Abstract
Bacteria owe much of their evolutionary success to the development of mechanisms that enable survival under harsh conditions; despite their importance, however, we have only recently begun to understand these survival strategies. Several of these mechanisms have been shown to target ribosomes, robustly blocking their ability to translate mRNAs into proteins and preserving their integrity in response to extreme environmental conditions. Now, two articles in The EMBO Journal reveal the structural basis of one such strategy, providing new insights and new questions regarding the mechanism and regulation of “ribosome hibernation”.
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Protein Biosynthesis & Quality Control; Structural Biology
Nowhere in nature is the capacity to endure extreme environmental conditions more central to survival than in the microbial world. In their native habitats, bacteria are constantly subjected to environmental insults—from temperature fluctuations to dehydration to nutrient deprivation to antibiotic attacks. Consistent with their evolutionary success, bacteria have developed numerous strategies for coping with the various types of stresses they continuously encounter. In response to metabolic stress and energy depletion, for example, the adenosine diphosphate (ADP)‐bound form of an adenosine triphosphatase (ATPase) regulatory translation factor known as energy‐dependent translational throttle A (EttA) binds to ribosomes and blocks their ability to catalyze protein synthesis (Boel et al, 2014; Chen et al, 2014). Similarly, in response to nutrient deprivation and other environmental stresses, ribosomes enter into a so‐called hibernating state that presumably inhibits protein synthesis (Wada et al, 1990). At the molecular level, ribosome hibernation involves the dimerization of two 70S ribosomes into a 100S ribosome dimer (disome). Now, in two articles in this issue, Beckert et al (2017) and Khusainov et al (2017) use cryo‐EM to determine the structure of 100S disomes from Bacillus subtilis and Staphylococcus aureus, respectively (Fig 1).
Figure 1. 100S disome structures and the mechanism of ribosome hibernation.
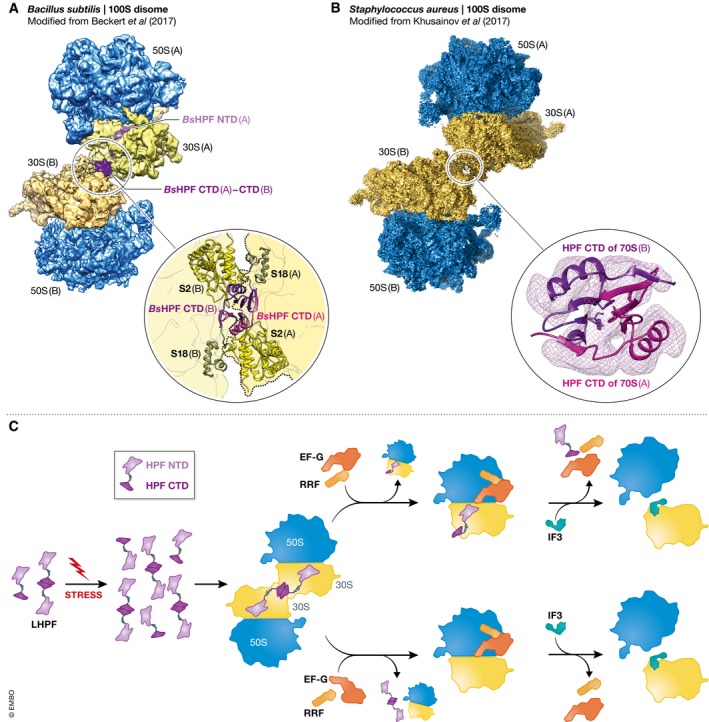
During ribosome hibernation, binding of lHPF to 70S ribosomes enables the formation of 100S disomes in (A) Bacillus subtilis and (B) Staphylococcus aureus. The insets depict the intermolecular contacts between the dimerized lHPF CTDs and, in the case of B. subtilis, additional components of the 30S subunits that constitute each 70S ribosome in the 100S disome. (C) A mechanistic model of ribosome hibernation highlighting several aspects of 100S disome formation and disassembly that remain to be investigated.
Early studies of ribosome hibernation revealed that amino acid and/or glucose deprivation in a subset of γ‐proteobacteria, including Escherichia coli, induces the expression of ribosome modulation factor (RMF), which, together with a short variant of hibernation promoting factor (sHPF), binds to 70S ribosomes, inhibiting protein synthesis and driving dimerization of the 70S ribosomes into 100S disomes. Structural studies of RMF‐ and sHPF‐bound 70S ribosomes (Polikanov et al, 2012) subsequently revealed that sHPF binds across the mRNA‐ and tRNA‐binding sites of the 30S subunit, suggesting that sHPF may inhibit protein synthesis by preventing mRNA and tRNAs from accessing the 70S ribosome. Contrasting with this, RMF binds the 30S subunit in a manner that could block translation initiation by preventing the Shine‐Dalgarno sequence of the mRNA from interacting with the 30S subunit. Moreover, RMF and sHPF seem to stabilize a conformation of the 30S subunit that enables it to interact with the 30S subunit of another RMF‐ and sHPF‐bound 70S ribosome, thereby forming a 100S disome.
Intriguingly, most bacteria lack an RMF homolog and encode a long variant of HPF (lHPF), leading to the discovery of a second mechanism of ribosome hibernation (Ueta et al, 2013). While the N‐terminal domain (NTD) of lHPF resembles sHPF, the C‐terminal domain (CTD) is only weakly related to RMF and the > 30 amino acid linker connecting the two domains is wholly unique to lHPF. It is the structural basis of this 1HPF‐mediated ribosome hibernation that is now addressed in the two articles in this issue (Fig 1A and B). Consistent with its strong homology to sHPF, Beckert et al (2017) and Khusainov et al (2017) find that the NTD of lHPF occupies the mRNA‐ and tRNA‐binding sites of the 30S subunit, thereby interfering with the binding of mRNA and tRNA to these sites in a manner that is indistinguishable from that observed for sHPF. The > 30 amino acid linker apparently passes through the mRNA channel of the 30S subunit, allowing the CTD of lHPF to exit the solvent accessible face of the 30S subunit near ribosomal protein uS2. In what is perhaps the most surprising discovery reported by both groups, the protruding CTD of the lHPF that is bound to one 30S subunit is observed to directly dimerize with the protruding CTD of the lHPF that is bound to the other 30S subunit, thereby resulting in a CTD‐CTD interaction that provides at least part of the physical basis for 100S disome formation. Whereas the 30S subunits within the S. aureus 100S disome apparently exhibit no additional intermolecular contacts beyond the CTD‐CTD interaction (Khusainov et al, 2017), the 30S subunits within the B. subtilis 100S disome exhibit additional intermolecular contacts (Beckert et al, 2017). Collectively, these results suggest that the CTD‐CTD interaction may provide a primary set of contacts that are universally present in the 100S disomes formed by lHPF, while the additional intermolecular interactions observed in the B. subtilis 100S disome provide a secondary set of contacts that may or may not be present in the 100S disomes that are formed by lHPF across bacterial species.
Both Beckert et al (2017) and Khusainov et al (2017) provide evidence that at least some fraction of lHPF can exist as a dimer under their experimental conditions. This raises the question of whether it is the monomeric or dimeric forms of free lHPF that first target ribosomes for inactivation and dimerization (Fig 1C). Likewise, it is not yet clear whether lHPF targets ribosomes during the initiation, elongation, termination, or ribosome recycling phases of the translation process. Thus, the exact mechanism by which free lHPF targets ribosomes for hibernation remains undefined.
Similarly, it remains unclear why ribosome hibernation involves dimerization of 70S ribosomes. The observation that bacteria have evolved at least two mechanisms for dimerizing 70S ribosomes (Ueta et al, 2013) suggests that 100S disome formation is an important aspect of ribosome hibernation. Consistent with this, deletion of RMF in E. coli causes ribosome degradation and cell death under stress conditions (Yamagishi et al, 1993; Niven, 2004), suggesting that 100S disome formation protects 70S ribosomes from degradation. As Beckert et al (2017) point out, 100S disome formation does not significantly decrease the amount of ribosomal RNA that would be exposed to degradation by non‐specific RNases, thereby suggesting that 100S disome formation might instead interfere with a specific ribosome degradation pathway. Indeed, the observation that 100S disomes formed via the RMF‐sHPF‐ and lHPF‐based mechanisms sterically occlude an overlapping region of the solvent accessible face of the 30S subunit (Kato et al, 2010; Ortiz et al, 2010; Polikanov et al, 2012; Beckert et al, 2017; Khusainov et al, 2017) is consistent with the possibility that a specific ribosome degradation pathway might target this region of the 30S subunit. It is notable that at least five of the bacterial species that lack a gene encoding RMF contain a gene encoding sHPF instead of lHPF. Since 70S ribosomes from these species presumably cannot form 100S disomes via the two known mechanisms, a third, as‐yet‐uncharacterized mechanism of 100S disome formation may exist.
Another important question is how 100S disomes are disassembled once environmental conditions become favorable again (Fig 1C). This may involve a variant of the canonical, RRF‐, EF‐G‐, and IF3‐mediated ribosome recycling mechanism, consistent with the observation by Khusainov et al (2017) that the 70S ribosomes in the S. aureus 100S disomes exhibit intersubunit dynamics resembling those of a ribosomal post‐termination complex that is typically targeted in the canonical ribosome recycling pathway. Moreover, the fact that RRF and EF‐G have been shown to recycle vacant E. coli ribosomes (Peske et al, 2005) suggests that such a mechanism should be tested for the recycling of 100S disomes. Assuming that 100S disomes are recycled in this manner, it would be interesting to investigate if recycling of the two constituent 70S ribosomes is regulated and coordinated. Are the dimerized lHPFs separated as a prerequisite for recycling the two 70S ribosomes? Are the two 70S ribosomes recycled individually or simultaneously, and independently or in a coupled manner? Is free lHPF targeted for degradation during disassembly so as to prevent re‐dimerization of 70S ribosomes?
The cryo‐EM structures of the lHPF‐bound B. subtilis and S. aureus 100S disomes reported here provide an essential framework for future studies that will help us elucidate the mechanism and regulation of ribosome hibernation. For example, knowing that 100S disome formation is facilitated by CTD‐mediated dimerization of lHPF suggests that kinetic studies of 100S disome formation using wild‐type and mutant lHPF should provide a time‐resolved sequence of events for lHPF dimerization, lHPF binding to 70S ribosomes, and 70S dimerization. Similarly, kinetic and structural studies aimed at addressing whether and how RRF, EF‐G, and/or IF3 drive the disassembly of 100S disomes should provide further insight into the mechanism and regulation of ribosome hibernation.
See also: B Beckert et al (July 2017) and I Khusainov et al (July 2017)
References
- Beckert B, Abdelshahid M, Schafer H, Steinchen W, Arenz S, Berninghausen O, Beckmann R, Bange G, Turgay K, Wilson DN (2017) Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J 36: 2061–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel G, Smith PC, Ning W, Englander MT, Chen B, Hashem Y, Testa AJ, Fischer JJ, Wieden HJ, Frank J, Gonzalez RL Jr, Hunt JF (2014) The ABC‐F protein EttA gates ribosome entry into the translation elongation cycle. Nat Struct Mol Biol 21: 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Boel G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL Jr, Hunt JF, Frank J (2014) EttA regulates translation by binding the ribosomal E site and restricting ribosome‐tRNA dynamics. Nat Struct Mol Biol 21: 152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Yoshida H, Miyata T, Maki Y, Wada A, Namba K (2010) Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18: 719–724 [DOI] [PubMed] [Google Scholar]
- Khusainov I, Vicens Q, Ayupov R, Usachev K, Myasnikov A, Simonetti A, Validov S, Kieffer B, Yusupova G, Yusupov M, Hashem Y (2017) Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. EMBO J 36: 2073–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niven GW (2004) Ribosome modulation factor protects Escherichia coli during heat stress, but this may not be dependent on ribosome dimerisation. Arch Microbiol 182: 60–66 [DOI] [PubMed] [Google Scholar]
- Ortiz JO, Brandt F, Matias VR, Sennels L, Rappsilber J, Scheres SH, Eibauer M, Hartl FU, Baumeister W (2010) Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ . J Cell Biol 190: 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Rodnina MV, Wintermeyer W (2005) Sequence of steps in ribosome recycling as defined by kineitc analysis. Mol Cell 18: 403–412 [DOI] [PubMed] [Google Scholar]
- Polikanov YS, Blaha GM, Steitz TA (2012) How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336: 915–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta M, Wada C, Daifuku T, Sako Y, Bessho Y, Kitamura A, Ohniwa RL, Morikawa K, Yoshida H, Kato T, Miyata T, Namba K, Wada A (2013) Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells 18: 554–574 [DOI] [PubMed] [Google Scholar]
- Wada A, Yamazaki Y, Fujita N, Ishihama A (1990) Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary‐phase Escherichia coli cells. Proc Natl Acad Sci USA 87: 2657–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M, Matsushima H, Wada A, Sakagami M, Fujita N, Ishihama A (1993) Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase‐ and growth rate‐dependent control. EMBO J 12: 625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]


