Abstract
The allantois-derived umbilical component of the chorio-allantoic placenta shuttles fetal blood to and from the chorion, thereby ensuring fetal-maternal exchange. The progenitor populations that establish and supply the fetal-umbilical interface lie, in part, within the base of the allantois, where the germ line is claimed to segregate from the soma. Results of recent studies in the mouse have reported that STELLA (DPPA-3, PGC7) co-localizes with PRDM1 (BLIMP1), the bimolecular signature of putative primordial germ cells (PGCs) throughout the fetal-placental interface. Thus, if PGCs form extragonadally within the posterior region of the mammal, they cannot be distinguished from the soma on the basis of these proteins. We used immunohistochemistry, immunofluorescence, and confocal microscopy of the mouse gastrula to co-localize STELLA with a variety of gene products, including pluripotency factor OCT-3/4, mesendoderm-associated T and MIXl1, mesendoderm- and endoderm-associated FOXa2 and hematopoietic factor Runx1. While a subpopulation of cells localizing OCT-3/4 was always found independently of STELLA, STELLA always co-localized with OCT-3/4. Despite previous reports that T is involved in specification of the germ line, co-localization of STELLA and T was detected only in a small subset of cells in the base of the allantois. Slightly later in the hindgut lip, STELLA+/(OCT-3/4+) co-localized with FOXa2, as well as with RUNX1, indicative of definitive endoderm and hemangioblasts, respectively. STELLA was never found with MIXl1. On the basis of these and previous results, we conclude that STELLA identifies at least five distinct cell subpopulations within the allantois and hindgut, where they may be involved in mesendodermal differentiation and hematopoiesis at the posterior embryonic-extraembryonic interface. These data provide a new point of departure for understanding STELLA’s potential roles in building the fetal-placental connection.
Keywords: Allantois, Brachyury, FOXa2, hindgut, MIXl1, OCT-3/4, primitive streak, primordial germ cells, Runx1, STELLA
INTRODUCTION
The allantois of placental mammals will become the umbilical cord, ensuring establishment of a continuous fetal-placental vasculature intimately associated with the maternal circulation. At one end, the allantois is connected to the embryo, where its vasculature becomes confluent with that of the fetus and yolk sac. At the other, the allantois fuses with the chorion, forming the chorio-allantoic labyrinth within the placenta and site of fetal-maternal exchange. While the source of the cells that build the fetal-umbilical connection, including the umbilical vasculature, hindgut endoderm, and canonically segregated primordial germ cells (PGCs), has long been thought to be located within the posterior embryonic primitive streak (Beddington, 1982; Beddington, 1981; Tam and Beddington, 1987), the latter’s contribution of descendant cells to the embryonic-extraembryonic interface is limited (Downs et al., 2009; Kinder et al., 1999; Mikedis and Downs, 2012; Tam and Beddington, 1987). Rather, as demonstrated in recent experiments (Downs, 2009; Downs et al., 2009; Inman and Downs, 2006a; Mikedis and Downs, 2012), the streak’s posterior end extends beyond the embryo and into the allantois, where it collaborates with the embryonic posterior primitive streak to provide distinct and overlapping descendant cell types to posterior tissues (Mikedis and Downs, 2012). This putative extraembryonic component of the primitive streak (XPS) is initially identifiable by juxtaposition of morphologically compact cells adjacent to posterior visceral endoderm (PVE), and robust localization of Brachyury (T; Downs et al., 2009). By headfold stages (~E7.75–8.0), the XPS expands, becoming the Allantoic Core Domain (ACD; Downs et al., 2009) that is essential for allantoic elongation and contributes to both the allantois itself (Downs et al., 2009) as well as to tissues encompassed by the fetal-placental interface (Mikedis and Downs, 2012).
Consistent with building the embryonic-extraembryonic interface, the ACD exhibits a variety of proteins involved in pluripotency (Downs, 2008; Mikedis and Downs, 2009, 2012), including those thought to signify the germ line (Chiquoine, 1954; Ginsburg et al., 1990; Lawson and Hage, 1994; reviewed in Leitch and Smith, 2013). Initially identified qualitatively on the basis of elevated alkaline phosphatase (AP) enzyme activity (Chiquoine, 1954), PGC progenitors are thought to enter the posterior primitive streak from the epiblast, congregate within the proximal end of the allantois, and there become segregated from the soma (Lawson and Hage, 1994). From the allantois, the PGCs then migrate as a segregated population into the gut as it forms (Lawson and Hage, 1994), and onto the genital ridges, populating the developing gonadal parenchyma (Chiquoine, 1954; reviewed in Mikedis and Downs, 2014). While clonal fate mapping, based on AP activity, has suggested that the site of PGC segregation from the soma lies within the base of the allantois (Lawson and Hage, 1994), rigorous evidence to show continuity via clonal fate mapping from the allantois to the gonads, or even to the hindgut, is absent from the literature (reviewed in Mikedis and Downs, 2014).
For more than six decades, cells within the PGC trajectory have been characterized largely by gene expression and protein localization. AP-positive putative PGCs were found to express several germline-specific “marker” gene products, one of which, STELLA (Saitou et al., 2002), is a demethylation-protective factor (Nakamura et al., 2007) required for pre-implantation development (Bortvin et al., 2004; Payer et al., 2003). However, its abrogation does not yield infertile progeny (Bortvin et al., 2004; Payer et al., 2003), and its role in this population remains unknown.
While the relationship between the PGCs and the allantois has generally been unappreciated, results of recent studies have demonstrated that STELLA protein localizes throughout the posterior region (Mikedis and Downs, 2012) and therefore appears to lack specificity in identifying a segregated germline. OCT-3/4 (Pou5f1), a POU domain transcription factor, has also been claimed to be specific to the germline, as its expression becomes restricted to the area of greatest tissue-nonspecific AP (TNAP) activity by embryonic day 8 in the base of the allantois (Scholer et al., 1990). Oct-3/4 also regulates Stella expression in maturing oocytes (Levasseur et al., 2008; Zuccotti et al., 2009). Like STELLA, however, OCT-3/4 localizes to myriad developing tissues of the mouse conceptus (Downs, 2008), including the primitive streak, where it maintains cell proliferation (DeVeale et al., 2013; Downs, 2008). Also, OCT-3/4, like STELLA, is associated with a variety of stem cell populations in mouse development (Garagna, 2009; Scholer, 1991) and pluripotency in embryonic stem cells in vitro (Sterneckert et al., 2012).
While STELLA+ descendants of ACD-derived cells colonize the hindgut, a component tissue of the PGC trajectory, ACD-derived STELLA is also found broadly throughout the fetal-umbilical junction and outside of the hindgut (Mikedis and Downs, 2012, 2013; reviewed in Mikedis and Downs, 2014). Whether the STELLA-positive cells of the ACD represent a pure segregated population, or whether they are subpopulations of cells that collectively build posterior tissues, including the PGCs, is obscure. For example, we have recently demonstrated that STELLA and PRDM1 (BLIMP1), a transcriptional repressor thought to regulate differentiation of progenitor cell populations, co-localize both within and outside of the canonical PGC trajectory (Mikedis and Downs, 2017). However, both STELLA and PRDM1 were also found independently of each other throughout the posterior region, including the allantois and hindgut, again calling into question the accuracy of expecting these proteins to specifically identify segregated putative PGCs.
These results suggest that a variety of molecularly distinct STELLA subpopulations may be present at the fetal-umbilical interface. To begin to identify these, we undertook systematic analysis by immunofluorescence (IF) and immunohistochemistry (IHC) during the presumed period when PGCs segregate within the allantois, then apparently leave this tissue to colonize the hindgut (~E7.5–E9.5). We paid particular attention to the allantois and hindgut, which are thought to encompass the PGC trajectory (McLaren, 2003) and examined a number of gene products, including T, MIXl1 (Pereira et al., 2011; Tada et al., 2005; Wolfe and Downs, 2014), and FOXa2 (HNF3β), a winged helix/forkhead transcription factor (Ang et al., 1993; Besnard et al., 2004) that can be employed as a specific marker of endoderm (Kubo et al., 2004). For reasons described above, we also co-localized STELLA and OCT-3/4, and we co-localized STELLA and Runx1 which identifies hemangioblasts (North et al., 1999) in the allantois and posterior region (Daane and Downs, 2011; Zeigler et al., 2006), and which has been shown to co-localize with PRDM1 (Mikedis and Downs, 2017).
Results of this study revealed distinct subpopulations of STELLA+ cells within the allantois, associated visceral endoderm, and hindgut that may play roles in mesendodermal differentiation of a variety of progenitor subpopulations at the fetal-umbilical junction.
MATERIALS AND METHODS
Mouse husbandry, dissections, embryo staging
All animals were treated in accordance with Public Health Service Policy on Humane Care and Use of Laboratory Animals (Public Law 99–158) as enforced by the University of Wisconsin-Madison. F2 conceptuses were obtained by mating inbred hybrids of B6CBAF1/J (The Jackson Laboratory, Bar Harbor, ME; Downs, 2006) or Runx1-LacZ heterozygous reporter males (Daane and Downs, 2011; North et al., 1999; Zeigler et al., 2006) with F1 females. Individual estrous females (Champlin et al., 1973) and stud males were placed together just before lights went off (13:00/01:00 or 21:00/09:00 lights off/on); copulation plugs were identified up to 12 hours later. Dissection and staging of conceptuses were as described by Downs and Davies (1993).
Staged conceptuses were fixed in 4% paraformaldehyde (PFA) for 2 hours at 4°C and sequentially rinsed at least three times in phosphate-buffered saline (PBS, Sigma-Aldrich, St. Louis, MO; Downs, 2008). Specimens were then dehydrated in increasing methanols/PBS through absolute methanol and stored at −20°C for at least three days. Prior to rehydration for protein localization, specimens were opened in the lateral yolk sac and amnion with the bladed bevel of a 27-gauge insulin needle to ensure reagent penetration.
Antibodies for immunofluorescence (IF) and immunohistochemistry (IHC)
Protein localization was evaluated by IF and IHC. Two primary antibodies raised in different species against STELLA were needed for co-localization: anti-STELLA AF2566 (goat polyclonal, raised against E. coli-derived recombinant full-length mouse STELLA/Dppa3; R&D Systems, Minneapolis, MN) used at 1:70 dilution (2.9μg/mL) for IF (Mikedis and Downs, 2012); and anti-STELLA sc-67249 (rabbit polyclonal, raised against full-length mouse STELLA; Santa Cruz Biotechnology, Santa Cruz, CA) used at 1:25-1:20 dilution (8–10μg/mL) for IF. Other primary antibodies included: anti-OCT-3/4 (goat polyclonal, raised against human N 1–134; sc-8268, Santa Cruz) used at 1:30 dilution (6.7μg/mL) for IF and 1:100 dilution (2μg/mL) for IHC (Downs, 2008); anti-T (goat polyclonal, raised against N-terminal 19 residues of human Brachyury; sc-17743, Santa Cruz) (Inman and Downs, 2006b) used at 1:25 dilution (8μg/mL) for IF; anti-MIXl1 (rabbit polyclonal, raised against mouse MIXl1 N-terminal residues 1–134; sc-98665, Santa Cruz) used at 1:25-1:20 dilution (8–10μg/mL) for IF; and at 1:100 dilution (2μg/mL) for IHC (Wolfe and Downs, 2014); anti-FOXa2 (HNF-3β; goat polyclonal, raised against C-terminus of mouse HNF-3β; sc-9187, Santa Cruz) used at 1:30 dilution (6.7μg/mL) for IF and 1:100 dilution (2μg/mL) for IHC (Kubo et al., 2004); and anti-RUNX1 (goat polyclonal, raised against N-terminus of mouse RUNX1; sc-8563, Santa Cruz) used at 1:25 dilution (8μg/mL) for IF and 1:100-1:50 dilution (2–4μg/mL) for IHC.
Secondary antibodies included: donkey anti-goat IgG Dylight 550 (Ab 96932, Abcam, Cambridge, MA) at 1:250 dilution (2μg/mL) for IF; donkey anti-rabbit IgG Dylight 650 (Ab 98501, Abcam) used at 1:250 dilution (2μg/mL) for IF; donkey anti-goat IgG-B (sc-2042, Santa Cruz) used at 1:500 dilution (0.8μg/mL) for IHC; and goat anti-rat IgG-B pre-adsorbed against mouse and human IgG (sc-2041, Santa Cruz) used at 1:500 dilution (0.8μg/mL) for IHC.
While the specificity of anti-STELLA AF2566 had been previously reported (Mikedis and Downs, 2012), anti-STELLA sc-67249 had not. To verify the specificity of sc-67249, IF specimens labeled with AF2566 were compared to stage-matched specimens labeled with sc-67249; the two antibodies exhibited identical localization patterns in posterior tissues at EB-9-somite (s) stages (data not shown). To verify the specificity of the sc-9187 FOXa2 antibody, we conducted IHC at 5s–7s stages using sc-9187 antibody compared with antibody pre-bound with its cognate peptide sequence (sc-9187P, Santa Cruz) and with IHC performed in the absence of primary antibody as described by Mikedis and Downs (2013). Specifically, the following was done in parallel on 5s–7s specimens: (1) 1:100 dilution of anti-FOXa2 primary antibody; (2) 1:100 dilution of anti-FOXa2 incubated in PBS + 5% donkey serum + 0.1% Triton X-100 (PBSST) for 8 hours at 4°C; (3) 1:100 dilution of anti-FOXa2 with 1:20 dilution of control peptide (CP) incubated in PBSST for 8 hours at 4°C; (4) 1:100 dilution of anti-FOXa2 with 1:10 dilution of CP incubated in PBSST for 8 hours at 4°C; and (5) PBSST without primary antibody were used as primary antibody solutions in IHC. No immunostaining signal was observed in the control peptide-treated (N=2) or minus-antibody (N=1) controls (data not shown). To verify the specificity of the sc-8563 RUNX1 antibody, we conducted IHC at 3–5s stages using sc-8563 antibody and its cognate peptide sequence (sc-8563P, Santa Cruz), as described above (N=5 specimens per treatment group), with no immunostaining signal observed in the control peptide-treated or minus-antibody controls (data not shown).
IF and confocal microscopy
Specimen rehydration was accomplished in graded methanols/PBS, followed by 3 × 5-minute washes in PBS. Blocking was performed in PBSST for 2 × 1h washes, then primary antibodies were added in PBSST and incubated with specimens at 4°C overnight. Excess primary antibody was removed by 5 × 1h PBSST washes at room temperature, then Dylight 550 and 650 secondary antibodies were added in PBSST and incubated with specimens at 4°C overnight. Excess secondary was removed by 5 × 1h PBSST washes at room temperature, followed by DAPI (4′,6-Diamidino-2-Phenylindole Dilactate, D3571, Invitrogen, Life Technologies, Grand Island, NY) at 1.8μg/mL in PBSST for 15 minutes at room temperature, then 3 × 30-minute PBSST washes.
Specimens were then positioned in fresh PBSST under a Nikon SMZ1500 dissection microscope (Nikon Instruments Inc., Melville, NY), and using a long glass scalpel (Beddington, 1987) and sharpened #5 forceps, the yolk sac, anterolateral visceral endoderm, ectoplacental cone, and anterior embryonic tissues were excised, leaving the posterior half of the embryo, posterior visceral endoderm (PVE), and allantois intact and exposed. Specimens were then placed in a drop of Aquamount compound (No. 13800, Lerner Laboratories, Pittsburgh, PA) on a gelatin-subbed glass slide, with the ventral surface (i.e., hindgut opening) oriented upward. A high performance glass coverslip (No. 1½, 170±5μm thickness, 18×18mm, 474030-9000-000, Zeiss, Lauda-Königshofen, Germany) was gently applied and slides were allowed to set overnight in the dark at room temperature.
Fluorescent imaging was performed on a Nikon A1R high-speed confocal system using 20× and 60× objectives. Z-series images were collected using 0.8–2μm steps, for a total depth of 40–50 μm, using Nikon Elements Confocal software. We standardized image acquisition such that signal from the Dylight 550 fluorophore appeared in red, and the Dylight 650 fluorophore appeared in green. Image processing was performed in Adobe Photoshop versions CS5 and CC (Adobe Systems Inc., San Jose, CA), utilizing red-green-blue channel isolation for preparation of single-color panels, as well as level and saturation adjustments and a sharpening filter.
As a negative control, 6s stage conceptuses were processed as above, omitting the primary antibodies and treating only with both fluorescent secondary antibodies prior to confocal microscopy. No cellular fluorescent signal was seen in the red or green channels, while DAPI nuclear signal was normal in all cells (data not shown).
IHC and histology
All conceptuses were immunostained in whole mount preparations as previously described (Downs, 2008). Runx1-LacZ specimens were X-gal-stained as previously described (Daane and Downs, 2011). After immunostaining, conceptuses were then re-fixed overnight in 4% PFA at 4°C, dehydrated and embedded in paraffin wax (Downs, 2008). Specimens were sectioned at a thickness of 6μm, and counterstained with Gill’s hematoxylin (Sigma-Aldrich, St. Louis, MO). Slides were viewed using a Nikon Microphot-SA compound microscope and photographed with an attached QImaging Retiga 2000R digital camera (QImaging, Surrey BC, Canada). Image processing was performed as described above. Specimens for morphology images (Fig. 1) were part of a previous study (Downs et al., 1998), though those selected for this manuscript have not previously been published.
Figure 1. Posterior sites of interest examined in this study.

All specimens were stained with hematoxylin/eosin (Downs et al., 1998) and oriented sagittally, with anterior to the left and posterior to the right. Upper right inset of each panel includes a mid-sagittal view of the whole conceptus, magnified in the main panel within the posterior region outlined by the grey box. A, early bud (EB) stage specimen with posterior region magnified to show the extraembryonic primitive streak (XPS) proximally within the bud of the allantois (al) and contiguous with the intraembryonic primitive streak (ips). The allantois-associated posterior visceral endoderm (aPVE) is indicated ventral to the XPS, and is contiguous with the more squamous intraembryonic posterior visceral endoderm (iPVE). B, 1-somite pair (1s) stage specimen with the compact allantoic core domain (ACD) indicated at the base of the allantois. C, 5s stage specimen magnified to show the early invagination of the hindgut (hg) and its protruding ventral endodermal edge, the hindgut lip (HGL). The distal extent of the hg is indicated (asterisk). Note that this specimen is oriented slightly obliquely, such that the head is largely absent from the whole conceptus (inset). D, 9s stage specimen magnified to illustrate the full length of the hg, its relationship to the HGL, and the mesenchyme distal to the hg (asterisk). E, F, Late headfold (LHF) stage mid-sagittal sections through the allantois immunostained for T (E) and STELLA (F). Boxed regions indicate the extent to which the ACD was sampled in the confocal analysis. Scale bar in F = 50μm in A–D, 351μm in A inset, 483μm in B inset, 525μm in C inset, 759μm in D inset, and 92.1 μm in E, F. Other abbreviations: am, amnion; nf, neural folds; x, exocoelom; ys, visceral yolk sac.
Numbers of specimens evaluated
Specimens were grouped by developmental stages into: early and late bud (EB-LB), early headfold through early somite (EHF-3s), early hindgut invagination (4s–7s), and late hindgut invagination (8s–12s). Table 1 summarizes the number of IF specimens analyzed by primary antibody, stage group, and genotype. In addition, the following numbers of specimens were examined for IHC: for FOXa2 IHC, wild type conceptuses were examined at EB-LB (N=2), EHF (N=1), 5s–6s (N=3) and 9s (N=1) and Runx1-lacZ heterozygotes were examined at 8s–12s (N=4). For RUNX1 IHC, wild type conceptuses were examined at 4s–7s (N=3) and 8–10s (N=3). For STELLA IHC, Runx1-lacZ heterozygotes were examined at 8s–12s (N=3).
Table 1.
Specimens analyzed by immunofluorescence
| Primary Antibody | Number of specimens analyzed
|
|||
|---|---|---|---|---|
| EB-LB | EHF-3s | 4s-7s | 8s-12s | |
| STELLA + OCT-3/4 | 2 | 3 | 8 | 8 |
| STELLA + T | 2 | 2 | 1 | 1 |
| STELLA + FOXa2 | 1 | 7 | 40 | 4 |
| STELLA + MIXl1 | 1 | 1 | 2 | 1 |
| STELLA + RUNX1 | 0 | 0 | 0 | 3 |
| MIXl1 + T | 0 | 2 | 0 | 1 |
| 1 Total STELLA | 6 | 13 | 15 | 17 |
| 1 Total T | 2 | 4 | 1 | 2 |
| 1 Total OCT-3/4 | 2 | 3 | 8 | 8 |
| 1 Total MIXl1 | 1 | 3 | 2 | 2 |
| 1 Total FOXa2 | 1 | 7 | 4 | 4 |
Totals reflect all specimens that included the indicated primary antibody, plus single-labeled specimens (i.e., that had not been co-localized with another protein).
RESULTS
Posterior tissues analyzed in this study
Throughout this study, we evaluated tissues associated with the PGC trajectory (Fig. 1 and reviewed in Mikedis and Downs, 2014), including the posterior primitive streak, which is partitioned into the allantoic component, initially the XPS (neural plate/pre- and allantoic bud stages) (Fig. 1A), and later the ACD (EHF-6s) (Fig. 1B, C) and the intraembryonic component (all stages, e.g., Fig. 1A, B), and developing hindgut endoderm (Fig. 1C, D). The XPS is 3–4 layers thick (Fig. 1A; Downs et al., 2009) and fully observable in the confocal microscope. However, the expanded ACD is about 6–10 cell layers thick from the headfold – 4s stages beginning at its site of contact with the aPVE (Fig. 1B; Downs et al., 2009; Inman and Downs, 2006b). Thus, in the confocal, the ventral 3–4 cell layers of the ACD were reliably assessed. This included much of the allantoic streak’s T population (Fig. 1E; Downs et al., 2009) and most, if not all of the STELLA population found in this ventral region at the bud - 2s stages (Fig. 1F; Mikedis and Downs, 2012). This population is thought to represent segregated PGCs (Lawson and Hage, 1994; Saitou et al., 2002). As putative PGCs are thought to enter the extraembryonic endoderm and travel through the hindgut during their journey as a lineage-restricted population (reviewed by McLaren, 2003), we included analysis of posterior visceral endoderm that spans the embryonic-extraembryonic interface, parsing it into allantois- and embryo-associated posterior visceral endoderm (headfold-3s stages) (Fig. 1A, B). We also examined the hindgut lip (4s–12s) (Fig. 1C, D), or caudal intestinal portal, and the invaginating hindgut itself (“early”, 4s–7s; “late”, 8s–12s) (Fig. 1C, D). At later stages, we evaluated the ACD/tailbud (7s–12s), as it encompasses a progenitor cell pool for building the tailbud (Tam and Tan, 1992; Wilson and Beddington, 1996), possibly derived, in part, from the ACD (Downs et al., 2009; not shown).
The method used for whole mount orientation of specimens for confocal microscopy (see Materials and Methods) allowed clear visualization of all of these tissues. Allantoic and embryonic mesoderm adjacent to the primitive streak could be identified by its proximity to landmarks including the XPS/ACD and node as well as its subjacent proximity to the posterior visceral endoderm in confocal Z-series. The amnion was excluded from IF evaluation, as it was technically unreliable to isolate for observation.
STELLA and OCT-3/4 co-localize but also identify two divergent posterior populations
STELLA and OCT-3/4 co-localized within the posterior region at all stages (EHF-12s) (Figs 2, 3). In all cases, all STELLA+ cells were OCT-3/4+, but OCT-3/4+ cells were initially found independent of STELLA (headfold stages, Fig. 2A–D; Fig. 3A–D). Where STELLA and OCT-3/4 co-localized in all tissues, STELLA was found throughout the nucleus, including the perinucleus and core, while OCT-3/4 appeared to be more concentrated in the core (e.g., Fig. 2H, L; Fig. 3H, L, V). STELLA often appeared independent of the nuclei (e.g., Fig. 2F, H); this is consistent with previous reports that STELLA is also found in the cytoplasm (Mikedis and Downs, 2012; Nakamura et al., 2007; Saitou et al., 2002). Because of its nuclear localization and nuclear export signals, the protein can be shuttled back and forth between the nucleus and cytoplasm (Nakamura et al., 2007; Saitou et al., 2002).
Figure 2. Localization of OCT-3/4 and STELLA in the allantoic core domain (ACD).

Red channel indicates OCT-3/4, green indicates STELLA, blue indicates DAPI. A–D, Late headfold (LHF) stage exhibits some STELLA+ cells co-localizing OCT-3/4 (white arrowheads, inset right cell) and other OCT-3/4+ cells lacking STELLA (black arrowheads, inset left cell). Dotted line indicates edges of proximal allantois. E–H, 6s stage exhibits all OCT-3/4+ cells co-localizing STELLA in the remnant of the ACD, distal to the invaginating hindgut (white arrowheads, inset). Dotted line indicates approximate site of embryonic-allantoic junction, determined by the site where the full width of the proximal allantois joins embryonic tissue. In this, the next set of panels, and in most of the subsequent figures, non-nuclear STELLA appears fragmented (e.g., arrow, F, H); we attribute these non-nuclear profiles to cytoplasmic STELLA, which has previously been described (see text). I–L, 9s stage illustrates persistent STELLA+/OCT-3/4+ cell cluster in the midline base of the allantois; the embryonic-allantoic junction is located below the bottom edge of the image. Occasional STELLA+/OCT-3/4+ cells can also be seen moving distally in the allantois from the cluster (white arrowhead, upper right inset; left inset, lower magnification of proximal allantoic midline. Scale bar in L = 20μm in A–L, 9μm in upper right insets A–L, 43μm in left insets I–L.
Figure 3. Localization of OCT-3/4 and STELLA within the visceral endoderm, hindgut lip, and hindgut.

Red channel indicates OCT-3/4, green indicates STELLA, blue indicates DAPI. AD, Late headfold (LHF) stage with focal plane at the allantois-associated posterior visceral endoderm (aPVE). OCT-3/4 and STELLA co-localizing cells are noted throughout the aPVE (white arrowheads, inset left cell), while some OCT-3/4+ cells are STELLA- in the aPVE (black arrowheads, inset right cell). E–L, 6s stage with focal plane at the visceral endoderm of the hindgut lip (HGL) in E–H and ventral hindgut endoderm in I–L. All STELLA+ cells co-localize OCT-3/4 and vice-versa within the HGL and hindgut (white arrowheads, insets). Dotted line indicates the ventral edge of the hindgut invagination. M–N, 7s stage IHC specimen, immunostained for OCT-3/4 and sectioned transversely, viewed just anterior to the site of splanchnopleure (spl) closure to form the hindgut (hg) (M) and at the site of hg closure (N). Punctate, darkly-staining OCT-3/4+ cells with large, prominent nuclei are localized to the spl (e.g., black arrowheads), while other, fainter OCT-3/4+ cells with smaller nuclei are found throughout the neural folds (nf) and subadjacent mesoderm (me; e.g., white arrowheads). O–V, 9s stage with focal plane at the HGL in O–R and ventral hg endoderm in S–V. Increased numbers of STELLA and OCT-3/4 co-localizing cells are noted throughout the HGL and ventral hg (white arrowheads, insets). Dotted line in O–R indicates the ventral edge of the hg invagination; in S–V it indicates lateral edges of the hg. Scale bar in V = 20μm in A–L, 23μm in M–N, 20μm in O–V, 9μm in insets A–L, S–V, 6.4μm in insets O–R. Other abbreviation: da, dorsal aorta.
Consistent with previous results (Downs, 2008; Mikedis and Downs, 2012), STELLA and OCT-3/4 were found within a broad domain of the primitive streak, including the ACD and embryonic posterior streak, and later the tailbud mesoderm (Fig. 2 and data not shown). By contrast, OCT-3/4 appeared independent of STELLA within the ACD and posterior embryonic primitive streak. Within the allantois, while some STELLA+/OCT-3/4+ cells remained within the ACD (Fig. 2A–L), some formed a file through the allantoic midline, reaching toward the distal region of the allantois (Fig. 2I–L), consistent with previous STELLA localization studies (Mikedis and Downs, 2012). The population of STELLA+/OCT-3/4+ cells associated with the ACD and allantoic midline was observed through the 12s stage (data not shown), and persisted within the tailbud mesoderm. Thus, a STELLA+/OCT-3/4+ population is a permanent feature of the allantois.
By contrast, and consistent with the claims that PGCs leave the allantois (Lawson and Hage, 1994) and with fate mapping the ACD (Downs, 2008; Mikedis and Downs, 2012), some headfold-stage STELLA+/OCT-3/4+ cells appeared to take up residence within the allantois-associated posterior visceral endoderm. Within the posterior visceral endoderm, OCT-3/4 was found both with and without STELLA (Fig. 3A–D and data not shown). By 2s, however, nearly all OCT-3/4+ cells of the allantois-associated posterior visceral endoderm co-localized STELLA (data not shown). From there, STELLA+/OCT-3/4+ cells were found within the hindgut lip by 6s stage (Fig. 3E–H), and within ventral hindgut endoderm (Fig. 3I–L). Intriguingly, immunostaining suggested that the OCT-3/4+ nuclei found within the hindgut lip were much larger than those in surrounding cells (Fig. 3M, N; Downs, 2008). During elongation of the hindgut, a dense population of STELLA+/OCT-3/4+ cells remained clustered within the hindgut lip and was not noted in any other visceral endoderm (Fig. 3O–R). Within the ventral hindgut endoderm, STELLA+/OCT-3/4+ cells appeared to increase dramatically, and spanned the longitudinal extent of the hindgut (Fig. 3S–V). STELLA+/OCT-3/4+ cells were not observed within the foregut, nor in posterior visceral endoderm anterior to the site of hindgut invagination through the 12s stage (data not shown).
Thus, while all STELLA+ cells spatiotemporally co-localized OCT-3/4 throughout the posterior region at all stages examined, OCT-3/4+/STELLA− cells dropped out of the posterior region by 2s, at which point OCT-3/4 was found only with STELLA. Thereafter, STELLA+/OCT-3/4+ cells localized to two topographically distinct posterior tissues, extraembryonic mesoderm (remnant ACD) and endoderm, which included extraembryonic visceral endoderm, the hindgut lip and hindgut endoderm.
A small population of STELLA+ cells transiently co-localizes T within the extraembryonic and embryonic components of the primitive streak
Because STELLA+/OCT-3/4+ cells are found within the ACD, which is defined by Brachyury (T) (Downs et al., 2009; Inman and Downs, 2006b; Mikedis and Downs, 2012), we next asked about STELLA’s relationship to T. Within the XPS (Fig. 4A–H), ACD (Fig. 4I–L), embryonic posterior streak (not shown), allantois-associated posterior visceral endoderm (Fig. 4M–P), embryonic streak-associated visceral endoderm (not shown), hindgut lip (not shown) and hindgut (not shown), STELLA+(/OCT-3/4+) and T generally formed separate cell populations (parentheses indicate inferred presence of OCT-3/4 in the STELLA+ cells, based on results in the previous section). However, additionally within the posterior primitive streak (XPS/ACD and embryonic posterior streak), 3 rare ACD cells were STELLA+(/OCT-3/4+)/T+, but only at the LHF stage (N=1; Fig. 4I–L; total ACD counted = 140 cells; STELLA+/T− = 7/140; T+/STELLA− = 130). The event of encountering T/STELLA protein co-localization was rare, transient, and limited to this developmental stage: specimens observed after LHF (i.e., through 2s; Table 1) no longer exhibited cells with T/STELLA co-localization in the ACD. T/STELLA co-localization was not found within any endoderm.
Figure 4. T and STELLA localization within the posterior region at EB and LHF stages.

Red channel indicates T, green indicates STELLA, blue indicates DAPI. All immunofluorescence specimens are oriented with posterior (i.e., allantois) upward and anterior downward. A–D, Early bud (EB) stage with focal plane at the extraembryonic primitive streak (XPS). Dotted line indicates location of allantoic bud, as identified in analysis of other planes in the Z-series. White box indicates approximate site of panels E–H. E–H, increased magnification of EB stage XPS, with occasional STELLA-T co-localizing cells (black arrowheads) and many STELLA+/T- cells (white arrowheads). I–L, Late headfold (LHF) stage with focal plane at the allantoic core domain (ACD). Dotted line indicates approximate site of embryonic-allantoic junction. Some cells co-localize STELLA and T (black arrowheads, lower inset) and other STELLA+ cells are T− (white arrowheads, upper inset). M–P, LHF stage with focal plane at the allantois-associated posterior visceral endoderm (aPVE). Dotted line indicates approximate site of embryonic-allantoic junction. STELLA and T are not noted to co-localize; white arrowhead indicates adjacent STELLA+ (upper) and T+ (lower) cells, magnified in inset. Scale bar in P = 20μm in A–D, I–P, 9μm in E–H, 6μm in insets I–P.
T identifies several distinct posterior cell populations
As studies on embryonic stem cells have indicated that T plays a role in mesendodermal fate (Izumi et al., 2007; Kubo et al., 2004) and that STELLA facilitates endodermal differentiation in human cells (Wongtrakoongate et al., 2013), we then asked whether T and/or STELLA identify distinct subpopulations with likely mesendodermal fate. For this, we asked whether T and STELLA co-localize with MIXl1 (headfold-8s stages). First, because T and MIXl1 proteins physically interact (Pereira et al., 2011; Pereira et al., 2012; Tada et al., 2005), and because Mixl1 is a regulatory target of T (Lolas et al., 2014), we evaluated the posterior region for co-localization of T and MIXl1. Beginning at headfold stages, a period during which MIXl1 was observed in the posterior expanse of the primitive streak and allantois (Wolfe and Downs, 2014), all MIXl1+ cells were T+ within the posterior primitive streak (XPS/ACD and embryonic posterior streak) and associated posterior visceral endoderm (Fig. 5A–D). By the late hindgut stages (e.g., 8s), MIXl1+/T+ co-localizing cells persisted in the ACD/presumptive tailbud mesoderm, and were limited to the posterior edge of the embryonic notochord. By contrast, at all stages examined, not all T+ cells contained MIXl1 (Fig. 5A–D and data not shown). T+/MIXl1− cells predominated in the hindgut’s dorsal component during hindgut elongation (e.g., 8s), consistent with prior localization of individual proteins (data not shown; Downs et al., 2009; Inman and Downs, 2006b; Wolfe and Downs, 2014) while a few MIXl1+/T− cells were also found in the dorsal hindgut (data not shown). We did not see STELLA co-localize with MIXl1+ (e.g., Fig. 5E–H, Table 1 and data not shown). Thus, STELLA+/(OCT-3/4+)/T+ and T+/MIXl1+ identify at least two distinct subpopulations within the primitive streak and tailbud.
Figure 5. Localization of T, MIXl1, and STELLA in the extraembryonic primitive streak (XPS) and allantoic core domain (ACD) at EB-EHF stages.

A–D, Early headfold (EHF) stage with focal plane at ACD. Red channel indicates T, green indicates MIXl1, blue indicates DAPI. T and MIXl1 co-localize in some cells (white arrowhead, inset left cell), while other T+ cells are MIXl1- (black arrowhead, inset right cell). E–H, Early bud (EB) stage with focal plane at XPS. Red channel indicates STELLA, green indicates MIXl1, blue indicates DAPI. STELLA and MIXl1 do not co-localize; white arrowhead indicates STELLA+ and black arrowhead indicates MIXl1+ cells, magnified in inset. Scale bar in H = 10μm in A–H, 3.5μm in insets A–D, 4.4μm in insets EH.
STELLA and FOXa2 co-localize in the hindgut lip
To further evaluate STELLA’s potential role in nascent hindgut endoderm, we examined its relationship to FOXa2. Consistent with previously described FOXa2 mRNA expression data (Ang et al., 1993) and previous protein localization data (Besnard et al., 2004), FOXa2 localized throughout posterior visceral endoderm associated with the embryo (Fig. 6A and data not shown). By the allantoic bud stage, FOXa2 was found in the allantois-associated posterior visceral endoderm and persisted there through headfold stages (Fig. 6A). Later, FOXa2 was found within the notochord and visceral and definitive hindgut endoderm (Fig. 6B–C). Thus, FOXa2 is found in sites of STELLA localization.
Figure 6. FOXa2 and STELLA localization in visceral and embryonic endoderm through 7s stage.
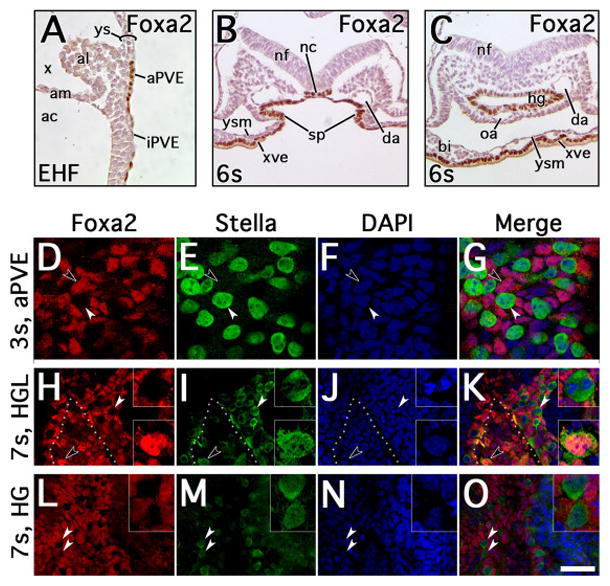
A, Early headfold (EHF) stage mid-sagittal section through posterior region, oriented with posterior upward, anterior downward, dorsal leftward and ventral rightward, illustrating the allantois (al), with immunohistochemical localization of FOXa2 to intraembryonic posterior visceral endoderm (iPVE) and allantois-associated posterior visceral endoderm (aPVE). B–C, 6s stage transverse sections through the posterior region anterior to splanchnopleure (sp) fusion to form the hindgut (B) and posterior to the site of hindgut (hg) invagination (C), oriented with dorsal upward and ventral downward. FOXa2 localizes to the notochord (nc), hg endoderm and yolk sac extraembryonic visceral endoderm (xve). D–O, immunofluorescent localization of FOXa2 (red) and STELLA (green) in the visceral endoderm and hg at 3s and 7s stages; blue channel indicates DAPI. D–G, 3s stage with focal plane at the aPVE, illustrating STELLA+ cells (white arrowheads) distinct from FOXa2+ cells (black arrowheads). H–K, 7s stage with focal plane at the visceral endoderm of the hindgut lip (HGL), illustrating some FOXa2+ cells co-localizing STELLA along the HGL (black arrowheads, bottom inset) while other STELLA+ cells of the nearby visceral endoderm exclude FOXa2 (white arrowheads, top inset). Dotted line indicates ventral edge of invaginating hg. L–O, 7s stage with focal plane within the hg; STELLA+ and FOXa2+ cells remain distinct (white arrowheads, inset). Scale bar in O = 42μm in A, 65μm in B–C, 10μm in D–G, 20μm in H–O, 6.8μm in insets H–O. Other abbreviations: ac, amniotic cavity; am, amnion; bi, blood island of yolk sac; da, dorsal aorta, nf, neural fold; oa, omphalomesenteric artery; x, exocoelom; ys, yolk sac; ysm, yolk sac mesoderm.
We next co-localized STELLA with FOXa2 via IF (EB-10s stages). Until hindgut formation (i.e., through 4s stage), STELLA and FOXa2 localized in separate populations throughout the posterior region (Fig. 6D–G and Table 2). Within the aPVE at 3s stage, non-co-localizing FOXa2+ and STELLA+ cells were counted in a ratio of approximately 2:1; no co-localizing FOXa2/STELLA cells were observed (Table 2). However, as the lateral splanchnopleure joined to form the ventral hindgut invagination (6s stage; e.g., Fig. 3M–N), FOXa2 and STELLA co-localized in approximately 1/3 of the cells counted at the nascent hindgut (Table 2). As the hindgut invagination progressed (7s–10s), a reproducible and sizeable population of these co-staining cells persisted within the hindgut lip (Fig. 6H–K), averaging approximately 20% of the cells counted in 6 specimens (Table 2). Conversely, while STELLA+/FOXa2− cells peppered the longitudinal extent of the hindgut, they were never observed to co-localize FOXa2 here (Fig. 6L–O and Table 2).
Table 2.
Relative frequencies of STELLA+ and FOXa2+ cells in endoderm based on immunofluorescence of co-stained specimens
| Stage (N) | Staining pattern | Average number of positive cells per specimen by site (% of all counted cells)
|
||
|---|---|---|---|---|
| aPVE | HGL | HG | ||
| Early bud (1) | STELLA+ / FOXa2− | 0 (0) | NA | NA |
| STELLA− / FOXa2+ | 3 (100) | NA | NA | |
| STELLA+ / FOXa2+ | 0 (0) | NA | NA | |
| Total | 3 (100) | NA | NA | |
| Headfold (2) | STELLA+ / FOXa2− | 37.5 (32) | NA | NA |
| STELLA− / FOXa2+ | 77.5 (68) | NA | NA | |
| STELLA+ / FOXa2+ | 0 (0) | NA | NA | |
| Total | 115 (100) | NA | NA | |
| 3s (5) | STELLA+ / FOXa2− | 49.6 (31) | NA | NA |
| STELLA− / FOXa2+ | 112.6 (69) | NA | NA | |
| STELLA+ / FOXa2+ | 0.0 (0) | NA | NA | |
| Total | 162.2 (100) | NA | NA | |
| 6s (1) | STELLA+ / FOXa2− | NA | 60 (32)1 | NA |
| STELLA− / FOXa2+ | NA | 67 (36)1 | NA | |
| STELLA+ / FOXa2+ | NA | 60 (32)1 | NA | |
| Total | NA | 187 (100)1 | NA | |
| 7–10s (6) | STELLA+ / FOXa2− | NA | 24.3 (15) | 60.5 (10) |
| STELLA− / FOXa2+ | NA | 104.8 (65) | 555.8 (90) | |
| STELLA+ / FOXa2+ | NA | 32.7 (20) | 0 (0) | |
| Total | NA | 161.2 (100) | 616.3 (100) | |
The hindgut invagination at 6s is early in its formation, morphologically not yet distinctly formed into HGL and HG; the definitive endoderm cells counted in this specimen were grouped as HGL.
Abbreviations: aPVE, allantois-associated posterior visceral endoderm; HG, hindgut; HGL, hindgut lip; NA, not applicable, as these features were not found at these stages; s, somite pairs.
Closer scrutiny of nuclear morphology in the DAPI channel clearly revealed mitotic events within the visceral endoderm, hindgut lip, and hindgut up to 7s. In all sites, many FOXa2+/STELLA− and STELLA+/FOXa2− daughter cells appeared to localize equivalent amounts of FOXa2 or STELLA signal, respectively (Fig. 7A–J). Similarly, in the hindgut lip, STELLA+/FOXa2+ cells distributed both STELLA and FOXa2 to their descendants (Fig. 71K–O). In two cases, however, (Fig. 7P–T), one daughter cell appeared to acquire FOXa2 and most of the STELLA signal, remaining within the hindgut lip, while the other daughter was predominantly FOXa2+/STELLA− and moving into the hindgut. Asymmetric distribution of STELLA and FOXa2 may have been difficult to discern as the hindgut lip was typically oriented transversely with respect to the confocal imaging plane.
Figure 7. Patterns of cell division among FOXa2+ and STELLA+ cells in the hindgut lip at 7s stage.

View of individual dividing cells within the hindgut lip at 7s stage; red channel indicates FOXa2, green indicates STELLA, blue indicates DAPI. A–D, FOXa2+/STELLA− cell at metaphase/anaphase with apparent symmetric FOXa2 localization to daughters. E, Schematic depiction of the cell of interest. F–I, STELLA+/FOXa2− daughter cells at telophase with apparent symmetric STELLA localization to daughters. J, Schematic depiction of the cell of interest. K–N, STELLA+/FOXa2+ cell at anaphase with apparent symmetric FOXa2 and STELLA localization to daughters. O, Schematic depiction of the cell of interest; orange coloration indicates STELLA/FOXa2 co-localization. P–S, STELLA+/FOXa2+ cell at telophase with apparent asymmetric localization of STELLA to one daughter and FOXa2 to both daughters. T, Schematic depiction of the cell of interest; orange coloration indicates STELLA/FOXa2 co-localization and pink indicates relatively fainter FOXa2 localization than in other positive cells. Scale bar in S = 3.9μm in A–D, 3.5μm in F–I, 3.9μm in K–N, 2.5μm in P–S.
Thus, a subset of STELLA+ cells appeared to transiently exhibit endodermal identity within the hindgut lip but not the hindgut itself, and appeared capable of producing hindgut endoderm. The STELLA+/(OCT-3/4+)/FOXa2+ cell population found within the hindgut lip, a site of putative PGCs, therefore appears to be involved in differentiation of a progenitor population into hindgut endoderm during a limited time period.
STELLA co-localizes with Runx1 within the hindgut lip and hindgut
Runx1 was previously found to localize to the visceral endoderm of the yolk sac (Daane and Downs, 2011; Zeigler et al., 2006). While the function of Runx1 in visceral endoderm is not known, visceral endoderm contributes to the definitive gut endoderm (Kwon et al., 2008) and thus, we wanted to know whether STELLA was excluded from the Runx1+ population. Co-localization of STELLA with Runx1 at the hindgut lip and hindgut (8s–12s; ~E9.0) revealed its presence both independent of and with Runx1 expression within the hindgut lip and ventral midline of the developing hindgut and omphalomesenteric artery (Fig. 8A). Moreover, FOXa2 also co-localized with Runx1 in both the yolk sac visceral endoderm and ventral hindgut (8s–12s) (Fig. 8B, C), further underscoring a role for FOXa2 in mesendodermal differentiation.
Fig. 8. Co-localization of STELLA and FOXa2 with Runx1 in the ventral hindgut.

A, 8s stage transverse section through hindgut, ventral toward bottom. STELLA (brown) co-localizes with Runx1 expression (blue, arrow) in cells of the ventral hindgut (hg), while other STELLA cells lack co-expression (arrowhead). B, C, 12s stage transverse sections through the hindgut lip (B) and hindgut (C), ventral toward bottom. FOXa2 (brown) also co-localizes with Runx1 in the ventral hg (arrows), while other FOXa2 cells appear devoid of Runx1 (arrowhead, C). Note that FOXa2 and Runx1 also co-localize within the extraembryonic visceral endoderm (xve). D, E, 4s- (D), 8s- (E) stage specimens (posterior, right) immunostained with the RUNX1 antibody, demonstrating appropriate localization to previously-reported sites in the Runx1:lacZ mouse line (Daane and Downs, 2011). Hematopoietic cells of the yolk sac blood islands (bi, arrowhead) and allantoic vessel of confluence (asterisk, arrow) are robustly positive for RUNX1; the xve of the yolk sac (ys), which was previously reported to express Runx1 (Zeigler et al., 2006), and the hg endoderm (arrow) exhibit cytoplasmic RUNX1. F–I, immunofluorescent localization of nuclear RUNX1 (red) and nuclear STELLA (green) at the 9s stage with focal plane within the hg; blue channel indicates DAPI. Arrowhead indicates a STELLA+ cell co-localizing with RUNX1 in the nucleus (magnified in inset). Dotted line indicates the ventral edge of the hg invagination. J–M, Same specimen, 9s, shows co-localization of STELLA and RUNX1 within the cytoplasmic dots (arrowheads), suggesting that the cytoplasmic versions of both proteins localize to similar components. Scale bar in M = 10μm in J–M, 15 μm in A, 25 μm in B, C, 40μm in D, 30 μm in E, 20 μm in F–I, and 9 μm in F–I, insets. Other abbreviations: al, allantois; oa, omphalomesenteric artery; va, vitelline artery, x, exocoelom.
To verify these co-localization patterns, we showed, by IHC, that RUNX1 was appropriately found in the yolk sac blood islands and vessel of confluence of the allantois (Fig. 8D), a hemogenic blood vessel that marks the site of the future fetal-placental arterial connection (Daane and Downs, 2011; Downs et al., 1998). Outside of hematopoietic cells, RUNX1 localized in a punctate manner to the cytoplasm of the yolk sac’s extraembryonic visceral endoderm, in accord with previous results (Zeigler et al., 2006), and with its possible function as a cytoplasmic attenuator of NF-KB signaling in the repression of myeloid tumors (Nakagawa et al., 2011). RUNX1 also co-localized to the hindgut endoderm itself (Fig. 8E). Within the hindgut endoderm, results of IF verified that STELLA co-localizes with RUNX1, both nuclear and cytoplasmic profiles (Fig. 8E–M).
Thus, together with the results of FOXa2 co-localization, STELLA appears to be found within yet another population of cells, this one destined to become part of the hematopoietic lineage. Together, these data provide further evidence for the presence of multiple distinct STELLA+ mesendodermal populations in the nascent hindgut region.
DISCUSSION
Contrary to previous claims (McLaren, 2003; Saitou et al., 2002), we could find no evidence that STELLA identifies a single molecularly homogenous population within the allantois and posterior endoderm of the gastrula, thought to be the sites of formation and migration of the mouse PGCs. Rather, here we have identified four molecularly distinct STELLA populations within canonical PGC-associated tissues: the allantois, associated visceral endoderm, and hindgut. Added to previous results (Mikedis and Downs, 2017), we can now describe five distinct STELLA+ populations in these tissues. We further showed that STELLA is found with gene products that are involved in mesendodermal cell differentiation, and concentrated along the axial midline. These subpopulations are summarized in Table 3.
Table 3.
STELLA subpopulations identified by gene product localization
| XPS/ACD | STELLA/OCT/PRDM11 STELLA/(OCT)/(PRDM1)/T2 |
| aPVE | STELLA/OCT/PRDM11 STELLA/OCT |
| Hindgut Lip/ Hindgut | STELLA/OCT/PRDM11 STELLA/OCT STELLA/OCT/FOXa23 STELLA/OCT/Runx1 |
| Other possible, but not yet proven, combinations4 | STELLA/OCT/PRDM1/Runx1 STELLA/OCT/FOXa2/PRDM1 STELLA/OCT-3/4/FOXa2/Runx1 STELLA/OCT-3/4/FOXa2/PRDM1/Runx1 |
Together with data in this study, we conclude that a STELLA/OCT/PRDM1 population is found in all putative PGC tissues (Mikedis and Downs, 2017).
STELLA invariably co-localized with PRDM1 in the allantoic component of the primitive streak (Mikedis and Downs, 2017) and co-localized in this study with T in just 3 cells in one late headfold-stage specimen.
STELLA/OCT/Foxa2 were found only in the hindgut lip, and not within the hindgut itself in this study.
These possible combinations are based on results here combined with those of previous studies (Mikedis and Downs, 2012, 2017).
Within the ACD, STELLA always co-localized with OCT-3/4, and rarely with T, the latter subpopulation of which was present only at the late headfold stage. We had previously shown that STELLA always co-localized with PRDM1 in the ACD (Mikedis and Downs, 2017). Thus, within the ACD, two populations, STELLA/OCT-3/4/PRDM1 and STELLA/OCT-3/4/(PRDM1)/T, were observed. The latter, albeit very rare, population is consistent with previous data showing that the putative PGCs utilize PRDM1, acting in part through T, to suppress somatic fates and ultimately up-regulate other PGC-associated gene products including STELLA (Aramaki et al., 2013). However, that study was conducted in epiblast-like cell (EpiLC) culture and thus the spatiotemporal pattern of posterior region cell populations exhibiting these proteins – and their rarity - was not elucidated. Our previous localization studies (Mikedis and Downs, 2012) were conducted at stages separated by 2–4 hours. We are confident that from the bud through headfold stages, when the PGCs are claimed to segregate from the soma (Lawson and Hage, 1994), most, if not all of the full complement of the allantois’s STELLA cell population and thus, the putative PGCs, was assessed. In the present study, most allantoic core domain STELLA-positive cells did not co-localize T. This finding appears to contradict the described role for T in specification of putative “PGCs” within the allantois (Aramaki et al., 2013). However, we note that our protein-based localization methodology differs from previously published transcript detection; it is possible that these proteins were present at levels below the threshold of detection of our antibody-based methods. Alternatively, the scant cells co-localizing T/STELLA we observed at LHF stage (Fig. 4I–L) could represent the “true” PGCs (Lawson and Hage, 1994), albeit in an unexpectedly restricted quantity and developmental timepoint.
From the ACD, the STELLA+/OCT-3/4+ cell populations diverged, with some remaining within the allantois and becoming subsumed into tailbud mesoderm, while others appeared within the axial posterior visceral endoderm beginning at the EHF stage, in accord with previous results (Downs, 2008; Downs et al., 2009; Mikedis and Downs, 2012). The fate of those STELLA+/OCT3/4+ cells that remained within the ACD/tailbud mesoderm is not known. We had previously shown that allantoic OCT-3/4 co-localizes with flk1 (Downs, 2008), a major receptor for vascular endothelial growth factor (VEGF) and present on angioblasts (Kataoka et al., 1997). A major function of the allantois is to create the umbilical vasculature, which may be hemogenic (Corbel et al., 2007; Zeigler et al., 2006). STELLA/OCT-3/4 may be involved in blood vessel formation, especially as STELLA co-localized with RUNX1 (Fig. 8), which is found in nascent hemangioblasts (North et al., 1999). Alternatively, these may be “ectopic PGCs,” implying that they have erred from the earliest point in the PGC trajectory and must be culled by apoptosis (reviewed in Mikedis and Downs, 2014). This hypothesis has been supported by findings of increased apoptosis in OCT4+ PGCs that fail to complete their expected migration trajectory out of the gut in embryonic slice culture at and after E10.5 (Stallock et al., 2003). However, the conclusion that these cells can be dismissed as a priori developmental errors has been recently questioned (Mikedis and Downs, 2014). In any event, as the ACD-derived STELLA+ cells persisted in the allantois/tailbud and therefore did not appear to die or enter the hindgut, they instead appear to represent a viable geographic population distinct from those found in endoderm.
While Oct-3/4 was found without STELLA prior to the 2s stage within the allantois-associated visceral endoderm, thereafter STELLA/OCT-3/4 cells formed a single population. STELLA was never found here with T, FOXa2, or MIXl1. By contrast, T was found alone and with MIXl1, while MIXl1 was also found independent of T. FOXa2 was not found here with any other protein. Previous results also place PRDM1 with STELLA in the visceral endoderm (Table 3; Mikedis and Downs, 2017). However, in that study, STELLA/(OCT-3/4) was also found independent of PRDM1 in this tissue. Thus, within visceral endoderm, STELLA/OCT-3/4 forms at least two distinct populations, STELLA/OCT-3/4 and STELLA/(OCT-3/4)/PRDM1, that may have translocated there from the ACD. Translocation of ACD-derived cells to the posterior visceral endoderm has been suggested by fate mapping the STELLA-positive ACD (Mikedis and Downs, 2012). Other data place OCT-3/4 in the visceral endoderm at comparable stages (Anderson et al., 2000; Downs, 2008). Alternatively, perhaps visceral endoderm produces STELLA and/or OCT-3/4 de novo; this is currently not known.
While the STELLA+/OCT-3/4+ cells embedded within visceral endoderm appeared consistent with the expected PGC trajectory, we were surprised to discover that a number of them reproducibly co-localized FOXa2 in the hindgut lip. The hindgut lip represents the ventral midline site of joining of posterolateral splanchnopleure (Daane and Downs, 2011) and, presumably, deployment of definitive endoderm from the visceral endoderm into the hindgut proper (Hara et al., 2009; Kwon et al., 2008). The action of elongating endoderm appears necessary for normal translocation of presumed PGCs into the hindgut (Hara et al., 2009); this site also contains cells exhibiting MIXl1, and may therefore harbor hindgut progenitor cells (Wolfe and Downs, 2014). FOXa2, which identifies embryo-associated visceral endoderm and embryonic endoderm (Ang et al., 1993; Besnard et al., 2004; Kubo et al., 2004) and is found in vitro in putative mesendodermal stem cells (Tada et al., 2005), did not co-localize in most endodermal cells in which we found STELLA. Rather, individual STELLA+/FOXa2− cells were interspersed with STELLA−/FOXa2+ endoderm within the hindgut lip. Since we see stable STELLA localization with OCT-3/4+ and FOXa2 within this putative stem cell reservoir for definitive endoderm during initial hindgut formation, we propose that some STELLA+ cells are deployed at the hindgut lip as hindgut progenitors. While functional studies supporting this role for STELLA in the mouse have not been reported to date, we note in vitro findings that human STELLA overexpression can drive up-regulation of endodermally-associated genes such as Sox17, Sox7 and α-fetoprotein in embryonic stem cells (ESCs; Wongtrakoongate et al., 2013). Granting that human ESC culture conditions differ from those of in vivo murine development, our finding suggesting STELLA contribution to definitive endoderm warrants further investigation. We have recently reported STELLA in the hindgut with and without PRDM1 (Mikedis and Downs, 2017), and here have found that it localizes with and without Runx1. Thus, in the hindgut lip/hindgut, four distinct STELLA populations have now been documented (Table 3).
Together, across the canonical PGC trajectory, we have identified five distinct STELLA subpopulations (Table 3). In addition, we have suggested the possibility of higher orders of subpopulations with these gene products, to be investigated in future studies. Of the subpopulations identified here and combined with those of a previous study (Mikedis and Downs, 2017), only one, STELLA/OCT-3/4/PRDM1, was consistently found within the allantois, aPVE, and hindgut lip and hindgut. All three of these gene products have been implicated in identifying the PGCs. However, whether this subpopulation is continuous with the germ line is not known. We conclude, based on these and previous results (Mikedis and Downs, 2014), that STELLA alone cannot be used as a marker of presumptive PGCs. However, the nuclei of STELLA/OCT-3/4+ cells within the endodermal component of the splanchnopleure appeared to be larger than those containing OCT-3/4 in non-splanchnopleure tissue. As a variety of factors regulate the size of the nucleus (Jevtic et al., 2014), perhaps signals that regulate nuclear size could be useful in future studies. For example, nuclear atypia is a common diagnostic and prognostic marker that, in some instances, distinguishes normal from malignant cells (Weiss, 2002).
Why have the so-called segregated allantoic PGCs never been fate-mapped and shown to end up in the gonads via a hindgut intermediate? These fate mapping experiments are currently technically impossible for two reasons. First, while individual cells of the gastrula can be labeled (Lawson et al., 1991), exactly which cells of the allantois should be labeled is not clear. Our results underscore cell heterogeneity in the base of the allantois. In addition, results of studies involving alkaline phosphatase as a “marker” of allantoic PGCs place the number of potential targets at approximately 40 cells (Ginsburg et al., 1990), but these cannot be distinguished from other cells of the ACD by morphology or biochemical methods in the living state. Second, even if a population of interest could be identified prospectively, currently available technologies for whole embryo culture cannot sustain fetal development until the gonads form (Cockroft, 1990).
At present, the only available option to demonstrate continuity of a specific STELLA population between the allantois and gonads is to exploit the inducible STELLA-CRE reporter, ensuring that the presumptive segregated PGC population is present within the ACD at the time of induction, and that descendants of these cells are found subsequently in sequence in the established PGC tissues: from visceral endoderm, to hindgut, and ultimately to gonads (Hirota et al., 2011; discussed in Mikedis and Downs, 2014). However, a major caveat with studies of this type is the uncertainty that STELLA populations will arise in the trajectory at advanced timepoints, rather than originating within the allantois.
Barring prospective evidence for a continuous lineage, the only other way to evaluate whether STELLA/PRDM1 cells represent a distinct and continuous population within these tissues was to ask whether a unique STELLA/PRDM1 signature is confined to the trajectory, whether that signature is found outside of PGC tissues (Mikedis and Downs, 2012, 2017), or whether diverse STELLA subpopulations exist within PGC tissues (this study). Even within the trajectory, STELLA and PRDM1 proteins were found independently of the other. Thus, STELLA and PRDM1 do not appear to be the molecular signature of PGCs, and PGCs cannot be distinguished from somatic cells. Alternatively, PGCs may not be confined to the allantois, visceral endoderm, and hindgut, but are found throughout the posterior region. Either way, these cells need to be rigorously fate-mapped to show continuity with the gonads.
CONCLUSIONS
Our data argue against all posterior STELLA+ cells being a segregated, homogeneous germline population that suppresses somatic identity within the emerging hindgut of the mouse gastrula. We have presented findings that lead us to question the accepted immutability of lineage-restricted, fate-determined PGCs in the mouse posterior region from E7.5–9.5. STELLA, together with OCT-3/4, appears to identify cells which may have more expansive developmental roles in formation of the allantois, hindgut, vasculature, and tailbud than previously suspected. We recognize that these findings challenge a long-held belief in the specification and journey of unipotent mammalian PGCs from allantois to endoderm to gonads, and note that recent authors in the field (e.g., Nikolic et al., 2016) have appeared unwilling to acknowledge prior literature that questions these beliefs (e.g., Mikedis and Downs, 2014). Nevertheless, as we continue to explore the events that build the posterior embryo and the fetal-umbilical junction, we maintain an open mind to the possibility that mouse PGC specification, if it truly occurs in the posterior region, occurs there at a later developmental timepoint, and not within the allantois.
HIGHLIGHTS.
STELLA co-localizes with mesendodermal proteins at the fetal-placental interface.
STELLA only rarely – and transiently – co-localizes with Brachyury.
STELLA is not a specific identifier of germ cell antecedents in the gastrula.
Acknowledgments
The authors gratefully acknowledge Lance Rodenkirch and the W.M. Keck Laboratory for Biological Imaging at the University of Wisconsin-Madison for technical assistance with confocal microscopy, Erin DeCloux, Lauren Wierenga, and Maria Mikedis for technical assistance, and Maria Mikedis for critical appraisal of the findings presented in this work. This study was supported by Grants from the March of Dimes (1-FY09-511) and National Institutes of Child Health and Development (R01 HD042706 and RO1 HD079481 to K.M.D.; T32 HD041921 to A.M.R). A.D.W. was further supported by NIH National Heart, Lung, and Blood Institute Grant “Research Training in Hematology” (T32 HL07899), and an NIH Extramural Pediatric Research Loan Repayment Program Grant (NOT-OD-11-086).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson R, Copeland TK, Scholer H, Heasman J, Wylie C. The onset of germ cell migration in the mouse embryo. Mechanisms of Development. 2000;91:61–68. doi: 10.1016/s0925-4773(99)00271-3. [DOI] [PubMed] [Google Scholar]
- Ang SL, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, Mochizuki Y, Hamakubo T, Kato Y, Shirahige K, Saitou M. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev Cell. 2013;27:516–529. doi: 10.1016/j.devcel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Beddington RS. An autoradiographic analysis of tissue potency in different regions of the embryonic ectoderm during gastrulation in the mouse. J Embryol Exp Morphol. 1982;69:265–285. [PubMed] [Google Scholar]
- Beddington RS. Isolation, culture and manipulation of post-implantation mouse embryos. In: Monk M, editor. Mammalian Development: A Practical Approach. IRL Press; Oxford: 1987. pp. 43–69. [Google Scholar]
- Beddington SP. An autoradiographic analysis of the potency of embryonic ectoderm in the 8th day postimplantation mouse embryo. J Embryol Exp Morphol. 1981;64:87–104. [PubMed] [Google Scholar]
- Besnard V, Wert SE, Hull WM, Whitsett JA. Immunohistochemical localization of Foxa1 and Foxa2 in mouse embryos and adult tissues. Gene Expr Patterns. 2004;5:193–208. doi: 10.1016/j.modgep.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Goodheart M, Liao M, Page DC. Dppa3 / Pgc7 / stella is a maternal factor and is not required for germ cell specification in mice. BMC Dev Biol. 2004;4:2. doi: 10.1186/1471-213X-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champlin AK, Dorr DL, Gates AH. Determining the stage of the estrous cycle in the mouse by the appearance of the vagina. Biol Reprod. 1973;8:491–494. doi: 10.1093/biolreprod/8.4.491. [DOI] [PubMed] [Google Scholar]
- Chiquoine AD. The identification, origin, and migration of the primordial germ cells in the mouse embryo. Anat Rec. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- Cockroft DL. Postimplantation Mammalian Embryos: A Practical Approach. IRL Press; Oxford: 1990. Dissection and culture of post-implantation mouse embryos. [Google Scholar]
- Corbel C, Salaun J, Belo-Diabangouaya P, Dieterlen-Lievre F. Hematopoietic potential of the pre-fusion allantois. Dev Biol. 2007;301:478–488. doi: 10.1016/j.ydbio.2006.08.069. [DOI] [PubMed] [Google Scholar]
- Daane JM, Downs KM. Hedgehog signaling in the posterior region of the mouse gastrula suggests manifold roles in the fetal-umbilical connection and posterior morphogenesis. Dev Dyn. 2011;240:2175–2193. doi: 10.1002/dvdy.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeale B, Brokhman I, Mohseni P, Babak T, Yoon C, Lin A, Onishi K, Tomilin A, Pevny L, Zandstra PW, Nagy A, van der Kooy D. Oct4 is required ~E7.5 for proliferation in the primitive streak. PLoS Genet. 2013;9:e1003957. doi: 10.1371/journal.pgen.1003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs KM. In vitro methods for studying vascularization of the murine allantois and allantoic union with the chorion. Methods Mol Med. 2006;121:241–272. doi: 10.1385/1-59259-983-4:239. [DOI] [PubMed] [Google Scholar]
- Downs KM. Systematic localization of Oct-3/4 to the gastrulating mouse conceptus suggests manifold roles in mammalian development. Developmental dynamics: an official publication of the American Association of Anatomists. 2008;237:464–475. doi: 10.1002/dvdy.21438. [DOI] [PubMed] [Google Scholar]
- Downs KM. The enigmatic primitive streak: prevailing notions and challenges concerning the body axis of mammals. Bioessays. 2009;31:892–902. doi: 10.1002/bies.200900038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- Downs KM, Gifford S, Blahnik M, Gardner RL. Vascularization in the murine allantois occurs by vasculogenesis without accompanying erythropoiesis. Development. 1998;125:4507–4520. doi: 10.1242/dev.125.22.4507. [DOI] [PubMed] [Google Scholar]
- Downs KM, Inman KE, Jin DX, Enders AC. The Allantoic Core Domain: new insights into development of the murine allantois and its relation to the primitive streak. Dev Dyn. 2009;238:532–553. doi: 10.1002/dvdy.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagna S. Role of Oct-4 during acquisition of developmental competence in mouse oocyte. Reprod BioMed Online. 2009;19:57–62. doi: 10.1016/s1472-6483(10)60284-2. [DOI] [PubMed] [Google Scholar]
- Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- Hara K, Kanai-Azuma M, Uemura M, Shitara H, Taya C, Yonekawa H, Kawakami H, Tsunekawa N, Kurohmaru M, Kanai Y. Evidence for crucial role of hindgut expansion in directing proper migration of primordial germ cells in mouse early embryogenesis. Dev Biol. 2009;330:427–439. doi: 10.1016/j.ydbio.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Hirota T, Ohta H, Shigeta M, Niwa H, Saitou M. Drug-inducible gene recombination by the Dppa3-MER Cre MER transgene in the developmental cycle of the germ cell lineage in mice. Biol Reprod. 2011;85:367–377. doi: 10.1095/biolreprod.110.090662. [DOI] [PubMed] [Google Scholar]
- Inman KE, Downs KM. Brachyury is required for elongation and vasculogenesis in the murine allantois. Development. 2006a;133:2947–2959. doi: 10.1242/dev.02454. [DOI] [PubMed] [Google Scholar]
- Inman KE, Downs KM. Localization of Brachyury (T) in embryonic and extraembryonic tissues during mouse gastrulation. Gene Expr Patterns. 2006b;6:783–793. doi: 10.1016/j.modgep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Izumi N, Era T, Akimaru H, Yasunaga M, Nishikawa S. Dissecting the molecular hierarchy for mesendoderm differentiation through a combination of embryonic stem cell culture and RNA interference. Stem Cells. 2007;25:1664–1674. doi: 10.1634/stemcells.2006-0681. [DOI] [PubMed] [Google Scholar]
- Jevtic P, Edens LJ, Vukovic LD, Levy DL. Sizing and shaping the nucleus: mechanisms and significance. Curr Opin Cell Biol. 2014;28:16–27. doi: 10.1016/j.ceb.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka H, Takakura N, Nishikawa S, Tsuchida K, Kodama H, Kunisada T, Risau W, Kita T, Nishikawa SI. Expressions of PDGF receptor alpha, c-Kit and Flk1 genes clustering in mouse chromosome 5 define distinct subsets of nascent mesodermal cells. Dev Growth Differ. 1997;39:729–740. doi: 10.1046/j.1440-169x.1997.t01-5-00009.x. [DOI] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Quinlan GA, Hadjantonakis AK, Nagy A, Tam PP. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development. 1999;126:4691–4701. doi: 10.1242/dev.126.21.4691. [DOI] [PubMed] [Google Scholar]
- Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ, Keller G. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell. 2008;15:509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Hage WJ. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp. 1994;182:68–84. doi: 10.1002/9780470514573.ch5. discussion 84–91. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- Leitch HG, Smith A. The mammalian germline as a pluripotency cycle. Development. 2013;140:2495–2501. doi: 10.1242/dev.091603. [DOI] [PubMed] [Google Scholar]
- Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH. Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes Dev. 2008;22:575–580. doi: 10.1101/gad.1606308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolas M, Valenzuela PD, Tjian R, Liu Z. Charting Brachyury-mediated developmental pathways during early mouse embryogenesis. Proc Natl Acad Sci U S A. 2014;111:4478–4483. doi: 10.1073/pnas.1402612111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- Mikedis MM, Downs KM. Collagen type IV and Perlecan exhibit dynamic localization in the Allantoic Core Domain, a putative stem cell niche in the murine allantois. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:3193–3204. doi: 10.1002/dvdy.22129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikedis MM, Downs KM. STELLA-positive subregions of the primitive streak contribute to posterior tissues of the mouse gastrula. Dev Biol. 2012;363:201–218. doi: 10.1016/j.ydbio.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikedis MM, Downs KM. Widespread but tissue-specific patterns of interferon-induced transmembrane protein 3 (IFITM3, FRAGILIS, MIL-1) in the mouse gastrula. Gene Expr Patterns. 2013 doi: 10.1016/j.gep.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikedis MM, Downs KM. Mouse primordial germ cells: a reappraisal. Int Rev Cell Mol Biol. 2014;309:1–57. doi: 10.1016/B978-0-12-800255-1.00001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikedis MM, Downs KM. PRDM1/BLIMP1 is Widely Distributed to the Nascent Fetal-Placental Interface in the Mouse Gastrula. Dev Dyn. 2017;246:50–71. doi: 10.1002/dvdy.24461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Shimabe M, Watanabe-Okochi N, Arai S, Yoshimi A, Shinohara A, Nishimoto N, Kataoka K, Sato T, Kumano K, Nannya Y, Ichikawa M, Imai Y, Kurokawa M. AML1/RUNX1 functions as a cytoplasmic attenuator of NF-kappaB signaling in the repression of myeloid tumors. Blood. 2011;118:6626–6637. doi: 10.1182/blood-2010-12-326710. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Nikolic A, Volarevic V, Armstrong L, Lako M, Stojkovic M. Primordial Germ Cells: Current Knowledge and Perspectives. Stem Cells Int. 2016;2016:1741072. doi: 10.1155/2016/1741072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JPC, Zahn D, Colledge WH, Carlton MBL, Nakano T, Surani MA. stella Is a Maternal Effect Gene Required for Normal Early Development in Mice. Current Biology. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Pereira LA, Wong MS, Lim SM, Sides A, Stanley EG, Elefanty AG. Brachyury and related Tbx proteins interact with the Mixl1 homeodomain protein and negatively regulate Mixl1 transcriptional activity. PLoS One. 2011;6:e28394. doi: 10.1371/journal.pone.0028394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LA, Wong MS, Mei Lim S, Stanley EG, Elefanty AG. The Mix family of homeobox genes--key regulators of mesendoderm formation during vertebrate development. Dev Biol. 2012;367:163–177. doi: 10.1016/j.ydbio.2012.04.033. [DOI] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Scholer HR. Octamania: the POU factors in murine development. Trends Genet. 1991;7:323–329. doi: 10.1016/0168-9525(91)90422-m. [DOI] [PubMed] [Google Scholar]
- Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. Embo J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallock J, Molyneaux K, Schaible K, Knudson CM, Wylie C. The pro-apoptotic gene Bax is required for the death of ectopic primordial germ cells during their migration in the mouse embryo. Development. 2003;130:6589–6597. doi: 10.1242/dev.00898. [DOI] [PubMed] [Google Scholar]
- Sterneckert J, Hoing S, Scholer HR. Concise review: Oct4 and more: the reprogramming expressway. Stem Cells. 2012;30:15–21. doi: 10.1002/stem.765. [DOI] [PubMed] [Google Scholar]
- Tada S, Era T, Furusawa C, Sakurai H, Nishikawa S, Kinoshita M, Nakao K, Chiba T, Nishikawa SI. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- Tam PP, Beddington RS. The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis. Development. 1987;99:109–126. doi: 10.1242/dev.99.1.109. [DOI] [PubMed] [Google Scholar]
- Tam PP, Tan SS. The somitogenetic potential of cells in the primitive streak and the tail bud of the organogenesis-stage mouse embryo. Development. 1992;115:703–715. doi: 10.1242/dev.115.3.703. [DOI] [PubMed] [Google Scholar]
- Weiss SW. Smooth muscle tumors of soft tissue. Adv Anat Pathol. 2002;9:351–359. doi: 10.1097/00125480-200211000-00004. [DOI] [PubMed] [Google Scholar]
- Wilson V, Beddington RSP. Cell fate and morphogenetic movement in the late mouse primitive streak. Mechanisms of Development. 1996;55:79–89. doi: 10.1016/0925-4773(95)00493-9. [DOI] [PubMed] [Google Scholar]
- Wolfe AD, Downs KM. Mixl1 localizes to putative axial stem cell reservoirs and their posterior descendants in the mouse embryo. Gene Expr Patterns. 2014;15:8–20. doi: 10.1016/j.gep.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongtrakoongate P, Jones M, Gokhale PJ, Andrews PW. STELLA facilitates differentiation of germ cell and endodermal lineages of human embryonic stem cells. PLoS One. 2013;8:e56893. doi: 10.1371/journal.pone.0056893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler BM, Sugiyama D, Chen M, Guo Y, Downs KM, Speck NA. The allantois and chorion, when isolated before circulation or chorio-allantoic fusion, have hematopoietic potential. Development. 2006;133:4183–4192. doi: 10.1242/dev.02596. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Merico V, Sacchi L, Bellone M, Brink TC, Stefanelli M, Redi CA, Bellazzi R, Adjaye J, Garagna S. Oct-4 regulates the expression of Stella and Foxj2 at the Nanog locus: implications for the developmental competence of mouse oocytes. Hum Reprod. 2009;24:2225–2237. doi: 10.1093/humrep/dep191. [DOI] [PubMed] [Google Scholar]


