Abstract
Small regulatory RNAs have major roles in many regulatory circuits in Escherichia coli and other bacteria, including the transition from planktonic to biofilm growth. We tested Hfq-dependent sRNAs in E. coli for their ability, when overproduced, to inhibit or stimulate biofilm formation, in two different growth media. We identify two mutually exclusive pathways for biofilm formation. In LB, PgaA, encoding an adhesion export protein, played a critical role; biofilm was independent of the general stress factor RpoS or CsgD, regulator of curli and other biofilm genes. The PgaA-dependent pathway was stimulated upon overproduction of DsrA, via negative regulation of H-NS, or of GadY, likely by titration of CsrA. In YESCA (Yeast Extract Casamino acids) media, biofilm was dependent upon RpoS and CsgD, but independent of PgaA; RpoS appears to indirectly negatively regulate the PgaA-dependent pathway in YESCA medium. Deletions of most sRNAs had very little effect on biofilm, although deletion of hfq, encoding an RNA chaperone, was defective in both LB and YESCA. Deletion of ArcZ, a small RNA activator of RpoS, decreased biofilm in YESCA; only a portion of this defect could be bypassed by overproduction of RpoS. Overall, sRNAs highlight different pathways to biofilm formation.
Introduction
In response to various stress conditions and hostile environments, microorganisms can form communities of surface-adherent cells embedded in a matrix called biofilm. Biofilms are ubiquitous and are able to form on a variety of surfaces, contaminating food, water sources, and medical devices (Donlan & Costerton, 2002). A more complete understanding of the regulatory mechanisms involved in biofilm formation and how they change with growth conditions may lead to new strategies for successfully controlling its synthesis and treating biofilm-associated infections. Biofilm synthesis is a complex process involving a multitude of gene regulatory pathways. Flagella play a role in bringing cells to surfaces, and has been implicated in other stages of biofilm formation as well; FlhDC acts as the master regulator for flagellar synthesis (Chevance & Hughes, 2008). Proteins, including curli amyloid fibers, and exopolysaccharides such as PGA [Polyβ-1,6-GlcNAc] contribute to the interaction of cells with surfaces, with each other, and contribute to formation of a matrix around the bacteria. The transcriptional regulator for curli synthesis is CsgD (Liu et al., 2014). PGA synthesis is activated by the transcriptional regulator NhaR (Goller et al., 2006) and translation is repressed by CsrA. CsrA is titrated by CsrB and CsrC, as well as the Hfq-dependent sRNA McaS (Wang et al., 2005, Jorgensen et al., 2013).
Small RNAs [sRNAs] are known to regulate genes involved in these networks and in the regulation of biofilm formation, with multiple sRNAs regulating expression of FlhDC and CsgD [reviewed in (Mika & Hengge, 2013, Mika & Hengge, 2014, Chambers & Sauer, 2013, Van Puyvelde et al., 2013)]. There are at least 90 sRNAs detected in Escherichia coli (Raghavan et al., 2011), and a large number of these require the chaperone protein Hfq (De Lay et al., 2013). Here, we focus on Hfq-dependent sRNAs; these sRNAs regulate their mRNA targets at the post-transcriptional level by complementary base-pairing (Updegrove et al., 2016).
Hfq has been shown to be required for biofilm formation (Monteiro et al., 2012, Bak et al., 2015), and is considered a master regulator of biofilm under various environmental conditions in S.enterica serovar Typhimurium (Monteiro et al., 2012). In addition, many of the Hfq-dependent sRNAs play important roles during various stress responses and in regulation of motility (Zhao et al., 2013, Chambers & Sauer, 2013, Thomason et al., 2012, Van Puyvelde et al., 2013) (De Lay & Gottesman, 2012) and curli synthesis (Boehm & Vogel, 2012), all pathways that are known to be important in the biofilm development process. Stress conditions such as changes in growth temperatures or pH, peroxide, high metal concentration, and biocides increase biofilm formation in E. coli and other enteric bacteria (Goller & Romeo, 2008); (Goller et al., 2006, Zhang et al., 2007). Three sRNAs, DsrA, ArcZ, and RprA play important roles in the induction of the general stress response controlled by the sigma factor RpoS (Battesti et al., 2011), while at least two others, GadY and SdsR, are members of the RpoS regulon (Frohlich et al., 2012, Opdyke et al., 2004).
Many biofilm studies have been performed using rich (LB) medium; a smaller portion of these studies have used other media such as colonization factor antigen (CFA) medium and yeast extract-Casamino Acids (YESCA) to study specific regulated genes (Kikuchi et al., 2005, Jorgensen et al., 2013) (Hung et al., 2013, Bordeau & Felden, 2014); (Thomason et al., 2012). Here, we overproduced sRNAs to probe the pathways important for biofilm formation in Escherichia coli in both LB and YESCA media. In preliminary experiments, we found that RpoS was needed for biofilm in YESCA, but not in LB, suggesting that a comparison would be useful. Our work demonstrates the complexity of biofilm formation, with different sRNAs affecting this process in different media.
Results
Multiple sRNAs Regulate the Expression of Biofilm
Using a crystal violet assay for the development of biofilm by E. coli K12, we investigated whether overexpressed sRNAs can positively or negatively affect biofilm formation. sRNAs were overexpressed using a library of plasmids, each expressing a different Hfq-dependent sRNA under the control of an inducible Plac promoter (Mandin & Gottesman, 2010). All of the Hfq-dependent sRNAs known at the time when this work was initiated were in the library, as was the Hfq-independent CsrB RNA. Strains harboring the vector control or sRNA plasmid were grown at 37°C, the plasmids were induced with IPTG, and cultures were incubated in microtiter plates at 25 °C with fresh LB-ampicillin or YESCA-ampicillin for 24–48 hrs. Growth was measured using the OD600 and biofilm levels were determined at OD550 by staining with 0.1% crystal violet. Those sRNAs with two-fold or greater effects, normalized to the level of biofilm in the plac vector control, are shaded in Fig. 1A and B.
Figure 1. Effects of overexpressed sRNAs and sRNA deletions on Biofilm A, B: Effect of sRNAs overexpressed from a plasmid library.
Overnight cultures of WT MG1655 harboring pBR-plac (vector) or a derivative expressing one of 29 different sRNAs from a lac-based promoter were grown at 25°C in microtiter dishes and assayed for biofilm formation as described in Materials and Methods. Cells were grown for (A) 24 hours of incubation in LB-Amp or (B) 48 hours in YESCA-Amp. The fold change relative to the strain harboring the vector control was calculated. The results are an average of three independent experiments, with error bars representing standard deviation. The shaded grey bars indicate over-expressed sRNAs that increased or decreased biofilm formation two-fold or more.
C, D: Effect of sRNA deletions. Strains containing sRNA and hfq deletions were grown overnight in LB media and tested for biofilm formation after (C) 24 hours incubation in LB or (D) 48 hours in YESCA at 25°C. Biofilm was measured and the fold change relative to the wild-type MG1655 control (black filled bars), set to 1, was calculated. The results are an average of three independent experiments, with error bars representing standard deviation. The grey bars represent a significant decrease in biofilm formation (of two-fold or greater). Strain names are listed in Table S1.
Plasmids expressing sRNAs DsrA, GadY, and MicF increased biofilm formation in the library screen. Strains overexpressing MicF increased biofilm formation (five-fold) only in YESCA (Fig. 1B). Multicopy GadY increased biofilm 7-fold, only in LB (Fig. 1A). Overexpression of DsrA led to a 5-fold increase in the levels of biofilm formation using both types of media (Fig. 1A, B). Overall, overproduction of many (19/28) Hfq-dependent sRNAs had significant effects on biofilm formation in at least one of the two different types of media.
In the initial screen, strains overexpressing RydC, SdsR, and MicC reduced biofilm formation in both LB and YESCA media (Fig. 1A, B). A larger set of sRNAs reduced biofilm 2-fold or more in YESCA medium but not in LB, while RybD reduced biofilm in LB but not YESCA (Fig. 1, compare A and B).
McaS has previously been shown to increase motility and flagella synthesis via activation of flhDC (Thomason et al., 2012). It directly represses translation of csgD but activates pgaA, causing increased biofilm formation in LB, CFA (colonization factor antigen), and YESCA media (Thomason et al., 2012), (Jorgensen et al., 2013). For reasons not currently understood, we did not observe this activation in either LB or YESCA; in fact, McaS repressed biofilm formation in YESCA (Fig. 1). Biofilm formation is, in part, activated via McaS interaction with CsrA, and the titration mechanism allows activation of pgaA, leading to biofilm production (Jorgensen et al., 2013). Consistent with a negative role for CsrA in biofilm formation, overproduction of CsrB, which also titrates CsrA (Wang et al., 2005, Weilbacher et al., 2003), significantly increased biofilm formation in both LB and YESCA media (Fig. 1A, B). CsrB is not an Hfq-dependent sRNA and was not a focus of this work. Because the McaS plasmid did stimulate biofilm in preliminary experiments (not shown) and stimulated pgaA translation in a recent experiment (see below), it seems most likely that the plasmid used in Fig. 1 had acquired a mutation. We cannot rule out loss or mutation of other plasmids in this assay; thus the list of sRNAs affecting biofilm may be an underestimate.
Deletions of each of eleven of these Hfq-dependent sRNAs were tested for biofilm formation; also included was a deletion of hfq and a deletion of csrB, encoding a negative regulator of CsrA (Fig. 1C, D). The deleted sRNAs included all of those that activated biofilm when overproduced (DsrA, GadY and MicF), as well as some of those that showed strong negative effects when overproduced (ChiX, SdsR, RydC, ArcZ, DicF, and MicC). Finally, McaS, previously implicated in biofilm formation, and RprA known, with ArcZ and DsrA, to activate RpoS translation, were also included. In both LB and YESCA, the deletion of hfq had the greatest effect, essentially abolishing biofilm formation (Fig. 1C, D), consistent with a previous study carried out in LB (Bak et al., 2015). Most of the other deletions had only modest effects, although deletion of mcaS reduced biofilm almost two-fold in LB, and deletion of arcZ, dsrA or gadY all reduced biofilm at least two-fold in YESCA. These differences between LB and YESCA reinforce the idea that different pathways are likely used in the different media.
Although a number of sRNAs had significantly decreased biofilm when overproduced (Fig. 1A, B), none of the deletions resulted in more biofilm under these conditions (RydC, ChiX and SdsR for instance, Fig. 1C, D). One explanation for this is that the effects on biofilm require significant overproduction, for instance to titrate Hfq. We note that ChiX is known to titrate Hfq (Ellis et al., 2015, Moon & Gottesman, 2011, Santiago-Frangos et al., 2016). If Hfq titration is the basis for the decrease in biofilm, we would not expect the chromosomal level of these sRNAs to significantly perturb Hfq availability. Alternatively, the target perturbed by overproduction of these sRNAs may not be rate-limiting for biofilm formation. For instance, if gene X is required for biofilm formation, but more X does not lead to more biofilm, overproduction of an sRNA that negatively regulates X may eliminate biofilm, but deletion of the gene for the same sRNA may not show a difference in biofilm levels. ArcZ, which negatively regulated biofilm formation when overproduced in YESCA (Fig. 1B), led to reduced biofilm when deleted, again only in YESCA medium (Fig. 1D). With the exception of ArcZ, discussed further below, the sRNAs that negatively regulated biofilm only when overproduced were not further investigated.
Here, we focused on four of these sRNAs, the three activators (DsrA, GadY, MicF) and one multicopy repressor, ArcZ. We start by focusing our attention on DsrA, which was a strong activator under both LB and YESCA conditions (Fig. 1).
DsrA Acts through H-NS to Promote Biofilm Formation
In E. coli, DsrA is a three stem-loop 87 nucleotide long sRNA that is known to regulate the translation of two global transcriptional regulators, H-NS and RpoS, by RNA-RNA interactions (Majdalani et al., 1998) (Gottesman, 2004) (Lease & Belfort, 2000). DsrA stimulates translation of RpoS, the master regulator of the general stress response; the region of interaction with the rpoS mRNA is in the first stem loop of DsrA (bold type in Fig. 2A) (Majdalani et al., 1998). Base pairing occurs upstream of the rpoS translational start site (nt −97 to −125 relative to the ATG; Fig. 2C). The global transcriptional repressor protein, H-NS is negatively regulated by DsrA, using the second stem loop (circled in Fig. 2A), with base pairing occurring just beyond the ATG in the coding region of hns (Fig. 2B) (Lease et al., 1998, Lease & Belfort, 2000).
Figure 2. Interaction of DsrA with downstream targets hns and rpoS.
(A) DsrA sequences necessary for interaction with hns are circled and for interaction with rpoS are shown in bold.
(B) DsrA and hns base pairing shows the DsrA pairing region with hns (Lease et al., 1998). The nucleotides in DsrA mutated to create DsrA*h and in hns to create hns*, designed to study the role of this interaction in biofilm formation, are shown above or below the sequences.
(C) Base pairing of DsrA upstream of the rpoS translational start is required to positively regulate rpoS. The nucleotides mutated in DsrA to create DsrA*r are shown above the line.
(D) Specificity of regulation in DsrA mutants.
Beta-galactosidase assays of hns and rpoS translational fusions (HL1061 and PM1409, see Table S1) were used to confirm the specificity of pDsrA mutations. pDsrA*r should interfere with pairing to rpoS and pDsrA*h should disrupt pairing with hns. WT pDsrA and each of the mutant plasmids were introduced into the fusion strains. Overnight cultures of each strain grown in LB ampicillin media were diluted 200-fold into fresh LB medium containing IPTG (100μM), ampicillin (100μg/ml), and arabinose (0.001%), and incubated at 37°C for 6 hours. The activity of the hns and rpoS fusions were determined using the Miller assay (Miller, 1992). The error bars are a representation of the standard deviation of three independent experiments.
Because different regions of DsrA are involved in regulation of rpoS and hns, it is possible to distinguish which targets of DsrA are important for a given phenotype, using appropriate mutations (Fig. 2). DsrA*h (Fig. 2B) should disrupt pairing with hns, while DsrA*r disrupts pairing with rpoS (Fig. 2C). The specificity of these mutants was tested on appropriate translational reporter fusions (Fig. 2D). DsrA over-expression significantly reduced hns-lacZ expression levels, while the DsrA specificity mutant pDsrA*h lost the ability to repress (Fig. 2D, compare second and third bars for hns-lacZ fusion). The DsrA specificity mutant pDsrA*r, designed to prevent base pairing with rpoS, continued to strongly repress the hns-lacZ fusion (Fig. 2D, fourth bar). As expected, the effects of these mutations were reversed for regulation of the rpoS-lacZ fusion, as expected, with DsrA*h still able to activate this fusion while DsrA*r was unable to (Fig. 2D, rpoS-lacZ fusion). These results confirm that DsrA regulation of hns and rpoS is independent, as previously seen (Majdalani et al., 1998), and provide tools for asking which of these effects contribute to DsrA regulation of biofilm.
Overexpression of wild-type DsrA transformed in the MG1655 background increased biofilm formation by 5-fold in LB (Fig. 3). A plasmid overexpressing DsrA*h did not stimulate biofilm formation, while expression of DsrA*r had an effect similar to expression of wild-type DsrA (Fig. 3). This result suggests that negative regulation of H-NS by DsrA may be sufficient for increased biofilm, and that stimulation of RpoS by DsrA is not necessary.
Figure 3. DsrA regulation of biofilm formation through hns pairing.
DsrA multi-copy plasmids were used to transform MG1655 and the chromosomal hns* derivative (SC144) to test the specificity of pDsrA*h on biofilm formation. Overnight cultures were diluted to an OD600 of ~0.05 in fresh LB containing ampicillin (100μg/ml) and IPTG (100μM). These strains were assayed for biofilm as measured by crystal violet staining after 24 hours incubation at 25°C. Biofilm values for each strain were normalized to the plac vector control. Error bars are a representation of the standard deviation of three independent experiments.
To confirm that it is DsrA pairing with hns that leads to increased biofilm, rather than another target of DsrA that pairs with this same region, we constructed a compensatory mutant, hns*, first tested it in the hns-lacZ fusion (Fig. 4) and then introduced this mutation into the chromosomal copy of hns. As seen in Fig. 4, right panel, wild-type DsrA and DsrA*r are unable to regulate hns*-lacZ; DsrA*h, which can pair with hns*, can repress it. Note that the basal level of expression of the hns*-lac fusion is reduced significantly, suggesting that this mutation (in the second to fourth codons of hns, Fig. 2B) reduces translation.
Figure 4. Multi-copy DsrA post-transcriptional regulation of hns by direct base- pairing.
Mutations in the hns translational fusion (hns*-lacZ, SC71) were tested for regulation by pDsrA, pDsrA*h, and pDsrA*r. Overnight cultures of each strain were diluted 200-fold into fresh LB medium containing IPTG (100μM), ampicillin (100μg/ml), arabinose (0.001%), and cultures were incubated at 37°C for 6 hours. The activity of the hns and hns* fusions were determined as described using the Miller assay (Miller, 1992). Error bars are a representation of the standard deviation of three independent experiments.
If DsrA is working only through its repression of hns, the plasmid overexpressing DsrA*h should stimulate biofilm formation only when it is able to pair with hns (in the hns* strain). This was what was observed (Fig. 3, right side); DsrA*h increased biofilm formation in the hns* strain relative to the plac vector control, while DsrA+ and DsrA*r (unable to regulate the hns* strain) did not. This experiment confirms that the increase in biofilm formation is specifically due to DsrA negative regulation of hns and not another target. The reduced HNS in the hns* strain did not have a significant effect on biofilm levels (compare level of biofilm in plac (vector control) lanes in MG1655 and hns* strain). This suggests that chromosomally-encoded DsrA regulation of hns is not important for the biofilm levels observed in wild-type cells, since the hns* allele should be resistant to endogenous DsrA. The lack of an effect of endogenous DsrA on biofilm in LB was confirmed by deletion of dsrA (Fig. 1C). In addition, the lower level of expression of the hns*-lacZ fusion suggests that reducing the levels of hns translation two-fold is not sufficient to increase biofilm.
A prediction of these results is that deletion of hns should also increase biofilm. This was confirmed for growth in LB (Fig. 5). Under these conditions, deletion of hns mimicked the effect of DsrA overproduction, consistent with H-NS negatively regulating biofilm. An rpoS deletion mutant was also included in the biofilm assay and no changes in biofilm were observed in LB (Fig. 5). Therefore, under our LB biofilm assay conditions, neither more RpoS (activation by DsrA) nor absence of RpoS affects biofilm levels. We return to the role of RpoS in YESCA medium later in the manuscript.
Figure 5. hns chromosomal deletion mutant increases biofilm formation.
MG1655, an rpoS deletion mutant (SC124), an hns deletion mutant (SC110), and the hns* mutant (SC144) were grown overnight and tested for biofilm formation in LB after 24 hours incubation at 25°C. Biofilm formation was measured as OD570/OD600 expressed as the fold change relative to the MG1655 control. The results are an average of three independent experiments, with error bars representing standard deviation.
DsrA and H-NS Targets and Biofilm
H-NS is known to repress multiple genes in E. coli (Bertin et al., 1994, Soutourina et al., 1999) (Chib & Mahadevan, 2012, Donato et al., 1997). We carried out epistasis experiments to try to identify possible critical H-NS targets for biofilm formation. A selected set of genes, each important for specific pathways implicated in biofilm formation, were deleted and tested for biofilm formation in the presence or absence of H-NS or in the cells overexpressing DsrA (Fig. 6). In LB, deletion of flhD, the regulator of flagellar synthesis, reduced the basal level of biofilm in hns+ cells (Fig. 6A, compare lane 4 to lane 1), while deletion of rpoS, csgD (regulator of curli synthesis and other genes associated with biofilm formation) or pgaA, encoding the first gene in the pgaABCD operon for synthesis and export of PGA adhesin, had no effect (Fig. 6A, lanes 2, 3, and 5). However, the increased level of biofilm in either Δhns or pDsrA cells was reduced to or below the level of WT cells by deletion of pgaA (Fig. 6A, compare lane 7 to lane 6 and lane 12 to lane 11). These results show that the increased biofilm levels observed when H-NS is either deleted or down-regulated by DsrA is dependent upon PGA.
Figure 6. H-NS regulation of biofilm.
Deletion mutants of known biofilm targets, either alone (SC135: ΔpgaA::kan; SC137: ΔcsgD::cat; SC138: ΔflhD::kan; SC124: ΔrpoS::tet), in Δhns strains (SC134: ΔpgaA Δhns::kan; SC133: ΔcsgD::cat hns::kan; SC136: ΔflhD hns::kan; SC122: ΔrpoS::tet Δhns::kan) or in the presence of an induced multicopy DsrA plasmid were assayed for biofilm formation. (A) Overnight cultures were grown and diluted in LB or, for cultures containing plasmids, were diluted to an OD600 concentration of ~0.05 in fresh LB containing ampicillin (100μg/ml) and IPTG (100μM), incubated in microtiter wells for 24 hours at 25°C or (B) in YESCA medium with IPTG and ampicillin as appropriate for 48 hours at 25°C, and then measured for biofilm by crystal violet staining as described in Materials and Methods. Biofilm formation was determined as OD570/OD600 and the error bars are a representation of the standard deviation of triplicate experiments.
In Actinobacillus pleuropneumoniae, H-NS represses the pgaA operon and thus regulates biofilm formation (Bosse et al., 2010). The regulation of pgaA by H-NS was examined in E. coli, initially using a fusion under the control of the pgaA promoters, that measures both transcription and translation of pgaA. In this strain, deletion of hns increased expression (Fig. S1C, compare lane 4 to lane 1), and also increased activity on Lactose MacConkey indicator plates (Fig. S1B, compare plac quadrant to plac in Fig. S1A). However, multicopy DsrA had a less dramatic effect, although it showed some induction, consistent with down-regulation of H-NS (Fig. S1A, S1C). DsrA specificity mutants tested on Lactose MacConkey indicator plates (Fig. S1A) were consistent with DsrA acting via repression of H-NS synthesis; pDsrA*h had no effect, and pDsrA*r was similar to wild-type DsrA (Fig. S1A). Therefore, pgaA is a direct or indirect target of H-NS, but it seems likely from these results that additional H-NS targets contribute to the higher biofilm formation in Δhns cells. We suggest that in hns+ cells, redundant pathways, possibly including PGA, contribute to the basal biofilm formation so that deletion of pgaA had little or no effect. This is consistent with reports that wild-type MG1655 does not produce much PGA (Itoh et al., 2008).
As noted above, RpoS was not needed for the basal biofilm levels in LB (Fig. 5, Fig. 6). If anything, lack of RpoS appeared to modestly promote biofilm levels in cells deleted for hns or expressing multicopy DsrA (Fig. 6A, compare lane 10 to lane 6 and lane 15 to lane 11). This is consistent with work suggesting that RpoS can repress biofilm formation in cells assayed in LB media (Ferrieres et al., 2009).
In two cases, the deletion of hns and overproduction of DsrA gave somewhat different results; we suggest that these reflect differences in the timing and contribution of motility to biofilm development. First, deletion of flhD reduced biofilm in the Δhns strain, but not in the strain overproducing DsrA. The ability of DsrA to overcome the deletion of flhD was still dependent on the ability to pair with hns (Fig. S2A). Second, multicopy DsrA, in strains deleted for pgaA, led to a total loss of the basal level of biofilm; this reduction of the basal level was not seen in cells deleted for hns (Fig. 6, compare lanes 2, 7, and 12). This result suggested that DsrA either negatively affects a target important for the basal level of biofilm or positively regulates a negative regulator of biofilm formation. DsrA is known to stimulate RpoS synthesis, so we asked if RpoS was (indirectly) interfering with biofilm formation in the ΔpgaA cells. We examined the ability of DsrA to repress biofilm in cells deleted for rpoS, pgaA, or a double mutant of pgaA and rpoS (Fig. S2B). DsrA suppressed biofilm in the ΔpgaA cells, as seen in Fig. 6; deletion of rpoS did not relieve this repression.
If biofilm levels were measured after 48 hours (rather than 24 hours) in LB, the dependence on flhD disappeared, and the repression by DsrA in cells deleted for pgaA also was lost (Fig. S2C). Note that the basal level of biofilm was somewhat lower at 48 hrs in LB (compare Fig. 6, lane 1 to Fig. S2C, lane 1). Deletion of flhD, encoding a master regulator for motility, decreased biofilm at 24 hours (Fig. 6, compare lane 4 to lane 1). However, after 48 hours, not only was the dependence on flhDC lost, but the basal level of biofilm was increased (Fig. S2C, compare lane 9 to lane 1). The low level of biofilm at 48 hr was no longer repressed by DsrA in the absence of pgaA (Fig. S2C, compare lane 20 to lane 17). This suggests that DsrA negatively regulates a process needed early but not late in biofilm formation, presumably slowing the process. One candidate would be motility. Therefore, it is possible that DsrA overproduction negatively regulates motility or something else under FlhDC control; this process would be critical, as is FlhDC, at the early stages of biofilm formation, but would, if anything, be detrimental at later times. We would also suggest that the ability of DsrA to stimulate PGA synthesis changes the pathway or kinetics of biofilm formation sufficiently to overcome the dependence on FlhDC. Differences in the kinetics of biofilm formation and thus the dependence on motility genes in cells in which H-NS is totally lost (Δhns), compared to that in cells in which H-NS is down-regulated by DsrA may explain why cells deleted for hns had different behavior (Fig. 6).
Overall, we conclude that, in LB, loss of H-NS, via deletion or via expression of DsrA, increases biofilm in a process that is fully dependent upon pgaA. This pathway of biofilm production is independent of RpoS and CsgD, and conditionally dependent upon FlhDC.
Biofilm Production in YESCA
Some sRNAs had effects on biofilm in LB but not in YESCA (GadY, in particular, stimulated only in LB) or in YESCA but not LB (MicF stimulated in YESCA but not in LB) (Fig. 1A, B). In addition, in previous studies, we had noted a requirement for RpoS for biofilm formation in YESCA (Parker & Gottesman, 2016), suggesting that different pathways might be responsible for biofilm formation in this media. Therefore, parallel experiments were done in YESCA.
Most strikingly, and consistent with our previous observations, deletion of rpoS or csgD, neither of which were required in LB, significantly reduced basal levels of biofilm formation in the MG1655 strain grown in YESCA (Fig. 6B, lanes 3 and 5). The defect in csgD cells could not be overcome by deletion of hns, or by overproduction of DsrA (Fig. 6B, lanes 8 and 13). In general, deletion of hns or overproduction of DsrA had much less effect in increasing biofilm in YESCA than in LB (compare Fig. 6B to Fig. 6A, lane 11 vs. lane 1). The increase in biofilm in cells deleted for hns was, as in LB, dependent upon pgaA (compare Fig. 6B, lanes 6 and 7). Deletion of flhD reduced biofilm, but not as much as in LB (compare lanes 4 to 1 in Fig. 6A and 6B). Interestingly, the deletion of hns fully suppressed the deletion of rpoS in YESCA, consistent with H-NS repression of important downstream targets of RpoS, and with a possible inhibitory effect of RpoS in the absence of H-NS.
Thus, when E. coli K12 grows in YESCA, biofilm formation switches from the RpoS-independent, PGA-dependent pathway seen in LB overexpressing DsrA to an RpoS-dependent, CsgD-dependent pathway.
Three sRNAs, DsrA, ArcZ, and RprA, are known to each activate translation of RpoS [reviewed in (Battesti et al., 2011)]. Consistent with the dependence of biofilm formation in YESCA on RpoS (Fig. 6B and 7B, lane 5 compared to lane 1), we note that deletion of arcZ or dsrA reduced biofilm formation specifically in YESCA medium (Fig. 1D). ArcZ negatively regulates multiple targets, in addition to activating RpoS (Mandin & Gottesman, 2010, Monteiro et al., 2012, Papenfort et al., 2009). Overproduced RpoS was fully able to complement the deletion of rpoS for biofilm formation (Fig. S3, compare rpoS::kan pRpoS, lane 10 to lane 6), but only partially suppressed deletion of arcZ (compare arcZ::zeo/pRpoS, lane 11 to lanes 5 and 9). While these other ArcZ targets important for biofilm development have not been identified, ArcZ has also been shown to be essential for curli-dependent biofilm formation in Salmonella, and this effect is not entirely due to effects on RpoS (Monteiro et al., 2012). ArcZ also negatively regulates the master regulator of the flagellar genes (De Lay & Gottesman, 2012). Therefore, ArcZ regulates multiple targets that contribute to biofilm formation in YESCA medium, one of which is likely RpoS.
Figure 7. Alternative biofilm pathways regulated by sRNAs.
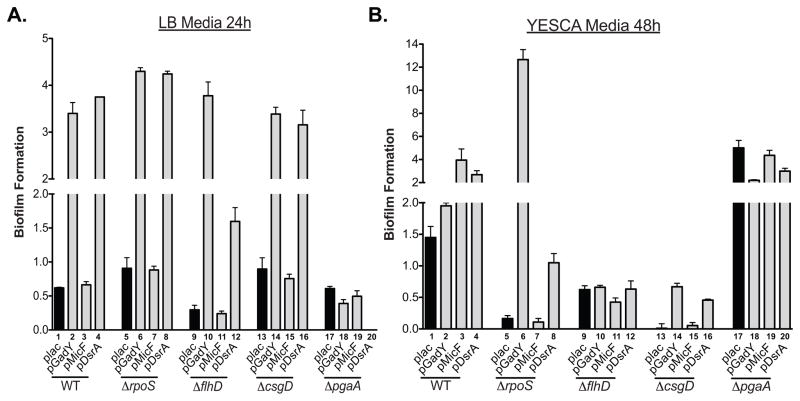
Single deletion mutants of known biofilm targets (WT: MG1655; ΔrpoS: SC124; ΔflhD: SC138; ΔcsgD: SC137; ΔpgaA: SC135) were transformed with the vector control plac and multicopy plasmids pGadY, pMicF, and pDsrA. Overnight cultures were diluted to an OD600 ~0.05 in fresh LB (A) or YESCA (B) containing ampicillin (100μg/ml) and IPTG (100μM). Biofilm assays were performed after (A) 24 hours incubation in LB or (B) 48 hours in YESCA media at 25°C. Biofilm formation was determined as OD570/OD600 and the error bars are a representation of the standard deviation of triplicate experiments.
Small RNAs Regulate Multiple Biofilm Targets
As noted above, different pathways are important for biofilm formation in LB and YESCA (Fig. 6), and different sRNAs stimulate biofilm in LB and YESCA (Fig. 1A, B). We compared the role of DsrA, discussed above, to two other sRNAs that were observed to increase biofilm, GadY, and MicF, in strains deleted for the various pathways (Fig. 7).
Consistent with the results in Fig. 6, DsrA stimulated biofilm formation in LB in a manner that was fully dependent upon pgaA, but was unaffected by csgD or rpoS (Fig. 7A, compare lanes 4, 8 and 16). Note that, as discussed for Fig. 6, DsrA led to loss of the basal level of biofilm in cells deleted for pgaA (Fig. 7A, compare lane 20 to lane 17).
GadY, like DsrA, stimulated biofilm formation in LB, dependent upon pgaA (Fig. 7A, lane 18 compared to lane 2), but was independent of rpoS and csgD (Fig. 7A, compare lane 6 and 14 to lane 2). Unlike DsrA, GadY did not repress biofilm formation in the absence of pgaA (Fig. 7A, compare the effect of DsrA, lane 20 to the effect of GadY, lane 18). GadY also activates expression of a pgaA-lacZ fusion, more than DsrA (Fig. S1A, C). However, GadY does not stimulate pgaA via repression of H-NS, since the stimulation by the GadY plasmid was seen even in cells deleted for hns (Fig. S1B and S1C).
We further examined how GadY might act on pgaA expression. McaS, an Hfq-dependent sRNA that regulates some targets by direct pairing, was found to positively regulate pgaA by titrating the translational repressor CsrA (Jorgensen et al., 2013). This induction was shown to be dependent on GGA sites within McaS and on CsrA binding sites close to the ribosome binding site within the pgaA leader (Jorgensen et al., 2013). We asked if GadY might act in a similar fashion, based on the presence of two GGA sites in this sRNA (underlined in Fig. 8A). First, GadY, activated translation of pgaA in two fusions, both of them driven by a pBAD promoter, and both of them activated by McaS as well (Fig. 8B). One of these fusions contains the full 234 nt leader for pgaA; the other contains only 30 nt of the leader. The fusion with the 30 nt leader has been shown to still be subject to CsrA repression (Jorgensen et al., 2013). Therefore, GadY activates translation, and at least some of the critical sites are close to the ribosome binding site. Second, mutations were made in each or both of the two GGA sites, changing the A to T. Both mutants were still able to repress a target that pairs with GadY, although with somewhat lower efficiency (Fig. S1D). Mutation of A34 to T reduced activation of a transcriptional and translational fusion (Fig. 8C) and of a translational fusion under control of the pBAD promoter (Fig. 8D). Inactivation of csrA (right hand panel in Fig. 8D) increased the expression of the fusion, as expected; in these cells, GadY stimulation was reduced from the 2.5× seen in the csrA+ host to less than fold; the A34T mutant was not significantly less effective (Fig. 8D, compare left, csrA+, and right, csrA−, panels). We note that this csrA mutant is not a complete null, and that both CsrB and McaS have been shown to show some, but reduced activation in this csrA- strain (Jorgensen et al., 2013). Therefore, we suggest that GadY, like McaS, activates biofilm formation via increased pgaA expression, and that this is due at least in part to the ability of a GGA site to titrate CsrA.
Figure 8. GadY activation of pgaA translation.
A. Sequence of GadY with mutations of GGA sequences (underlined) tested shown below the line. The terminator stem is overlined, as is the short stem around the first GGA sequence. B. Strains carrying pgaA-lacZ translational fusions, each driven by a pBAD promoter, with either the full leader of 234 nt (GS0568; left panel) or truncated to a 30 nt leader (GS0641; right panel) were transformed with a vector, pGadY or pMcaS plasmids, grown to an OD600 ~ 0.2 in LB/amp at 37C before addition of 0.1M IPTG and 0.2% Ara; assayed in triplicate. Note the difference in the y axis for the two fusions. C. Lactose MacConkey indicator plate with NM750, carrying a pgaA-lacZ transcriptional and translational fusion, transformed with vector, GadY, and mutant derivatives of GadY. D. Isogenic csrA+ (GS0644) and csrA::kan (GS0645) strains, each carrying the pBAD-pgaA-lacZ translational fusion with a full leader were transformed with vector control, pGadY, and GadY mutant plasmids, grown and assayed as in Fig. 8B. Note the different y-axis scales for these two strains.
In YESCA medium, overexpression of GadY did not significantly stimulate biofilm formation (Fig. 7B, compare lane 2 to lane 1), consistent with it acting on the pgaA-dependent pathway that is not used in YESCA. Strikingly, however, GadY overexpression fully suppressed the need for RpoS, giving biofilm levels that were significantly higher than in the ΔrpoS host (Fig. 7B, compare lane 5 to lane 6). It seemed likely that biofilm in this case, in the absence of RpoS, was again pgaA-dependent. This was tested by measuring the ability of GadY to stimulate biofilm formation in YESCA in cells deleted for both rpoS and pgaA (Fig. S4). Basal levels of biofilm were again decreased in the rpoS::tet mutant, and were similarly low in the pgaA::kan rpoS::tet double mutant. However, while multicopy DsrA or GadY could stimulate biofilm in the rpoS mutant, they did not in the pgaA rpoS double mutant. These results suggest that normally in YESCA, biofilm is independent of PgaA and dependent upon RpoS, and that conditions that might be expected to increase the PgaA-dependent pathway are somehow blocked by RpoS. Thus, there is no additive effect, at least in our crystal violet staining assay, of expressing GadY in an rpoS+ csgD+ strain, and in some experiments in which the PGA pathway should be activated in YESCA, for instance in the absence of H-NS, there is more biofilm in the absence of RpoS than with it (Fig. 6B, compare lane 6 to 10). This suggests that one or more products of the RpoS regulon are responsible for blocking the PGA pathway. A similar, but less striking, stimulation of biofilm is seen in YESCA in cells deleted for csgD (Fig. 7B, compare lanes 14 and 16 to lane 13), but not for cells deleted for flhD (Fig. 7B, compare lanes 10 and 12 to lane 9).
MicF stimulated biofilm formation only in a wild-type strain in YESCA (Fig. 1A, B, Fig. 7, compare lane 3 to lane 1 in A and B), suggesting it is working via the CsgD/RpoS pathway. It behaved similarly to the vector control in all of the epistasis experiments in Fig. 7. Therefore, unlike DsrA or GadY, MicF does not change the pathway for biofilm formation but instead may increase the efficiency with which these cells use the RpoS and CsgD pathway for biofilm formation. MicF represses the major outer membrane porin OmpF, as well as the Lrp and CpxR regulators in E. coli (Mizuno et al., 1984, Delihas & Forst, 2001) (Holmqvist et al., 2012) (Lee & Gottesman, 2016). In Salmonella, MicF overexpression decreased expression of the mRNA for bssS, a negative regulator of biofilm (Domka et al., 2006) (Corcoran et al., 2012). All of these genes are possible relevant targets of MicF in our experiments; we did not further investigate what the downstream target(s) of MicF might be.
Discussion
There are many genes, including those encoding sRNAs, that regulate biofilm formation; in this study we have examined the ability of overproduced sRNAs to affect biofilm formation under both LB and YESCA growth conditions and found that they were significantly different. When sRNAs were overproduced, both positive and negative regulators of biofilm were identified. We focused on the subset of Hfq-dependent sRNAs that increased biofilm formation, as measured by crystal violet staining, when overproduced. These were then tested in epistasis experiments to identify the pathways they acted on. Consistent with a role for sRNAs, deletion of hfq abolished biofilm formation in both LB and YESCA (Fig. 1C, D), as previously found by others using cells grown in LB (Bak et al., 2015). The defect in hfq mutants was more severe than effects of deleting any single one of the sRNAs studied, likely suggesting that combinations of sRNAs are involved. In addition, hfq mutants have global changes in gene expression and slow growth rates that may lead to more complex effects on biofilm formation.
Our results show two of the alternative pathways that E. coli K12 can use in establishing biofilms, and how changing the expression of an sRNA regulator can shift the bacteria from one pathway to another. While deletion of the sRNAs had only modest effects under our laboratory conditions (Fig. 1C, D), overproduction of sRNAs allowed us to highlight these alternative pathways, and emphasized the complex cross talk between sRNAs and transcriptional regulators. This is outlined in Fig. 9 and discussed further below.
Figure 9. Alternative pathways for Biofilm formation.
Two pathways for biofilm formation were defined in this paper. Pathway 1, seen here with cells grown on LB, is dependent on PgaA and independent of RpoS or CsgD. Pathway 2, seen here with cells grown on YESCA, is dependent upon RpoS and CsgD but is independent of PgaA. Increased biofilm in LB in the presence of high levels of DsrA or GadY (blue arrows) is via Pathway 1. DsrA stimulates biofilm by down-regulation of H-NS; H-NS negatively regulates pgaA expression. GadY stimulates translation of the pgaA operon by titrating CsrA, a known negative regulator of pgaA. Both H-NS and GadY may have other targets that contribute to Pathway 1 function expression. In YESCA media, biofilm via Pathway 2 is dependent upon RpoS and CsgD and not on PgaA. High levels of MicF increase biofilm expression using this pathway; the direct MicF targets are not yet known. High levels of DsrA or GadY are only able to significantly stimulate PgaA-dependent biofilm formation in YESCA in the absence of RpoS (see text), suggesting that RpoS or something dependent upon RpoS inhibits this pathway (red bar from RpoS to PgaA-dependent biofilm).
Basal Levels of Biofilm Differ in LB and YESCA
Requirements for the basal level of biofilm were defined by effects of mutants in various pathways implicated in biofilm formation, in the absence of the sRNA. By this definition, FlhD, the master regulator of flagellar synthesis, is needed in both LB and YESCA (Fig. 6 and 7) for the full level of biofilm. None of the other tested mutants decreased basal levels of biofilm in LB, but mutants in both rpoS, encoding the stationary/stress sigma factor, and csgD, encoding the master regulator for curli synthesis and other genes involved in biofilm formation, abolished biofilm in YESCA (Fig. 6B, compare lanes 3 and 5 to lane 1; Fig. 7B, compare lanes 5 and 13 to lane 1). Therefore, we suggest that in LB the low basal level of biofilm may use multiple redundant pathways. In YESCA medium, CsgD and RpoS played important roles for the basal level of biofilm, while PgaA did not (Fig. 6, 7), consistent with previous reports for a minor role of PGA in wild-type E. coli K12 (Itoh et al., 2008) (Fig. 9).
sRNAs stimulate a PGA-dependent pathway of biofilm formation in LB
In LB, increased biofilm was seen in the presence of multicopy DsrA or multicopy GadY or in the absence of H-NS; this increased biofilm was dependent upon pgaA, encoding the polysaccharide beta-1, 6 N-acetyl-D-glucosamine outer membrane porin (Fig. 6, 7). This pathway, which was independent of both rpoS and csgD, is shown as Pathway 1 in Fig. 9.
Our results show that DsrA stimulates biofilm specifically by negatively regulating hns translation (Fig. 9). DsrA stimulates biofilm only when it can pair with hns (Fig. 3), and deletion of hns mimicked the effect of overproducing DsrA in most experiments (Fig. 6). H-NS repression of pgaA transcription is likely to contribute to this increase (Fig. S1), but the pga operon is likely not the only target of H-NS. For instance, H-NS has complex effects on motility, including effects on cyclic-di-GMP regulation (Kim & Blair, 2015). We do not currently know if H-NS repression of pgaA transcription is direct and/or indirect, for instance, via repression of an activator of pgaA, such as NhaR (Dover et al., 1996); (Goller et al., 2006). Finally, under the growth conditions used here (LB, 25°C), chromosomally-encoded DsrA did not contribute significantly to biofilm formation (Fig. 1C, Fig. 3), although it seems possible that there are growth conditions under which DsrA repression of H-NS may be important. We note that deletion of dsrA did have a biofilm-deficient phenotype in YESCA (Fig. 1D), likely due to the role of DsrA in stimulating RpoS translation.
A previous study demonstrated increased adhesion, a critical process for biofilm formation, upon inactivation of hns (Landini & Zehnder, 2002), although the role of PGA in adhesion was not investigated in that paper. Studies in A. pleuropneumoniae reported that hns mutants led to increased biofilm formation (Dalai et al., 2009, Bosse et al., 2010), consistent with our findings in E. coli, and that H-NS specifically repressed the pga operon and the expression of the PGA polysaccharide matrix (Bosse et al., 2010). Our results support a similar pathway for biofilm in E. coli K12, at least in rich (LB) medium, shown both by increased biofilm (Fig. 5, 6) and increased expression of a pgaA-lacZ fusion in the absence of H-NS (Fig. S1B, C). Still to be explained, however, is why multicopy DsrA was less effective than deletion of hns in increasing expression of this fusion (Fig. S1). Possibly the promoter for pgaA is a particularly sensitive target for H-NS, and residual H-NS present when DsrA is overexpressed was still sufficient for some pgaA repression. Intriguingly, microarray analysis of an hns mutant in A. pleuropneumoniae suggested that, unlike the large number of genes regulated by H-NS in E. coli (Hommais et al., 2001), only the pgaA operon was up-regulated in an hns mutant (Bosse et al., 2010).
In contrast to our findings, in another study screening many sRNAs in E. coli, overexpression of DsrA reduced biofilm levels rather than increasing them, and deletion of dsrA modestly increased biofilm (Bak et al., 2015). The biofilm assays in that study were performed using a 12 hour incubation period, at 30°C (Bak et al., 2015), compared to the 24 hours at 25°C that we used. Although we have not fully explored this difference, we note that these investigators also saw a decrease in motility when DsrA was overproduced. It seems likely that with these short incubation periods, inhibition of motility may be sufficient to explain their observations. As discussed above, at 24 hours, deletion of flhD (master regulator for flagellar synthesis) lowered basal levels of biofilm significantly (Fig. 6, compare lane 1 to lane 4, Fig. 7, compare lane 9 to lane 1, Fig. S2A), while after 48 hours the same deletion increased, rather than decreased biofilm levels (Fig. S2C, compare lane 9 to lane 1). Bak et al detected little effect of DsrA on csgD-lacZ, flhD′-’lacZ or pgaA’-’lacZ translational fusions (Bak et al., 2015), consistent with our findings that DsrA is likely to work primarily via silencing of H-NS, a transcriptional repressor. However, when pgaA was absent, we observed DsrA repression of biofilm in LB (Fig. 6, 7, Fig. S2B); this effect disappeared after 48 hours (Fig. S2C). We suggest that this reflects negative regulation of motility by DsrA, and this may also explain the repression by DsrA noted by Bak et al (Bak et al., 2015).
GadY increased PgaA-dependent biofilm (Fig. 6, 7); this can be explained by its ability to increase expression of a pgaA-lacZ fusion (Fig. S1). While this fusion reflects both transcriptional and translational regulation of pgaA, our further work suggests that GadY stimulates pgaA translation (Fig. 8B), and may do this by titrating CsrA, a negative regulator of pgaA (Wang et al., 2005). The stimulation of pgaA is dependent on a GGA sequence, contained in a short hairpin, within GadY, can stimulate pgaA translation even when most of the leader is deleted, and is partially suppressed by a csrA mutant (Fig. 8). This pattern is similar to that observed for McaS, another Hfq-dependent sRNA that has been shown to activate pgaA via CsrA titration (Jorgensen et al., 2013). Therefore, there may be more bifunctional sRNAs that link the Hfq and CsrA regulons than first thought. Bak et al, who found inhibition of biofilm with DsrA overproduction (discussed above), did not find any effect of GadY on biofilm levels, although they did report that GadY repressed swarming and activated translation of pgaA (Bak et al., 2015). Possibly these two effects balance out in their experiments. Our findings, in addition to others, suggest that GadY activation of biofilm in LB is primarily via translational activation of pgaA.
RpoS stimulates CsgD dependent biofilm in YESCA medium
Pathway 2 in Fig. 9 depicts the dependence of biofilm in YESCA on both RpoS and CsgD. CsgD, known to be involved in adhesion via production of curli and biofilm formation, and is a modulator of a subset of genes in the rpoS regulon (Gualdi et al., 2007). csgD transcription is directly controlled by RpoS (Battesti et al., 2011, Ogasawara et al., 2010). The requirement for RpoS in YESCA (Fig. 6B, lane 5 vs. lane 1, Fig. 7B, lane 5 vs. lane 1) is likely at least in part due to its role in regulating csgD expression (Hammar et al., 1995). Deletions of ArcZ or DsrA, both positive regulators of RpoS translation, reduced biofilm formation in YESCA, but not in LB (Fig. 1C, D), consistent with their effect on RpoS levels, although ArcZ probably has other targets in this pathway (Fig. S3).
Consistent with our findings, others have found a role for curli in YESCA, but not in LB. Uropathogenic E. coli (UPEC) strain UT189 was shown to produce a curli and cellulose dependent morphology on YESCA agar and form curli-dependent biofilms when grown in YESCA medium (Lim et al., 2012, DePas et al., 2013); other studies have shown that biofilms developed in LB are not curli dependent (Cegelski et al., 2009).
When MicF was overexpressed, biofilm increased, but unlike the effects of GadY and DsrA, this biofilm was still fully dependent upon RpoS and CsgD (Fig. 7B) and is only seen in YESCA (Fig. 1B, Fig. 7B). Therefore, it seems likely that MicF, by an as yet undefined pathway, specifically stimulates Pathway 2.
Interference between Pathway 1 and Pathway 2
Pathway 1 and 2 appear to be mutually exclusive. In our experiments, RpoS interferes with PgaA-dependent (Pathway 1) biofilm production in YESCA (Fig. 9). This conclusion is based on the inability of GadY or DsrA to stimulate biofilm in YESCA unless cells also carry a deletion of rpoS (Fig. 7B, lanes 5–8); that biofilm was dependent upon pgaA (Fig. S4). The mechanism of this inhibition remains to be determined. A negative effect of RpoS on biofilm formation was previously reported, with different strains and under somewhat different growth conditions (Corona-Izquierdo & Membrillo-Hernandez, 2002).
Other work supports an “either-or” regulation of these two pathways. For instance, the sRNA McaS stimulates the PgaA pathway by titrating CsrA (Jorgensen et al., 2013), but represses csgD (Jorgensen et al., 2012). We would predict that further work will uncover further mechanisms for down-regulating one pathway while up-regulating the other.
Negative Regulation of Biofilm by sRNA Overproduction
While we did not investigate the basis for negative regulation by many sRNAs (Fig. 1A, B), we can imagine a number of ways for sRNAs to repress biofilm. CsgD, clearly important for biofilm formation in YESCA, has been reported to be negatively regulated by six sRNAs (Thomason et al., 2012) (Bordeau & Felden, 2014), four of which (McaS, RydC, RprA, and OmrB) we saw as biofilm repressors in Fig. 1B. However, RydC is likely to have another important biofilm target, since it also inhibited biofilm in LB, which is not dependent on csgD (Fig. 1A). We note that there was not a good correlation of sRNAs that inhibited motility in TB agar (De Lay & Gottesman, 2012) with those that inhibited biofilm (Fig. 1A, B), suggesting that this is not the major pathway for inhibiting biofilm. ChiX, a negative regulator of biofilm in YESCA (Fig. 1B), is known to effectively titrate Hfq (Moon & Gottesman, 2011, Ellis et al., 2015) and thus overproducing this sRNA may partially mimic the effect of deleting Hfq. It is interesting that ChiX negative regulation was much stronger in YESCA than in LB, suggesting a particularly sensitive Hfq-dependent target for biofilm formation in YESCA, possibly translation of RpoS.
In conclusion, several Hfq-dependent sRNAs are capable of perturbing biofilm synthesis pathways. The variation between different media highlights the redundancy between pathways and the ways in which the cell coordinates alternative regulatory circuits. Our work reinforces previous findings suggesting that positive and negative regulation by RpoS has a role in which pathway cells use.
Experimental Procedures
Growth conditions
Strains were grown aerobically at 37°C in LB (10g tryptone, 5g yeast extract, 10g NaCl per liter) overnight or until stationary phase. Biofilm formation was performed in 96-well microtiter plates using LB or YESCA (10g casamino acids and 1g yeast extract per liter) media (Epstein et al., 2009). Ampicillin (100μg/ml), Kanamycin (25μg/ml), Arabinose (.01%), isopropyl-β-D-thogalactopyranoside (IPTG) (100μM), and 5-bromo- 4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) (20 μg/ml) were used where needed.
Strains and Plasmids
The strains used in this study are listed in Table S1. Translational fusions in this study were engineered using lambda red recombination (Yu et al., 2000). The hns*-lacZ fusion was designed by moving the hns-mut1 DNA oligonucleotide into PM1205 (Supplementary Table S1 and S2) and selecting on minimal glycerol X-gal plates containing sucrose plates to create strain SC71. Chromosomal hns* (SC144) was designed by first PCR amplifying the ccdB-kan cassette from NM570 genomic DNA, using primers Kn-hnsmut-ccdBfor and Kn-hnsmut-ccdBrev. Lambda red recombination was used to recombine the PCR fragment into MG1655 containing mini-lambda::tet (NM1100) (Yu et al., 2000) (Court et al., 2003), and the recombinants were selected using 1% glucose-kanamycin agar plates at 32°C. This hns::kan ccdB chromosomal deletion, in which the toxin CcdB is expressed from a PBAD promoter (Tripathi et al., 2012) was confirmed using primers (ab05- Δhns::km forward and ab06- Δhns::km reverse). In a second step, the hns::kan ccdB cassette was further replaced by the hns-mut1 oligonucleotide (hns*) as described (Court et al., 2003). The hns* strain was counter-selected using 1% arabinose agar plates and confirmed by loss of kanR and sequencing.
The pgaA-lacZ fusion present in NM750 was constructed using a standard PCR (ABI) reaction using pgaA_trnsltnl.F and pgaA_trnsltnl.R primers and MG1655 genomic DNA as template; the region of interest was designed to include the promoter, upstream elements of the promoter, as well as the 5′ UTR, to the ATG. This region extended from −502 relative to ATG or −268 relative to the start of transcription to the ATG. This PCR fragment also contained 40 extra nucleotides on either end that are homologous to the region of insertion in NM580, namely to the zeoR marker on the 5′ end and to the lacZ ORF on the 3′end. This PCR fragment was recombineered into electrocompetent λ-Red proficient NM580 cells, recovered overnight on the bench in LB-1% glucose and plated on LB-1% arabinose for counter-selection. Recombinants were screened for KanS and checked by PCR for the correct insertion and sequenced to confirm. The resulting strain was named NM750. This strain was then transduced with P1 to hns::kan to yield NM751 (Table S1).
Deletion-substitutions of dsrA (NM607) and arcZ (NM665) were created by introducing PCR products from NM1201 with the appropriate primers (dsrA-zeo.F and dsrA-zeo.F for ΔdsrA7::zeo and arcZ-zeo.F and arcZ-zeo.R for arcZ::zeo) into NM1100 by recombineering, selecting for zeomycin resistance.
The plasmids used in this study are derived from the pBR-plac vector (Guillier & Gottesman, 2006) (Table S1). Mutations in the pDsrA plasmid vector were constructed using Quik Change Lightning Site-Directed Mutagenesis Kit (Stratagene). The pDsrA plasmid was used to generate mutations in the region important for pairing with the rpoS 5′ UTR (pDsrA*r) using primers DsrA*rmut2for and DsrA*rmut2rev; mutations in the pairing regions important for hns (pDsrA*h) was previously constructed (Lee & Gottesman, 2016)(Table S1). Small RNA plasmids were introduced into strains using TSS transformation (Chung & Miller, 1988) and deletion mutants were constructed in the chromosome using P1 transduction.
Mutagenesis of pGadY was performed using the QuickChange kit from Agilent (Santa Clara, CA) according to the manufacturer’s specifications. Primer sets used for these reactions were gadY_a34t/gadY_a34t-as and gadY_a60t/gadY_a60t-as. Plasmids were sequenced to confirm the mutations.
Biofilm assay
Strains used for biofilm experiments were grown in LB (with Ampicillin at 100 μg/ml where appropriate) at 37°C overnight or until stationary phase, using well-aerated tubes in a roller drum rotating at 250rpm. Cultures were diluted to a final OD600 concentration of ~0.05 in fresh LB or YESCA, LB+amp (100μg/ml)+IPTG (100μM), or YESCA+amp (100μg/ml)+IPTG (100μM). 200μl of each culture was aliquoted into separate wells in a 96-well plate. The wells on the edge of the 96-well plate were avoided as they generate more variance. LB or YESCA media was used as negative controls. Microtiter plates were carefully wrapped using parafilm and placed in a 25°C incubator without shaking. After 24hrs or 48hrs, the plates were removed and the planktonic cell growth (OD600) was measured using a SpectraMax Plus microplate reader. Planktonic cells were removed using a multi-channel pipette without disturbing the biofilm area and individual wells were washed twice using 200μl of dH2O. To stain for biofilm, 220μl of 0.1% crystal violet solution was added to each well for 10 min and then washed twice with 200μl of dH2O and once with 300μl of dH2O. The plates were allowed to dry for 10 min at 37°C prior to dissolving the stained biofilm with 230μl of 20% Acetone/80% Ethanol solution for 10 minutes at room temperature. Biofilm levels were measured using the OD570 normalized by the OD600 (planktonic cell growth). This assay does not provide insight into the structure of the biofilm and it is possible that staining with crystal violet of cells in the two pathways studied here is not equivalent. Because cell growth was very slow in the YESCA medium, we report the 48 hr rather than the 24 hr results for YESCA. For LB, the amount of biofilm was seen to decrease at 48 hr (compare WT plac cells in Fig. S2C to Fig. S2A and B or Fig. 7A) and the dependence upon flhD disappeared (Fig. S2C).
β-Galactosidase assays
Overnight cultures of hns-lacZ (HL1061), hns*-lacZ (SC71), and rpoS-lacZ (PM1409) containing plac, pDsrA, pDsrA*h, pDsrA*r plasmids were diluted 200-fold into fresh LB medium containing IPTG (100μM), ampicillin (100μM), and arabinose (0.001%), and incubated at 37°C in a roller drum for 6 hours. The cultures were then removed, and a β-galactosidase assay was performed as described by Miller (Miller, 1992). PgaA-lacZ (NM750) and Δhns pgaA-lacZ (NM751) transcriptional/translational fusions were grown overnight in LB and diluted 1:1000 in LB medium or in LB ampicillin (100μM) for strains containing plasmids. GS0568, GS0641, GS0644 and GS0645 were grown and assayed similarly, except that 0.2% arabinose was added to the culture medium after dilution to induce the pBAD promoter. Cultures were grown in a shaking water bath at 37°C and strains with plasmids were induced at OD600 = 0.3 using 100μM IPTG. Samples were taken at OD600 1.5–1.8 and assayed for activity using the Miller assay (Miller, 1992).
Supplementary Material
Acknowledgments
We thank members of the Gottesman and Storz labs for comments on the work and the manuscript. We thank G. Storz (NICHD) for strains used in this work. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The authors do not have any conflicts of interest to report.
Footnotes
Author contributions
SC, ND, AP, NM and SG designed the study, SC, ND, AP and NM developed materials, carried out and analyzed the data, SG, AP, NM and SC wrote the manuscript.
References
- Bak G, Lee J, Suk S, Kim D, Young Lee J, Kim KS, Choi BS, Lee Y. Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli. Scientific reports. 2015;5:15287. doi: 10.1038/srep15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annual review of microbiology. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin P, Terao E, Lee EH, Lejeune P, Colson C, Danchin A, Collatz E. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. Journal of bacteriology. 1994;176:5537–5540. doi: 10.1128/jb.176.17.5537-5540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A, Vogel J. The csgD mRNA as a hub for signal integration via multiple small RNAs. Molecular microbiology. 2012;84:1–5. doi: 10.1111/j.1365-2958.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- Bordeau V, Felden B. Curli synthesis and biofilm formation in enteric bacteria are controlled by a dynamic small RNA module made up of a pseudoknot assisted by an RNA chaperone. Nucleic acids research. 2014;42:4682–4696. doi: 10.1093/nar/gku098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse JT, Sinha S, Li MS, O’Dwyer CA, Nash JH, Rycroft AN, Kroll JS, Langford PR. Regulation of pga operon expression and biofilm formation in Actinobacillus pleuropneumoniae by sigmaE and H-NS. Journal of bacteriology. 2010;192:2414–2423. doi: 10.1128/JB.01513-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nature chemical biology. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JR, Sauer K. Small RNAs and their role in biofilm formation. Trends in microbiology. 2013;21:39–49. doi: 10.1016/j.tim.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FFV, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chib S, Mahadevan S. Involvement of the global regulator H-NS in the survival of Escherichia coli in stationary phase. Journal of bacteriology. 2012;194:5285–5293. doi: 10.1128/JB.00840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CT, Miller RH. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic acids research. 1988;16:3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran CP, Pdkaminski D, Papenfort K, Urban JH, Hinton JCD, Vogel J. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Molecular microbiology. 2012;84:428–445. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- Corona-Izquierdo FP, Membrillo-Hernandez J. A mutation in rpoS enhances biofilm formation in Escherichia coli during exponential phase of growth. FEMS microbiology letters. 2002;211:105–110. doi: 10.1111/j.1574-6968.2002.tb11210.x. [DOI] [PubMed] [Google Scholar]
- Court DL, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, Sawitzke J, Sharan SK. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- Dalai B, Zhou R, Wan Y, Kang M, Li L, Li T, Zhang S, Chen H. Histone-like protein H-NS regulates biofilm formation and virulence of Actinobacillus pleuropneumoniae. Microbial pathogenesis. 2009;46:128–134. doi: 10.1016/j.micpath.2008.11.005. [DOI] [PubMed] [Google Scholar]
- De Lay N, Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Molecular microbiology. 2012;86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. The Journal of biological chemistry. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delihas N, Forst S. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. Journal of molecular biology. 2001;313:1–12. doi: 10.1006/jmbi.2001.5029. [DOI] [PubMed] [Google Scholar]
- DePas WH, Hufnagel DA, Lee JS, Blanco LP, Bernstein HC, Fisher ST, James GA, Stewart PS, Chapman MR. Iron induces bimodal population development by Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2629–2634. doi: 10.1073/pnas.1218703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J, Lee J, Wood TK. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Applied and environmental microbiology. 2006;72:2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato GM, Lelivelt MJ, Kawula TH. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. Journal of bacteriology. 1997;179:6618–6625. doi: 10.1128/jb.179.21.6618-6625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical microbiology reviews. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover N, Higgins CF, Carmel O, Rimon A, Pinner E, Padan E. Na+-induced transcription of nhaA, which encodes an Na+/H+ antiporter in Escherichia coli, is positively regulated by nhaR and affected by hns. J Bacteriol. 1996;178:6508–6517. doi: 10.1128/jb.178.22.6508-6517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Trussler RS, Haniford DB. Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. Molecular microbiology. 2015;96:633–650. doi: 10.1111/mmi.12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein EA, Reizian MA, Chapman MR. Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. Journal of bacteriology. 2009;191:608–615. doi: 10.1128/JB.01244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrieres L, Thompson A, Clarke DJ. Elevated levels of sigma S inhibit biofilm formation in Escherichia coli: a role for the Rcs phosphorelay. Microbiology. 2009;155:3544–3553. doi: 10.1099/mic.0.032722-0. [DOI] [PubMed] [Google Scholar]
- Frohlich KS, Papenfort K, Berger AA, Vogel J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic acids research. 2012;40:3623–3640. doi: 10.1093/nar/gkr1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goller C, Wang X, Itoh Y, Romeo T. The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcripiton, required for the production of the biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. Journal of bacteriology. 2006;188:8022–8032. doi: 10.1128/JB.01106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goller CC, Romeo T. Environmental influences on biofilm development. Curr Top Microbiol Immunol. 2008;322:37–66. doi: 10.1007/978-3-540-75418-3_3. [DOI] [PubMed] [Google Scholar]
- Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annual review of microbiology. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Gualdi L, Tagliabue L, Landini P. Biofilm formation-gene expression relay system in Escherichia coli: modulation of sigmaS-dependent gene expression by the CsgD regulatory protein via sigmaS protein stabilization. Journal of bacteriology. 2007;189:8034–8043. doi: 10.1128/JB.00900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Molecular microbiology. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Molecular microbiology. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Holmqvist E, Unoson C, Reimegard J, Wagner EGH. A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Molecular microbiology. 2012;84:414–427. doi: 10.1111/j.1365-2958.2012.07994.x. [DOI] [PubMed] [Google Scholar]
- Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Molecular microbiology. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, Heuser J, Chapman MR, Hadjifrangiskou M, Henderson JP, Hultgren SJ. Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio. 2013;4:e00645–00613. doi: 10.1128/mBio.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF, 3rd, Romeo T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen MG, Nielsen JS, Boysen A, Franch T, Moller-Jensen J, Valentin-Hansen P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Molecular microbiology. 2012;84:36–50. doi: 10.1111/j.1365-2958.2012.07976.x. [DOI] [PubMed] [Google Scholar]
- Jorgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. Dual function of the McaS small RNA in controlling biofilm formation. Genes & development. 2013;27:1132–1145. doi: 10.1101/gad.214734.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiology and immunology. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- Kim EA, Blair DF. Function of the histone-Like Protein H-NS in motility of Escherichia coli: Multiple regulatory roles rather than direct action at the flagellar motor. J Bacteriol. 2015;197:3110–3120. doi: 10.1128/JB.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini P, Zehnder AJ. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. Journal of bacteriology. 2002;184:1522–1529. doi: 10.1128/JB.184.6.1522-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Gottesman S. sRNA roles in regulating transcriptional regulators: Lrp and SoxS regulation by sRNAs. Nucleic acids research. 2016 doi: 10.1093/nar/gkw358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JY, May JM, Cegelski L. Dimethyl sulfoxide and ethanol elicit increased amyloid biogenesis and amyloid-integrated biofilm formation in Escherichia coli. Applied and environmental microbiology. 2012;78:3369–3378. doi: 10.1128/AEM.07743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Niu H, Wu S, Huang R. CsgD regulatory network in a bacterial train-altering biofilm formation. Emerg Microbes Infect. 2014;3 doi: 10.1038/emi.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. The EMBO journal. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int, J Mol Sci. 2013;14:4560–4579. doi: 10.3390/ijms14034560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA biology. 2014;11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press; Plainview, NY: 1992. [Google Scholar]
- Mizuno T, Chen MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proceedings of the National Academy of Sciences of the United States of America. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro C, Papenfort K, Hentrich K, Ahmad I, Le Guyon S, Reimann R, Grantcharova N, Romling U. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA biology. 2012;9:489–502. doi: 10.4161/rna.19682. [DOI] [PubMed] [Google Scholar]
- Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Molecular microbiology. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology. 2010;156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. Journal of bacteriology. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JCD, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Molecular microbiology. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- Parker A, Gottesman S. Small RNA Regulation of TolC, the Outer Membrane Component of Bacterial Multidrug Transporters. Journal of bacteriology. 2016;198:1101–1113. doi: 10.1128/JB.00971-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome research. 2011;21:1487–1497. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Frangos A, Kavita K, Schu DJ, Gottesman S, Woodson SA. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E6089–E6096. doi: 10.1073/pnas.1613053113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. Journal of bacteriology. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason MK, Fontaine F, De Lay N, Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Molecular microbiology. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Dewan PC, Barua B, Varadarajan R. Additional role for the ccd operon of F-plasmid as a transmissible persistence factor. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12497–12502. doi: 10.1073/pnas.1121217109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove TB, Zhang A, Storz G. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol. 2016;30:133–138. doi: 10.1016/j.mib.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Puyvelde S, Steenackers HP, Vanderleyden J. Small RNAs regulating biofilm formation and outer membrane homeostasis. RNA biology. 2013;10:185–191. doi: 10.4161/rna.23341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Molecular microbiology. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Molecular microbiology. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XS, Garcia-Contreras R, Wood TK. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. Journal of bacteriology. 2007;189:3051–3062. doi: 10.1128/JB.01832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Koestler BJ, Waters CM, Hammer BK. Post-transcriptional activation of a diguanylate cyclase by quorum sensing small RNAs promotes biofilm formation in Vibrio cholerae. Molecular microbiology. 2013;89:989–1002. doi: 10.1111/mmi.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.