Abstract
Vertical sleeve gastrectomy (VSG) produces sustainable weight loss, remission of type 2 diabetes (T2D), and improvement of nonalcoholic fatty liver disease (NAFLD). However, the molecular mechanisms underlying the metabolic benefits of VSG have remained elusive. According to our previous results, diet-induced obesity induces epigenetic modifications to chromatin in mouse liver. We demonstrate here that VSG in C57BL/6J wild-type male mice can reverse these chromatin modifications and thereby impact the expression of key metabolic genes. Genes involved in lipid metabolism, especially omega-6 fatty acid metabolism, are up-regulated in livers of mice after VSG while genes in inflammatory pathways are down-regulated after VSG. Consistent with gene expression changes, regulatory regions near genes involved in inflammatory response displayed decreased chromatin accessibility after VSG. Our results indicate that VSG induces global regulatory changes that impact hepatic inflammatory and lipid metabolic pathways, providing new insight into the mechanisms underlying the beneficial metabolic effects induced by VSG.
Introduction
A dramatic increase in the prevalence of obesity over the last few decades has led to a worldwide epidemic. In the United States alone, one-third of adults are classified as obese (BMI > 30)1. Obesity is a risk factor for many other diseases, including type 2 diabetes (T2D), cardiovascular diseases, and many types of cancer2. Conventional treatments for obesity such as exercise and dietary changes are often insufficient leading to regaining weight. Pharmacological options are limited and patients often experience severe side effects3. Bariatric surgeries, including vertical sleeve gastrectomy (VSG), Roux-en-Y gastric bypass and adjustable gastric band, have shown sustainable weight loss and remission of T2D4. VSG is not only an effective treatment for obesity and T2D, but also has been shown to significantly improve nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH)5, 6. Obese mice that receive VSG have reduced hepatic steatosis prior to weight loss5. Despite these benefits, bariatric surgeries are invasive and are associated with the risk of mortality and nutritional deficiencies4. Furthermore, not all patients sustain the weight reduction7. In order to develop safer and more effective therapeutic approaches for the treatment of these diseases, it is important to better understand the molecular mechanisms by which bariatric surgeries exert these beneficial metabolic effects.
There is increasing evidence that metabolic diseases are associated with epigenetic dysregulation. We have previously demonstrated that diet-induced obesity leads to chromatin modifications in the liver of mice8 and these can persist even upon the removal of an obesogenic diet9. Alteration of promoter methylation of metabolic genes has been observed in muscle tissue from obese patients, and these changes can be reversed after surgery-induced weight loss10. Analysis using liver biopsies of NAFLD and normal individuals revealed NAFLD-specific DNA methylation and transcriptional changes for genes involved in metabolism and insulin signaling. These NAFLD-specific DNA methylation signatures were shown to be partially reversible after bariatric surgery11.
To investigate the molecular mechanisms underlying the beneficial metabolic effects induced by VSG, we performed genome-wide profiling of chromatin accessibility and gene expression in C57BL/6J wild-type male mice after VSG surgery and compared these profiles to mice with a sham surgery. We demonstrate that diet-induced changes in gene expression and chromatin accessibility can be reversed by VSG.
Results
Reversible chromatin accessibility changes after VSG
We resected liver tissue from wild-type male C57BL/6J mice that underwent Sham or VSG surgery (Fig. 1A). As a control, we used livers from age-matched C57BL/6J mice fed on a control standard chow without surgery (Fig. 1A). As described previously, VSG mice had significant weight loss compared to Sham mice over the course of the study12, although VSG mice still weighed more than age-matched control-fed mice (Fig. 1B). Mice that underwent VSG showed reversible changes of liver lipid storage (Fig. 1C) and triglyceride (TG) levels (Fig. 1D). H&E staining showed more lipid droplets in the liver of Sham mice as compared to Control, and the number of lipid droplets decreased in the VSG mice compared to Sham mice (Fig. 1C). Similarly, the hepatic TG levels in Sham mice significantly increased when compared to Control mice (Fig. 1D). In VSG mice, although the hepatic TG levels were higher than Control, they were significantly decreased compared to Sham mice (Fig. 1D). Though the overall weight of VSG mice was greater than Control mice, lipid accumulation and triglyceride levels in liver were reversed by VSG. We next sought to determine the molecular changes associated with these phenotypic changes.
Figure 1.

Characteristics of C57BL/6J mice after vertical sleeve gastrectomy (VSG). (A) Scheme for mice feeding and surgery for Control, Sham, and VSG group. (B) Body weight (grams) over the course of the 14 weeks for Control, Sham, and VSG group. Error bars represent standard error of the mean from 9 to 12 mice in each group. (**Indicates p < 0.01) Sham and VSG weights are from Ding, L. et al.12. (C) Hematoxylin and eosin (top) and Oil Red O (bottom) staining from livers of Control, Sham and VSG mice. (D) Liver triglyceride (milligrams/g) levels of Control, Sham, and VSG mice. Values are mean ± S.D. (n = 5 per group, **Indicates p < 0.01).
To understand the regulatory changes induced by VSG, we profiled genome-wide chromatin accessibility in livers of mice that underwent VSG or Sham procedures, as well as Control mice (Fig. 1A). To evaluate the genome-wide signatures of chromatin accessibility in response to VSG, we performed hierarchical clustering on the union set of accessible chromatin sites across the three conditions (Fig. 2A). Overall, the clustering analysis indicated that the chromatin accessibility profiles of mice that underwent VSG were more similar to Control mice than the mice that underwent Sham procedures (Fig. 2A), indicating that chromatin modifications associated with diet-induced obesity are largely reversed by VSG.
Figure 2.

Reversible chromatin accessibility changes after VSG. (A) Heatmap of relative read counts at each peak of chromatin accessibility, sorted by relative fold change between Sham and Control group. The dendrogram above the heatmap shows the distance between Control, Sham, and VSG group characterized by read counts at each site. (B) Enriched mouse phenotype terms for chromatin sites that are less accessible in VSG mice compared to Sham. (C and D) Examples of reversible genes and accessible chromatin sites at the Cyp2c29 (C) and Cd14 (D) loci.
In order to understand the molecular networks involved in the VSG response, we began by focusing on regions of the genome that displayed variable accessibility between livers of VSG and Sham mice. Based on GREAT analysis13, sites with reduced chromatin accessibility after VSG were enriched for inflammatory phenotypes (Fig. 2B), as well as lipid metabolic diseases (Supplementary Fig. 1). Sites with increased chromatin accessibility after VSG were not enriched for any terms by GREAT analysis. Examples of sites of variable chromatin accessibility and nearby genes are shown in Fig. 2C and D.
Inflammatory and lipid metabolic genes are altered in mouse liver after VSG
After focusing on putative regulatory regions involved in the VSG response, we next wanted to examine the transcriptional response to VSG. We profiled gene expression by RNA sequencing (RNA-seq) using livers from Control, Sham and VSG mice (Fig. 1A). As with the FAIRE-seq analysis, we focused on genes that were differentially expressed between the VSG and Sham mice. We identified 34 differentially expressed genes between Sham and VSG mice (FDR 10%; Supplementary Table 1). Up-regulated genes in VSG compared to Sham were enriched for metabolic pathways, including retinol and linoleic acid metabolism (Fig. 3A and B). In contrast, genes down-regulated in VSG compared to Sham were enriched for inflammatory and defense response (Fig. 3C and D). This result is consistent with chromatin accessibility changes between VSG and Sham mice, for which sites with reduced accessibility were enriched for inflammatory pathways (Fig. 2B). Using a less stringent threshold to identify differentially expressed genes, we found 355 differentially expressed genes between Sham and VSG mice (p-value < 0.05; Supplementary Table 2). With this threshold, up-regulated genes in VSG were still enriched for fatty acid metabolism pathways, and down-regulated genes were still enriched for immune-related pathways (Supplementary Fig. 2), consistent with results obtained using the more stringent threshold (Fig. 3B and D).
Figure 3.

Inflammatory and lipid metabolic genes are altered in mouse liver after VSG. (A) Line plot of relative gene expression for genes up-regulated in VSG compared to Sham mice. (B) Enriched KEGG Pathway terms for genes up-regulated in VSG compared to Sham mice. (C) Line plot of relative gene expression for genes down-regulated in VSG compared to Sham mice. (D) Enriched Gene Ontology Biological Process terms for genes down-regulated in VSG compared to Sham mice.
When we considered the expression levels of genes of Control mice, 68% (23/34) of the genes differentially expressed between Control and Sham showed “reversible” gene expression trends (blue lines in Fig. 3A and red lines in Fig. 3C). Specifically, several cytochrome P450 (CYP450) genes were down-regulated in Sham compared to Control mice, and reversed in VSG mice (Fig. 3A). Similarly, several immune-related genes, such as Cd14 (Fig. 2D) and secretory leukocyte peptidase inhibitor (Slpi), were up-regulated in Sham compared to Control mice, and reversed in VSG mice (Fig. 3C). We also identified differentially expressed genes between Control and Sham mice (FDR 10% DESeq2; Supplementary Table 3). The down-regulated genes in Sham compared to Control mice are enriched for metabolic pathways, including linoleic acid metabolism (Supplementary Fig. 3A), which were up-regulated in VSG compared to Sham (Fig. 3B). The up-regulated genes in Sham compared to Control mice were enriched for inflammatory and immune response (Supplementary Fig. 3B), which were down-regulated in VSG compared to Sham mice (Fig. 3D). This correspondence of pathways further indicates that VSG can reverse diet-induced transcriptome changes.
Omega-6 metabolism altered in liver after VSG
Several VSG-induced differentially expressed genes were from the CYP450 superfamily. The CYP450 superfamily of proteins act as mono-oxygenases for the synthesis and degradation of endogenous compounds such as fatty acids14. Several Cyp genes, including Cyp2c29, Cyp2c50, Cyp2c55, Cyp3a25, and Cyp3a59 (Supplementary Fig. 4A), are down-regulated in Sham compared to Control and reversed in VSG. We also noticed the high variability of the FPKM values of different Cyp genes, which is consistent with a previous report of diverse expression levels of Cyp gene in liver15. We further validated the VSG-induced expression changes of three Cyp genes with RT-qPCR (Fig. 4A).
Figure 4.

Altered omega-6 metabolism by VSG. (A) Relative expression levels of Cyp2b29 Cyp3a11, and Cyp2c55 in Sham and VSG compared to Control mice liver. Expression levels were defined by RT-qPCR. (*Indicates p < 0.05, **Indicates p < 0.01) (B) Representative chromatogram of arachidonic acid and linoleic acid from standards and liver tissues of Sham and VSG mice. Numbers in parentheses represent peak area. (C) Peak area from mass spectrometry of arachidonic acid and linoleic acid in Sham and VSG mice liver (*Indicates p < 0.05, **Indicates p < 0.01; t test).
These Cyp genes we identified as altered in VSG are involved in the metabolism of linoleic acid (LA) and arachidonic acid (AA), two polyunsaturated omega-6 fatty acids. Given the expression changes of lipid metabolic enzymes, we further examined the changes in lipid profiles from Sham and VSG mice by mass spectrometry. We observed reduced levels of TGs in VSG compared to Sham mice (Supplementary Fig. 5A), which is consistent with the measurement by the TG kit (Fig. 1D). We also observed reduced levels of diglyceride (DG) in VSG compared to Sham mice (Supplementary Fig. 5A). The level of total free fatty acids (FFA) in liver was the same between Sham and VSG mice (Supplementary Fig. 5B). However, the levels of both LA and AA were reduced in livers of VSG compared to Sham mice (Fig. 4B and C), which is consistent with the result of increased expression of LA and AA metabolic genes in VSG compared to Sham mice (Fig. 4A). These results indicate that VSG induces changes of omega-6 fatty acid metabolism, although the lipid composition in the diet was the same between Sham and VSG mice12.
Another Cyp gene altered by VSG is Cyp7a1, a key rate-limiting enzyme for bile acid synthesis from cholesterol16. Cyp7a1 is up-regulated in VSG compared to both Sham and Control mice (Supplementary Fig. 4B). Previous studies have demonstrated increased bile acid in both mouse and human after VSG5, 17, 18. The increased levels of Cyp7a1 expression may be an explanation of the bile acid change after VSG.
Regulatory factors altered by VSG
To identify potential upstream regulators for the differentially expressed genes, we first used Ingenuity Pathway Analysis (IPA) and identified several nuclear receptors and inflammatory factors involved with the gene expression changes after VSG (Fig. 5A). The most enriched upstream regulators were proinflammatory cytokines, TNF-α and IFN-γ19. These factors correspond to the VSG-induced regulatory changes and differentially expressed genes in inflammatory response pathways (Figs 2B and 3D).
Figure 5.
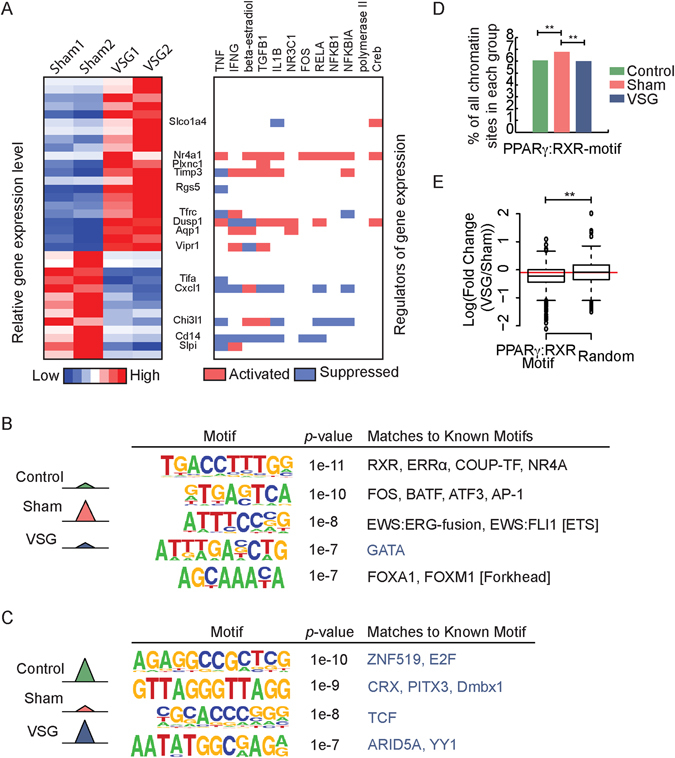
Upstream regulators altered by VSG. (A) Heatmap of differentially express genes between Sham and VSG (left), and upstream regulators from Ingenuity Pathways Analysis (right). (B and C) Identified motifs from reversible chromatin sites that are more (B) or less (C) accessible in Sham compared to Control and VSG. Numbers in parentheses represent p-values of enrichment of motif occurrence in the given set of sequences compared to background, and the percentage of sequences with the motif. (D) Percentage of accessible chromatin site in each group that contained PPARγ:RXR motif (**Indicates p < 0.01, Fisher’s exact test). (E) Fold change (VSG vs Sham) of FAIRE read counts within chromatin sites contain PPARγ:RXR motifs or random chromatin sites (**Indicates p < 0.01, Wilcoxon’s rank-sum test).
To identify specific TFs potentially contributing to gene expression changes, we performed de novo motif analysis on the sets of chromatin sites with reversible accessibility changes in VSG mice (Fig. 5B,C). The top enriched motif for sites more accessible in Sham and reversed in VSG matched with several nuclear receptors, including retinoid X receptor (RXR) (Fig. 5B). RXR forms a heterodimer with liver X receptors (LXRs), and can act as either a repressor or activator depending on the availability of ligands20. LXR-RXR plays a crucial role in lipid homeostasis, and has been shown to regulate Cyp7a1 20, 21. RXR can also heterodimerize with PPARγ, a key regulator of lipid metabolism (Supplementary Fig. 6A)22. To further investigate this, we scanned the whole genome for PPARγ:RXR motif sites and found that the accessible chromatin sites in Sham mice contained significantly more PPARγ:RXR motif sites compared to Control and VSG mice (Fig. 5D). Examining the fold-change of FAIRE-seq read counts further validated the reduction of chromatin accessibility at PPARγ:RXR motif in VSG compared to Sham mice (Fig. 5E). We also checked the PPARγ expression from the different groups. Both isoforms of PPARγ were up-regulated in Sham compared to Control mice, and VSG mice maintained the up-regulated expression of PPARγ (Supplementary Fig. 6B). The induction of PPARγ expression in Sham mice was consistent with previous reports of PPARγ upregulation in fatty liver23, 24. However, the consistent expression level of PPARγ in VSG mice does not explain the reduced accessibility at the PPARγ:RXR motif. One possible explanation for the VSG-induced chromatin variation at PPARγ:RXR binding sites could be availability of ligands. Fatty acids, such as linoleic acid, can activate PPAR25, which may explain the altered accessibility of PPARγ:RXR binding sites after VSG.
We also searched for enriched motifs at sites less accessible in Sham mice that were reversed in VSG mice. We noticed that many of these sequences were GC rich. Indeed, 59% (103/175) of these sites are at CpG islands. Consistent with this, the de novo identified motifs also showed high GC content (Fig. 5C), and matched known motifs have lower matching scores, indicating less confident motif matches (Fig. 5B,C: matched motifs that scored less than 0.7 are colored in blue, while the ones with a score higher than 0.8 are in black).
Discussion
This study has uncovered molecular pathways and regulatory factors altered in mouse liver after VSG surgery. Our results indicate that inflammatory genes and corresponding regulatory factors are suppressed after VSG, while cytochrome P450-mediated fatty acid metabolic pathways are upregulated after VSG. We further demonstrated that diet-induced changes of gene expression and chromatin accessibility can be reversed by VSG, consistent with the improvement of metabolic phenotypes associated with NAFLD.
The overall body weight for mice that underwent VSG was significantly lower than that of Sham mice, consistent with previous studies on the effectiveness of VSG in persistent weight loss4, 12. However, mice that underwent VSG still weighed more than age-matched Control mice 14 weeks post surgery. It may be that a longer period of tracking would show a more significant body weight change. However, within the current experimental design, VSG mice showed reversible hepatic lipid accumulation and triglyceride levels, which further indicates that the beneficial effects of VSG can be independent of overall weight5.
Strikingly, one of the most variable pathways altered by VSG involved the Cyp genes, which play an important role in lipid metabolism in liver. The upregulation of these genes after VSG was further validated by the reduced levels of linoleic acid in livers of VSG compared to Sham mice. Interestingly, previous studies have demonstrated that dietary conjugated linoleic acid reduces weight gain and fat mass in rodents, although there have been conflicting reports on its effects in humans26. Recent studies also show that excessive linoleic acid intake may contribute to chronic inflammation in obese patients27. The Sham and VSG mice we used here were fed with the same diet and food intake was similar between the two groups for a majority of the time after surgery12. Our results therefore, indicates an alteration of metabolic regulations without the differences of overall change of dietary linoleic acid. In addition, previous studies have shown that polyunsaturated fatty acids, including linoleic acid and arachidonic acid, play important roles in energy balance28, 29, although the exact molecular mechanisms remain unclear. This is consistent with the increased energy expenditure in VSG mice we observed previously12.
In the post-operative period following VSG, several studies have observed changes in bile acid (BA) composition30–32. Besides their functions in lipid absorption and digestion in the intestine, BAs can also act as hormones, interacting with the G-protein-coupled BA receptor 1 (GPBAR-1, also known as TGR5) and the nuclear farnesoid X receptor (FXR)33. A recent study demonstrated that the beneficial effects of VSG disappear in FXR knockout mice17. We previously demonstrated that TGR5 is necessary for VSG-induced weight loss, insulin sensitizationing, and improvements of fatty liver12. A separate study recently demonstrated that TGR5 involved in VSG-induced improvement of glucose homeostasis34. We observed the elevated expression of Cyp7a1, which encodes a bile acid synthesis enzyme, in livers of mice that underwent VSG compared to Sham and Control. The increased levels of Cyp7a1 expression may explain the changes in BA after VSG, although a previous study, with different time frames, did not observe the same trend5.
In summary, we have shown that VSG modulates a specific group of genes, and corresponding regulatory elements to exert its beneficial metabolic effects. This study may provide novel insight into the future development of effective therapies for obesity, T2D, and NAFLD.
Research Design And Methods
Ethics Statement
All the procedures within this study were performed following guidelines from National Institutes of Health. All animal study were approved by the City of Hope Institutional Animal Care and Use Committee (IACUC#14031). All methods were carried out in accordance with the relevant guidelines and regulations.
Animal and diets
Male C57BL/6J mice were obtained from the Jackson Laboratory and maintained on a standard chow diet ad libitum and a standard 12 h:12 h light/dark cycle until 8 weeks of age. Mice at this age were then given a high-fat diet (Research Diets D12492, 60 kcal% saturated fat) for 14 weeks. These mice were then randomly divided into 2 groups, followed by VSG or Sham surgery. These mice were maintained on the same high-fat diet for 14 weeks after surgery except the recovery during the immediate postoperative period. The mice in the Control group were maintained on standard chow for 28 weeks. During the post-surgery period, the body weights of all mice and food intake were measured weekly. At the end of the experiment, all mice were sacrificed and livers were harvested. Oil Red O and Hematoxylin and eosin (H&E) staining was performed on liver sections by the Pathology Core at the City of Hope using standard procedures.
Triglycerides were assayed by GPO-PAP method (Nanjing Jiancheng Bioengineering Institute). The color intensity of the red dye product is measured photometrically at 510 nm, and is directly proportional to the triglyceride concentration (mmol/L). After adjustment with the protein concentration (gprot/L) in each sample, the reported units of measurement of liver triglycerides are in mmol/g. The Bio-Rad protein concentration assay kit was used for protein measurement.
VSG surgery
After anaesthetization with isoflurane, the lateral ~80% of the stomach was resected, as previous described17. For the Sham surgery group, the stomach was similarly isolated and then subjected to pressure. After the surgery, mice were maintained on a liquid (Osmolite One Cal) diet during the 4-day recovery period. All animals were transitioned onto regular pellets of the designated diet after the recovery period.
FAIRE-seq
Formaldehyde Isolation of Regulatory Elements (FAIRE) was performed as described previously8. FAIRE DNA from two mice of each condition was barcoded and sequenced on the HiSeq 2500 to generate 100 × 100-bp paired-end reads. Sequenced reads were aligned to the mouse mm9 reference genome using bowtie1 and only reads that could be mapped to a unique genomic location were retained35. Mapped reads were filtered to exclude improperly paired reads and PCR duplicates. Wiggle tracks were generated for visualization on the UCSC genome browser36.
To identify accessible chromatin “peaks” from FAIRE-seq reads, F-seq was used with default parameters and a 250 bp feature length37. Irreproducible discovery rate (IDR) analysis was then used to find reproducible peaks between two replicates38. A union set of peaks among different conditions was generated by the mergeBed function of BEDTools39. To assess global chromatin accessibility across different conditions, read counts for all peaks were normalized40, and the conditions were hierarchically clustered with Cluster 3.041 and viewed with TreeView42. To identify peaks with variable accessibility between different conditions, we utilized DESeq2 (p-value < 0.01)43.
RNA-seq
RNA was extracted from the same livers (two replicates) used for FAIRE using TRIzol (Invitrogen). Ribosomal RNA was depleted from total extracted RNA (Epicenter Ribo-ZeroTM magnetic kit, MRZH11124). Eluted RNA was sequenced with Illumina protocols on a HiSeq 2500 to generate 50-bp reads. Sequenced reads were aligned to the mouse mm9 reference genome with TopHat244. Transcript expression was quantified by HTSeq45, and DESeq2 was utilized to identify differentially expressed genes. The identification of differentially expressed genes by DEseq2 included adjustment for inter-replicate variations. Genes that differentially expressed in Control vs Sham and Sham vs VSG groups in common were defined as reversible genes. Transcript abundance was quantified as fragments per kilobase of transcript per million fragments mapped (FPKM) using Cufflinks46.
Quantitative real-time PCR
Quantitative Real-Time PCR (qRT-PCR) were performed to validate the expression change of selected genes using additional four liver samples of each condition. Total RNA was extracted from frozen liver tissue using Tri Reagent (Molecular Research Center, Inc), followed by cDNA synthesis using SuperScript First-Strand Synthesis System (Bioland Scientific LLC). qRT-PCR was then performed using SYBR Green qPCR Supermix (Applied Biosystems). The mRNA expression for each gene was normalized to M36B417. Results are shown as relative mRNA level according to 2ˆ(–delta delta CT)47. Primer sequences are available in Supplementary Table 4.
Molecular pathway and motif analysis
Enriched gene ontology terms and KEGG pathways for differentially expressed genes were identified using DAVID (v6.7)48. Bonferroni-corrected p-values < 0.1 were considered statistically significant. Gene interaction networks were generated by submitting differentially expressed genes to the Ingenuity Pathways Analysis (IPA) (www.ingenuity.com)
To characterize the enriched biological functions of sites of accessible chromatin, we used genomic coordinates of accessible chromatin sites as input for Genomic Regions Enrichment of Annotations Tool (GREAT)13. Region-gene associations were defined using default parameters and significant associations for “Mouse Phenotype” and “Disease Ontology” were included. Only terms with a false discovery rate (FDR) < 0.05 were reported.
De novo motif discovery was carried out using HOMER (version 4.8) with default parameters49. Motifs were matched to known motifs with higher scores being better matches. STORM50 was further used to scan for occurrences of the PPARγ:RXR motif genome wide. Putative binding sites were defined by motif occurrences within accessible chromatin regions identified in C57BL/6J mouse liver, similar to what has been reported before51.
Lipid extraction and Mass spectrometry
Livers from additional three mice of each condition was used for lipid extraction followed by mass spectrometry. Mouse liver tissue was flash frozen in liquid nitrogen immediately after sacrifice, then a portion of 100 mg was transferred to tubes containing glass beads and homogenized at 4 °C for 6 min in 0.25 mL PBS containing 2 mM EDTA and 100 μM t-butylated hydroxytoluene52. The homogenate was extracted using 0.75 mL of chloroform/methanol (2:1), concentrated in a nitrogen evaporator then dissolved in a volume of buffer B (10% acetonitrile in isopropanol containing 10 mM ammonium formate), proportional with the initial tissue weight for analysis.
The lipids (10 µL injection) were separated with a Dionex Ultimate 3000 UHPLC on a Phenomenex Kinetex 2.1 × 100 mm C18 column with 1.7 µm particle size with a gradient of isopropanol/acetonitrile/water over 25 min from 60% buffer A (40% acetonitrile in 10 mM ammonium formate in water) to 100% buffer B over 15 min then 4 min at 100% B53. Lipids eluting from the UHPLC column were measured with a Thermo Orbitrap Fusion mass spectrometer operated in DDA in either positive or negative ion mode, scanning from 210–1410 at 120000 resolution with a 1 sec duty cycle during which the maximum number of precursor ions were selected by the quadrupole, with an isolation width of 1.4 Da, for HCD fragmentation and their product ion spectra subsequently measured in the Orbitrap at 30000 resolution. The data was then analyzed by in silico product ion database search using Thermo Lipid search 4.1.
Statistical analysis
Data in this study is presented as mean ± standard deviation or standard error of the mean. Statistical tests were performed using Student’s t test or one way ANOVA, and p < 0.05 was considered statistical significant.
Electronic supplementary material
Acknowledgements
We would like to acknowledge Debbie Thurmond and members of the Huang and Schones laboratories for helpful discussions and comments. We thank Gabriel Gugiu for performing lipidomics analysis. This work was supported by Schaeffer Endowed Fund, John Hench Fund, National Cancer Institute 2R01CA139158 (WH), Transform Medicine Innovation Foundation of Shanghai Jiao Tong University School of Medicine 15ZH2001 (JT) and the Fund of the Key Laboratoty of Stem Cell Biology of Chinese Academy of Sciences No.201601 (JT). Research reported in this publication included work performed in the Integrative Genomics Cores supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572.
Author Contributions
J.D., J.T., D.E.S. and W.H. designed the study. J.D., J.T., L.D., C.T., B.X., S.S. performed experiments and analyzed data. J.D., J.T., D.E.S. and W.H. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Juan Du and Jingyan Tian contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05349-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jingyan Tian, Email: tianjypaper@163.com.
Dustin E. Schones, Email: dschones@coh.org
Wendong Huang, Email: WHuang@coh.org.
References
- 1.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of Obesity and Trends in the Distribution of Body Mass Index Among US Adults, 1999-2010. Jama-Journal of the American Medical Association. 2012;307:491–497. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- 2.Pi-Sunyer X. The medical risks of obesity. Postgrad Med. 2009;121:21–33. doi: 10.3810/pgm.2009.11.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palermo A, et al. Oral anti-diabetic drugs and fracture risk, cut to the bone: safe or dangerous? A narrative review. Osteoporosis International. 2015;26:2073–2089. doi: 10.1007/s00198-015-3123-0. [DOI] [PubMed] [Google Scholar]
- 4.Stefater MA, Wilson-Perez HE, Chambers AP, Sandoval DA, Seeley RJ. All bariatric surgeries are not created equal: insights from mechanistic comparisons. Endocr Rev. 2012;33:595–622. doi: 10.1210/er.2011-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myronovych A, et al. Vertical Sleeve Gastrectomy Reduces Hepatic Steatosis While Increasing Serum Bile Acids in a Weight-Loss-Independent Manner. Obesity. 2014;22:390–400. doi: 10.1002/oby.20548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karcz WK, et al. Influence of Sleeve Gastrectomy on NASH and Type 2 Diabetes Mellitus. J Obes. 2011;2011:765473. doi: 10.1155/2011/765473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGrice M, Don Paul K. Interventions to improve long-term weight loss in patients following bariatric surgery: challenges and solutions. Diabetes Metab Syndr Obes. 2015;8:263–274. doi: 10.2147/DMSO.S57054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung A, et al. Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet. J Biol Chem. 2014;289:23557–23567. doi: 10.1074/jbc.M114.581439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung, A., Trac, C., Du, J., Natarajan, R. & Schones, D. E. Persistent chromatin modifications induced by high fat diet. J Biol Chem, doi:10.1074/jbc.M115.711028 (2016). [DOI] [PMC free article] [PubMed]
- 10.Barres R, et al. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep. 2013;3:1020–1027. doi: 10.1016/j.celrep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Ahrens M, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Ding L, et al. Vertical sleeve gastrectomy activates GPBAR-1/TGR5 to sustain weight loss, improve fatty liver, and remit insulin resistance in mice. Hepatology. 2016;64:760–773. doi: 10.1002/hep.28689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honkakoski P, Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J. 2000;347:321–337. doi: 10.1042/bj3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furukawa M, et al. Cytochrome P450 gene expression levels in peripheral blood mononuclear cells in comparison with the liver. Cancer Science. 2004;95:520–529. doi: 10.1111/j.1349-7006.2004.tb03243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorbek G, Lewinska M, Rozman D. Cytochrome P450s in the synthesis of cholesterol and bile acids–from mouse models to human diseases. FEBS J. 2012;279:1516–1533. doi: 10.1111/j.1742-4658.2011.08432.x. [DOI] [PubMed] [Google Scholar]
- 17.Ryan KK, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509:183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinert RE, et al. Bile acids and gut peptide secretion after bariatric surgery: a 1-year prospective randomized pilot trial. Obesity (Silver Spring) 2013;21:E660–668. doi: 10.1002/oby.20522. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Soloski MJ, Diehl AM. Dietary factors alter hepatic innate immune system in mice with nonalcoholic fatty liver disease. Hepatology. 2005;42:880–885. doi: 10.1002/hep.20826. [DOI] [PubMed] [Google Scholar]
- 20.Hong C, Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov. 2014;13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 21.Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. Journal of Clinical Investigation. 2006;116:607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen R, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panasyuk G, et al. PPARgamma contributes to PKM2 and HK2 expression in fatty liver. Nat Commun. 2012;3:672. doi: 10.1038/ncomms1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue M, et al. Increased expression of PPARgamma in high fat diet-induced liver steatosis in mice. Biochem Biophys Res Commun. 2005;336:215–222. doi: 10.1016/j.bbrc.2005.08.070. [DOI] [PubMed] [Google Scholar]
- 25.Ahmadian M, et al. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang YW, Jones PJH. Conjugated linoleic acid and obesity control: efficacy and mechanisms. International Journal of Obesity. 2004;28:941–955. doi: 10.1038/sj.ijo.0802641. [DOI] [PubMed] [Google Scholar]
- 27.Naughton SS, Mathai ML, Hryciw DH, McAinch AJ. Linoleic acid and the pathogenesis of obesity. Prostaglandins & Other Lipid Mediators. 2016;125:90–99. doi: 10.1016/j.prostaglandins.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Clarke SD. Polyunsaturated fatty acid regulation of gene transcription: a mechanism to improve energy balance and insulin resistance. The British journal of nutrition. 2000;83(Suppl 1):S59–66. doi: 10.1017/s0007114500000969. [DOI] [PubMed] [Google Scholar]
- 29.Choque B, Catheline D, Rioux V, Legrand P. Linoleic acid: between doubts and certainties. Biochimie. 2014;96:14–21. doi: 10.1016/j.biochi.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Patti ME, et al. Serum bile acids are higher in humans with prior gastric bypass: potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring) 2009;17:1671–1677. doi: 10.1038/oby.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerhard GS, et al. A role for fibroblast growth factor 19 and bile acids in diabetes remission after Roux-en-Y gastric bypass. Diabetes Care. 2013;36:1859–1864. doi: 10.2337/dc12-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakatani H, et al. Serum bile acid along with plasma incretins and serum high-molecular weight adiponectin levels are increased after bariatric surgery. Metabolism. 2009;58:1400–1407. doi: 10.1016/j.metabol.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap–bile acids in metabolic control. Nat Rev Endocrinol. 2014;10:488–498. doi: 10.1038/nrendo.2014.60. [DOI] [PubMed] [Google Scholar]
- 34.McGavigan AK, et al. TGR5 contributes to glucoregulatory improvements after vertical sleeve gastrectomy in mice. Gut. 2015 doi: 10.1136/gutjnl-2015-309871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kent, W. J. et al. The human genome browser at UCSC. Genome Res12, 996–1006, doi:10.1101/gr.229102. Article published online before print in May 2002 (2002). [DOI] [PMC free article] [PubMed]
- 37.Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li QH, Brown JB, Huang HY, Bickel PJ. Measuring Reproducibility of High-Throughput Experiments. Annals of Applied Statistics. 2011;5:1752–1779. doi: 10.1214/11-AOAS466. [DOI] [Google Scholar]
- 39.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hicks SC, Irizarry R. A. quantro: a data-driven approach to guide the choice of an appropriate normalization method. Genome Biol. 2015;16:117. doi: 10.1186/s13059-015-0679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 43.Du J, et al. Chromatin variation associated with liver metabolism is mediated by transposable elements. Epigenetics & chromatin. 2016;9:28. doi: 10.1186/s13072-016-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu F, et al. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 45.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian JY, et al. Role and mechanism of rosiglitazone on the impairment of insulin secretion induced by free fatty acids on isolated rat islets. Chinese medical journal. 2006;119:574–580. [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 49.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schones DE, Smith AD, Zhang MQ. Statistical significance of cis-regulatory modules. BMC Bioinformatics. 2007;8:19. doi: 10.1186/1471-2105-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gugiu BG, et al. Identification of oxidatively truncated ethanolamine phospholipids in retina and their generation from polyunsaturated phosphatidylethanolamines. Chemical Research in Toxicology. 2006;19:262–271. doi: 10.1021/tx050247f. [DOI] [PubMed] [Google Scholar]
- 53.Castro-Perez JM, et al. Comprehensive LC-MSE Lipidomic Analysis using a Shotgun Approach and Its Application to Biomarker Detection and Identification in Osteoarthritis Patients. Journal of Proteome Research. 2010;9:2377–2389. doi: 10.1021/pr901094j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


