Abstract
Whole-genome sequencing was performed for Lactobacillus parakefiri JCM 8573T to confirm its hitherto controversial taxonomic position. Here, we report its first reliable reference genome. Genome-wide metrics, such as average nucleotide identity and digital DNA-DNA hybridization, and phylogenomic analysis based on multiple genes supported its taxonomic status as a distinct species in the genus Lactobacillus. The availability of a reliable genome sequence will aid future investigations on the industrial applications of L. parakefiri in functional foods such as kefir grains.
Keywords: Lactobacillus parakefiri, taxonomy, lactic acid bacteria, whole-genome sequence
Type strains hold significant positions in bacterial nomenclature: the taxonomic affiliation of any other isolate is identified on the basis of comparison with the type strains. For example, an isolate showing a DNA-DNA hybridization (DDH) similarity of more than 70% to the type strain of a certain species is considered to belong to the same species. The DDH value of 70%, a classical threshold described nearly 30 years ago, still remains for the gold standard for species boundary, although it has been partly supplemented by the use of 16S rRNA gene sequence similarity [1]. However, recently, the use of genome-wide metrics, such as average nucleotide identity (ANI) and digital DDH (dDDH), has been proposed as a replacement for the laborious process of DDH. ANI is calculated from the mean identity of homologous regions between two genomes, and the cutoff value of 95% is widely acknowledged as the threshold to delineate two species [2, 3]. The dDDH value is also based on the sequence alignment between a given pair of genomes, as implemented by the Genome-to-Genome Distance Calculator (GGDC, http://ggdc.dsmz.de), which infers an in-silico analogue of DDH with confidence intervals [4]. These sequence-based methods are reproducible and scalable and thus may be applied in the detection and validation of taxonomic mislabeling in public sequence databases [5,6,7]. In our previous study, we used ANI to assess the taxonomic status of 718 publicly available genomes of the genus Lactobacillus, the largest group of lactic acid bacteria, with over 180 known species [8]. We found mislabeling of organism names in several genomes, possibly caused by taxonomic misidentification or human errors such as sample mix-ups during experimental procedures or data handling. Moreover, our results suggested that even genomes for type strains might be erroneous in several species, including L. parakefiri and L. homohiochii.
L. parakefiri is a heterofermentative lactic acid bacterium described by Takizawa et al. in 1994, with strain GCL 1731T (= JCM 8573T = DSM 10551T = ATCC 51648T) designated as its type strain [9]. It was originally isolated from kefir grains and, together with another kefir-isolated species, L. kefiri, belongs to the L. buchneri group [10, 11]. The complex compositions and mechanisms of kefir grain microbiota have been studied extensively because of the health benefits associated with kefir ingestion [12, 13]. Recently, conflicting views were reported by two research groups for L. parakefiri on the basis of genomic analyses, resulting in controversies about the taxonomic status of L. parakefiri. Zheng et al. suggested that L. parakefiri is a later heterotypic synonym of L. kefiri [14], whereas Sun et al. acknowledged it as the species with the largest genome in the genus Lactobacillus [15]. In both studies, the draft genome was reconstructed independently using the raw sequencing reads for DSM 10551T deposited under the accession numbers SRR1151226 and ERR433484, respectively, in the Sequence Read Archive (SRA). The reconstructed genome from ERR433484 was also deposited in the International Nucleotide Sequence Database Collaboration (INSDC) under the accession number AZEN01. Our preliminary assessment suggested that the genome AZEN01 was contaminated with L. kefiri. Indeed, housekeeping genes that normally exist in single copies, such as pheS and rpoA, were found in duplicate, with one matching the sequence of L. parakefiri deposited in the public sequence database and the other matching that of L. kefiri. Therefore, the only publicly available genome for the type strain of L. parakefiri does not serve as a reference for taxonomic studies. In this study, we obtained a type strain of L. parakefiri, JCM 8573T, from the Japan Collection of Microorganisms (JCM) and reanalyzed its taxonomic status by conducting whole-genome sequencing.
Genomic DNA of JCM 8573T was extracted from cells cultured in de Man, Rogosa, and Sharpe broth (Difco) at the mid-logarithmic phase and then purified using Qiagen Genomic-tip 500/G gravity-flow, anion-exchange tips and a Qiagen Genomic DNA Buffer Set with lysozyme (Sigma) and proteinase K (Qiagen), according to the manufacturer’s instructions. Whole-genome sequencing using a 300-bp pair-end Illumina MiSeq system yielded 5,829,866 reads, which corresponds to approximately 700-fold coverage. De novo assembly was performed using the Platanus_B assembler (version 1.1.0) with the default settings after preprocessing the raw reads to remove low-quality bases and adapter sequences using Platanus_trim (version 1.0.7) [16]. The draft genome was annotated using the DDBJ Fast Annotation and Submission Tool (DFAST, http://dfast.nig.ac.jp). We obtained a draft genome that consisted of 161 contigs with an estimated genome size of 2,493,412 bp, which was comparable with those of other species in the L. buchneri subgroup. The genome completeness and contamination values were also calculated using CheckM (version 1.0.5) by inspecting for the presence/absence of single-copy gene markers conserved in the genus Lactobacillus [17]. For comparison, two data sets of raw reads, SRR1151226 and ERR433484, were downloaded from the SRA and assembled de novo in the same manner. The genome statistics are summarized in Table 1. Our data showed a high completeness value and a low contamination level. Contrarily, the data from SRR1151226 and ERR433484 showed high contamination values (14.20% and 99.35%, respectively) and high ANI values (97.9% and 91.7%, respectively) against the publicly available draft genome of L. kefiri DSM 20587T (accession number: AYYV01), supporting our previous findings of contamination in the sequencing data deposited in the public databases. The different genome sizes and contamination values implied different extents of contamination in the SRR1151226 and ERR433484 data. We identified one copy each of rpoA, pheS, and recA genes in the genome of JCM 8573T, whose nucleotide sequences exactly matched the ones reported for L. parakefiri LMG 15133T (accession numbers: AM087851, AM263510.1, and AJ621665, respectively), while showing only 92%, 84%, and 82% nucleotide identity with those of L. kefiri LMG 9480T (AM087840, AM263508, and AJ621650, respectively). This strongly suggests that the strain JCM 8573T belongs to a species distinct from L. kefiri. Notably, full-length 16S rRNA genes could not be identified in the draft genome. Bacterial genomes generally harbor multiple copies of rDNA regions, and such repetitive sequences make genome assembly difficult, often yielding collapsed or fragmented contigs for such regions [18]. We found that several contigs contained fragmented 16S rDNA gene sequences, which showed nearly 100% identity with the ones deposited in the public databases for L. parakefiri.
Table 1. Genome statistics of Lactobacillus parakefiri from different data sources.
| Data source /Strain | Total size/bp | Contigs | GC % | Completeness % | Contamination % | ANI against L. kefiri DSM 20587T |
|
|---|---|---|---|---|---|---|---|
| BDGB01 /JCM 8573T | 2,493,412 | 161 | 43.6 | 98.71 | 0.81 | 79.7% | Newly sequenced |
| SRR1151226 /DSM 10551T | 2,928,489 | 456 | 41.9 | 98.06 | 14.20 | 97.9% | Newly reconstructed |
| ERR433484 /DSM 10551T | 4,903,546 | 318 | 42.6 | 99.03 | 99.35 | 91.7% | Newly reconstructed |
| AZEN01 /DSM 10551T | 4,872,317 | 421 | 42.6 | 99.03 | 98.07 | 99.9% | INSDC data, reconstructed from ERR433484 |
The draft genome sequence of L. parakefiri JCM 8573 T was deposited in the INSDC under the accession numbers BDGB01000001–BDGB01000161. The raw sequencing reads were also deposited in the SRA under the accession number DRR064132.
Next, ANI and GGD calculation were performed between L. parakefiri JCM 8573T and each type strain of the 13 species in the L. buchneri subgroup. Genomes included in the analysis were obtained from the DFAST Archive of Genome Annotation (DAGA) [8]. ANI was calculated using the modified pyani script provided at https://github.com/widdowquinn/pyani, and the dDDH values were calculated using the GGDC web service (http://ggdc.dsmz.de). All the ANI values were less than the 95% cutoff line used to delineate two species [2], with the highest value being 84.92% against L. buchneri DSM 20057T. All the dDDH values were also well below 70%. Together, these results (summarized in Table 2) support the phylogenetic position of L. parakefiri as a distinct species.
Table 2. Average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values between L. parakefiri JCM 8573T and each type strain of closely related species.
| Strain | Data source | ANI | dDDH |
|---|---|---|---|
| L. buchneri DSM 20057T | GCA_001434735.1 | 84.92% | 29.4% |
| L. curieae CCTCC M 2011381T | GCA_000785105.1 | 72.24% | 20.1% |
| L. diolivorans DSM 14421T | GCA_001434255.1 | 74.87% | 21.5% |
| L. farraginis DSM 18382T | GCA_001435875.1 | 73.85% | 20.6% |
| L. hilgardii DSM 20176T | GCA_001434655.1 | 74.05% | 20.9% |
| L. kefiri JCM 5818T | GCA_001311745.1 | 79.71% | 23.1% |
| L. kisonensis DSM 19906T | GCA_001434135.1 | 75.16% | 20.9% |
| L. otakiensis DSM 19908T | GCA_001434145.1 | 80.62% | 23.6% |
| L. parabuchneri DSM 5707T | GCA_001435315.1 | 78.33% | 21.7% |
| L. parafarraginis DSM 18390T | GCA_001435895.1 | 74.30% | 21.9% |
| L. rapi DSM 19907T | GCA_001436255.1 | 75.12% | 19.8% |
| L. senioris DSM 24302T | GCA_001436555.1 | 71.30% | 19.3% |
| L. sunkii DSM 19904T | GCA_001435575.1 | 79.57% | 22.7% |
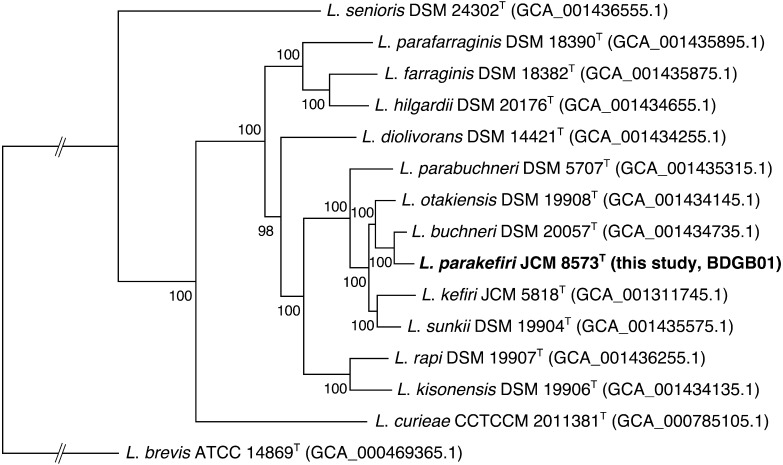
Figure 1 shows a phylogenetic tree constructed based on the conserved genes among the L. buchneri groups. The protein sequences of 14 type strains in the L. buchneri group and L. brevis DSM 20054T (as an outgroup) were subjected to analysis with the GET_HOMOLOGUES software (version 1.3) [19], and 465 single-copy orthologous genes shared by all the strains were selected for phylogenetic reconstruction. The amino acid sequences within each cluster were aligned using MUSCLE (version 3.8.31) [20]. Poorly aligned or divergent regions were trimmed using Gblocks [21], and conserved regions were then concatenated using FASconCAT-G [22]. A partitioned maximum likelihood analysis was performed to construct the phylogenetic tree with RAxML (version 8.1.22) [23] using the best-fit evolutionary models predicted for each alignment by ProtTest [24]. The number of bootstrap replicates was set at 1,000. All the species were well separated with high bootstrap support values. The most closely related species of L. parakefiri is L. buchneri, which is in agreement with the ANI and dDDH results as well as the phylogenetic tree constructed based on the 16S rRNA gene sequences [25], although the tree topology is slightly different.
Fig. 1.
Maximum likelihood phylogenetic tree of lactobacilli in the Lactobacillus buchneri subgroup constructed based on multiple alignments of protein sequences of 465 single-copy genes.
L. brevis is included an the outgroup. The numbers at the internal nodes represent the bootstrapping values (1,000 replications).
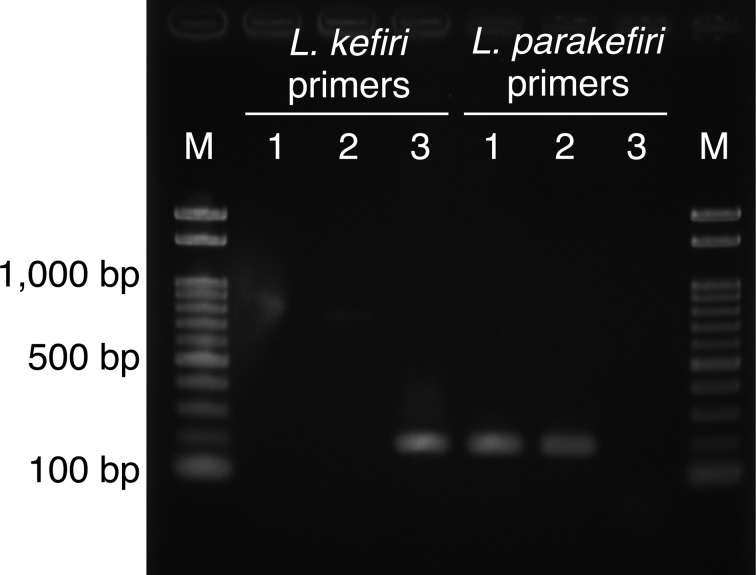
All these results confirmed the taxonomic position of L. parakefiri and indicated no contamination of L. kefiri in the type strain deposited in JCM. According to the cell history of JCM 8573T (http://www.straininfo.net/strains/54579) [9], it was directly deposited in the JCM from a subculture of the original isolate, GCL 1731T, and then transferred to other culture collections, including the German Collection of Microorganisms and Cell Cultures (DSMZ). Hence, the JCM strain is most unlikely to contain contamination. To examine whether the DSMZ strain contains contamination, we also obtained the cell line of L. parakefiri DSM 10551T. We randomly picked 16 colonies from cells cultured on de Man, Rogosa, and Sharpe broth and sequenced their 16S rRNA genes using the primers described previously [26]. All sequences were identical to that of L. parakefiri JCM 8573T (accession number: LC096211). In addition, PCR amplification of partial pheS genes was conducted by using two sets of primers, each of which were specific for L. parakefiri and L. kefiri. Briefly, genomic DNAs of all tested strains were prepared as described above. Primers for specific amplification of partial pheS genes of L. parakefiri or L. kefiri were designed: for L. parakefiri, LPKF, AGACCAGCGACTTAAAGGCA, and LPKR, CCAACGTTTCTCGATGATTC, which amplified a 186-bp fragment, and for L. kefiri, LKF, CAAACCAACGACTTGAAGAAG, and LKR, CAAGCTTTTGGCGTTGATCG, which amplified a 187-bp fragment. The reactions were performed with a GeneAmp PCR System 9700 (Thermo Fisher Scientific) using EmeraldAmp PCR Master Mix (TaKaRa) according to the manufacturer’s instructions. The reaction was initiated by denaturation for 30 sec at 98°C, followed by 40 cycles of 10 sec at 98°C, 30 sec at 60°C, and 30 sec at 72°C. As shown in Fig. 2, an amplified product was obtained from the DSMZ sample only when the L. parakefiri-specific primers were used, indicating no evidence of contamination with L. kefiri. Therefore, the reason for contamination in both the SRR1151226 and ERR433484 data, for which DSM 10551T was used in genome sequencing, remains unclear.
Fig. 2.
Amplification products obtained from a pheS PCR assay using the L. kefiri- and L. parakefiri-specific primers.
Lanes: M, Gene Ladder 100 (Wako Pure Chemical Industries, Ltd.); 1, L. parakefiri DSM 10551T; 2, L. parakefiri JCM 8573T; 3, L. kefiri JCM 5818T.
In this study, we report the first reliable genome for L. parakefiri. The genome contains 2,444 protein-coding sequences (CDSs). It has been reported that L. parakefiri produces gas from glucose but not from gluconate [9]. This unusual trait is likely due to the frameshift mutation that resulted in a premature stop codon in the coding region of gluconate permease, which is located adjacent to the gluconate kinase gene. In other heterofermentative species, including L. buchneri and L. kefiri, the two genes are thought to take roles in uptake and phosphorylation of gluconate, constituting an initial stage of its metabolic pathway. Other metabolic gene contents of L. parakefiri are similar to those of its most closest relative, L. buchneri. They share homologues of both lactaldehyde dehydrogenase and lactaldehyde reductase involved in the anaerobic lactate degradation to acetate and 1,2-propanediol. This characteristic property of L. buchneri is important for the dairy industry, because acetate enhances the aerobic stability of silage after exposure to air by preventing the growth of molds and yeasts [27, 28]. JCM 8573T also possesses two genes responsible for the conversion of serine to cysteine (serine O-acetyltransferase and cysteine synthase A), which differentiates it from L. buchneri and L. kefiri that do not possess these genes. Interestingly, as many as 10 of the predicted CDSs were shared by L. kefiranofaciens subsp. kefirgranum, a rather distantly related homofermentative lactobacillus, with a nucleotide identity of more than 99%. This number exceeds that for the more closely related L. kefiri (6 CDSs) detected under the same condition. L. kefiranofaciens subsp. kefirgranum was first isolated from kefir grains together with L. parakefiri by the same authors [9]. The high nucleotide identity may indicate the recent acquisition of these genes in similar biological environments. These include genes annotated as major facilitator superfamily transporter, nicotinate-nucleotide pyrophosphorylase, or hypothetical protein. It will be interesting to speculate how these genes have affected the coevolution of the two species and how they function in the microbiome of the kefir grain. The genome information we provide here may be used as a reference, especially for taxonomic studies, as well as for biotechnological applications of kefir-grain microbial flora.
Acknowledgments
Funding
This study was supported in part by MEXT KAKENHI (grant number 221S0002) and NIG-JOINT (grant number 2016-54).
We thank Ms. M. Ezure and Ms. H. Ikezawa for technical support and helpful discussions. Computational analysis was performed on the NIG supercomputer at the Research Organization of Information and Systems (ROIS).
References
- 1.Wayne LG, Brenner DJ. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37: 463–464. [Google Scholar]
- 2.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91. [DOI] [PubMed] [Google Scholar]
- 3.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Federhen S, Rosselló-Mora R, Klenk HP, Tindall BJ, Konstantinidis KT, Whitman WB, Brown D, Labeda D, Ussery D, Garrity GM, Rita R, Colwell NH, Graf J, Parte A, Yarza P, Goldberg B, Sichtig H, Karsch-Mizrachi I, Clark K, McVeigh R, Pruitt KD, Tatusova T, Falk R, Turner S, Madden T, Kitts P, Kimchi A, Klimke W, Agarwala R, DiCuccio M, Ostell J. 2016. Meeting report: GenBank microbial genomic taxonomy workshop (12–13 May, 2015). Stand Genomic Sci 11: 15. [Google Scholar]
- 6.Figueras MJ, Beaz-Hidalgo R, Hossain MJ, Liles MR. 2014. Taxonomic affiliation of new genomes should be verified using average nucleotide identity and multilocus phylogenetic analysis. Genome Announc 2: e00927–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, Kyrpides NC, Pati A. 2015. Microbial species delineation using whole genome sequences. Nucleic Acids Res 43: 6761–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanizawa Y, Fujisawa T, Kaminuma E, Nakamura Y, Arita M. 2016. DFAST and DAGA: web-based integrated genome annotation tools and resources. Biosci Microbiota Food Health 35: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takizawa S, Kojima S, Tamura S. 1994. Lactobacillus kefirgranum sp. nov. and Lactobacillus parakefir sp. nov., two new species from kefir grains. Int J Syst Bacteriol 44: 435–439. [Google Scholar]
- 10.Kandler O, Kunath P. 1983. Lactobacillus kefir sp.nov., a component of the microflora of Kefir. Syst Appl Microbiol 4: 286–294. [DOI] [PubMed] [Google Scholar]
- 11.Salvetti E, Torriani S, Felis GE. 2012. The Genus Lactobacillus: a taxonomic update. Probiotics Antimicrob Proteins 4: 217–226. [DOI] [PubMed] [Google Scholar]
- 12.Hamet MF, Londero A, Medrano M, Vercammen E, Van Hoorde K, Garrote GL, Huys G, Vandamme P, Abraham AG. 2013. Application of culture-dependent and culture-independent methods for the identification of Lactobacillus kefiranofaciens in microbial consortia present in kefir grains. Food Microbiol 36: 327–334. [DOI] [PubMed] [Google Scholar]
- 13.Vardjan T, Mohar Lorbeg P, Rogelj I, Čanžek Majhenič A. 2013. Characterization and stability of lactobacilli and yeast microbiota in kefir grains. J Dairy Sci 96: 2729–2736. [DOI] [PubMed] [Google Scholar]
- 14.Zheng J, Ruan L, Sun M, Gänzle M. 2015. A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl Environ Microbiol 81: 7233–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Z, Harris HMB, McCann A, Guo C, Argimón S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O’Sullivan O, Ritari J, Douillard FP, Paul Ross R, Yang R, Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H, O’Toole PW. 2015. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6: 8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, Kohara Y, Fujiyama A, Hayashi T, Itoh T. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 24: 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanizawa Y, Tohno M, Kaminuma E, Nakamura Y, Arita M. 2015. Complete genome sequence and analysis of Lactobacillus hokkaidonensis LOOC260T, a psychrotrophic lactic acid bacterium isolated from silage. BMC Genomics 16: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79: 7696–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577. [DOI] [PubMed] [Google Scholar]
- 22.Kück P, Longo GC. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool 11: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pot B, Felis GE, De Bruyne K, Tsakalidou E, Papadimitriou K, Leisner J, Vandamme P. 2014. The genus Lactobacillus, pp. 249–353. In Lactic Acid Bacteria: Biodiversity and Taxonomy First edition, Holzapfel WE, Wood BJB (eds.), John Wiley & Sons, Ltd., Hoboken. [Google Scholar]
- 26.Tohno M, Kitahara M, Irisawa T, Ohmori H, Masuda T, Ohkuma M, Tajima K. 2015. Lactobacillus mixtipabuli sp. nov. isolated from total mixed ration silage. Int J Syst Evol Microbiol 65: 1981–1985. [DOI] [PubMed] [Google Scholar]
- 27.Oude Elferink SJ, Krooneman J, Gottschal JC, Spoelstra SF, Faber F, Driehuis F. 2001. Anaerobic conversion of lactic acid to acetic acid and 1, 2-propanediol by Lactobacillus buchneri. Appl Environ Microbiol 67: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danner H, Holzer M, Mayrhuber E, Braun R. 2003. Acetic acid increases stability of silage under aerobic conditions. Appl Environ Microbiol 69: 562–567. [DOI] [PMC free article] [PubMed] [Google Scholar]