterinarians to their household members. PLoS ONE. 2014;9:e100823. doi: 10.1371/journal.pone.0100823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Witte W, Cuny C. Emergence and spread of cfr-mediated multiresistance in staphylococci: an interdisciplinary challenge. Future Microbiol. 2011;6:925–931. doi: 10.2217/fmb.11.69. [DOI] [PubMed] [Google Scholar]
- 62.Brennan GI, et al. The emergence and spread of multiple livestock-associated clonal complex 398 methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains among animals and humans in the Republic of Ireland, 2010–2014. PLoS ONE. 2016;11:1–11. doi: 10.1371/journal.pone.0149396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


