Abstract
The study was designed to isolate and characterize Pseudomonas spp. from sugarcane rhizosphere, and to evaluate their plant- growth- promoting (PGP) traits and nitrogenase activity. A biological nitrogen-fixing microbe has great potential to replace chemical fertilizers and be used as a targeted biofertilizer in a plant. A total of 100 isolates from sugarcane rhizosphere, belonging to different species, were isolated; from these, 30 isolates were selected on the basis of preliminary screening, for in vitro antagonistic activities against sugarcane pathogens and for various PGP traits, as well as nitrogenase activity. The production of IAA varied from 312.07 to 13.12 μg mL−1 in tryptophan supplemented medium, with higher production in AN15 and lower in CN20 strain. The estimation of ACC deaminase activity, strains CY4 and BA2 produced maximum and minimum activity of 77.0 and 15.13 μmoL mg−1 h−1. For nitrogenase activity among the studied strains, CoA6 fixed higher and AY1 fixed lower in amounts (108.30 and 6.16 μmoL C2H2 h−1 mL−1). All the strains were identified on the basis of 16S rRNA gene sequencing, and the phylogenetic diversity of the strains was analyzed. The results identified all strains as being similar to Pseudomonas spp. Polymerase chain reaction (PCR) amplification of nifH and antibiotic genes was suggestive that the amplified strains had the capability to fix nitrogen and possessed biocontrol activities. Genotypic comparisons of the strains were determined by BOX, ERIC, and REP PCR profile analysis. Out of all the screened isolates, CY4 (Pseudomonas koreensis) and CN11 (Pseudomonas entomophila) showed the most prominent PGP traits, as well as nitrogenase activity. Therefore, only these two strains were selected for further studies; Biolog profiling; colonization through green fluorescent protein (GFP)-tagged bacteria; and nifH gene expression using quantitative real-time polymerase chain reaction (qRT-PCR) analysis. The Biolog phenotypic profiling, which comprised utilization of C and N sources, and tolerance to osmolytes and pH, revealed the metabolic versatility of the selected strains. The colonization ability of the selected strains was evaluated by genetically tagging them with a constitutively expressing GFP-pPROBE-pTetr-OT plasmid. qRT-PCR results showed that both strains had the ability to express the nifH gene at 90 and 120 days, as compared to a control, in both sugarcane varieties GT11 and GXB9. Therefore, our isolated strains, P. koreensis and P. entomophila may be used as inoculums or in biofertilizer production for enhancing growth and nutrients, as well as for improving nitrogen levels, in sugarcane and other crops. The present study, to the best of our knowledge, is the first report on the diversity of Pseudomonas spp. associated with sugarcane in Guangxi, China.
Keywords: antibiotic gene, genetic diversity, GFP, Pseudomonas, nifH, sugarcane, Biolog
Introduction
Sugarcane (Saccharum officinarum L.) is one of the most important industrial agricultural crops, being cultivated in over 110 tropical and subtropical countries and, providing a source of sugar, renewable energy, and biomaterials (Fischer et al., 2012). The main sugarcane producing country in the world market is Brazil, and the next major producers are India, China and Thailand (FAO, 2016). More than fifty diseases are caused by plant pathogens in sugarcane (Croft and Magarey, 2000; Rao et al., 2002) with 10–15% of sugar being lost due to such diseases. Among them, red rot, smut, wilt, and pineapple diseases caused by fungi, and ratoon stunting disease caused by bacteria, are found to cause considerable yield loss (Viswanathan and Rao, 2011). Sugarcane is a long duration economical crop, so it requires large amounts of plant nutrients i.e., N, P, K, as well as of other micro nutrients. An abundant supply of nitrogen is required for the early stages of plant growth. However, in many countries, farmers apply even higher doses of fertilizers, chemicals, and pesticides to sugarcane to promote early growth and development and to increase yields. Although, N fertilizer use is comparatively low in Brazil (~50 kg N ha−1), other countries average ~120–300 kg N ha−1 with extreme rates in excess of 700 kg N ha−1 (Robinson et al., 2011). However, a higher dose of fertilizer not only raises the production cost, but also causes serious environmental pollution (Herridge et al., 2008; Li and Yang, 2015). It may have negative and unpredictable effects on the environment, and contribute to the pollution of soil, water, and natural areas.
There is vast microbial flora available globally, and microbes are found in all types of soils, such as sands, deserts, and soils of volcanic origin, and in bogs and moors, snow covered soils, sediments, and semi-aquatic ecosystems, and on rocks (Manoharachary and Mukerji, 2006). There is a clear incentive to exploit this microbial diversity and to isolate and develop functional microbes that can be used, in effect, as targeted fertilizers as an alternative to traditional fertilizer applications. Here, we focused on nitrogen-fixing bacterial genera that are often found in large populations in rhizospheric soils and that exhibit general disease-suppression and PGP traits. In principle, biological nitrogen fixation (BNF) promises an alternative approach to plant N fertilizer requirements (Xing et al., 2006, 2015). Some Brazilian sugarcane varieties are capable of obtaining substantial nitrogen from the soil through BNF (Lima et al., 1987; Urquiaga et al., 1992, 2012). Studies using long-term N balances, 15N natural abundance, and 15N isotope dilution methods have shown that some sugarcane cultivars can obtain a significant amount of their nitrogen requirements in this way (Urquiaga et al., 1992, 2012), but the bacteria responsible remains unknown (Boddey et al., 1995; James and Olivares, 1998; James, 2000).
A diverse array of bacteria, including species of Azoarcus, Azospirillum, Arthrobacter, Azotobacter, Bacillus, Burkholderia, Erwinia, Enterobacter, Gluconacetobacter, Herbaspirillum seropedicae, Klebsiella, Kosakonia, Paenibacillus, Pantoea, Pseudomonas, Stenotrophomonas, Serratia, and Xanthomonas are among the main plant growth promoting rhizobacteria used to promote the growth of several crops, including sugarcane (Somers et al., 2004; Bhattacharyya and Jha, 2012; Carvalho et al., 2014; Rafikova et al., 2016; Xing et al., 2016; Solanki et al., 2017). Strains of some bacterial genera, e.g., Azotobacter, Bacillus, Enterobacter, Pseudomonas, Serratia, and Azospirillum are already being used as biofertilizers for enhancing the growth and yield of crops, as well as for maintaining soil fertility (De Souza et al., 2015). Nitrogen-fixing microorganisms play an important role both in the soil and in plants. Many plant-associated rhizobacteria are recognized for their PGP ability, for their capacity to increase disease resistance, and for their of phytohormones under various stress conditions. However, the use of inoculated microbial activity requires observation of the efficiency and colonization rate to track and identify the inoculated strain within the host plant. We used a popular marker gene that encodes green fluorescent protein (GFP), which was easily detected in cell samples by using confocal microscopy (Unge et al., 1999). Confocal laser scanning electron microscopy (CLSEM), in combination with GFP is a powerful tool for studying plant-microbe interactions (Chi et al., 2004; Liu et al., 2006). N2-fixing microorganisms contain dinitrogenase, one of the subunits of which is encoded by nifH, the detection of nifH mRNA indicates the presence of N2-fixing bacteria, as well as indicating N2-fixation in plants (Young, 1992). Quantitative real-time polymerase chain reaction (qRT-PCR) has been found to be a powerful approach for quantification of an active N2-fixing population within a multifarious community (Wallenstein, 2004). The undeviating contribution of plant-inhabiting diazotrophs (Azospirillum spp., Rhizobium spp., etc.) to nitrogen-fixation and nitrogen-uptake in cucumber has been estimated by quantifying the nifH gene copy number using qRT-PCR (Juraeva et al., 2006).
Researchers have explored and focused on identifying N2-fixing Pseudomonas associated with plants to increase crop production, reduce harmful chemicals and protect the soil and environment. Pseudomonas spp. belong to the family Pseudomonadaceae, which contains a large number of species and is divided into subdivisions (Mehnaz, 2011). Pseudomonas fluorescens and Pseudomonas putida are very well-known and well-studied species of this genus that have been used as inoculums to promote plant growth. Some earlier reports on the isolation of Pseudomonas from sugarcane are: Pseudomonas spp. (Li and Macrae, 1991; Antwerpen et al., 2002; Magnani et al., 2010), P. aeruginosa (Viswanathan et al., 2003), P. aurantiaca (Mehnaz et al., 2009b), P. fluorescens (Viswanathana and Samiyappan, 2002; Mendes et al., 2007; Mehnaz et al., 2009a), P. putida (Viswanathana and Samiyappan, 2002; Mehnaz et al., 2009a), and P. reactans (Mehnaz et al., 2010).
In this study, Pseudomonas strains were isolated from the rhizosphere of sugarcane plants grown in the field in Guangxi, China. We specifically focused on the diversity of Pseudomonas spp. and the major objectives were: (1) to investigate the antagonistic ability of Pseudomonas spp. isolated from sugarcane against pathogens; (2) to evaluate their PGP traits as well as nitrogenase activities in order to use them further as bio-fertilizers; (3) to use polymerase chain reaction (PCR) and qRT-PCR based techniques to detect the nifH gene, characterize antibiotic genes, and assess their genetic diversity through BOX, ERIC and REP-PCR; (4) to analyse 16S rRNA gene sequences for effective identification of Pseudomonas spp.; (5) to test the utilization of numerous sources of carbon, and nitrogen, as well as tolerance to osmolytes and different pH conditions and (6) to investigate the interaction mechanisms between the sugarcane plant and selected potential strains through a GFP technique. As for the available literature, to the best of our knowledge, this is the first report on nitrogen-fixing Pseudomonas koreensis and Pseudomonas entomophila isolated from sugarcane in China.
Materials and methods
Locations and collection of soil sampling site and properties
The study area is Nanning City, Guangxi Autonomous Region in South China. It has a warm with an average temperature of 21.7°C and humid subtropical climate. Summers are hot and the average temperature is 25 (lowest), 33°C (highest) in July, and winters are the coldest, 10°C in January. The average annual rainfall is between 1,000–2,800 mm and precipitation is 1,372 mm. It is situated between 22°49′1.21″ N latitude and 108°21′59.55″ E longitude, and elevation is 79.51 m.
Soil samples were randomly collected from sugarcane fields that have highly fertile soils. Five healthy plants were sampled using a sterile auger from different locations in a sterile specimen container and immediately transported to the laboratory. In all cases, soil samples were taken from 2 to 20 cm layers in April 2015. The soil particles attached to roots were carefully collected after uprooting plants and mixed well. Root debris was removed by sieving through 2 mm mesh. Samples were stored at 4°C for further studies and processed within 24 h of collection. The pulverized soil samples were used for analysis of physico-chemical properties.
Media and growth conditions of bacterial strains
We selected four different enrichment media (Ashbey's medium, Yeast Mannitol Agar, LGI, and Dworkin and Foster salts minimal medium) for the isolation of bacteria from soil samples; all the media contained some component that permitted the growth of specific types of nitrogen-fixing bacteria. A universal nutrient agar (NA) medium was also used for the isolation of all types of bacterial strains (Table S1). Ten grams of soil from each sample were separately suspended in 90 mL of saline water (0.85% of NaCl) in a flask and placed on an orbital shaker (at 100 rpm) at 30 ± 2°C for 1 h.
In vitro test of plant-growth-promoting attributes
The growth promotion traits of all bacterial isolates were evaluated by performing standard protocol for the estimation of indole acetic acid (IAA), P-solubilization, siderophore, hydrogen cyanide (HCN) and ammonia production according to Glickmann and Dessaux (1995), Brick et al. (1991), Schwyn and Neilands (1987), Lorck (1948) and Dey et al. (2004), respectively. For P-solubilization, plates containing Pikovskaya's media amended with tri-calcium phosphate were observed for clearing or solubilisation zones around the colonies. For siderophore production the Chrome Azurol S (CAS) medium was prepared by mixing 25 mL of solution A (composition in gL−1: 60.5 mg CAS was dissolved in 50 mL of distilled water and 10 mL iron (III) solution; 1 mM FeCl3.6H2O, 10 mm HCl). This solution was slowly added to 72.9 mg hexadecyl-trimethyl ammonium bromide (HDTMA) dissolved in 40 mL of water. The resultant dark medium was autoclaved and 75 mL of solution B (Nutrient agar) after autoclaving separately mixed at about 40–50°C and chromazurol sulphonate agar plate has been prepared. The bacterial strains were spot inoculated on chromazurol sulphonate agar plate medium and incubated at 28 ± 2°C for 2–6 days. After incubation of plates siderophore production was assayed by the change in the color of the medium from blue to orange haloes zone formation.
Antifungal activity
All isolates were evaluated for their in vitro antifungal activity by dual culture assays on NA + potato dextrose agar (1:1) plate against the plant pathogens, Ustilago scitaminea and Ceratocystis paradoxa according to Singh et al. (2013, 2014). The strains exhibiting more than 50% inhibition in mycelial growth were considered as promising antagonists.
Nitrogen fixation by acetylene reduction assay (ARA)
Nitrogen-fixing ability of all isolate was tested by using ARA previously described by Hardy et al. (1968). All bacterial isolate was inoculated in a 25 mL flask containing 10 mL semi solid JNFb medium (Table S1) and bacteria were grown at 30 ± 2°C for 3 days. Five Percent air from the tubes was replaced by acetylene through a syringe, incubated for 12 h, 0.5 mL gas was withdrawn from the tube, and ethylene formation was analyzed through a gas chromatograph (GC-17A, Shimadzu, Kyoto, Japan) with a flame ionization detector and a column filled with DB-1701 (Agilent, Santa Clara, USA).
1-Aminocyclopropane-1-Carboxylate (ACC) deaminase activity
Screening for ACC deaminase activity of all isolates was done based on their ability to use ACC as a sole nitrogen source, a trait that is consequence of the activity of the enzyme ACC deaminase. All the isolates were grown in 10 mL of LB broth medium incubated at 30 ± 2°C at 120 rpm for 24–36 h. The cells were harvested by centrifugation at 10,000 rpm for 5 min and washed twice with sterile 0.1 M Tris-HCl (pH 7.5). And it was spotted on petri plates of the modified nitrogen free Dworkin and Foster (DF) medium (Jacobson et al., 1994). Plates without ACC were used as negative control and those with ACC (3 mM) or (NH4)2SO4 (0.2% w/v) plates were used as positive control. The plates were incubated at 30 ± 2°C for 3–5 days. The isolates had ability to grow on ACC plates confirmed that it possessed ACC deaminase activity.
Quantitative estimations of ACC deaminase activities of the bacterial isolates were determined for selected isolates according to the procedures described by Honma and Shimomura (1978) with a standard curve of α-ketobutyrate ranging between 0.01 and 1.0 μ mole. The absorbance was measured at 540 nm (Shimadzu UV-1800, Japan). Enzyme activity was expressed as μmol mg−1 protein h−1.
Genomic DNA extraction
Extraction of total genomic DNA from all the selected bacterial strains was performed using a DNA isolation kit (CWBIO, Beijing, China). After extraction, the quantity, integrity, and quality of the DNA obtained were checked by 0.8% (wt/vol) agarose gel electrophoresis, followed by staining in ethidium bromide, and visualization under UV light. The extracted DNA was further quantified using a nano-photometer (Pearl, Implen-3780, USA).
PCR amplification of 16S rRNA gene
To amplify the 16S rRNA gene, polymerase chain reaction (PCR) was performed from the genomic DNA of strains using a universal primer pair for pA-F and pH-R (Table 1). The PCR program for 16S rRNA gene included initial denaturation at 95°C for 5 min, 30 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 1 min and final extension cycle at 72°C for 5 min. Amplified fragments were checked and purified by using PCR purification kit BioFlux (Hangzhou, China) and then sequenced at Sangon Biotech (Shanghai, China).
Table 1.
PCR primers used for identification, functional genes amplifications and nifH gene expression.
| Gene | Primer | Sequence (5′ → 3′) | Product size | References |
|---|---|---|---|---|
| 16S | pA-F | AGAGTTTGATCCTGGCTCAG | 1,300–1,500 bp | Edwards et al., 1989 |
| pH-R | AAGGAGGTGATCCAGCCGCA | |||
| NifH | Pol-F | TGCGAYCC-SAARGCBGACTC | 360 bp | Poly et al., 2001 |
| Pol-R | ATSGCCATCATYTCRCCGGA | |||
| PhCA | PhCA-F | TTGCCAAGCCTCGCTCCAAC | 1,150 bp | Raaijmakers et al., 1997 |
| PhCA-R | CCGCGTTGTTCCTCGTTCAT | |||
| PRN | Prn-F | GGGGCGGGCCGTGGTGATGGA | 786 bp | Souza and Raaijmakers, 2003 |
| Prn-R | YCCCGCSGCCTGYCTGGTCTG | |||
| HCN | HCN-F | ACTGCCAGGGGCGGATGTGC | 587 bp | Ramette et al., 2003 |
| HCN-R | ACGATGTGCTCGGCGTAC | |||
| BOX | BOX A1R | CTACGGCAAGGCGACGCTGACG | 50–5,000 bp | Rademaker and de Bruijn, 1997 |
| ERIC | ERIC-1 | ATGTAAGCTCCTGGGGATTCAC | 50–5,000 bp | |
| ERIC-2 | AAGTAAGTGACTGGGGTGAGCG | |||
| REP | REP-F | IIIICGICGICATCIGGC | 50–5,000 bp | |
| REP-R | ICGICTTATCIGGCCTAC | |||
| Q-RT-PCR PRIMER | ||||
| NifH | Pol-F | TGCGAYCC-SAARGCBGACTC | Poly et al., 2001 | |
| Pol-R | ATSGCCATCATYTCRCCGGA | |||
| Glyceraldehyde 3-phosphate dehydrogenase | GAPDH-1 | CTCTGCCCCAAGCAAAGATG | Niu et al., 2013 | |
| GAPDH-2 | TGTTGTGCAGCTAGCATTG | |||
Phylogenetic analysis
To perform molecular phylogenetic analysis and evolutionary relationship analysis, the 16S rRNA gene sequences of the isolated Pseudomonas strains were compared with reference strain sequences deposited in the National Center for Biotechnology Information (NCBI) GenBank public database. The sequences were aligned by ClustalW (Saitou and Nei, 1987) and the phylogenetic tree was reconstructed using MEGA software version 7.0 (Kumar et al., 2016) and unweighted pair group method with arithmetic mean (UPGMA) (Sneath and Sokal, 1973) in a Kimura two-parameter model (Tamura et al., 2004). To obtain the confidence values, the gaps were treated by pairwise deletions and bootstrap analysis was carried out by the method of Felsenstein (1985) using 1,000 pseudoreplications.
Amplification, cloning, and sequencing of the nifH gene
Isolated DNA template from all 30 selected strains were used to amplify a conserved region of the nifH gene fragment by PCR, according to the method of Poly et al. (2001) using the primers PolF and PolR (Table 1).
Detection of antibiotic genes
Genomic DNA of Pseudomonas strains was used for the PCR amplification of genes involved in the biosynthesis of three different types of antibiotics genes:- phenazine-1-carboxylic acid (PhCA) (Raaijmakers et al., 1997), pyrrolnitrin (PRN) (Souza and Raaijmakers, 2003), and hydrogen cyanide (HCN) (Ramette et al., 2003) respectively (Table 1). The antibiotics PhCA and PRN are broad-spectrum antibiotics produced by several strains of Pseudomonas, which play an important role in the suppression of multiple plant pathogenic fungi, whereas HCN is a broad-spectrum antimicrobial compound that play a key role in the biological control of root diseases by many plant-associated Pseudomonas isolates.
Genetic diversity studies
The genetic PCR fingerprinting was carried out using repetitive consensus with BOX-PCR (based on primers targeting the highly conserved repetitive DNA sequences of the BOXA subunit of the BOX element), ERIC-PCR (based on primers targeting the highly conserved enterobacterial repetitive intergenic consensus) and REP-PCR (based on primers targeting the repetitive extragenic palindromic sequence). The genomic fingerprints were obtained as described by Rademaker and de Bruijn (1997) to determine phylogenetic relatedness and sequences of different primers (Table 1). All PCR reactions were carried out in Peltier Thermal Cycler BIORAD. PCR amplifications were performed in a 25 μL reaction volume, the reaction mixture and conditions are given in Table S2. Amplification was analyzed by 1.6% agarose gels electrophoresis containing 0.5 μg mL−1 ethidium bromide. A low range ladder (TaKaRa, Dalian, China) was used as molecular size marker. The gels were visualized and gel images were documented in Bio-Rad gel documentation system. The profiles generated by genetic diversity analysis were compared by calculating Jaccard's similarity coefficient for each pairwise comparison and dendrogram was constructed from the similarity matrix by the unweighted pairgroup method with arithmetic average (UPGMA) using NTSYS pc, version 2.02h. All the experiment was carried out in three replicates.
BIOLOG(R) phenotypic assays
Assays of utilization of potential carbon (C), nitrogen (N), and tolerance to different osmotic and pH conditions were tested using BIOLOG Phenotype Micro-Array™ plates GENIII, PM3B, PM9, and PM10 (Biolog Inc., Hayward, CA). The number of all the possible conditions was assayed in the four different types of microplates. GENIII plates were used to study C sources metabolism, and PM3B plates to assess N metabolism sources, respectively. In addition, PM9 and PM10 plates were used to test the growth under various stress conditions and different pH (Bochner, 2009; Mazur et al., 2013). Two isolates (CY4 and CN11) were grown at 30 ± 2°C on LB agar medium and then suspended in an inoculation fluid (IF) after washing to get the transmittance of 90–98% according to procedure. A 100 mL cell suspension was then transferred into the 96 wells of all Micro-Plate, and then incubated at 30 ± 2°C for 48 h to allow the phenotypic fingerprint to form. During incubation there is an increased respiration in the wells and cells can utilize different sources and grow. Increased the respiration causes reduction of the tetrazolium dye and forming a purple color. After incubation the readings were obtained using automated BIOLOG(R) Micro-Station Reader according to the instructions of the manufacturer.
Green fluorescent protein technique
Plasmid transformation
The Pseudomonas strains (CY4 and CN11) resistant to ampicillin (40 μg mL−1) were chosen as recipients for genetic tagging with GFP-pPROBE-pTetr-OT. Both the strains were sensitive to kanamycin (100 μg mL−1). Plasmid pPROBE-pTetr-OT containing the GFP and kanamycin genes expressed under the control of a Tetr promoter was introduced by biparental mating using donor strain E. coli TG1. Plasmid pPROBE-pTetr-OT is a derivative of plasmid pBBR1, which is a small (2.6 kb), broad-host range plasmid and stably maintained in a number of gram (+) and (−) bacteria. The recipient strains and donor strains were mixed at a ratio of 1:2. An aliquot (100 μL) of this mixture was spread onto LB agar. After overnight incubation of the plates at 30°C, the bacteria were washed off the plates and suitable dilutions of the cultures were plated onto selective media containing ampicillin (40 μg mL−1) and kanamycin (100 μg mL−1). Ex-conjugants showing green fluorescence under UV illumination were selected for further study.
Inoculation of micro-propagated sugarcane plantlets
Sugarcane micro-propagated plantlets were inoculated with Pseudomonas strains and GFP-tagged Pseudomonas strain/pPROBEpTetr-TT as described by Oliveira et al. (2002). Five individual rooted plantlets were transferred into a glass bottle of 300 mL capacity containing 50 mL of liquid one-tenth MS medium (sucrose and basal salt mixture) (Reis et al., 1999). Two days after plant transfer, the media free of clear microbial contamination were inoculated with bacterial suspension, providing an initial bacterial cell numbers of approximately 2.0 × 105 mL−1 medium. Plantlets without inoculation were prepared as control. Plantlets were grown in a growth chamber at 30°C with a 14 h photoperiod at a 60 μ moL m−2 s−1 photon flux density.
Laser scanning confocal microscopy (olympus SXZ16)
Three days after inoculation, sugarcane tissue culture plantlets (inoculated and un-inoculated) were taken out from the tubes and sugarcane plantlets were washed with autoclaved distilled water. After cut into small pieces, the root and stem tissues were mounted on bridge slide with 10% (v/v) glycerol. Whole root parts and optical sections of the root and stem pieces were observed with a Leica DMI 6000 microscope attached to a Leica TCS SP5 laser scanning confocal microscope (Leica Microsystems, Mannheim, Germany). GFP fluorescence was detected with an emission band from 500 nm to 530 nm while root auto fluorescence was detected by adjusting the band width from 600 nm to 800 nm depending on the intensity of auto fluorescence (Lin et al., 2012).
RNA extraction and cDNA synthesis
Total RNA was extracted using Trizol reagent (Tiangen, Beijing, China), according to the manufacturer's instructions. DNA contamination of the RNA was removed by DNase I (Promega, USA). The extracted RNA was further quantified using a Nano photometer (Pearl, Implen-3780, USA). Synthesis of first-strand cDNA from the RNA was carried out using the Prime-ScriptTM RT Reagent Kit (TaKaRa, Dalian, China).
Quantitative real-time polymerase chain reaction
Expression patterns of the target nifH gene during plant-microbe interaction were studied in a greenhouse experiment for the selected strains (CY4 and CN11) in two sugarcane varieties (GT11 and GXB9) at 90 and 120 days, as well as for a control. Leaf samples of both sugarcane varieties were used as the experimental material. The relative expression of the target genes was calculated as the expression level of the inoculated sample minus the level of the control at each corresponding time point. Each qRT-PCR experiment was conducted in triplicate. qRT-PCR was carried out with SYBR Premix Ex Tap™ II (TaKaRa, Japan) in Real Time PCR Detection System (Bio-Rad, USA). Reactions were carried out in a final volume of 20 μL, which contained 10 μL SYBR Premix Ex Tap™ II, 2 μL template (10 × diluted cDNA), 0.8 μL of each 10 μM primer and 6.4 μL ddH2O. PCR with distilled water as the template was performed as a control. The primer sequences for nifH are presented in Table 1. The qRT-PCR program was as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 20 s (Niu et al., 2013). Melting curve analysis was conducted at the end of amplification to confirm the specificity of the reaction. The 2−ΔΔCt method was used to quantify the relative gene expression (Livak and Schmittgen, 2001).
Statistical analysis
Experimental data was analyzed using standard analysis of variance (ANOVA) followed by Duncan's multiple range test (DMRT). Standard errors were calculated for all mean values. Differences were considered significant at the p ≤ 0.05 level. All biochemical experiments were performed in triplicate, and the results were expressed as mean values. OrigiPro 9.1 (2013) software was used for the principle component analysis (PCA).
Results
Analysis of chemical and trace elements composition in the soil
The soil texture was analyzed, and characterized as a medium loam. The pH varied from a low of 5.99 to a high of 6.70, and the electrical conductivity of the soil samples varied from 111.0 to 72.8 μS cm−1. The chemical analysis of the soil revealed that the total amounts of nitrogen (N), phosphorus (P) and potassium (K) were 0.30, 0.43, and 13.81 g kg−1, respectively, at the time of sampling, (April 2015); calcium and magnesium levels were 784.6 and 147.6 mg kg−1, respectively. The amounts of trace elements (mg kg−1) were Fe, 106.14; Mn, 85.22; Zn, 7.49; B, 0.42; , 136.67; Cl−, 30.64.
Isolation and PGP potential
A total of 350 bacterial strains were isolated from the sugarcane rhizospheric soil samples. Of these, 100 strains were selected on basis of morphology. These selected strains were further tested by in vitro screening for antifungal activity against sugarcane pathogens (U. scitaminea and C. paradoxa), and for selected PGP traits, as well as for nitrogenase activity. PGP activities were evident from the ability of the selected isolates to produce plant hormones. Of these, only 30 strains were selected that showed various different PGP traits, such as P-solubilization, or siderophore, ammonia, HCN, ACC, or indole-3-acetic acid IAA production, or acetylene reduction activity for molecular identification. In the case of phosphate solubilization, out of the 30 isolates, 26 (87%) isolates showed halo zone formation on Pikovskaya's agar plates, confirming their ability that solubilized the tricalcium phosphate in the medium. An in vitro siderophore production assay revealed that 20 (66.67%) of the strains were able to produce siderophores, 40% of the strains were able to produce ammonia, and 43.33% were able to produce HCN. In the case of qualitative screening of ACC-utilizing bacterial strains, only 18 (60%) exhibited ACC deaminase activity, the strains consumed 3 mM ACC in DF-ACC medium after 48 h incubation at 30 ± 2°C and the color in the DF-ACC medium containing the bacterial strains appeared weaker compared with the non-inoculated medium. In the quantitative estimation of ACC deaminase activity, all strains ranged from 77.0 to 15.13 μmoL mg−1 h−1, with a maximum activity in CY4 and a minimum in the BA2 strain (Table 3). Twenty-one isolates showed antagonistic activity against sugarcane pathogens. Fourteen isolates were antagonistic to U. scitaminea (46.66%), and eleven were antagonistic to C. paradoxa (36.67%) (Table 2).
Table 2.
In vitro biochemical characterization of antagonistic Pseudomonas species on the plant growth promotion traits isolated from sugarcane.
| Culture code | Phosphate | Siderophore | Ammonia | HCN | ACC | Antifungal activity | |
|---|---|---|---|---|---|---|---|
| Ustilago scitaminea | Ceratocystis paradoxa | ||||||
| CoY1 | ++ | +++ | − | − | + | ++ | − |
| CoA5 | ++ | − | ++ | − | − | + | ++ |
| CoA6 | ++ | +++ | ++ | − | − | + | ++ |
| AY1 | ++ | ++ | − | − | − | ++ | − |
| AY2 | +++ | +++ | − | − | + | − | − |
| AA8 | ++ | − | − | +++ | − | ++ | + |
| AN15 | − | +++ | ++ | ++ | − | − | +++ |
| BY2 | ++ | +++ | − | ++ | + | ++ | − |
| BY3 | +++ | − | − | − | − | ++ | − |
| BY10 | +++ | − | − | − | + | − | ++ |
| BA2 | ++ | − | − | − | + | ++ | − |
| BA4 | +++ | − | − | − | − | ++ | − |
| BA7 | +++ | − | +++ | − | − | − | − |
| BA12 | +++ | +++ | − | ++ | + | ++ | − |
| BA13 | +++ | − | +++ | − | − | − | + |
| BA17 | ++ | +++ | − | ++ | + | − | − |
| BN6 | ++ | +++ | +++ | +++ | − | − | − |
| BN7 | ++ | +++ | +++ | +++ | − | − | − |
| CY4 | ++ | ++ | +++ | ++ | + | + | − |
| CY7 | ++ | ++ | − | − | + | + | ++ |
| CA5 | ++ | +++ | +++ | +++ | + | − | − |
| CA7 | +++ | +++ | − | +++ | + | − | +++ |
| CN1 | +++ | ++ | − | − | + | − | − |
| CN2 | − | ++ | +++ | − | + | − | − |
| CN3 | +++ | − | − | − | + | + | − |
| CN9 | +++ | +++ | − | ++ | − | − | + |
| CN11 | +++ | +++ | +++ | +++ | + | − | + |
| CN12 | +++ | − | − | − | + | − | +++ |
| CN15 | − | +++ | − | +++ | + | − | − |
| CN20 | − | ++ | ++ | − | + | + | − |
+, showing low activity; ++, moderate activity; +++, strong activity; −, showing no activity.
Phosphate and Siderophore activity of the bacterial isolates 3 mm or greater clear zone of inhibitions on suitable medium after 3–5 days of incubation at 30 ± 2°C.
Antifungal activity by dual culture plate measured as zone of inhibition after 3–5 days of incubation at 26 ± 2°C.
Biosynthesis of IAA showed differences among the strains, which are summarized in Table 3. The quantitative production of IAA varied from 312.07 to 13.12 μg mL−1 in tryptophan supplemented medium, and a higher level of production was recorded in strain AN15, and a lower one in CN20. In the case of medium without tryptophan, the maximum IAA production was observed from the strain BN7 (23.24 μg mL−1) and the minimum from CoA5 (12.92 μg mL−1). Nitrogen fixation efficiency of all strains was estimated by the acetylene reduction assay (ARA) under laboratory conditions. The data revealed considerable variability in the nitrogenase activity among the studied strains, which ranged from 108.30 to 6.16 μmoL C2H2 h−1 mL−1. On average, under our experimental conditions, the strain CoA6 fixed higher amounts and AY1 fixed lower amounts of nitrogen as compared to the other strains (Table 3).
Table 3.
Screening of different Pseudomonas species for IAA, ARA, and ACC deaminase activity.
| Culture Code | IAA (μg mL−1) | ARA (μmoL C2H2h−1 mL−1) | ACC (μmoL mg−1 h−1) | |
|---|---|---|---|---|
| A-Tryptophane | P-Tryptophane | |||
| CoY1 | 21.84a | 118.12e | 13.61ijk | 30.32ghi |
| CoA5 | 12.92k | 130.86c | 18.41g | – |
| CoA6 | 18.92bc | 153.31b | 108.30a | – |
| AY1 | 15.27fgh | 76.19f | 6.16n | – |
| AY2 | 14.23g−k | 128.35d | 17.46gh | 44.23d |
| AA8 | 14.23g−k | 20.63h | 22.32ef | – |
| AN15 | 13.90g−k | 312.07a | 25.91bcd | – |
| BY2 | 17.44cde | 17.43ij | 22.34ef | 32.92fgh |
| BY3 | 16.47ef | 16.32ijk | 25.71b−e | – |
| BY10 | 13.84g−k | 14.23kl | 29.26b | 19.04k |
| BA2 | 16.38ef | 16.37ijk | 19.30fg | 15.13k |
| BA4 | 17.17de | 17.40ij | 27.08bcd | – |
| BA7 | 13.92g−k | 13.74kl | 25.59cde | – |
| BA12 | 15.38fg | 15.37jkl | 26.04bcd | 36.49ef |
| BA13 | 14.72g−j | 14.92jkl | 18.12g | – |
| BA17 | 15.14fgh | 15.08jkl | 23.83de | 60.78b |
| BN6 | 15.21fgh | 15.21jkl | 17.34gh | – |
| BN7 | 23.13a | 23.12g | 26.08bcd | – |
| CY4 | 20.25b | 310.63a | 26.33bcd | 77.00a |
| CY7 | 14.90f−i | 14.90jkl | 28.30bc | 24.33j |
| CA5 | 13.84g−k | 13.84kl | 8.49lmn | 25.97ij |
| CA7 | 14.51g−k | 14.36kl | 10.82kl | 52.92c |
| CN1 | 13.62h−k | 13.59l | 6.87mn | 32.72fgh |
| CN2 | 13.32ijk | 13.26l | 13.62ijk | 29.68hi |
| CN3 | 13.99g−k | 14.06kl | 18.41g | 34.99fg |
| CN9 | 23.24a | 23.31g | 14.24hij | – |
| CN11 | 23.00a | 23.26g | 16.89ghi | 33.17fgh |
| CN12 | 18.16cd | 18.29i | 11.11jkl | 29.64hi |
| CN15 | 13.77g−k | 13.77kl | 9.71lm | 40.23de |
| CN20 | 13.12jk | 13.12l | 18.58g | 15.62k |
| SEM | 0.499 | 0.785 | 1.082 | 1.479 |
| CD (P = 0.05) | 1.413 | 2.224 | 3.063 | 4.252 |
| CV (%) | 5.3 | 2.5 | 8.6 | 7.3 |
Means followed by same letter within a row are not significantly different (P ≤ 0.05) according to Duncan's Multiple Range Test (DMRT). SEM standard error of the difference between means, CD, critical difference; CV, coefficient of variation.
Molecular identification and phylogenetic analysis
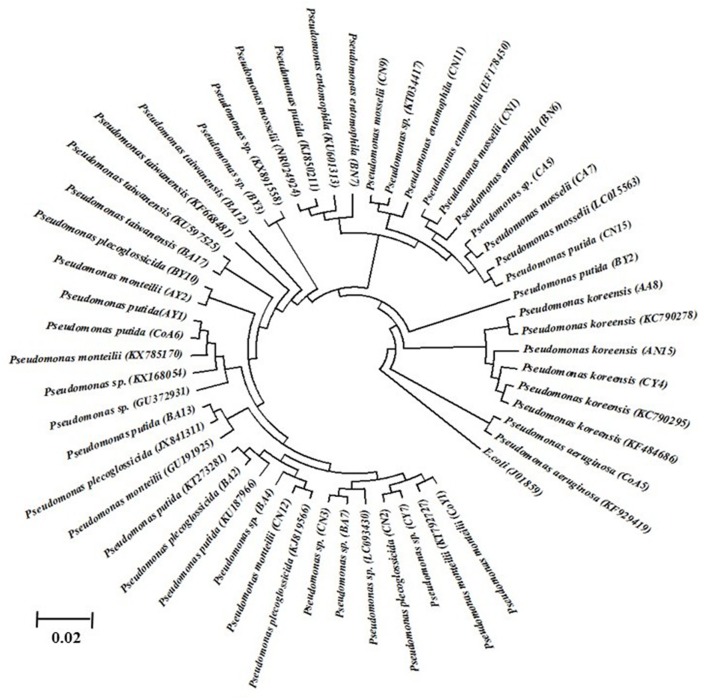
Identification of all isolates was performed based on partial 16S rRNA gene sequencing. The strain sequences obtained were compared using the BlastN tool, with NCBI GenBank database nucleotide sequences and similarity values ≥97% being obtained. The results are suggestive that all the isolates belonged to different species of Pseudomonas, i.e., P. monteilii (3), P. aeruginosa (1), P. putida (5), P. koreensis (3), P. spp. (7), P. plecoglossicida (3), P. taiwanensis (2), P. entomophilla (3), and P. mosselii (3), and the nucleotide sequence data have been submitted to the NCBI GenBank database (Table 4). These DNA sequences were aligned and used to reconstruct a phylogenetic tree, possessing five clusters with 1,000 bootstrap samplings with representative strains of related taxa, as shown in Figure 1.
Table 4.
Identification of putative plant growth-promoting bacterial strains isolated from sugarcane based on 16S rDNA sequence and NCBI GenBank databases.
| Isolates | 16S rRNA gene | % Similarity | Accession number match | No. of nucleotides | Accession numbers |
|---|---|---|---|---|---|
| CoY1 | Pseudomonas monteilii | 97 | KX785170 | 1,501 | KY460972 |
| CoA5 | Pseudomonas aeruginosa | 99 | KF929419 | 1,438 | KY460973 |
| CoA6 | Pseudomonas putida | 99 | KC952984 | 1,440 | KY460974 |
| AY1 | Pseudomonas putida | 99 | KU187966 | 1,414 | KY460975 |
| AY2 | Pseudomonas monteilii | 99 | LC015566 | 1,383 | KY460976 |
| AA8 | Pseudomonas koreensis | 99 | KC790278 | 1,424 | KY460977 |
| AN15 | Pseudomonas koreensis | 99 | KC790275 | 1,446 | KY460978 |
| BY2 | Pseudomonas putida | 98 | KT759148 | 1,040 | KY460979 |
| BY3 | Pseudomonas sp. | 99 | KX891558 | 1,162 | KY460980 |
| BY10 | Pseudomonas plecoglossicida | 99 | LT671913 | 1,396 | KY460981 |
| BA2 | Pseudomonas plecoglossicida | 97 | JQ976892 | 1,436 | KY460982 |
| BA4 | Pseudomonas sp. | 99 | GU372931 | 1,433 | KY460983 |
| BA7 | Pseudomonas sp. | 99 | LC093430 | 1,486 | KY460984 |
| BA12 | Pseudomonas taiwanensis | 99 | KU597525 | 1,417 | KY460985 |
| BA13 | Pseudomonas putida | 99 | KT273281 | 1,458 | KY460986 |
| BA17 | Pseudomonas taiwanensis | 99 | KU597525 | 1,433 | KY460987 |
| BN6 | Pseudomonas entomophila | 99 | KU601313 | 1,386 | KY460988 |
| BN7 | Pseudomonas entomophila | 99 | KU601313 | 1,409 | KY460989 |
| CY4 | Pseudomonas koreensis | 99 | KF484686 | 1,444 | KY460990 |
| CY7 | Pseudomonas sp. | 99 | GU372931 | 1,429 | KY460991 |
| CA5 | Pseudomonas sp. | 98 | KP126777 | 1,445 | KY460992 |
| CA7 | Pseudomonas mosselii | 98 | KU550148 | 1,353 | KY460993 |
| CN1 | Pseudomonas mosselii | 99 | LC015563 | 1,419 | KY460994 |
| CN2 | Pseudomonas plecoglossicida | 99 | KY006168 | 1,404 | KY460995 |
| CN3 | Pseudomonas sp. | 99 | KX168054 | 1,460 | KY460996 |
| CN9 | Pseudomonas mosselii | 98 | LN995691 | 1,490 | KY460997 |
| CN11 | Pseudomonas entomophila | 98 | KU601313 | 1,398 | KY460998 |
| CN12 | Pseudomonas monteilii | 98 | KX785170 | 1,514 | KY460999 |
| CN15 | Pseudomonas putida | 98 | KJ850211 | 1,410 | KY460100 |
| CN20 | Pseudomonas sp. | 97 | KT034417 | 1,448 | KY460101 |
Figure 1.
The 16S rDNA phylogenetic tree of Pseudomonas isolates from sugarcane. The evolutionary history was inferred using the UPGMA method. Bootstrap values of 1,000 replications are indicated as percent confidence values for particular branching. All positions containing gaps and missing data were eliminated. Sequences indicated in code were determined in this study. The E. coli was used as an out group.
Detection of nifH and antibiotic genes
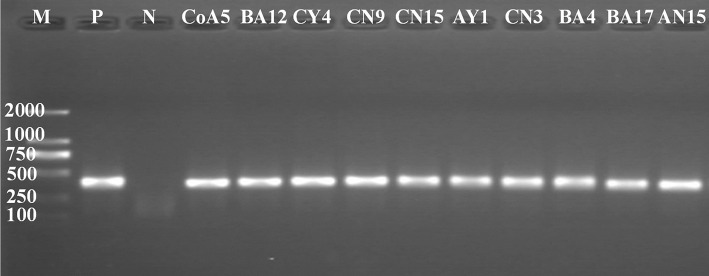
PCR products of the correct size were amplified from the total genomic DNA extracted from all strains. Of these, 10 strains were positive for nifH gene amplification, producing an amplified fragment of about 360 bp (Figure 2); these strains were CoA5, BA12, CY4, CN9, CN15, AY1, CN3, BA4, BA17, and AN15. The All positive strains were selected and used to establish nifH clone libraries. A total of ten clones selected from each strain were analyzed by sequencing. All clones were sequenced, analyzed, and identified by BlastN. All sequences were related to the partial nifH gene. The clones obtained had similarity levels that varied from 90 to 100% and the NCBI GenBank accession numbers are KY508382–KY508391.
Figure 2.
PCR-amplification of nifH gene from genomic DNA of Pseudomanas species. M, molecular size marker from 100 to 2,000 bp. P is a positive control (Klebsiella verticola- DX120E) and N is a negative control (Water). M, molecular size marker (100 bp–2 kb), (Takara).
Amplifications of three genes of antibiotic biosynthesis; (involved in biocontrol activity in the biosynthesis of PhCA, PRN, and HCN) were used in this study. Only four strains showed the PhCA related amplified band size of 1.15 kb, and five showed PRN related gene amplification at around 786 bp. In the case of the HCN related gene, nine strains showed positive amplification at 587 bp. All the amplified products were purified and sequenced. The antibiotic related gene sequences were analyzed and identified using the BlastN program, but only the HCN related sequences submitted to the NCBI GenBank database, under accession numbers KY508373–KY508381. On the basis of the above results, we selected two strains (CY4 and CN11) for the study of substrate richness using different Biolog plates (carbon, nitrogen, and osmolytes), and for study of the plant-microbe interaction mechanism by means of GFP.
Genotypic diversity
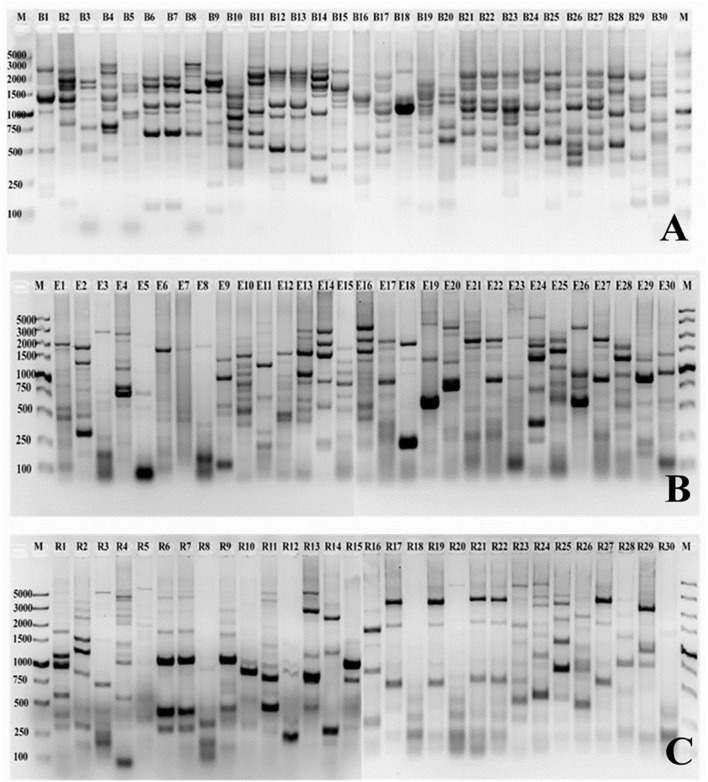
In the present study, the genetic diversity fingerprints of all the selected strains isolated from sugarcane rhizosphere were investigated through BOX, ERIC, and REP-PCR fingerprints. A number of polymorphic bands ranging between 50 bp and about 5 kb were observed, and the DNA fingerprints were clearly differentiated from each other. Nearly all the selected strains showed high-quality DNA fingerprint profiles generated with each primer set (Figure 3). The fingerprint patterns of the thirty isolates generated by BOX, ERIC, and REP-PCR were complex, producing a large number of polymorphic bands of variable intensity. Differences among strains were assessed visually, on the basis of the banding patterns of PCR products. BOX PCR generated a total of 268 bands ranging from 150 bp to 4 kb for all the 30 selected Pseudomonas strains studied. The maximum numbers of bands (12) were observed for CN1 and CN20 while the lowest (5) was found in the case of BN7 (Figure 3A). A total of 229 bands were identified by ERIC-PCR for all the Pseudomonas strains, and the band sizes ranged from 50 bp to 4 kb. CoY1, CoA5, BY10, BA17, CA5, and CN15 showed the highest number of bands (11), while AN15 showed the lowest number of bands (3) (Figure 3B). For REP-PCR, 210 bands were identified, of approximately 50 bp to 5 kb, and faint bands were also frequently observed. The highest number of bands (12) was observed for AY1, while the lowest (4) was found in the case of BN7 and CN11 (Figure 3C). In the case of all these PCRs, BOX-PCR fingerprints revealed a high genotypic diversity for all the Pseudomonas strains.
Figure 3.
PCR finger printing patterns from genomic DNA of Pseudomonas strains isolated from sugarcane. The BOX, ERIC and REP patterns are shown in (A–C), respectively. B1-B30 (BOX), E1-E30 (ERIC), and R1-R30 (REP) is a strain codes used in this study. A 12–15 μL of each product is loaded onto a 1.5% agarose gel. M, molecular size marker (100 bp–5 kb), low range DNA ruler (Takara). Strain codes: 1. CoY1, 2. CoA5, 3. CoA6, 4. AY1, 5. AY2, 6. AA8, 7. AN15, 8. BY2, 9. BY3, 10. BY10, 11. BA2, 12. BA4, 13. BA7, 14. BA12, 15. BA13, 16. BA17, 17. BN6, 18. BN7, 19. CY4, 20. CY7, 21. CA5, 22. CA7, 23. CN1, 24. CN2, 25. CN3, 26. CN9, 27. CN11, 28. CN12, 29. CN15, and 30. CN20.
To determine the relatedness of the isolates, a dendrogram was reconstructed based on BOX, ERIC, and REP-PCR fingerprint bands (Figure 4), and the data were analyzed by using Jaccard similarity coefficients and the neighbor-joining clustering method based on pair-wise similarity coefficients with UPGMA. The dendrogram generated through BOX PCR fingerprints revealed two major clusters, one containing five strains and the other containing 25 strains (Figure 4A). The clustering was clear, and represented 28 distinct clusters of Pseudomonas spp. In the ERIC-PCR, all 30 Pseudomonas spp. showed two major clusters containing 20 and 10 strains (Figure 4B), all grouped into different clusters. As with BOX and ERIC-PCR, the dendrogram generated based on the REP-PCR fingerprints also showed two major clusters, containing one and 29 strains (Figure 4C) respectively, and all 30 strains were grouped into 28 different clusters of Pseudomonas spp.
Figure 4.
Cluster analyses of (A) BOX, (B) ERIC, and (C) REP-PCR fingerprints showing the genotypic diversity of Pseudomonas species isolated from sugarcane. Dendrogram was obtained from the similarity coefficient calculations and clustering was done using unweighted pair-grouping method based on arithmetic averages (UPGMA) using NTSYS software and the Jaccard coefficient.
Substrate utilization patterns
Selected strains were tested for metabolic potential with several compounds as sole sources for carbon (C), using GNIII and nitrogen (N) PM3B micro-plates. Strain tolerance of osmotic stress was examined with PM9 micro-plates, and metabolic activity over a wide range of pH, 3.5–10, was determined by using PM10 micro-plates. The Biolog system was used to detect the biochemical, physiological, and chemical sensitivity of strains on the basis of substrate richness (Table S3). The results indicated that strain CY4, followed by CN11, showed the highest utilization of carbon sources, i.e., sugars (28.17%), carboxylic acids (18.30%), hexose acids (12.68%), and amino acids (9.86%), and the highest chemical sensitivity (86.95%) (Figure 5).
Figure 5.
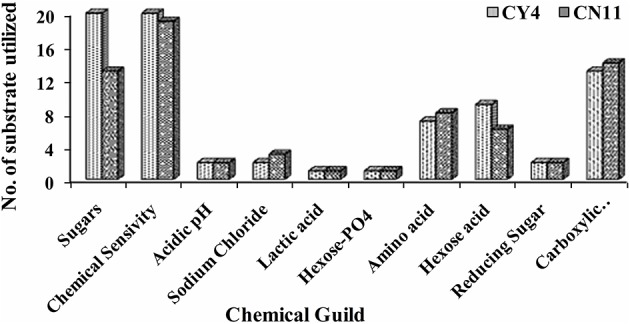
Metabolic differences among CY4 and CN11 in presence of sugars, chemical sensivity, acidic pH, sodium chloride, lactic acid, hexose PO4, amino acid, hexose acid, reducing sugar, and carboxylic acid using BIOLOG Phenotype Micro-Array™ plates GNIII.
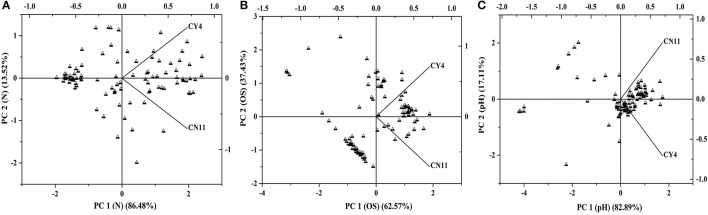
A qualitative analysis of metabolic differentiation between the selected strains was performed through principal component analysis (PCA), using PM3B, PM9, and PM10 (Figure 6 and Table S4). The main principle for the grouping of every substrate group to the individual component was the relationship between different metabolites and strain utilization. For these studies, separate evaluations of nitrogen, osmolytes, and pH were performed. A scatter plots diagram of PCA showed the nitrogen of different components accounted for 86.48% of the metabolic variation in PC1 for the selected strains (Figure 6A). The osmolytes accounted for 62.57% of the variance and the pH for 82.89% of the variance observed in PC1 (Figures 6B,C). Based on the data obtained through the Biolog analysis, the diversity index was measured and was evident for the selected strains, which showed high metabolic diversity (Table 5).
Figure 6.
The scatterplots of selected two strains Pseudomonas koreensis (CY4) and Pseudomonas entomophila (CN11) were analyzed through principle component analysis (PCA) under the different treatments by using the BIOLOG(R) micro-plates (A) nitrogen, (B) osmolytes, and (C) pH.
Table 5.
Substrate richness diversity indices calculated for nitrogen, osmolytes and pH through Biolog for selected strains Pseudomonas koreensis (CY4) and Pseudomonas entomophila (CN11).
| Index measure | Diversity indices through Biolog Micro-Plates | |||||
|---|---|---|---|---|---|---|
| Strain CY4 | Strain CN11 | |||||
| Nitrogen | Osmolytes | pH | Nitrogen | Osmolytes | pH | |
| PM3B | PM9 | PM10 | PM3B | PM9 | PM10 | |
| Dominance D | 0.01507 | 0.01634 | 0.01178 | 0.01488 | 0.01137 | 0.01131 |
| Simpson 1-D | 0.9849 | 0.9837 | 0.9882 | 0.9851 | 0.9886 | 0.9887 |
| Shannon H | 4.327 | 4.193 | 4.46 | 4.33 | 4.499 | 4.501 |
| Evenness e∧H/S | 0.7891 | 0.6899 | 0.9006 | 0.7912 | 0.9368 | 0.9385 |
| Brillouin | 1.357 | 2.177 | 2.729 | 1.311 | 2.072 | 2.522 |
| Menhinick | 10.71 | 8.977 | 7.258 | 11.29 | 7.911 | 7.339 |
| Margalef | 25.58 | 21.56 | 19.31 | 26.51 | 20.86 | 19.64 |
| Equitability J | 0.9481 | 0.9187 | 0.9771 | 0.9487 | 0.9857 | 0.9861 |
| Fisher alpha | 0 | 282 | 87.25 | 0 | 119.6 | 90.32 |
| Berger Parker | 0.01244 | 0.01749 | 0.01143 | 0.01384 | 0.01358 | 0.01169 |
Colonization of sugarcane plants by GFP-tagged bacteria
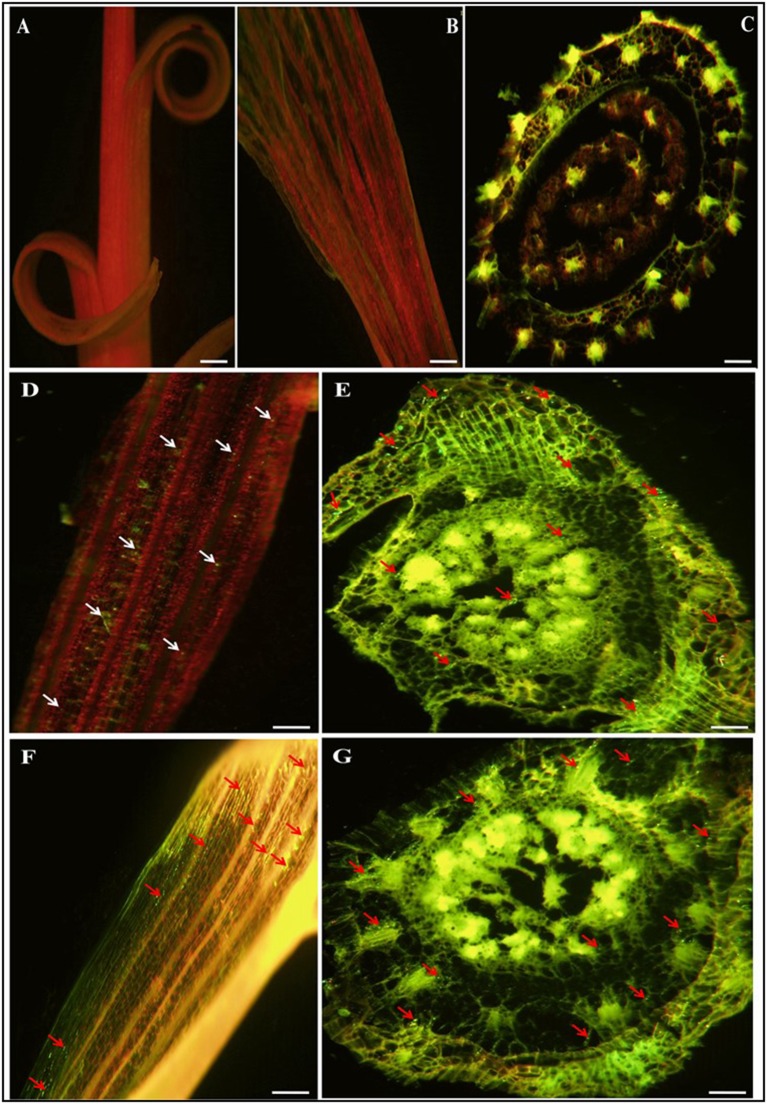
Visualization of root colonization was carried out by CLSEM. This technique facilitated study of the interaction of the selected potential strains in sugarcane. Bacterial colonization in the internal tissues of plants was observed in almost all parts of the sugarcane plant. After 72 h of incubation with the inoculated strains (CY4 and CN11), it was observed that the bacterial cell density had increased, and green fluorescent cells were distributed throughout the plant organs, including roots, stems, and leaves (Figure 7). No fluorescent bacteria were detected by CLSEM in the uninoculated plants served as a control (Figures 7A–C). In roots, the GFP-tagged bacteria were observed by CLSEM to colonize mostly mature root hair zones. Many of the cells were over shadowed by the green fluorescence light emitted from the cell walls of the epidermal cells, endodermal, xylem vessels, and junction sites between the primary and lateral root zones, which were excited by the blue light of the fluorescence microscope (Figures 7E,G). In the case of leaves, colonization of GFP-tagged bacteria was clearly observed through the green auto-fluorescence emitted as small dots in all plant parts (Figures 7D,F), in the case of roots, the cells emitted fluorescence in the maturation zone and within the body of the root, detected by CLSEM in transvers hand-cut optical sections.
Figure 7.
Confocal laser scanning micrographs images showing gfp-tagged strains (CY4 and CN11) colonized in and on roots and leaves of sugarcane micropropagated plantlets GT11 (variety). (A–C) is control sugarcane plantlets parts i.e., stem, leaf and root, without inoculated strains. Confocal microscopic images (D–G) present inoculated bacterial GFP fluorescence (500–530 nm) in green dots and auto-fluorescence in everywhere in leaf and root. Arrow heads point indicates bacterial cells present in a single or grouped of bacteria. (D,E) represents CY4 and (F,G) is CN11 strain. Bars present 50 μm.
nifH gene expression determined by qRT-PCR
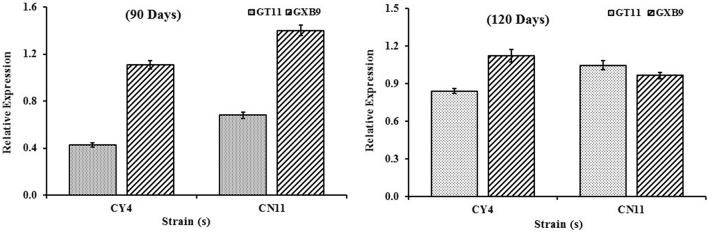
The nifH gene expression pattern for two sugarcane varieties (GT11 and GXB9) inoculated with CY4 and CN11 strains at 90 and 120 days was studied (Figure 8). The results showed that, using total RNA extracted from sugarcane leaf samples, nifH gene expression was positively detected by means of qRT-PCR. The highest expression of the nifH gene was recorded at 90 days, in GXB9 inoculated with strain CN11.
Figure 8.
qRT-PCR analysis of the nifH expression patterns in sugarcane varieties (GT11 and GXB9) and potent strains (Pseudomonas koreensis-CY4 and Pseudomonas entomophila-CN11) interaction. Data were normalized to the GAPDH expression level. All data points (with the deduction of their controls) are the means ± SE (n = 3).
Discussion
In this study, soil samples from sugarcane were used for the isolation of PGP nitrogen-fixing bacteria. Guangxi is the major sugarcane producing province in South China, and more than 60% of the total sugar production is from this area (Li and Yang, 2015). In China, the nitrogen application rate is very high, that is about 500–700 kg ha−1 annually for commercial sugarcane production and this is several times higher than in Brazil and other countries (Li et al., 2015). Higher application of nitrogen fertilizers not only raises the production cost, but also has an adverse effect on the environment and on soil health. Root-associated microbes have a positive effect on soil nutrient availability for plants (Glick et al., 1994). PGPR increases root surface area, increasing nutrient uptake and improving plant production (Mantelin and Touraine, 2004). Therefore, PGPR is an alternative method, the use of chemicals, for sugarcane nutrition because this crop requires large quantities of nitrogen fertilizer for growth and development. A number of nitrogen-fixing microbes have been reported in sugarcane (Xing et al., 2006, 2015; Mehnaz et al., 2009a; Lin et al., 2012; Solanki et al., 2017). Pseudomonas spp. are important PGPRs used as biofertilizer, and are able to enhance crop yield by direct and indirect mechanisms (Walsh et al., 2001), in addition to their characteristics of antibiotic production, phosphate solubilization, siderophore, IAA, HCN, and ammonia production (Ahemad and Khan, 2012a,b), nitrogen fixation, ACC deaminase activity, plant hormone production, and biological control. Of all the strains isolated in the present study, only 30 were selected on the basis of various different PGP traits, and nitrogenase activity.
Phosphorus is one of the most important plant nutrients and it greatly affects the growth of plants (Wang et al., 2009). In soil, P is highly insoluble and is therefore unavailable to plants. Phosphate-solubilizing microorganisms increase the performance of plants by providing them with soluble phosphorus. Phosphate solubilization has already been reported for different strains of Pseudomonas frederiks-bergensis (Zeng et al., 2016). In this study, we found that 26 (87%) isolates were positive for phosphate solubilization to convert insoluble tricalcium phosphate to the soluble form. Another important PGPR trait, which may indirectly influences plant growth, is the production of siderophores. Microorganisms play an important role in several essential biological processes and developed specific mechanisms for the assimilation of iron by production of low molecular weight iron-chelating compounds siderophores, which transport this element into their cells (Schwyn and Neilands, 1987; Arora et al., 2013). In the present investigation, 20 (66.67%) strains displayed siderophores production ability; siderophore production by Pseudomonas spp. is well known (Liu et al., 2011; Luo, 2014). HCN production by bacterial strains is important for diseases suppression and protection of plants from fungal diseases. HCN production by P. fluorescens strain CHA0 was related to biocontrol ability and root colonization; for example, suppression of tobacco black root rot caused by Thielaviopsis basicola (Voisard et al., 1989; Laville et al., 1992). The results of qualitative test of HCN production showed that 43.33% of the bacteria were capable of producing HCN. Volatile compounds, such as ammonia produced by a number of rhizobacteria, have been reported to play an important role in biocontrol (Brimecombe et al., 2001). Our results showed that 40.0% of the Pseudomonas strains were able to produce ammonia and might therefore play an important role in biocontrol activity. A previous isolate, P. fluorescens BAM-4, from semi-arid soil, is a potential biocontrol agent against M. phaseolina I mung bean and P. aeruginosa RM-3 showed lysis of several pathogenic fungi (Minaxi and Saxena, 2010a,b). To test strains for antifungal activity, we screened against two sugarcane pathogens (U. scitaminea and C. paradoxa); against these antagonistic activity was observed in 46.66 and 33.33% of strains, respectively (Table 2).
The role of ACC deaminase has been documented as one of the major mechanisms of PGP bacteria in promoting root and plant growth (Glick et al., 2007). Inoculation with ACC-deaminase containing bacteria promotes root growth of developing seedlings of various crops (Zahir et al., 2003). Inoculation with rhizobacteria having ACC deaminase activity resulted in the development of a better root system, which subsequently had a positive effect on shoot growth (Glick et al., 1998; Belimov et al., 2002). The screening of bacterial isolates obtained from sugarcane with DF medium and DF-ACC medium showed that 60% of the bacteria contained ACC deaminase, when grown on medium containing ACC as the sole nitrogen source. Most ACC-utilizing bacterial isolates are known to belong to genus Burkholderia and genus Pseudomonas (Blaha et al., 2006; Onofre-Lemus et al., 2009). All the selected bacterial strains showed ACC deaminase activity ranging from 77.0 to 15.13 μmoL mg−1 h−1, and higher activity was observed in strain CY4. In this study, we found that all bacterial strains were able to produce IAA in the range of 312.07–13.12 μg mL−1. The potential of the bacterial strains to produce IAA is indicative of their capability to be used as growth hormones or growth regulators. The results for nitrogen fixation as determined by ARA indicated that a large population of sugarcane-associated nitrogen-fixing bacterial strains is present in the soil and may be beneficial in improving the nitrogen level of sugarcane. The selected isolates were evaluated for their nitrogen-fixation efficiency, which was highly variable, ranging from 108.30 to 6.16 μmoL C2H2 h−1 mL−1. The incidence of nitrogen fixation in Pseudomonas spp. has been long debated, but recently several such Pseudomonas strains have been identified (Mirza et al., 2006). Several studies have also shown that nitrogenase producing bacteria can be isolated from sugarcane (Xing et al., 2006; Mehnaz et al., 2010; Lin et al., 2012; Solanki et al., 2017).
In this paper, we have also described a molecular approach for analyzing nitrogen-fixing genes in pure cultures isolated from sugarcane. Several primer have been used to amplify nifH genes, but after comparison of the sequences obtained from the nifH clones, we found the primer sets were is suitable for this study. All strains showed nitrogen-fixing activity by ARA in N-free medium, but the nifH gene was amplified only in 10 strains. nifH is one of the earliest characterized and best known functional genes (Rosado et al., 1998), and its amplification using degenerate primers is a useful tool for confirming nitrogen-fixation potential (Zehr and Capone, 1996). On the other hand, if amplification does not occur using the primers, this is not proof that strains are not proficient in nitrogen fixation, because the gene may show diverse nucleotide sequences between species and even within the same species (Zehr et al., 2003). An important objective of this study was to evaluate all the strains for antibiotic biosynthetic gene targets, because the production of various antimicrobial compounds is one of the biocontrol that helps to degrade pathogen cell walls and it is also an important factor for disease suppression (Sasirekha et al., 2013).
In the present study, we have demonstrated that repetitive extragenic sequences such as BOX, ERIC and REP are present in the genome of Pseudomonas bacteria. There little information published regarding the use of PCR to study the genetic diversity of Pseudomonas isolated from sugarcane. A method that was described for bacterial fingerprinting by examining strain specific banding patterns obtained from PCR amplification of repetitive DNA elements presented entire bacterial genomes (Versalovic et al., 1991). This technique is useful in the classification and differentiation of strains in many Gram-positive and Gram-negative bacteria, and also proved to be a powerful tool for initially screening within the strains. REP patterns are generally less complex than BOX and ERIC patterns, but all could give good discrimination at the strain level for the strains isolated from sugarcane (Figure 3). Our results also support a previous study in P. aeruginosa (Dawson et al., 2002), and the technique proved to be effective for evaluating the diversity of nitrogen-fixing bacterial strains.
Of all the strains, only two CY4 and CN11, were selected for further studies, such as Biolog profiling, GFP localization, and nifH gene expression, through qRT-PCR in sugarcane. Metabolic profiling of the selected isolates was performed using Biolog microplates, comprising analyses of the utilization of nutritional compounds (carbon and nitrogen), as well as tolerance to osmolytes and different pH conditions. The metabolic assets of an organism could contribute toward a particular adaptation and therefore might provide valuable information about bacteria supportive for root colonization (Mazur et al., 2013). The patterns of phenotypes obtained were suggestive that the selected strains were capable of utilizing a variety of metabolic substrates (Figure 5). Previously, it was suggested that more metabolically useful strains were more successful competitors in host plant nodulation (Wielbo et al., 2007). Remarkably, in our studies, the more metabolically diverse strain was CY4, rather than CN11. Through metabolic profiling, tolerance to osmolytes and different pH conditions was studied, using diversity indices such as Simpson 1-D, Shannon H, Evenness e∧H/S, Brillouin and Equitability J, and these methods revealed the substrate richness of the selected strains. Phenotypic profiling is important for understanding genotype differences, stress responses, media composition, and changes in environmental conditions for microorganisms (Chojniak et al., 2015).
We also investigated the effect of the selected Pseudomonas strains CY4 and CN11 on sugarcane by genetically tagging them with GFP-pPROBE-pTetr-OT and observing the level of colonization in plantlets. Both Pseudomonas strains colonized the whole plantlets as for roots, stems and leaves when inoculated separately. Uninoculated sugarcane plantlets served as the control and the plantlets did not show any appearance of fluorescence after 3 and 5 days of inoculation. This result showed that there was no fluorescence from whole plantlet tissues, but after comparison of strains CY4 and CN11, appeared that strain CY4 was a better colonizer for sugarcane plantlets. CLSEM and GFP have previously been used to demonstrate the colonization pattern of Bacillus megaterium in rice (Liu et al., 2006), Klebsiella pneumoniae in maize (Chelius and Triplett, 2000), Rhizobium sp. and Burkholderia sp. in rice (Singh et al., 2009), and Microbacterium sp. in sugarcane (Lin et al., 2012). This study shows that the GFP technique can be used effectively to evaluate competitive colonization capability for more than one plant-associated bacterium (Singh et al., 2009), or to select a potent strain isolated from different crops.
In, previous studies, it was considered that there were no nitrogen-fixing strains in the genus Pseudomonas (Setten et al., 2013). In fact, the inability of Pseudomonas spp. to fix nitrogen had been proposed as an important taxonomic character (Young, 1992; Anzai et al., 2000; Solanki et al., 2017). However, recent studies have confirmed that some strains belonging to the genus Pseudomonas sensu stricto, such as P. stutzeri A1501, P. stutzeri DSM4166, P. azotifigens 6HT33bT and Pseudomonas sp. K1, have the capability to fix nitrogen (Mehnaz, 2011; Setten et al., 2013; Solanki et al., 2017). In our study, the selected Pseudomonas strains CY4 and CN11 also showed nifH gene expression in sugarcane, detected through qRT-PCR. Therefore, the presence of the nifH gene is indicative of the existence of diazotrophs, and the expression of the nifH gene is suggestive of the occurrence of BNF (Akter et al., 2014). The qRT-PCR technique is advantageous because of its high sensitivity and specificity, and detection of mRNA has been reported in ecological samples (Noda et al., 1999; Brown et al., 2003).
Conclusion
To the best of our knowledge, this study is the first systematic report that has provided an awareness of the bacterial genus Pseudomonas associated with sugarcane rhizosphere. We have isolated bacteria that show useful activities in phosphate solubilization, siderophore production, ACC deaminase activity, and IAA-production, as well as N2-fixing activity and disease management. These features are measured as important PGP traits and have been found to be effective in improving the growth and nitrogen content of sugarcane plants. The strains CY4 (P. koreensis) and CN11 (P. entomophila) turned out to be very efficient in terms of enhancing the growth and development of plants, and disease control, as well as having nitrogenase activity. These organisms have greater potential to be used as biofertilizer due to their properties of nitrogen fixation, phytohormone production, and biocontrol capability. Assessment of these strains in field trials is now required, to determine their efficiency in plant growth promotion under field conditions. The inoculation of PGPR isolates may be an imminent development biofertilizer applications, for sustainable crop production, in reducing environmental pollution, and in biological agri-business.
Author contributions
HL, RS, LY, and YL conceived and proposed the idea and drafted the manuscript. HL, RS, PS, QS, and YX carried out the experiments and conducted data analysis. All authors have read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This present study was supported by the National Natural Science Foundation of China (31471449, 31171504, 31101122), the National High Technology Research and Development Program (“63” Program) of China (2013AA102604), Guangxi Special Funds for Bagui Scholars and Distinguished Experts (2013), Guangxi Natural Science Foundation (2011GXNSFF018002, 2012GXNSFDA053011, 2013NXNSFAA019073) and Guangxi Academy of Agriculture Sciences Fund (GNK2014YD01, GNKB2014021). Fund for Guangxi Innovation Teams of Modern Agriculture Technology (gjnytxgxcxtd-03-01) and Fund of Guangxi Academy of Agricultural Sciences (2015YT02).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01268/full#supplementary-material
References
- Ahemad M., Khan M. S. (2012a). Effect of fungicides on plant growth promoting activities of phosphate solubilizing Pseudomonas putida isolated from mustard (Brassica compestris) rhizosphere. Chemosphere. 86, 945–950. 10.1016/j.chemosphere.2011.11.013 [DOI] [PubMed] [Google Scholar]
- Ahemad M., Khan M. S. (2012b). Evaluation of plant growth promoting activities of rhizobacterium Pseudomonas putida under herbicide-stress. Ann. Microbiol. 62, 1531–1540. 10.1007/s13213-011-0407-2 [DOI] [Google Scholar]
- Akter Z., Pageni B. B., Lupwayi N. Z., Balasubramanian P. M. (2014). Biological nitrogen fixation and nifH gene expression in dry beans (Phaseolus vulgaris L.). Can. J. Plant Sci. 94, 203–212. 10.4141/cjps2013-200 [DOI] [Google Scholar]
- Antwerpen T. V., Rutherford R. S., Vogel J. L. (2002). Assessment of sugarcane endophytic bacteria and rhizospheric Burkholderia species as antifungal agents. Proc. S. Afr. Sug. Technol. Assoc. 76, 301–304. [Google Scholar]
- Anzai Y., Kim H., Park J. Y., Wakabayashi H., Oyaizu H. (2000). Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 4, 1563–1589. 10.1099/00207713-50-4-1563 [DOI] [PubMed] [Google Scholar]
- Arora N. K., Tewari S., Singh R. (2013). Multifaceted plant-associated microbes and their mechanisms diminish the concept of direct and indirect PGPRs, in Plant Microbe Symbiosis; Fundamentals and Advances, ed Arora N. K. (Lucknow: Springer; ), 411–449. [Google Scholar]
- Belimov A. A., Safronova V. I., Mimura T. (2002). Response of spring rape (Brassica napus L. var. Oleifera) to inoculation with plant growth promoting rhizobacteria containing 1-aminocyclopropane-1-carboxylate deaminase depends on nutrient status of the plant. Can. J. Microbiol. 48, 189–199. 10.1139/w02-007 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya P. N., Jha D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World. J. Microbiol. Biotechnol. 28, 1327–1350. 10.1007/s11274-011-0979-9 [DOI] [PubMed] [Google Scholar]
- Blaha D., Prigent-Combaret C., Mirza M. S., Moenne-Loccoz Y. (2006). Phylogeny of the 1-amino cyclopropane-1-carboxylic acid deaminase encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol. Ecol. 56, 455–470. 10.1111/j.1574-6941.2006.00082.x [DOI] [PubMed] [Google Scholar]
- Bochner B. R. (2009). Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 33, 191–205. 10.1111/j.1574-6976.2008.00149.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddey R. M., de Oliveira O. C., Urquiaga S., Reis V. M., de Olivares F. L., Baldani V. L., et al. (1995). Biological nitrogen fixation associated with sugar cane and rice: contributions and prospects for improvement. Plant Soil. 174, 195–209. 10.1007/BF00032247 [DOI] [Google Scholar]
- Brick J. M., Bostock R. M., Silverstone S. E. (1991). Rapid in situ assay for indole acetic acid production by bacteria immobilized on nitrocellulose membrane. Appl. Environ. Microbiol. 57, 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimecombe M. J., De Liej F. A., Lynch J. M. (2001). The effect of root exudates on rhizosphere microbial populations, in The Rhizosphere, eds Pinton R., Varanini Z., Nannipieri P. (New York, NY: Marcel Dekker; ), 95–140. [Google Scholar]
- Brown M. M., Friez M. J., Lovell C. R. (2003). Expression of nifH genes by diazotrophic bacteria in the rhizosphere of short form Spartina alterniflora. FEMS Microbiol. Ecol. 43, 411–417. 10.1111/j.1574-6941.2003.tb01081.x [DOI] [PubMed] [Google Scholar]
- Carvalho T. L. G., Balsemao-Pires E., Saraiva R. M., Ferreira S. G., Hemerly A. S. (2014). Nitrogen signalling in plant interactions with associative and endophytic diazotrophic bacteria. J. Exp. Bot. 65, 5631–5642. 10.1093/jxb/eru319 [DOI] [PubMed] [Google Scholar]
- Chelius M. K., Triplett E. W. (2000). Immuno localization of dinitrogenase reductase produced by Klebsiella pneumoniae in association with Zea mays L. Appl. Environ. Microbiol. 66, 783–787. 10.1128/AEM.66.2.783-787.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi F., Shen S. H., Chen S. F., Jing Y. X. (2004). Migration of Azospirillum brasilense Yu62 from root to stem and leaves inside rice and tobacco plants. Acta. Bot. Sin. 46, 1065–1070. [Google Scholar]
- Chojniak J., Wasilkowski D., Płaza G., Mrozik A., Brigmon R. (2015). Application of Biolog microarrays techniques for characterization of functional diversity of microbial community in phenolic-contaminated water. Int. J. Environ. Res. 9, 785–794. [Google Scholar]
- Croft B. J., Magarey R. C. (2000). Pachymetra root rot, in A Guide to Sugarcane Diseases, eds Rott P., Bailey R. A., Comstock J. C., Croft B. J., Saumtally A. S. (Montpellier: CIRAD/ISSCT, CIRAD Publication Services; ), 126–130. [Google Scholar]
- Dawson S. L., Fry J. C., Dancer B. N. (2002). A comparative evaluation of five typing techniques for determining the diversity of fluorescent pseudomonads. J. Microbiol. Methods 50, 9–22. 10.1016/S0167-7012(02)00003-9 [DOI] [PubMed] [Google Scholar]
- De Souza R., Ambrosini A., Passaglia L. M. P. (2015). Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 38, 401–419. 10.1590/S1415-475738420150053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R., Pal K. K., Bhatt D. M., Chauhan S. M. (2004). Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth promoting rhizobacteria. Microbiol. Res. 159, 371–394. 10.1016/j.micres.2004.08.004 [DOI] [PubMed] [Google Scholar]
- Edwards U., Rogall T., Blöcker H., Emde M., Böttger E. C. (1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2016). Top Sugarcane Production: Food and Agriculture Organization of the United Nations. Rome: FAO. [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39, 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Fischer D., Pfitzner B., Schmid M., Simoes-Araujo J. L., Reis V. M., Pereira W., et al. (2012). Molecular characterisation of the diazotrophic bacterial community in uninoculated and inoculated field-grown sugarcane (Saccharum sp.). Plant Soil 356, 83–99. 10.1007/s11104-011-0812-0 [DOI] [Google Scholar]
- Glick B. R., Jacobson C. B., Pasternak J. J. (1994). Partial purification and characterization of ACC deaminase from the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2. Can. J. Microbiol. 40, 1019–1025. 10.1139/m94-146 [DOI] [Google Scholar]
- Glick B. R., Penrose D. M., Li J. (1998). A model for the lowering of plant ethylene concentration by plant growth-promoting bacteria. J. Theor. Biol. 190, 63–68. 10.1006/jtbi.1997.0532 [DOI] [PubMed] [Google Scholar]
- Glick B. R., Todorovic B., Czarny J., Cheng Z., Duan J., McConkey B. (2007). Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant. Sci. 26, 227–242. 10.1080/07352680701572966 [DOI] [Google Scholar]
- Glickmann E., Dessaux Y. (1995). A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 61, 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R. W. F., Holsten R. D., Jackson E. K., Burns R. C. (1968). The acetylene ethylene assay for N2 fixation: laboratory and field evaluation. Plant Physiol. 43, 1185–1207. 10.1104/pp.43.8.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge D. F., Peoples M. B., Boddey R. M. (2008). Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 3, 1–18. 10.1007/s11104-008-9668-3 [DOI] [Google Scholar]
- Honma M., Shimomura T. (1978). Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 42, 1825–1831. [Google Scholar]
- Jacobson C. B., Pasternak J. J., Glick B. R. (1994). Partial purification and characterization of 1-aminocyclopropane-1-carboxylate deaminase from the plant growth promoting rhizobacterium Pseudomonas putida GR12-2. Can. J. Microbiol. 40, 1019–1025. 10.1139/m94-162 [DOI] [Google Scholar]
- James E. K. (2000). Nitrogen fixation in endophytic and associative symbiosis. F. Crop. Res. 65, 197–209. 10.1016/S0378-4290(99)00087-8 [DOI] [Google Scholar]
- James E. K., Olivares F. L. (1998). Infection and colonization of sugarcane and other graminaceous plants by endophytic diazotrophs. Crit. Rev. Plant Sci. 50, 77–119. 10.1016/S0735-2689(98)00357-8 [DOI] [Google Scholar]
- Juraeva D., George E., Davranov K., Ruppel S. (2006). Detection and quantification of the nifH gene in shoot and root of cucumber plants. Can. J. Microbiol. 52, 731–739. 10.1139/w06-025 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laville J., Voisard C., Keel C., Maurhofer M., Defago G., Haas D. (1992). Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. U.S.A. 89, 1562–1566. 10.1073/pnas.89.5.1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. P., Macrae I. C. (1991). Specific association of diazotrophic Acetobacers with sugarcane. Soil Biol. Biochem. 23, 999–1002. 10.1016/0038-0717(91)90181-I [DOI] [Google Scholar]
- Li Y. R., Zhou X. Z., Yang L. T. (2015). Biological nitrogen fixation in sugarcane and nitrogen transfer from sugarcane to cassava in an intercropping system. Inter. J. Sci. Nature. 6, 214–218. [Google Scholar]
- Li Y. R., Yang L. T. (2015). Sugarcane agriculture and sugar industry in China. Sugar Tech. 17, 1–8. 10.1007/s12355-014-0342-1 [DOI] [Google Scholar]
- Lima E., Boddey R. M., Dobereiner J. (1987). Quantification of biological nitrogen fixation associated with sugar cane using a 15N-aided nitrogen balance. Soil Biol. Biochem. 19, 165–170. 10.1016/0038-0717(87)90077-0 [DOI] [Google Scholar]
- Lin L., Guo W., Xing Y., Zhang X., Li Z., Hu C., et al. (2012). The actinobacterium Microbacterium sp. 16SH accepts pBBR1-based pPROBE vectors, forms biofilms, invades roots, and fixes N2 associate d with micropropagated sugarcane plants. Appl. Microbiol. Biotechnol. 93, 1185–1195. 10.1007/s00253-011-3618-3 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhao H., Chen S. (2006). Colonization of maize and rice plants by strain Bacillus megaterium C4. Curr. Microbiol. 52, 186–190. 10.1007/s00284-005-0162-3 [DOI] [PubMed] [Google Scholar]
- Liu Y. P., Teng S. S., Zhao L. (2011). Identification of a siderophore-producing bacterium Pseudomonas putida A3 and its growth- promoting effects on cucumber seedlings. J. Plant Nutrit. Fertil. Sci. 17, 1507–1514. [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lorck H. (1948). Production of hydrocyanic acid by bacteria. Physiol. Plant 1, 142–146. 10.1111/j.1399-3054.1948.tb07118.x [DOI] [Google Scholar]
- Luo L. J. (2014). Isolation and Identification of Endophytic Bacteria in Guangxi Sugarcane Wild Germplasms and Endogenous Rhizobia in Sugarcane Intercropping Beans. Master Thesis, Guangxi University. [Google Scholar]
- Magnani G. S., Didonet C. M., Cruz L. M., Picheth C. F., Pedrosa F. O., Souza E. M. (2010). Diversity of endophytic bacteria in Brazilian sugarcane. Genet. Mol. Res. 9, 250–258. 10.4238/vol9-1gmr703 [DOI] [PubMed] [Google Scholar]
- Manoharachary C., Mukerji K. G. (2006). Microbial activity in the Rhizosphere, in Soil Biology, eds Mukerji K. G., Manoharachary C., Singh J. (Berlin; Heidelberg: Springer-Verlag; ), 7. [Google Scholar]
- Mantelin S., Touraine B. (2004). Plant growth-promoting bacteria and nitrate availability impacts on root development and nitrate uptake. J. Exp. Bot. 55, 27–34. 10.1093/jxb/erh010 [DOI] [PubMed] [Google Scholar]
- Mazur A., Stasiak G., Wielbo J., Koper P., Kubik-Komar A., Skorupsk A. (2013). Phenotype profiling of Rhizobium leguminosarum bv. trifolii clover nodule isolates reveal their both versatile and specialized metabolic capabilities. Arch. Microbiol. 195, 255–267. 10.1007/s00203-013-0874-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnaz S. (2011). Plant growth-promoting bacteria associated with sugarcane in Bacteria in Agrobiology, ed Maheshwari D. K. (Berlin; Heidelberg: Crop Ecosystems, Springer-Verlag; ), 165–187. [Google Scholar]
- Mehnaz S., Baig D. N., Jamil F., Weselowski B., Lazarovits G. (2009a). Characterization of a phenazine and hexanoyl homoserine lactone producing Pseudomonas aurantiaca strain PB-St2, isolated from sugarcane stem. J. Microbiol. Biotechol. 19, 1688–1694. 10.4014/jmb.0904.04022 [DOI] [PubMed] [Google Scholar]
- Mehnaz S., Baig D. N., Lazarovits G. (2010). Genetic and phenotypic diversity of plant growth promoting rhizobacteria isolated from sugarcane plants growing in Pakistan. J. Microbiol. Biotechnol. 20, 1614–1623. 10.4014/jmb.1005.05014 [DOI] [PubMed] [Google Scholar]
- Mehnaz S., Weselowski B., Mufti F. A., Zahid S., Lazarovits G., Iqbal J. (2009b). Isolation, characterization and effect of fluorescent pseudomonads on micropropagated sugarcane. Can. J. Microbiol. 55, 1007–1011. 10.1139/W09-050 [DOI] [PubMed] [Google Scholar]
- Mendes R., Pizzirani-Kleiner A. A., Araujo W. L., Raaijmakers J. M. (2007). Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 73, 7259–7267. 10.1128/AEM.01222-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaxi and Saxena J. (2010a). Characterization of Pseudomonas aeruginosa RM-3 as a potential biocontrol agent. Myco Path. 170, 181–193. 10.1007/s11046-010-9307-4 [DOI] [PubMed] [Google Scholar]
- Minaxi and Saxena J. (2010b). Disease suppression and crop improvement in moong beans (Vignaradiata) through Pseudomonas and Burkholderia strains isolated from semi-arid region of Rajasthan, India. J. Biocontrol. 55, 799–810. 10.1007/s10526-010-9292-z [DOI] [Google Scholar]
- Mirza M. S., Mehnaz S., Normand P., Prigent-Combaret C., Moenne-Loccoz Y., Bally R., et al. (2006). Molecular characterization and PCR-detection of a nitrogen-fixing Pseudomonas strain promoting rice growth. Biol. Fertil. Soil. 43, 136–170. 10.1007/s00374-006-0074-9 [DOI] [Google Scholar]
- Niu J. Q., Wang A. Q., Huang J. L., Li Y. R., Yang L. T. (2013). Cloning and expression analysis of a soluble acid invertase gene (SoSAI1) of sugarcane. Sci. Agric. Sin. 46, 5248–5260. 10.3864/j.issn.0578-1752.2013.24.019 [DOI] [Google Scholar]
- Noda S., Ohkuma M., Usami R., Horikoshi K., Kudo T. (1999). Culture-independent characterization of a gene responsiblefor nitrogen fixation in the symbiotic microbial community in the gut of the termite Neotermes koshunensis. Appl. Environ. Microbiol. 65, 4935–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A. L. M., Urquiaga S., Dobereiner J., Baldani J. I. (2002). The effect of inoculating endophytic N2 -fixing bacteria on micropropagated sugarcane plants. Plant Soil 242, 205–215. 10.1023/A:1016249704336 [DOI] [Google Scholar]
- Onofre-Lemus J., Hernandez-Lucas I., Girard L., Caballero-Mellado J. (2009). ACC (1-amino cyclopropane-1-carboxylate) deaminase activity, a widespread trait in Burkholderia species, and its growth-promoting effect on tomato plants. Appl. Environ. Microbiol. 75, 6581–6590. 10.1128/AEM.01240-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poly F., Monrozier L. J., Bally R. (2001). Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152, 95–103. 10.1016/S0923-2508(00)01172-4 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J. M., Weller D. M., Thomashow L. S. (1997). Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 63, 881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademaker J. L. W., de Bruijn F. J. (1997). Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer assisted pattern analysis, in DNA Markers: Protocols, Applications and Overviews, eds Caetano-Anolles G., Gresshoff P. M. (New York, NY: John Wiley; ), 151–171. [Google Scholar]
- Rafikova G. F., Korshunova T., Yu Minnebaev L. F., Chetverikov S. P., Loginov O. N. (2016). A new bacterial strain, Pseudomonas koreensis IB-4, as a promising agent for plant pathogen biological control. Microbiology 85, 333–341. 10.1134/S0026261716030115 [DOI] [Google Scholar]
- Ramette A., Frapolli M., Defago G., Moenne-Loccoz Y. (2003). Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Mol. Plant Microb. Interact. 16, 525–535. 10.1094/MPMI.2003.16.6.525 [DOI] [PubMed] [Google Scholar]
- Rao G. P., Viswanathan R., Singh S. B. (2002). Current situation of sugarcane diseases in India in Sugarcane Crop Management, eds Singh S. B., Rao G. P., Easwaramoorthy S. (Houstan, TX: SCI Tech Publishing LLC; ), 734. [Google Scholar]
- Reis V. M., Olivares F. L., de Oliveira A. L. M., dos Reis J. F. B., Baldani J. I., Dobereiner J. (1999). Technical approaches to inoculate micro-propagated sugarcane plants with Acetobacter diazotrophicus. Plant Soil. 206, 205–211. 10.1023/A:1004436611397 [DOI] [Google Scholar]
- Robinson N., Brackin R., Vinall K., Soper F., Holst J., Gamage H., et al. (2011). Nitrate paradigm does not hold up for sugarcane. PLoS ONE 6:e19045 10.1371/journal.pone.0019045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A. S., Duarte G. F., Seldin L., Elsas J. D. V. (1998). Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR amplified gene fragments. Appl. Environ. Microbiol. 64, 2770–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sasirekha B., Srividya S., Shankar B. S. (2013). Molecular detection of antibiotic related genes from Pseudomonas aeruginosa FP6, an antagonist towards Rhizoctonia solani and Colletotrichum gloeosporioides. Turk. J. Biol. 37, 289–295. 10.3906/biy-1207-56 [DOI] [Google Scholar]
- Schwyn B., Neilands J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- Setten L., Soto G., Mozzicafreddo M., Fox A. R., Lisi C., Cuccioloni M., et al. (2013). Engineering Pseudomonas protegens Pf-5 for nitrogen fixation and its application to improve plant growth under nitrogen-deficient conditions. PLoS ONE 8:e63666. 10.1371/journal.pone.0063666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M. K., Kushwaha C., Singh R. K. (2009). Studies on endophytic colonization ability of two upland rice endophytes, Rhizobium sp. and Burkholderia sp., using green fluorescent protein reporter. Curr. Microbiol. 59, 240–243. 10.1007/s00284-009-9419-6 [DOI] [PubMed] [Google Scholar]
- Singh R. K., Kumar D. P., Solanki M. K., Singh P., Srivastva A., Kumar S., et al. (2013). Optimization of media components for chitinase production by chickpea rhizosphere associated Lysinibacillus fusiformis B-CM18. J. Basic Microb. 53, 451–460. 10.1002/jobm.201100590 [DOI] [PubMed] [Google Scholar]
- Singh R. K., Kumar D. P., Solanki M. K., Singh P., Srivastava S., Srivastva A. K., et al. (2014). Multifarious plant growth promoting characteristics of chickpea rhizosphere associated Bacilli help to suppress soil-borne pathogens. Plant Growth Regul. 73, 91–101. 10.1007/s10725-013-9870-z [DOI] [Google Scholar]
- Sneath P. H. A., Sokal R. R. (1973). Numerical Taxonomy. San Francisco, CA: Freeman. [Google Scholar]
- Solanki M. K., Wang Z., Wang F.-Y., Li C.-N., Lan T.-J., Singh R. K., et al. (2017). Intercropping in sugarcane cultivation influenced the soil properties and enhanced the diversity of vital diazotrophic bacteria. Sugar Tech. 19, 136–147. 10.1007/s12355-016-0445-y [DOI] [Google Scholar]
- Somers E., Vanderleyden J., Srinivasan M. (2004). Rhizosphere bacterial signalling: a love parade beneath our feet. Crit. Rev. Microbiol. 30, 205–240. 10.1080/10408410490468786 [DOI] [PubMed] [Google Scholar]
- Souza J. T., Raaijmakers J. M. (2003). Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microbiol. Ecol. 43, 21–34. 10.1111/j.1574-6941.2003.tb01042.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei M., Kumar S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unge A., Tombolini R., Molbak L., Jansson J. K. (1999). Simultaneously monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-lux AB marker system. Appl. Enviorn. Microbiol. 65, 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquiaga S., Cruz K. H. S., Boddey R. M. (1992). Contribution of nitrogen fixation to sugarcane: nitrogen-15 and nitrogen-balance estimates. Soil Sci. Soc. Am. J. 56, 105–114. 10.2136/sssaj1992.03615995005600010017x [DOI] [Google Scholar]
- Urquiaga S., Xavier R. P., de Morais R. F., Batista R. B., Schultz N., Leite J. M., et al. (2012). Evidence from field nitrogen balance and 15N natural abundance data for the contribution of biological N2 fixation to Brazilian sugarcane varieties. Plant Soil 356, 5–21. 10.1007/s11104-011-1016-3 [DOI] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831. 10.1093/nar/19.24.6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan R., Rajitha R., Sundar A. R., Ramamoorthy V. (2003). Isolation and identification of endophytic bacterial strains isolated from sugarcane stalks and their in vitro antagonism against red rot pathogen. Sugar Tech. 5, 25–29. 10.1007/BF02943760 [DOI] [Google Scholar]
- Viswanathan R., Rao G. P. (2011). Disease scenario and management of major sugarcane diseases in India. Sugar Tech. 13, 336–353. 10.1007/s12355-011-0102-4 [DOI] [Google Scholar]
- Viswanathana R., Samiyappan R. (2002). Induced systemic resistance by fluorescent pseudomonads against red rot disease of sugarcane caused by Colletotrichum falcatum. Crop Prot. 21, 1–10. 10.1016/S0261-2194(01)00050-3 [DOI] [Google Scholar]
- Voisard C., Keel C., Haas D., Defago G. (1989). Cyanide production by Pseudomonas fuorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8, 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallenstein M. D. (2004). Effect of Increased Nitrogen Deposition on Forest Soil Nitrogen Cycling and Community Structure. Ph.D. Dissertation, Duke University, Durham, NC. [Google Scholar]
- Walsh U. F., Morrissey J. P., O'Gara F. (2001). Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr. Opin. Biotechnol. 12, 289–295. 10.1016/S0958-1669(00)00212-3 [DOI] [PubMed] [Google Scholar]
- Wang X., Wang Y., Tian J., Lim B. L., Yan X., Liao H. (2009). Over expressing at PAP15 enhances phosphorus efficiency in soybean. Plant Physiol. 151, 233–240. 10.1104/pp.109.138891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielbo J., Marek-Kozaczuk M., Kubik-Komar A., Skorupska A. (2007). Increased metabolic potential of Rhizobium spp. is associated with bacterial competitiveness. Can. J. Microbiol. 53, 957–967. 10.1139/W07-053 [DOI] [PubMed] [Google Scholar]
- Xing Y.-X., Wei C.-Y., Mo Y., Yang L.-T., Huang S.-L., Li Y.-R. (2016). Nitrogen-fixing and plant growth-promoting ability of two endophytic bacterial strains isolated from sugarcane stalks. Sugar Tech. 18, 373–379. 10.1007/s12355-015-0397-7 [DOI] [Google Scholar]
- Xing Y. X., Yang L. T., Huang S. L., Li Y. R. (2006). Identification of a new nitrogen fixing endo-bacterium strain isolated from sugarcane stalk. Sugar Tech. 8, 49–53. 10.1007/BF02943741 [DOI] [Google Scholar]
- Xing Y., Yang L., Huang S., Li Y. (2015). A new nitrogen fixing endo-bacterium strain isolated from sugarcane stem, in Proceedings of International Symposium on Technologies to improve Sugar Productivity in Developing Countries, ed Guilin P. R. (Beijing: China Agricultural Science; ), 487–490. [Google Scholar]
- Young J. P. W. (1992). Phylogenetic classification of nitrogen fixing organisms in Biological Nitrogen Fixation, Vol. 1544, eds Stacey G., Burris R. H., Evans H. J. (New York, NY: Chapman & Hall; ), 43–86. [Google Scholar]
- Zahir Z. A., Muhammad A., Frankenberger W. T. J. (2003). Plant growth promoting rhizobacteria: applications and perspectives in agriculture. Adv. Agron. 81, 97–168. 10.1016/S0065-2113(03)81003-9 [DOI] [Google Scholar]
- Zehr J. P., Capone D. G. (1996). Problems and promises of assaying the genetic potential for nitrogen fixation in the marine environment. Microb. Ecol. 32, 263–281. 10.1007/BF00183062 [DOI] [PubMed] [Google Scholar]
- Zehr J. P., Jenkins B. D., Short S. M., Steward G. F. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5, 539–554. 10.1046/j.1462-2920.2003.00451.x [DOI] [PubMed] [Google Scholar]
- Zeng Q., Wu X., Wen X. (2016). Identification and characterization of the rhizosphere phosphate-solubilizing bacterium Pseudomonas frederiksbergensis JW-SD2, and its plant growth-promoting effects on poplar seedlings. Ann. Microbiol. 66, 1343–1354. 10.1007/s13213-016-1220-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.