Abstract
High-affinity antibody production through the germinal centre (GC) response is a pivotal process in adaptive immunity. Abnormal development of follicular helper T (TFH) cells can induce the GC response to self-antigens, subsequently leading to autoimmunity. Here we show the transcriptional repressor Capicua/CIC maintains peripheral immune tolerance by suppressing aberrant activation of adaptive immunity. CIC deficiency induces excessive development of TFH cells and GC responses in a T-cell-intrinsic manner. ETV5 expression is derepressed in Cic null TFH cells and knockdown of Etv5 suppresses the enhanced TFH cell differentiation in Cic-deficient CD4+ T cells, suggesting that Etv5 is a critical CIC target gene in TFH cell differentiation. Furthermore, we identify Maf as a downstream target of the CIC–ETV5 axis in this process. These data demonstrate that CIC maintains T-cell homeostasis and negatively regulates TFH cell development and autoimmunity.
Follicular helper T (TFH) cells promote germinal centre (GC) response for efficient generation of protective antibodies during humoral immunity. Here the authors show that deficiency of the translational repressor, Capicua/CIC, enhances TFH differentiation and GC responses potentially via the derepression of Etv5.
The germinal centre (GC) response is one of the most elegant processes in adaptive immunity and produce antibodies that have high affinity to antigens. In follicles, B cells that recognize antigens proliferate and form GCs. GCs are expanded by the proliferation of GC B cells and polarize into two microenvironments, the dark zone and the light zone1,2,3. GC B cells cycle between these two zones. In the dark zone, GC B cells rapidly proliferate and undergo somatic hypermutation, which enables antibody diversification and affinity maturation. In the light zone, GC B cells are selected on the basis of antigen affinity, undergo immunoglobulin class-switch recombination, and eventually give rise to antibody-secreting plasma cells or memory B cells1,2,3. During the GC response, several types of immune cell collaborate with B cells in the follicles, where follicular helper T (TFH) cells have an instrumental function. TFH cells facilitate the selection and maturation of high-affinity GC B cells by multiple rounds of cognate interaction with B cells in the light zone; these interactions provide the selected B cells with crucial signals for survival and re-entry into the dark zone3. Because TFH cells have an important function in the generation of isotype-switched and affinity-maturated antibodies, dysregulation of TFH cell development and function is closely associated with immunodeficiency-related pathogenesis or antibody-mediated autoimmune diseases including systemic lupus erythematosus4,5,6.
Differentiation of TFH cells is initiated by the interaction of naïve T cells with dendritic cells (DCs), which, together with environmental factors, including cytokines, triggers expression of the chemokine receptor CXCR5 on DC-primed T cells7,8. The surface expression of CXCR5 enables T cells to migrate into B-cell follicles7,8. T cells that are targeted to enter B-cell follicles upregulate expression of the transcriptional repressor BCL6 and express an intermediate level of typical TFH molecules (for example, CXCR5, PD-1, ICOS and SAP) at the junction between T-cell and B-cell zone9,10. At this stage, developing TFH cells interact with cognate B cells and differentiate into GC TFH cells that express high levels of TFH molecules, such as PD-1 and CXCR5 (ref. 9). BCL6 as a master transcription factor for TFH cell differentiation11,12,13 and BLIMP1 as an antagonist of BCL6 (ref. 11), plus several other transcription factors, help orchestrate TFH cell differentiation by exerting either a positive or negative effect, depending on the cellular context8. Among these factors, MAF (also known as c-MAF) was identified as a positive regulator of TFH cell differentiation in mice and humans. Maf deficiency decreases the frequency of CD4+CXCR5+ T cells in mice14. MAF regulates expression of Il21 in mouse TFH cells and, in conjunction with BCL6, MAF induces expression of CXCR4, CXCR5, PD-1, ICOS and IL-21 in human TFH cells14,15,16. Moreover, MAF expression is induced in CD4+ T cells by ICOS co-stimulation14 or by IL-6, an important cytokine for initiating TFH cell differentiation15.
Capicua/CIC is a transcriptional repressor that is evolutionarily conserved from cnidarians to mammals17, and it exists in short (CIC-S) and long (CIC-L) isoforms17. In mammals, CIC interacts with Ataxin-1/ATXN1 (ref. 18), of which polyglutamine (polyQ)-expanded form causes spinocerebellar ataxia type-1 (SCA1) neuropathogenesis, and its haploinsufficiency alleviates SCA1 progression19. Loss of the ATXN1–CIC complex results in hyperactivity, impaired learning and memory, and abnormal maturation and maintenance of upper-layer cortical neurons in mice20. CIC also suppresses the progression of several types of cancer21,22,23. CIC target genes that are critical for regulation of cancer progression include PEA3 group genes, ETV1/ER81, ETV4/PEA3 and ETV5/ERM, which are frequently overexpressed and promote tumourigenesis and metastasis in various types of cancer cell21,24,25,26. In vivo functions of CIC have been reported in studies of Cic hypomorphic (Cic-L−/−) mice19. These mice die within 4 weeks after birth and have defects in lung alveolarization27 and bile acid homeostasis28. However, CIC is expressed in most mouse tissues27,28, so it may also have other important physiological functions. CIC levels are relatively high in the thymus27,28 and in immune cells including T and NK cells (http://www.humanproteomemap.org)29. Moreover, our previous study shows that genes associated with immune activation and autoimmune diseases, such as SLE, asthma and thyroiditis, are significantly upregulated in the liver of Cic-L−/− mice28. These findings suggest that CIC may regulate immune responses and immune disorders.
Here we investigate CIC functions in immune system using various immune cell-specific Cic null mice. Our study finds previously unrecognised functions of CIC in regulation of T-cell activation and GC responses and suggests CIC as a key transcription factor in the suppression of autoimmunity.
Results
Autoimmunity in immune cell-specific Cic null mice
To investigate in vivo requirement of CIC in homeostasis and functions of immune cells, we generated haematopoietic lineage cell-specific Cic null mice (Cicf/fVav1-Cre) by crossing mice carrying floxed Cic alleles (Cicf/f) with Vav1-Cre mice (Supplementary Fig. 1a), and characterized them at 9–12 weeks of age. Development of lymphocytes in primary lymphoid organs was largely normal (Supplementary Fig. 1b,c), although an increase in proportion of thymic CD4+FOXP3+ regulatory T (Treg) cells was observed (Supplementary Fig. 2).
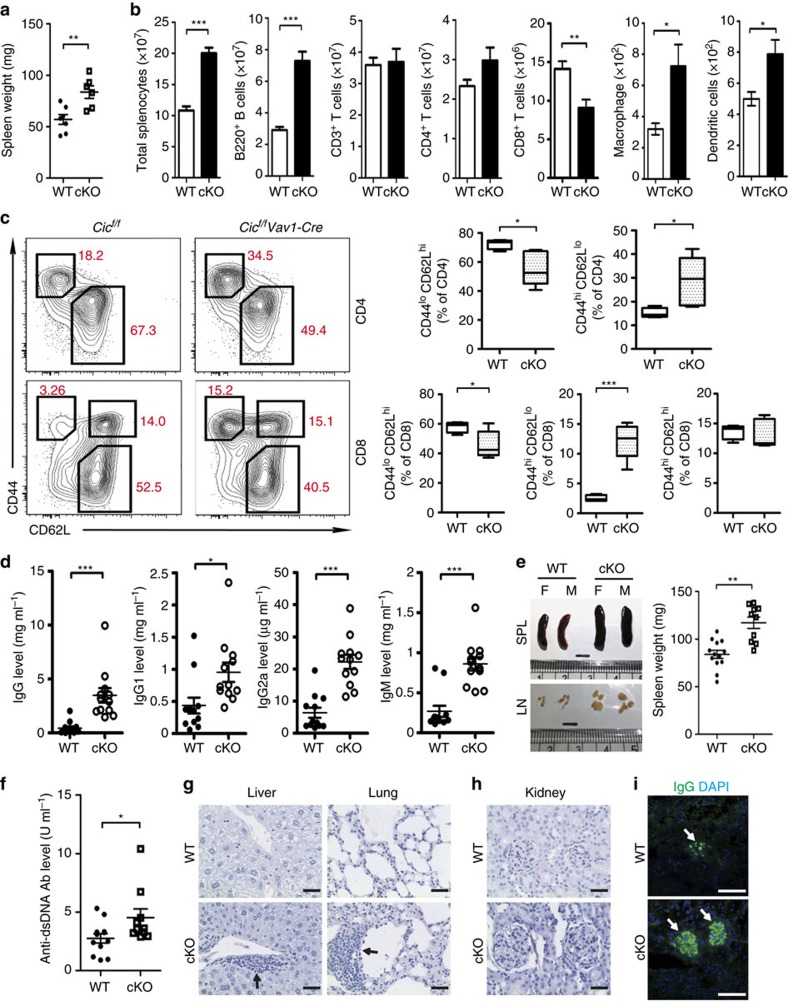
Strikingly, all Cicf/fVav1-Cre mice developed enlarged secondary lymphoid organs, including spleens (Fig. 1a), mainly due to proliferation of B220+ B cells (Fig. 1b). In addition, the number of CD8+ T cells was significantly decreased, whereas that of macrophages and DCs was increased in spleen of Cicf/fVav1-Cre mice (Fig. 1b). The expression levels of several T-cell co-stimulatory ligands, such as CD80, CD86, CD40 and ICOSL, on the surface of DCs were comparable between Cicf/f (WT) and Cicf/fVav1-Cre mice (Supplementary Fig. 3). Cicf/fVav1-Cre mice had a higher frequency of effector/memory-phenotype (CD44hiCD62Llo) cells for both CD4+ and CD8+ subsets, compared with wild-type (WT) mice (Fig. 1c). Cic-deficient CD4+CD44+ T cells showed elevated expression of cell surface co-stimulatory molecules such as ICOS, PD-1, OX40 and GITR (Supplementary Fig. 4). Flow cytometry analysis on cytokine expression profiles revealed that the frequency of IFNγ+, TNF+, IL-21+, IL-13+ and IL-22+ CD4+ T cells was significantly increased in spleen of Cicf/fVav1-Cre mice (Supplementary Fig. 5a). Cicf/fVav1-Cre mice also had a higher frequency of CCR4−CXCR3+ (TH1-related) and CCR4+CCR6− (TH2-related) CD4+ T cells30 (Supplementary Fig. 5b). Thus, loss of CIC spontaneously activates T cells, resulting in accumulation of effector/memory cells with mixed T helper (TH) phenotypes in mice. On the other hand, Cic null naïve CD4+ T cells differentiated into each TH subset with similar efficiency to WT cells under in vitro polarizing conditions (Supplementary Fig. 6), suggesting that CIC might not be a crucial factor for each TH subset differentiation.
Figure 1. Lymphoproliferative autoimmunity in immune cell-specific Cic null mice.
(a,b) Spleen weights (a) and the numbers of total splenocytes, B220+ B, CD3+ T, CD4+ T, CD8+ T, macrophages (CD11b+CD11c−F4/80+) and dendritic cells (CD11b+CD11c+MHCII+) (b) in 10-week-old Cicf/f (WT) and Cicf/fVav1-Cre (cKO) mice. (c) FACS analysis showing the proportion of CD44loCD62Lhi naïve T cells and CD44hiCD62Llo effector/memory T cells in spleen of Cicf/f and Cicf/fVav1-Cre mice. All data are representative of two independent experiments. Data are presented as box-and-whisker plots. n=4–5 per each genotype. (d) Serum levels of total IgG, IgG1, IgG2a and IgM in 12-week-old Cicf/f and Cicf/fVav1-Cre mice were measured by ELISA. (e) Images of dissected spleens (SPL) and draining lymph nodes (LN) from Cicf/f and Cicf/fVav1-Cre mice at the age of 12 months. The graph shows average weights of spleens from the Cicf/f and Cicf/fVav1-Cre mice. M, male; F, female. (f) Serum levels of anti-dsDNA antibody in the 12-month-old Cicf/f and Cicf/fVav1-Cre mice were measured by ELISA. The graphs in a,b,d–f show data as mean±s.e.m. and each dot in graphs represents an individual mouse. *P<0.05, **P<0.01 and ***P<0.001 (two-tailed two-sample unequal variance Student t-test). (g) Immune cell infiltration in liver and lung from the 12-month-old Cicf/fVav1-Cre mice. Tissue sections were stained with haematoxylin and eosin (H&E). Arrows indicate immune cell infiltrates. (h) H&E-stained kidney sections showing glomerulonephritis in the 12-month-old Cicf/fVav1-Cre mice. (i) Immunostaining for IgG deposition in kidney glomeruli of the 12-month-old Cicf/fVav1-Cre mice. Representative images from two independent experiments are shown. (g–i) Scale bars, 100 μm.
Cicf/fVav1-Cre mice had a higher proportion of CD4+CD25−FOXP3+ T cells in spleen, but a comparable proportion of CD4+CD25+FOXP3+ T cells, compared with WT mice (Supplementary Fig. 7a). Expression levels of surface molecules that are critical for controlling Treg cell homeostasis and function, such as GITR, CTLA-4, CD103 and GARP (refs 31, 32), were comparable between WT and Cic null CD4+ FOXP3+ T cells regardless of CD25 expression (Supplementary Fig. 7b). Consistent with this result, WT and Cic-deficient CD4+CD25+ Treg cells had a comparable suppressive activity on IL-2 secretion and proliferation of responding CD4+CD25− T cells (Supplementary Fig. 7c,d). The numbers of CD8+CD25+ FOXP3+ and CD8+ICOSL+CXCR5+ (Qa-1-restricted CD8+ Treg cells)33,34 T cells were also comparable between WT and Cicf/fVav1-Cre mice (Supplementary Fig. 7e,f). These results, together with the increased frequency of thymic Treg cells (Supplementary Fig. 2), indicate that the T cell hyperactivation in Cicf/fVav1-Cre mice was not due to defects in Treg cell compartment.
Because the abnormal activation of T cells and the increased frequency of CD4+CD25−FOXP3+ T cells are often associated with autoimmunity35,36, we assessed whether Cicf/fVav1-Cre mice display autoimmune phenotypes with age. The Cicf/fVav1-Cre mice developed hyperglobulinemia at 12 weeks of age (Fig. 1d), and exhibited several typical autoimmune attributes at around 1 year of age, including enlarged secondary lymphoid organs (Fig. 1e), increased serum levels of anti-dsDNA antibody (Fig. 1f), infiltration of immune cells into tissues (Fig. 1g), glomerulonephritis (Fig. 1h) and IgG deposition in kidney glomeruli (Fig. 1i). Together, these findings demonstrate that CIC is indispensable for suppression of lymphoproliferative autoimmunity.
Spontaneous induction of GC responses by CIC deficiency
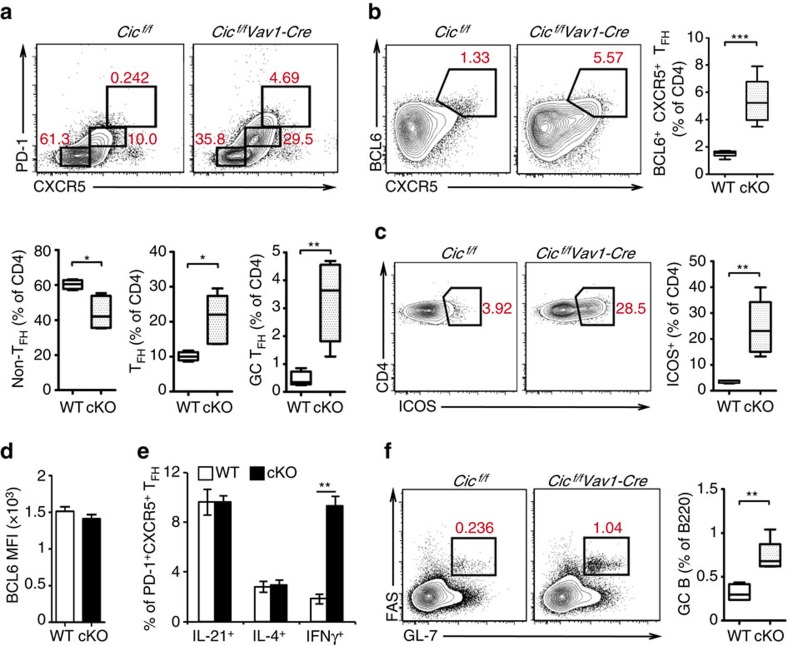
Increased frequencies of TFH-like cells are often observed in patients with autoimmune diseases including SLE6, and studies of Roquinsan mice show that overexpression of IFNγ and ICOS, a co-stimulatory molecule highly expressed in TFH cells37,38, promotes TFH cell development, which in turn spontaneously induces a lupus-like autoimmune syndrome39,40. It is also known that IL-21 and IL-4 are key cytokines that TFH cells express during the GC response41. Because Cicf/fVav1-Cre mice had the increased T-cell surface expression of ICOS, the increased proportions of CD4+IFNγ+, CD4+IL-21+ and CD4+IL-4+ T cells, and the autoimmune-like symptoms similar to those in Roquinsan mice42, we set out to determine whether the TFH subset was elevated in Cicf/fVav1-Cre mice. The frequencies of TFH (PD-1intCXCR5int) and GC TFH (PD-1hiCXCR5hi) cells were significantly increased in the spleen of 9-week-old Cicf/fVav1-Cre mice at the expense of non-TFH cells (Fig. 2a). The increased TFH cell frequency in Cicf/fVav1-Cre mice was confirmed by flow cytometry analysis of CD4+BCL6+CXCR5+ T cells (Fig. 2b). Consistent with these results, the proportion of CD4+ICOS+ T cells was also markedly increased (Fig. 2c). On the other hand, levels of BCL6 in PD-1+CXCR5+ TFH cells were comparable between WT and Cicf/fVav1-Cre mice (Fig. 2d). The frequency of IFNγ+ TFH cells was substantially increased in Cicf/fVav1-Cre mice, whereas that of IL-21+ and IL-4+ TFH cells was comparable between WT and Cicf/fVav1-Cre mice (Fig. 2e). Because the primary function of TFH cells is to help B cells form GC reactions, we further analysed GC B (B220+Fas+GL-7+) cells. The proportion of GC B cells was significantly higher in Cicf/fVav1-Cre mice than in WT mice (Fig. 2f). Taken together, these data indicate that CIC is required for suppression of spontaneous induction of GC reactions.
Figure 2. Increased proportion of TFH and GC B cells in Cicf/fVav1-Cre mice.
(a–c,f) Flow cytometry analyses of non-TFH, TFH and GC TFH cells (a), CD4+BCL6+CXCR5+ TFH cells (b), CD4+ICOS+ T cells (c) and GC B cells (f) in spleen from 9-week-old Cicf/f (WT) and Cicf/fVav1-Cre (cKO) mice. All data are representative of three independent experiments with n=4–5 mice per group in each experiment. Numbers adjacent to outlined areas indicate per cent of each cell population among splenic CD4+ or B220+ cells. Data are presented as box-and-whisker plots. *P<0.05, **P<0.01 and ***P<0.001 (two-tailed two-sample unequal variance student t-test). (d) Mean fluorescence intensity (MFI) of BCL6 expression in CD4+PD-1+CXCR5+ TFH cells from 9-week-old Cicf/f and Cicf/fVav1-Cre mice. n=3 mice per each genotype. (e) Flow cytometry analysis of IL-21, IL-4 or IFNγ-expressing TFH cells in spleen of 9-week-old Cicf/f and Cicf/fVav1-Cre mice. The proportions of each cytokine-expressing TFH cells are presented. (d,e) n=3 mice per each genotype. The bar graphs show data as mean±s.e.m. **P<0.01 (two-tailed two-sample unequal variance Student t-test).
T-cell-intrinsic functions of CIC
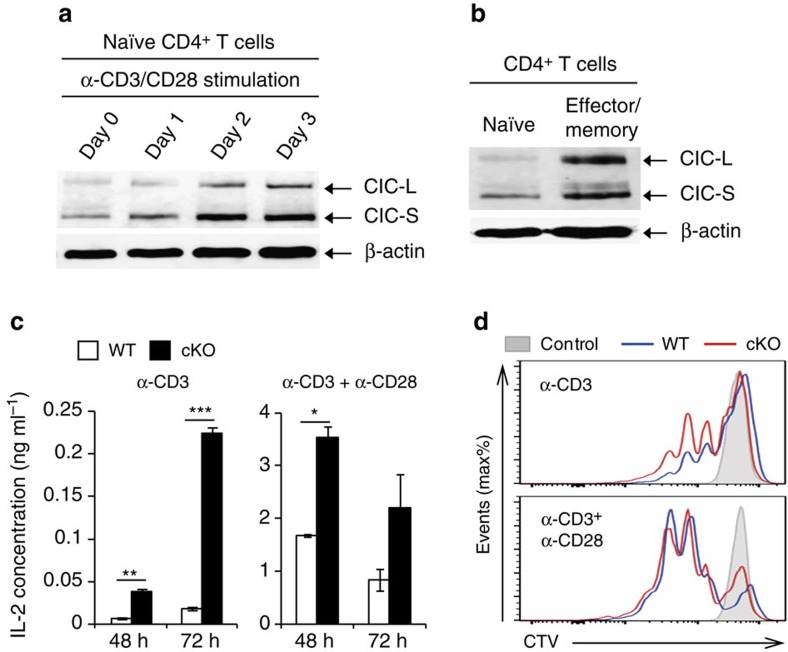
Because several abnormalities in T cells were observed in Cicf/fVav1-Cre mice, we next examined CIC in T cells. CIC protein levels gradually increased along with the activation of CD4+ T cells by anti-CD3 and anti-CD28 antibodies (Fig. 3a), and were higher in effector/memory CD4+ T cells than in naïve CD4+ T cells (Fig. 3b). Cic-deficient CD4+ T cells secreted IL-2 and proliferated more efficiently than WT T cells when stimulated with anti-CD3 (Fig. 3c,d), suggesting that CIC negatively regulates an activation signal that is mediated by T-cell receptors (TCRs). However, this difference was largely abrogated by addition of anti-CD28 (Fig. 3c,d), indicating that a strong co-stimulatory signal through CD28 overrides the enhanced TCR response in Cic-deficient T cells.
Figure 3. CIC negatively regulates T-cell activation.
(a,b) Increased CIC expression in activated CD4+ T cells. Western blot analysis for CIC levels in CD4+ T cells during T-cell activation by α-CD3/CD28 antibodies (a) and in sorted naïve (CD44loCD62Lhi) and effector/memory (CD44hiCD62Llo) CD4+ T cells (b). (c) ELISA of IL-2. WT and Cic-deficient (cKO) CD4+CD25−CD44loCD62Lhi naïve T cells were stimulated with plate-bound anti-CD3 (1.0 μg ml−1) in the presence (right) or absence (left) of plate-bound anti-CD28 (2.0 μg ml−1). The supernatants were taken 48 h and 72 h after stimulation and subjected to ELISA for IL-2 concentration. n=4 per each sample. Error bars indicate s.e.m. *P<0.05, **P<0.01 and ***P<0.001 (two-tailed two-sample unequal variance Student t-test). (d) In vitro T-cell proliferation assay. Naïve CD4+ T cells purified from pooled spleens and lymph nodes of Cicf/f and Cicf/fVav1-Cre mice were labelled with CTV dye and stimulated with plate-bound anti-CD3 (1.0 μg ml−1) in the presence (right) or absence (left) of plate-bound anti-CD28 (2.0 μg ml−1). The cells were analysed 72 h after stimulation. Data are representative of three independent experiments. Shaded area: unstimulated control, blue line: Cicf/f, red line: Cicf/fVav1-Cre.
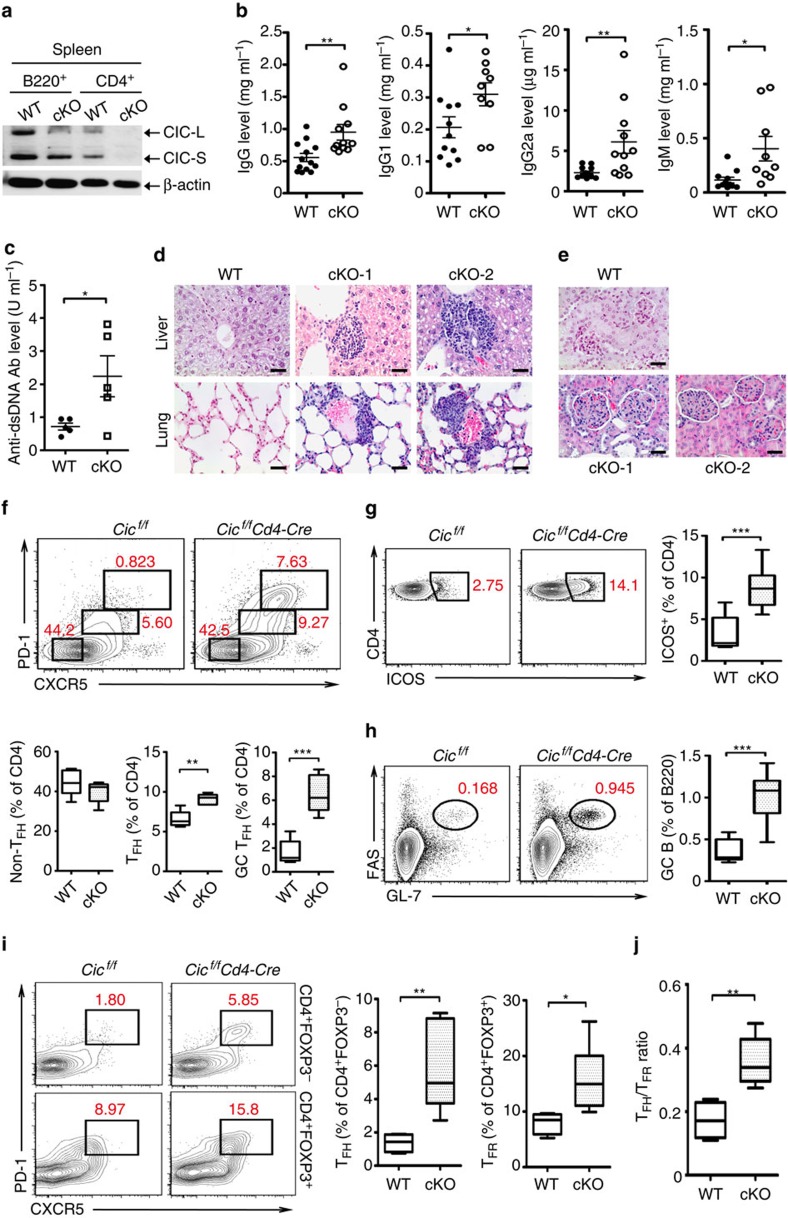
To determine the in vivo consequences of CIC deficiency in T cells, we generated mice with Cic deletion specific to T cells (Cicf/fCd4-Cre) (Fig. 4a). Development of T cells in thymus was largely normal in Cicf/fCd4-Cre mice (Supplementary Fig. 8a). Given that CIC deficiency induced autoimmunity and that Cic null CD4+ T cells more robustly responded to TCR stimulation than WT cells in vitro (Fig. 3c,d), we assessed thymic negative selection, which is a critical process that removes autoreactive T cells in thymus43, in WT and Cicf/fCd4-Cre mice. TCR-induced apoptosis comparably occurred in WT and Cic null double positive (DP) thymocytes (Supplementary Fig. 8b), suggesting that TCR-induced negative selection might not be affected by CIC deficiency. Consistent with this finding, WT and Cic null CD4+ thymocytes exhibited a comparable induction of Nur77, which has been implicated in negative selection through its ability to convert BCL2 into a proapoptotic molecule44,45, in response to TCR stimulation (Supplementary Fig. 8c).
Figure 4. Spontaneous induction of TFH cell differentiation and GC response in T-cell-specific Cic null mice.
(a) Western blot analysis showing T-cell-specific ablation of CIC expression in Cicf/fCd4-Cre mice. B220+ B and CD4+ T cells were purified from spleen of 12-week-old Cicf/f (WT) and Cicf/fCd4-Cre (cKO) mice. (b) Levels of total IgG, IgG1, IgG2a and IgM in sera from 12-week-old Cicf/f and Cicf/fCd4-Cre mice. (c) Serum levels of anti-dsDNA antibody in 14.5-month-old Cicf/f and Cicf/fCd4-Cre mice were measured by ELISA. (b,c) The graphs show data as mean±s.e.m. and each dot in graphs represent an individual mouse. *P<0.05 and **P<0.01 (two-tailed two-sample unequal variance Student t-test). (d) Immune cell infiltration in liver and lung from 14.5-month-old Cicf/fCd4-Cre mice. Tissue sections were stained with H&E. (e) H&E-stained kidney sections showing glomerulonephritis in 14.5-month-old Cicf/fCd4-Cre mice. (d,e) Representative images from two Cicf/fCd4-Cre mice (cKO-1 and cKO-2) are shown. Scale bars, 100 μm. (f–i) Flow cytometry analyses of non-TFH, TFH and GC TFH cells (f), CD4+ICOS+ T cells (g), GC B cells (h) and CD4+FOXP3−PD-1+CXCR5+ TFH and CD4+FOXP3+PD-1+CXCR5+ TFR cells (i) in spleen from 12-week-old Cicf/f and Cicf/fCd4-Cre mice. All data are representative of three independent experiments with n=4–5 mice per group in each experiment. Numbers adjacent to outlined areas indicate per cent of each cell population among splenic CD4+ or B220+ cells. *P<0.05, **P<0.01 and ***P<0.001 (two-tailed two-sample unequal variance Student t-test). (j) Quantification of the ratio TFH/TFR from the experiments as in Fig. 3i. **P<0.01 (two-tailed two-sample unequal variance Student t-test).
On the other hand, as observed in the Cicf/fVav1-Cre mice, hyperglobulinemia, T-cell hyperactivation and increased populations of CD25−FOXP3+−, TH1-type and TH2-type CD4+ T cells in spleen occurred in Cicf/fCd4-Cre mice at 12 weeks of age (Fig. 4b, Supplementary Fig. 9). These mice also showed several systemic autoimmune phenotypes with age, including increased serum levels of anti-dsDNA antibody (Fig. 4c), infiltration of immune cells into tissues (Fig. 4d) and glomerulonephritis (Fig. 4e). Moreover, the proportions of TFH, GC TFH, CD4+ICOS+ T and GC B cells were significantly higher in Cicf/fCd4-Cre mice than in WT mice (Fig. 4f–h). The frequency of IFNγ-expressing TFH cells was increased in the spleen of Cicf/fCd4-Cre mice (Supplementary Fig. 10a). Because IFNγ drives IgG2a class-switching in B cells during GC reactions40,46,47, we further assessed IgG2a+ GC B cells. As expected, the frequency and the number of IgG2a+ GC B cells were also increased in Cicf/fCd4-Cre mice (Supplementary Fig. 10b). Taken together, these findings demonstrate that the spontaneous inductions of T-cell activation, GC response and systemic autoimmunity were due to loss of CIC in T cells.
Follicular regulatory T (TFR) cells, defined as CD4+ FOXP3+PD-1+CXCR5+ cells, are a specialized subset of regulatory T cells that inhibit antibody production48,49,50. Because the proportion of CD4+FOXP3+ T cells increased in Cic-deficient mice (Supplementary Figs 7a and 9b), we determined whether TFR cells were increased in Cicf/fCd4-Cre mice. As with CD4+FOXP3−PD-1+CXCR5+ TFH cells, CD4+FOXP3+PD-1+CXCR5+ TFR cells were also significantly overrepresented in the spleen of Cicf/fCd4-Cre mice compared with WT mice (Fig. 4i). Importantly, the ratio TFH/TFR, which is a critical factor that dictates the magnitude of antibody production50, was also increased in the Cicf/fCd4-Cre mice (Fig. 4j), consistent with the observation that GC B cells and serum immunoglobulin levels were elevated in Cicf/fCd4-Cre mice (Fig. 4b,h).
Normal T-cell homeostasis in Treg-specific Cic null mice
Although the suppressive activity toward conventional T cells and expression profiles of Treg cell-associated surface molecules were comparable between WT and Cic-deficient CD4+ Treg cells (Supplementary Fig. 7b–d), the frequency of CD4+CD25−FOXP3+ T cells was substantially increased in Cic-deficient mice (Supplementary Figs 7a and 9b). To directly address the function of CIC in development and function of FOXP3+ Treg cells, we generated FOXP3+ Treg cell-specific Cic null (Cicf/fFoxp3-YFP-Cre) mice by crossing Cicf/f mice with Foxp3-YFP-Cre mice51. The ablation of Cic alleles in Foxp3-expressing (YFP+) cells did not affect formation of CD25+FOXP3+ and CD25−FOXP3+ CD4+ T cells in both thymus and spleen (Supplementary Fig. 11a–c). The proportion of naïve and effector/memory cells for both CD4+ and CD8+ subsets, TFH, TFR and GC B cells was also comparable between Cic+/+Foxp3-YFP-Cre (WT) and Cicf/fFoxp3-YFP-Cre mice (Supplementary Fig. 11d–f). These results suggest that CIC deficiency in FOXP3+ Treg cells does not cause defects in development and function of FOXP3+ Treg cells.
Since the FOXP3+ Treg cell-specific Cic deletion did not promote formation of CD4+CD25−FOXP3+ T-cell population in spleen (Supplementary Fig. 11c), we examined expression of FOXP3 in conventional CD4+ T cells from WT and Cicf/fCd4-Cre mice. Interestingly, the proportion of FOXP3-expressing naïve (CD45RBhiCD44loCD62Lhi) CD4+ T cells was markedly increased in the spleen of Cicf/fCd4-Cre mice, compared with WT mice (Supplementary Fig. 12a). Consistent with this result, CD4+CD25−FOXP3+ T cells in Cicf/fCd4-Cre mice were composed of both CD44loCD62Lhi and CD44hiCD62Llo populations, while those in WT mice were mainly CD44hiCD62Llo cells (Supplementary Fig. 12b). We also determined the origin of CD4+CD25−FOXP3+ T cells in Cicf/fCd4-Cre mice by flow cytometry analysis for expression of Helios and Neurophilin-1/NRP1, both of which are markers for thymus-derived Treg (tTreg) cells52,53,54. The frequency of Helios−NRP1− cells for both CD25+FOXP3+ and CD25−FOXP3+ T-cell subsets was significantly increased in the spleen of Cicf/fCd4-Cre mice (Supplementary Fig. 12c), demonstrating that the increased frequency of CD4+CD25−FOXP3+ T cells in Cicf/fCd4-Cre mice was due to the expansion of peripherally-induced Treg (pTreg) cells. Taken together, our data suggest that CIC deficiency induces FOXP3 expression in naïve CD4+ T cells, thereby increasing the population of CD4+CD25−FOXP3+ T cells in the periphery.
Regulation of TFH cell differentiation by the CIC–ETV5 axis
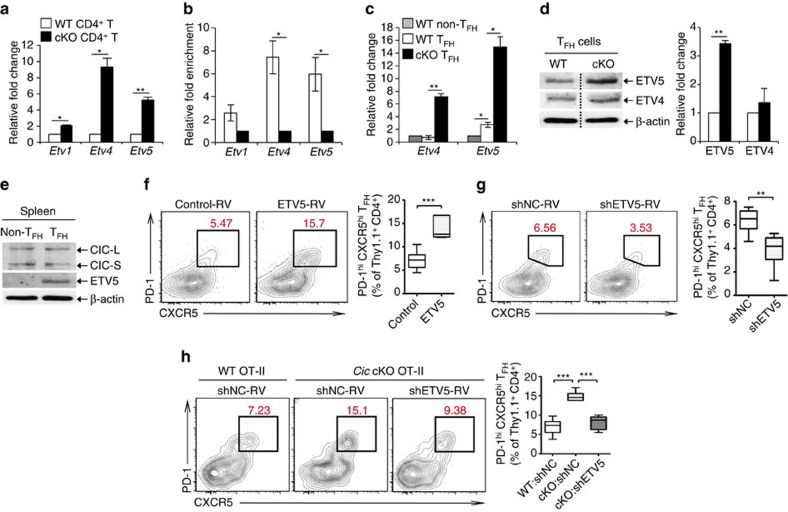
To understand the molecular mechanism mediating the CIC deficiency-induced increase in TFH cells, we set out to identify CIC target genes in CD4+ T cells. We analysed gene expression profiles in naïve and anti-CD3/CD28 antibody-activated CD4+ T cells from WT and Cicf/fCd4-Cre mice by RNA sequencing. Among the genes significantly upregulated (log2(fold change)>1.5; P<0.05) in Cic-deficient CD4+ T cells, only Etv4 and Etv5 were upregulated irrespective of the status of T-cell activation (Supplementary Data 1). Both Etv4 and Etv5 have been well-characterized as CIC target genes in various tissues19,22,24,27. qRT-PCR analysis for mRNA levels of Pea3 group genes (Etv1, Etv4 and Etv5) (Fig. 5a) and quantitative PCR (qPCR) analysis for Pea3 group gene promoter regions in total CIC-associated DNA fragments (Fig. 5b) verified that Etv4 and Etv5 are direct targets of CIC in CD4+ T cells.
Figure 5. De-repression of Etv5 promotes TFH cell differentiation in Cic-deficient CD4+T cells.
(a) qRT-PCR analysis for levels of Pea3 group genes in splenic CD4+ T cells from Cicf/f (WT) and Cicf/fCd4-Cre (cKO) mice. n=3 per each genotype. (b) Chromatin immunoprecipitation (ChIP)-qPCR analysis showing CIC promoter occupancy of Etv4 and Etv5 in CD4+ T cells. CD4+ T cells purified from spleen of Cicf/f and Cicf/fCd4-Cre mice were used. n=3 per each genotype. (c) qRT-PCR analysis for levels of Etv4 and Etv5 in sorted CD4+PD-1−CXCR5− non-TFH and CD4+PD-1+CXCR5+ TFH cells. Splenocytes from 5 to 6 Cicf/f mice or 3 to 4 Cicf/fCd4-Cre mice were pooled and subjected to cell sorting. Three independent experiments were conducted. (d) Western blot analysis for levels of ETV4 and ETV5 in sorted splenic CD4+PD-1+CXCR5+ TFH cells. The dash lines on blot images indicate cropping without image manipulation to either side. Three independent experiments were conducted. The graphs in a–d show data as mean±s.e.m. *P<0.05 and **P<0.01 (two-tailed two-sample unequal variance Student t-test). (e) Western blot analysis for levels of CIC and ETV5 in sorted non-TFH and TFH cells from spleen of WT C57BL/6 (B6) mice. The images are representative of two independent experiments. (f) Thy1.1+ OT-II cells infected with control or ETV5-expressing retrovirus were adoptively transferred to Thy1.2+ B6 recipient mice. Eight days after immunization with NP-OVA in alum, donor cells were analysed for TFH cell differentiation using flow cytometry. Control-RV, n=12; ETV5-RV, n=9. ***P<0.001 (two-tailed two-sample unequal variance Student t-test). (g) Thy1.1+ OT-II cells infected with control (shNC) or Etv5 shRNA (shETV5) expressing retrovirus were subjected to adoptive transfer experiment. Eight days after immunization, donor cells were analysed for TFH cell differentiation. shNC, n=6; shETV5, n=8. **P<0.01 (two-tailed two-sample unequal variance Student t-test). (h) WT and Cic cKO OT-II cells were prepared from spleens of Thy1.1+ OT-II Cicf/f and Thy1.1+ OT-II Cicf/fCd4-Cre mice, respectively. WT OT-II cells infected with shNC retrovirus and Cic cKO OT-II cells infected with shNC or shETV5-expressing retrovirus were transferred into Thy1.2+ B6 recipient mice. Seven days after immunization with NP-OVA in alum, donor cells were analysed for TFH cell differentiation. WT:shNC, n=9; cKO:shNC, n=6; cKO:shETV5, n=6. ***P<0.001 (two-tailed two-sample unequal variance Student t-test).
We then examined the levels of Etv4 and Etv5 in CD4+PD-1+CXCR5+ TFH cells from WT and Cicf/fCd4-Cre mice by qRT-PCR analysis. Consistent with the previous result (Fig. 5a), levels of both genes were markedly increased in Cic-deficient TFH cells (Fig. 5c), indicating that their expression was derepressed in TFH cells in the absence of CIC. Interestingly, however, only ETV5 expression, but not ETV4 expression, was substantially increased in Cic-deficient TFH cells at the protein level (Fig. 5d), suggesting that ETV4 expression is tightly controlled in TFH cells at the posttranscriptional level. We also found that ETV5 levels were higher in TFH cells than in non-TFH cells (Fig. 5c,e), whereas CIC levels in the two T-cell populations were comparable (Fig. 5e), implying that ETV5 might be critical for differentiation or function, or both, of TFH cells. To further explore this possibility, we performed adoptive transfer experiments using OT-II cells, which express ovalbumin (OVA)-specific TCRs55. Thy1.1+OT-II cells were infected with control or ETV5-expressing retrovirus (Supplementary Fig. 13a) and adoptively transferred to Thy1.2+ recipient mice. Immunization of the host mice with 4-hydroxyl-3-nitrophenyl (NP)-OVA in alum resulted in generation of a higher percentage of TFH cells (Fig. 5f), demonstrating that ETV5 promotes TFH cell differentiation. Consistent with this result, knockdown of ETV5 in OT-II cells by shRNA against Etv5 (shETV5) suppressed TFH cell differentiation (Fig. 5g). Lastly, we conducted the adoptive transfer experiment to examine whether the increased TFH cells in Cic mutant mice was due to derepression of ETV5. WT and Cic-deficient OT-II cells infected with control or shETV5-expressing retrovirus were adoptively transferred to Thy1.2+ recipient mice. Seven days after immunization with NP-OVA in alum, the donor OT-II cells were analysed for TFH cell differentiation by flow cytometry. As expected from previous results (Figs 2a,b and 4f), TFH cell differentiation was more prominent with Cic-deficient donor OT-II cells than with WT OT-II cells (Figs 5h and 6f). Strikingly, such enhanced TFH cell differentiation of Cic-deficient OT-II cells was blunted when ETV5 expression was knocked-down with shETV5 (Fig. 5h, Supplementary Fig. 13b). Altogether, these data demonstrate that the CIC–ETV5 axis is crucial for regulation of TFH cell differentiation.
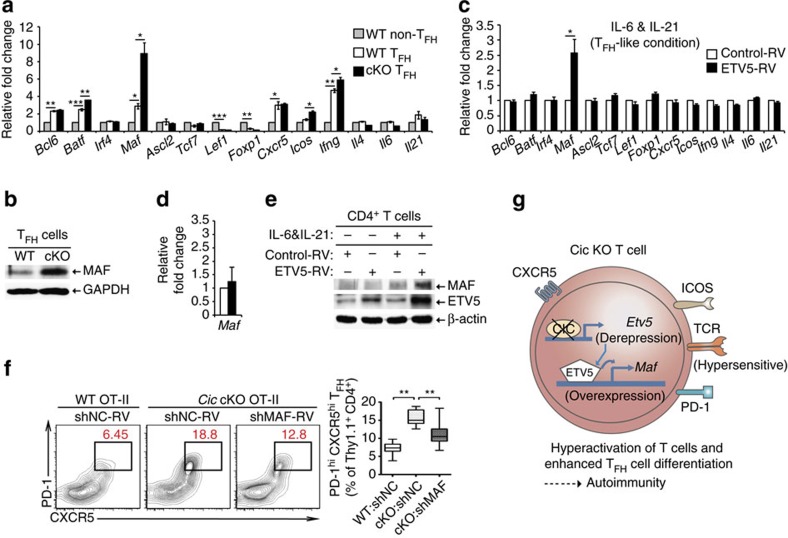
Figure 6. Maf is a critical downstream target of the CIC–ETV5 axis in the process of TFH cell differentiation.
(a) qRT-PCR analysis for levels of TFH-related genes in sorted non-TFH and TFH cells from Cicf/f and Cicf/fCd4-Cre mice. Splenocytes from 5 to 6 Cicf/f mice or 3 to 4 Cicf/fCd4-Cre mice were pooled and subjected to cell sorting. Three independent experiments were performed. (b) FACS-sorted splenicTFH cells from Cicf/f and Cicf/fCd4-Cre mice were subjected to western blot analysis for MAF expression. The images are representative of two independent experiments. (c) qRT-PCR analysis of TFH-related genes in CD4+ T cells infected with control or ETV5-expressing retrovirus. The cells were incubated in the presence of IL-6 and IL-21, and re-stimulated with anti-CD3 for 2 h prior to RNA extraction. Four independent experiments were performed. (d) qRT-PCR analysis for Maf levels in CD4+ T cells infected with control or ETV5-expressing retrovirus in the absence of IL-6 and IL-21. Before RNA extraction, the cells were activated with anti-CD3 for 2 h. Four independent experiments were performed. (a,c,d) Error bars indicate s.e.m. *P<0.05, **P<0.01 and ***P<0.001 (two-tailed two-sample unequal variance student t-test). (e) Western blot analysis for MAF levels in CD4+ T cells infected with control or ETV5-expressing retrovirus. The cells were cultured in the presence or absence of IL-6 and IL-21, and then re-stimulated with anti-CD3 for 12 h. The images are representative of two independent experiments. (f) WT Thy1.1+ OT-II cells infected with control retrovirus and Cic null Thy1.1+ OT-II cells infected with control or Maf shRNA (shMAF) expressing retrovirus were transferred into Thy1.2+ B6 recipient mice. Seven days after immunization with NP-OVA in alum, the Thy1.1+ OT-II cells were analysed for TFH cell differentiation using flow cytometry. WT:shNC, n=11; cKO:shNC, n=9; cKO:shMAF, n=8. **P<0.01 (two-tailed two-sample unequal variance Student t-test). (g) Schematic illustration on how CIC deficiency induces T-cell activation, TFH cell differentiation and autoimmunity. CIC deficiency in T cells makes the TCR response hypersensitive, thereby inducing T-cell activation. During TFH cell differentiation, de-repression of Etv5 induces Maf expression in Cic null TFH cells, subsequently promoting TFH cell differentiation. These T-cell abnormalities could contribute to autoimmunity in the Cic-deficient mice.
Maf as a critical ETV5 target in TFH cell differentiation
ETV5 function in TFH cell development has not been investigated. To comprehensively understand how the CIC–ETV5 axis regulates differentiation of TFH cells, we sought to identify ETV5 target genes that could be involved in this process. We initially analysed expression profiles of several TFH-related genes in non-TFH and TFH cells from WT and Cicf/fCd4-Cre mice. Bcl6, Batf, Maf, Cxcr5 and Ifng were significantly increased, whereas Lef1 and Foxp1 were decreased in WT TFH cells compared with WT non-TFH cells (Fig. 6a). Importantly, Batf, Maf, Icos and Ifng were significantly upregulated in Cic-deficient TFH cells compared with WT TFH cells; the increase was greatest in Maf, which promotes TFH cell differentiation and GC reactions14,16 (Fig. 6a). We confirmed the overexpression of MAF in Cic-deficient TFH cells at the protein level (Fig. 6b).
We also examined which TFH-related genes are regulated by ETV5 in CD4+ T cells. Anti-CD3/CD28 antibody-activated CD4+ T cells were infected with control or ETV5-expressing retrovirus and cultured in the presence of IL-6 and IL-21 (TFH-like condition)56. Among the genes tested, only Maf levels were substantially increased by ETV5 overexpression (Fig. 6c). Consistent with this, it is known that Maf is a direct target gene of ETV5 in ocular lens cells57. We confirmed that ETV5 directly bound to Maf promoter in IL-6-treated CD4+ T cells by chromatin immunoprecipitation (ChIP)-qPCR analysis (Supplementary Fig. 14). In contrast, in the absence of IL-6 and IL-21, ETV5 overexpression did not significantly increase levels of Maf (Fig. 6d), suggesting that ETV5 might induce expression of Maf during TFH cell development. These results were also confirmed by western blot analysis (Fig. 6e). Because IL-6 and IL-21 can activate STAT3 pathway, we examined whether activation of STAT3 is required for ETV5-mediated induction of Maf expression. Treatment with AG490 (inhibitor of JAK2-STAT3 pathway)58 and Stattic (inhibitor for STAT3 dimerization)59 completely suppressed the induction of Maf expression by ETV5 in CD4+ T cells incubated under TFH-like condition (Supplementary Fig. 15a,b). However, treatment with Stattic did not affect ETV5 promoter occupancy of Maf (Supplementary Fig. 15c). These results indicate that STAT3 activity is required for ETV5-mediated induction of Maf expression, but dispensable for ETV5 binding to Maf promoter.
Finally, we examined whether the upregulation of MAF expression contributed to the enhanced TFH cell differentiation in Cic-deficient T cells by the adoptive transfer experiment. WT and Cic null OT-II cells transduced with control or Maf shRNA (shMAF)-expressing retrovirus were adoptively transferred to Thy1.2+ recipient mice. Seven days after immunization with NP-OVA in alum, the donor OT-II cells were analysed for TFH cell differentiation by flow cytometry. The enhanced TFH cell differentiation of Cic-deficient OT-II cells was indeed significantly alleviated by knockdown of Maf (Fig. 6f, Supplementary Fig. 13b). Taken together, these data suggest that the derepression of Etv5 increased Maf expression in Cic-deficient TFH cells, and subsequently promoted TFH cell differentiation and GC responses.
Discussion
In this study, we examined a previously unrecognised function of CIC in the immune system. Our findings demonstrate that CIC is a key transcriptional repressor that maintains peripheral immune homeostasis and restrains TFH cell differentiation, thereby suppressing autoimmunity (Fig. 6g). CIC deficiency causes lymphoproliferative autoimmunity in mice, which is accompanied by augmented T-cell responses and increased frequencies of TFH and GC B cells in secondary lymphoid organs. Given that excessive formation of TFH cells is often observed in patients with autoimmune diseases5 and that autoimmunity in Roquinsan mice is reduced when TFH cell development is suppressed either by removing one allele of Bcl6, by deleting SAP (Sh2d1a), or by dampening IFNγ signalling40,60, spontaneous induction of the GC response might be a primary cause of the onset of autoimmune-like symptoms in Cic mutant mice.
The immune cell-specific Cic null mice shared many phenotypes including hyperglobulinemia, T-cell hyperactivation, accumulation of effector/memory cells with TH1 and TH2 phenotypes, systemic autoimmunity and increased proportions of CD4+FOXP3+CD25− T, TFH and GC B cells with the T-cell-specific Cic null mice, demonstrating that T-cell-intrinsic functions of CIC are crucial for maintenance of T-cell homeostasis and suppression of spontaneous induction of the GC response and autoimmunity. A specific loss of CIC in B cells, from which antigen presentation and co-stimulatory signals are required for differentiation and maintenance of TFH cells8, did not promote TFH cell differentiation in mice (Supplementary Fig. 16), accentuating the T-cell-intrinsic requirement of CIC in regulation of TFH cell development. However, most phenotypes were more prominent in the immune cell-specific Cic null mice than in the T-cell-specific Cic-deficient mice; this difference implies that T-cell-extrinsic mechanisms also contributed in part to those phenotypes. To comprehensively understand the function of CIC in maintenance of peripheral immune tolerance, functions of CIC in other types of immune cell, such as DCs and B cells, need to be further determined.
Our study provided a molecular basis of how CIC deficiency promotes TFH cell differentiation. We determined that ETV5 is a critical CIC target that promotes TFH cell differentiation. Among Pea3 group genes, Etv4 and Etv5 are major CIC target genes in CD4+ T cells. Although mRNA levels of both genes increased greatly in the Cic null TFH cells, only ETV5 expression increased significantly at the protein level, indicating that ETV4 levels are tightly controlled in TFH cells by posttranscriptional regulation. ETV5 expression also seems to be regulated at the posttranscriptional level in the Cic null TFH cells, because the increase in ETV5 expression by CIC deficiency was greater at the mRNA level (∼5.5-fold) than at the protein level (∼3.4 fold). One possible mechanism of posttranscriptional control of PEA3 group transcription factor levels in TFH cells is proteasomal degradation mediated by E3 ubiquitin ligase COP1 (also known as RFWD2). COP1-mediated degradation of PEA3 group transcription factors is involved in regulation of prostate cancer progression61, β-cell insulin secretion62 and lung-branching morphogenesis63, but the function of COP1 in TFH cell development has not been determined. Because Rfwd2 is expressed in T lymphocytes64, one interesting follow-up would be to examine whether COP1 regulates TFH cell differentiation through degradation of ETV5 or other PEA3 group transcription factors in T cells.
We identified Maf as an important downstream target gene of the CIC–ETV5 axis in the process of TFH cell differentiation. To identify genes that may be regulated by ETV5 during TFH cell differentiation, we examined 14 genes that encode transcription factors, cytokines or cell surface molecules, all of which are critically involved in regulation of TFH cell development. Strikingly, ETV5 overexpression induced only Maf expression in CD4+ T cells. The finding that TFH cell differentiation of the Cic null OT-II cells was suppressed by knockdown of Maf further demonstrates that the CIC–ETV5 axis regulates TFH cell differentiation via MAF. However, in the Cic-deficient OT-II cells, Maf RNAi suppressed TFH cell differentiation less efficiently than did Etv5 RNAi; this comparison suggests that, in addition to Maf, other ETV5 target genes may contribute to promotion of TFH cell differentiation. Moreover, Cic-deficient TFH cells exhibited the enhanced expression of IFNγ, which is critical for TFH cell differentiation40, but not regulated by MAF14. This result also implies that Maf may not be the only ETV5 target gene that contributes to promotion of TFH cell differentiation. To improve our understanding of how ETV5 promotes TFH cell development, studies of the function of ETV5 in TFH cells, such as a genome-wide identification of ETV5 targets and their function in TFH cells, should be conducted.
Given that ETV5 upregulated Maf expression only in the presence of IL-6 and IL-21, which are cytokines that induce TFH cell development56, and that Maf levels were increased in Cic null TFH cells, but in neither naïve nor anti-CD3/CD28-activated Cic-deficient CD4+ T cells (Supplementary Data 1), ETV5 might specifically regulate Maf expression during TFH cell development. Consistent with this inference, Maf levels were comparable between WT and Etv5 deficient TH17 cells65. However, ETV5 facilitates differentiation of TH17 cells65 and of IL-17-producing γδ effector cells66 by activating Il17 expression. ETV5 also promotes IL-9 production in TH9 cells by binding and recruiting histone acetyltransferases to the Il9 locus67. These findings, together with our data, suggest that ETV5 participates in differentiation processes of several TH subtypes by regulating a different set of genes in each TH subset.
Our study is the first demonstration that the CIC–ETV5 axis is critical for regulation of TFH cell development. This axis also contributes to cancer progression22,23,24; therefore, dysregulation of this regulatory axis could contribute to pathogenesis of both cancer and antibody-mediated autoimmune diseases. In this regard, CIC and ETV5 themselves or their target genes that commonly mediate pathogenesis of both diseases could be evaluated as molecular targets for treatment of both autoimmune diseases and cancers.
Methods
Mice
All mice were maintained on a C57BL/6 background. Vav1-Cre (ref. 68), Cd4-Cre (ref. 69), Foxp3-YFP-Cre (ref. 51), Cd19-Cre (ref. 70) and OT-II (ref. 55) mice have been described previously. The mouse with a loxP-flanked Cic allele was generated by mating Cictm1a(KOMP)Wtsi to Actin-FLP mice (Stock No: 003800). The Cictm1a(KOMP)Wtsi allele had been obtained as ES cells (Clone No. EPD0285_3_F07) from EUCOMM and injected into C57BL/6 blastocysts. Animals were maintained in a specific pathogen-free animal facility under standard 12 h light/12 h dark cycle. Mice were fed standard rodent chow and water ad libitum. Three-to-thirteen mice that were used in individual experiments were assigned randomly to the experimental groups. Male and female mice were also randomly assigned to the experimental groups. Blinding was not possible in most animal experiments. All procedures were approved by the Pohang University of Science and Technology Institutional Animal Care and Use Committee.
Enzyme-linked immunosorbent assay
Ninety six-well ELISA plates were pre-coated with 2 μg ml−1 anti-mouse Ig (1010-01, Southern Biotechnology) at 4 °C overnight. On the next day, the plates were washed with washing buffer (phosphate-buffered saline (PBS) with 0.05% Tween 20) and blocked with blocking buffer (washing buffer with 2% bovine serum albumin (BSA)) for 1–2 h at room temperature (RT); then 50 μl of diluted serum and standards were added and incubated for 2 h at RT. The plates were washed and incubated for 1 h at RT with secondary antibodies conjugated with horseradish peroxidase. For total IgG, goat anti-mouse IgG (H+L) (pooled antisera from goats hyperimmunized with mouse IgG, 1:3,000 diluted, 1031-05) was used; for IgG1, goat anti-mouse IgG1 (pooled antisera from goats hyperimmunized with mouse IgG1, 1:3,000 diluted, 1070-05) was used; for IgG2a, goat anti-mouse IgG2a (pooled antisera from goats hyperimmunized with mouse IgG2a paraproteins, 1:1,000 diluted, 1080-05) was used; and for IgM, goat anti-mouse IgM (pooled antisera from goats hyperimmunized with mouse IgM, 1:2,000 diluted, 1020-05) was used. The plates were washed at least five times, then 50 μl of TMB substrate (SurModics, TMBM-1000-01) was added and incubated for 20 min in the dark. Finally, 50 μl of stop solution (1 M H2SO4) was added and the plate was read at 450 nm. The serum concentration was calculated according to the standard curve generated. Serum anti-dsDNA concentration was measured using mouse anti-dsDNA ELISA Kit (Shibayagi, AKRDD-061) according to the manufacturer’s instruction. For IL-2 ELISA, naïve CD4+ T cells were activated with plate-bound anti-CD3 (1.0 μg ml−1, 145-2C11, BD) in the presence or absence of plate-bound anti-CD28 (2.0 μg ml−1, 37.51, BD), then cell supernatants were collected 48 and 72 h after stimulation. Briefly, plates pre-coated with 2 μg ml−1 anti-IL-2 (14-7022, eBioscience) were washed and blocked as described above, then 50 μl of diluted samples and standards were added and incubated for 2 h at RT. The plates were washed, then incubated sequentially with biotin-conjugated anti-IL-2 (13-7021) for 1.5 h, avidin-HRP (18-4100) for 30 min, TMB substrate for 15 min, and 1 M H2SO4 as a stop solution.
Tissue histology
Liver, lung and kidney tissues obtained from 12-month-old Cicf/f and Cicf/fVav1-Cre mice or 14.5-month-old Cicf/f and Cicf/fCd4-Cre mice were fixed in 10% formalin, then embedded in paraffin before sectioning. The tissues were cut into 5-μm sections (Leica RM2245), then sections were deparaffinized and dehydrated by using xylene, 100% ethanol and 95% ethanol sequentially. They were washed in distilled water, then stained with haematoxylin (Sigma, HHS32) and eosin (Sigma, HT110132). After H&E staining, × 200 images of specimens were obtained using an Olympus CX31 light microscope.
Immunofluorescence staining of IgG deposition
Kidney tissues from 12-month-old Cicf/f and Cicf/fVav1-Cre mice were snap-frozen in OCT medium (Sakura Finetek, 4583), then cut into 10-μm sections (Leica CM3050S). The kidney sections were air-dried for 24 h, then washed in PBS (pH 7.4) to remove fixation compound. Diluted anti-mouse CD16/CD32 antibodies (eBioscience, 14-0161) were treated for Fc blocking and incubated for 30 min at RT. After washing, the kidney sections were stained with anti-IgG antibody conjugated to fluorescein isothiocyanate (FITC) (1:500 diluted, Sigma, F0257) and 4′,6-diamidino-2-phenylindole to reveal the IgG complexes and the nuclei, respectively. The specimens were washed in PBS, then covered in mounting medium (Dako, C0563), and × 400 images of specimens were obtained using an Olympus IX82-ZDC2 fluorescence microscope.
Plasmids and retrovirus production
The amino-terminal FLAG-tagged coding sequence (CDS) of mouse Etv5 was amplified by PCR, initially cloned into T-blunt vector (SolGent), and then verified by sequencing. A confirmed Etv5 CDS fragment digested by XhoI/HpaI was sub-cloned into MigR1 retroviral vector (MigR1-ETV5-GFP). The shRNA expression vectors for knockdown of mouse Etv5 and Maf were generated using MSCV-LTRmiR30-PIG (LMP) vector (Open Biosystems) according to the manufacturer’s instruction. The target sequences were as follows. For shETV5: 5′-ACCCGAGAGACTGGAAGGCAAA-3′ and for shMAF: 5′-AAGATATAACCTGCAAGCATAT-3′.
Viruses were generated by transient co-transfection of Platinum-E (Plat-E) retroviral packaging cell line (Cell Biolabs) with the cloned retroviral vectors and pCL-Eco helper plasmid (Imgenex). Briefly, 0.5–0.8 × 106 plat-E cells were plated in 6-well plates. On the next day, the cells were transfected with 1.2 μg of retroviral vector and 0.8 μg of pCL-Eco using FuGENE HD transfection reagent (E2311, Promega). Retrovirus-containing supernatants were collected 48 h later and frozen at −80 °C.
Flow cytometry and cell sorting
Single-cell suspensions of spleens and thymuses were prepared and surface-stained in FACS buffer (PBS+1.5% fetal bovine serum (FBS)) with monoclonal antibodies. The following antibodies were obtained from eBioscience, BD PharMingen or BioLegend: anti-CD4 (1:100 diluted, GK1.5), anti-CD8 (1:100 diluted, 53-6.7), anti-CD3 (1:100 diluted, 17A2), anti-CD11b (1:100 diluted, M1/70), anti-CD11c (1:100 diluted, N418), anti-MHCII (1:100 diluted, AF6-120.1), anti-F4/80 (1:100 diluted, BM8), anti-CD44 (1:100 diluted, IM7), anti-CD62L (1:100 diluted, MEL-14), anti-CD25 (1:50 diluted, PC61), anti-PD-1 (1:50 diluted, J43), anti-GITR (1:100 diluted, DTA-1), anti-OX40 (1:100 diluted, OX-86), anti-ICOS (1:100 diluted, 7E.17G9), anti-B220 (1:200 diluted, RA3-6B2), anti-IgM (1:100 diluted, II/41), anti-CD43 (1:100 diluted, R2/60), anti-GL-7 (1:250 diluted, GL7), biotinylated anti-mouse FAS (1:200 diluted, Jo2), anti-CD40 (1:100 diluted, 1C10), anti-CD80 (1:100 diluted, 16-10A1), anti-CD86 (1:100 diluted, GL1), anti-ICOSL (1:100 diluted, HK5.3), anti-PDCA-1 (1:100 diluted, eBio927), anti-CCR4 (1:100 diluted, 2G12), anti-CCR6 (1:100 diluted, 140706), anti-CXCR3 (1:150 diluted, CXCR3-173), anti-CTLA-4 (1:100 diluted, UC10-4B9), anti-CD103 (1:100 diluted, 2E7), anti-GARP (1:100 diluted, YGIC86), anti-IgG2a (1:100 diluted, m2a-15F8), anti-CD304 (Neuropilin-1; 1:50 diluted, 3E12) and APC-conjugated streptavidin (1:100 diluted, eBioscience). For CXCR5 staining, tertiary staining was used11. Briefly, the cells were sequentially incubated with following reagents: rat anti-CXCR5 (1:100 diluted, 2G8) for 1 h, biotinylated anti-rat IgG (1:400 diluted, eBioscience) for 30 min, and then APC- or PerCP-Cy5.5-labelled streptavidin (1:100 diluted, eBioscience) with other surface antibodies. For intracellular staining, cells were fixed and permeabilized with ‘Foxp3 staining buffer set’ (00-5523, eBioscience) following the manufacturer’s protocol, then stained with anti-Foxp3 (1:75 diluted, MF23, BD), anti-BCL6 (1:50 diluted, 7D1), anti-T-bet (1:100 diluted, 4B10), anti-GATA3 (1:100 diluted, TWAJ), anti-RORγt (1:100 diluted, B2D), anti-Nur77 (1:100 diluted, 12.14) and anti-HELIOS (1:100 diluted, 22F6). For cytokine staining, total splenocytes were stimulated with PMA and ionomycin in the presence of Golgi-stop (554724, BD) and Golgi-plug (555029, BD) for 5 h, and then the intracellular staining of cytokines was performed using antibodies for IFNγ (1:100 diluted, XMG1.2), IL-2 (1:100 diluted, JES6-5H4), IL-17A (1:100 diluted, TC11-18H10), TNF (1:200 diluted, MP6-XT22), IL-21 (1:100 diluted, mhalx21), IL-4 (1:50 diluted, 11B11), IL-9 (1:100 diluted, RM9A4), IL-13 (1:100 diluted, eBio13A) and IL-22 (1:100 diluted, Poly5164). The stained cell samples were analysed using either a CantoII flow cytometer (BD Biosciences) or a LSRFortessa flow cytometer (BD Biosciences). Data were analysed by FlowJo software (Tree Star). MoFlo-XDP (Beckman Coulter) was used for cell sorting. The sorted populations were >98.5% pure. In FACS plots, isotype controls for each of antibodies were used for separating negative and positive populations and all gates were based on this method. All FACS sorting/gating strategies are described in Supplementary Fig. 17.
In vitro T-cell proliferation assay
Naïve CD4+ T cells were purified to >95% purity from pooled spleens and lymph nodes of Cicf/f and Cicf/fVav1-Cre mice by using a CD4+ negative selection method (Stem Cell Technologies) for CD4+CD25−CD44loCD62Lhi cells. The cells were labelled with 5 μM Cell Trace Violet (CTV, Invitrogen, C34557) in pre-warmed PBS. Labelling was performed by incubating in a 37 °C incubator for 20 min and stopped by adding five times the original staining volume of culture medium (containing ≥1% protein). The cells were incubated for at least 10 min to allow the CTV reagent to undergo acetate hydrolysis. The cells were stimulated with plate-bound anti-CD3 (1.0 μg ml−1, 145-2C11, BioXcell) and anti-CD28 (2.0 μg ml−1, 37.51, BioXcell), then collected 72 h after stimulation. The samples were analysed using the LSRFortessa flow cytometer. Data were analysed using FlowJo software (Tree Star).
In vitro T-cell differentiation assay
Naïve CD4+ T cells (CD4+CD25−CD44loCD62Lhi) were stimulated with plate-bound anti-CD3 and anti-CD28 in medium supplemented as follows: for TH0 differentiation, anti-IL-4 (10 μg ml−1) and anti-IFNγ (10 μg ml−1); for TH1 differentiation, IL-12 (10 ng ml−1) and anti-IL-4 (10 μg ml−1); for TH2 differentiation, IL-4 (10 ng ml−1) and anti-IFNγ (10 μg ml−1); for TH17 differentiation, IL-1β (20 ng ml−1), IL-6 (20 ng ml−1), TGF-β (2 ng ml−1), anti-IFNγ (10 μg ml−1) and anti-IL-4 (10 μg ml−1). The samples were analysed using the LSRFortessa flow cytometer, and data were analysed using FlowJo software.
In vitro Treg cell suppression assay
CD4+CD25− CD44lo T cells were prepared from CD45.1 C57BL/6 mice. The cells were labelled with CTV and used as responder cells (Tresp). CD4+CD25+ Treg cells were isolated from Cicf/f and Cicf/fCd4-Cre mice by the MoFlo-XDP (Beckman Coulter). Tresp cells were mixed with an equal number of irradiated splenocytes (3000 Rad) from Rag1−/− mice and different numbers of CD4+CD25+ Treg cells and incubated in round bottom 96-well tissue culture plates with anti-CD3 (1 μg ml−1). Seventy-two hours later, CD45.1+ CD4+ T cells were gated and proliferation was analysed based on CTV dilution. For detection of IL-2 secretion, CD4+CD25+ Treg cells from Cicf/f and Cicf/fCd4-Cre mice were used at a fixed concentration of 1 × 105 cells per well and co-cultured with CD4+CD25− Tresp cells in 96-well plates. The cells were stimulated with plate-bound anti-CD3 (1 μg ml−1) and anti-CD28 (1 μg ml−1), then supernatants were collected 72 h after stimulation. The samples were subjected to ELISA for IL-2 concentration.
Thymic negative selection assay
In vitro thymic negative selection assay was performed as described previously45. Briefly, DP thymocytes were prepared from Cicf/f and Cicf/fCd4-Cre mice using the MoFlo-XDP and stimulated with anti-CD3 (20 μg ml−1) and anti-CD28 (50 μg ml−1) for 24 h to evaluate TCR-induced apoptosis. Cell death was assessed by staining for Annexin V according to the manufacturer’s protocol (BD, 556419). To investigate the induction of Nur77 expression after TCR cross-linking, thymocytes from Cicf/f and Cicf/fCd4-Cre mice were stimulated with anti-CD3 (5 μg ml−1) and anti-CD28 (10 μg ml−1) for 6 h. The samples were analysed using the LSRFortessa flow cytometer, and data were analysed using FlowJo software.
Western blot analysis
Single-cell suspensions of spleen, lymph nodes and thymus from Cicf/f and Cicf/fVav1-Cre mice were used as total immune cells in lymphoid organs. Naïve CD4+ T cells were activated by anti-CD3 and anti-CD28 and collected on days 0 (unstimulated), 1, 2 and 3. B220+ B and CD4+ T cells from Cicf/f and Cicf/fCd4-Cre mice were prepared using MACS (130-049-501 and 130-104-454, Miltenyi Biotec) according to the manufacturer’s protocol. Naïve CD4+ T, effector/memory CD4+ T, TFH and non-TFH cells were purified from total CD4+ T cells by FACS sorting using the MoFlo-XDP. Splenic CD19+ B and CD3+ T cells were prepared from Cicf/f and Cicf/fCd19-Cre mice by FACS sorting using the MoFlo-XDP. Total lysates from 0.2 to 1.0 × 106 cells of each cell population were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE). The primary antibodies used are as follows. anti-CIC (1:500 diluted, homemade)28, anti-ETV4 (1:1,000 diluted, 10684-1-AP, ProteinTech), anti-ERM/ETV5 (1:1,000 diluted, ab102010, Abcam), anti-MAF (1:750 diluted, sc-7866, Santa Cruz Biotechnology), anti-β-actin (1:2,000 diluted, sc-47778, Santa Cruz Biotechnology) and anti-GAPDH (1:2,000 diluted, sc-32233, Santa Cruz Biotechnology). Raw western blot images are presented in Supplementary Figs 18 and 19.
Retroviral transduction and cell transfers
OT-II Thy1.1 CD4+ T cells were purified from whole splenocytes by negative selection using EasySep (StemCell), then re-suspended in T-cell medium (RPMI-1640+10% FBS, supplemented with 2.05 mM L-Glutamine, 100 U ml−1 Penicillin/Streptomycin and 55 mM BME). The purified OT-II T cells were stimulated in 24-well plates pre-coated with 2 μg ml−1 anti-CD3 (145-2C11, BD) and anti-CD28 (37.51, BD) in the presence of rmIL-2 (50 U ml−1). The OT-II T cells were transduced with control retrovirus (MigR1-GFP) or ETV5-expressing retrovirus (MigR1-ETV5-GFP) at 24 and 48 h after in vitro stimulation. After a total of 72 h stimulation, the cells were transferred into new six-well plates with rmIL-2 for 24 h, and then transferred again into new six-well plates in the presence of rmIL-7 for 24 h prior to cell sorting. For shRNA experiments, after in vitro stimulation and double transduction with shNC, shETV5 or shMAF expressing retrovirus for 72 h, the cells were transferred into six-well plates with rmIL-2 for 12 h and then transferred into new six-well plates with rmIL-7 for 12 h.
For adoptive transfer experiments, 0.2–0.5 × 106 OT-II T cells infected with retroviruses were transferred into C57BL/6 Thy1.2 recipient mice by tail-vein injection. Three days later, mice were immunized intraperitoneally with 200 μg of NP-OVA (9006-59-1, Sigma) in 200 μl of alum. Seven or eight days after immunization, the donor cells were analysed for TFH differentiation by flow cytometry.
Cell culture
Naïve (CD4+CD25−CD44loCD62Lhi) CD4+ T cells were isolated by negative selection using EasySep (StemCell), then activated with plate-bound anti-CD3 (145-2C11, BD) and anti-CD28 (37.51, BD) under neutral (anti-IFNγ, anti-IL-4, anti-TGFβ) or TFH-like (anti-IFNγ, anti-IL-4, anti-TGFβ, rmIL-6, rmIL-21) condition. The cells were transduced with retrovirus (MigR-GFP or MigR-ETV5-GFP) at 24 h and at 36 h after in vitro stimulation. Two days after culture, the cells were rested for 24 h in the presence of rmIL-7, then re-stimulated with pre-coated anti-CD3 for 2 h for RNA samples and for 12 h for protein samples.
For STAT3 inhibitor treatment experiments, naïve CD4+ T cells were polarized under TFH-like condition as described above. The cells were transduced with retrovirus (MigR1-GFP or MigR1-ETV5-GFP) at 24 h after in vitro stimulation, then treated with AG490 (Tokyo Chemical, 50 μM) for 16 h or Stattic (Santa Cruz Biotechnology, 20 μM) for 12 h. Two days after culture, the cells were rested for 24 h. The cells were collected for RNA samples.
ChIP and qPCR
Chromatin immunoprecipitation was performed as previously described28. Chromatin from 5 × 106 CD4+ T cells were crosslinked in 1.5% formaldehyde for 12 min with constant shaking, quenched cross-linking by adding 1.5 M glycine for 10 min, and then rinsed with cold PBS twice. After centrifugation for 5 min, the pellet was resuspended in Buffer A (100 mM Tris pH 9.4, 1 × protease inhibitor cocktail (11836170001, Roche)), Buffer 1 (10 mM HEPES, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton-X, 1 × protease inhibitor cocktail), and Buffer 2 (10 mM HEPES, 0.2 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 1 × protease inhibitor cocktail), successively. T-cell lysate was resuspended in 200 μl of nuclei lysis buffer (5 mM Tris pH 8.1, 10 mM EDTA, 1% SDS, 1 × protease inhibitor cocktail) and sonicated. After microcentrifugation, the supernatant was pre-cleared with protein G agarose (16-266, Millipore) in dilution buffer (16.7 mM Tris pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton-X, 1 × protease inhibitor) for 1–2 h. Six micrograms of anti-CIC antibody (homemade)28 were added to chromatin samples and incubated overnight at 4 °C. The chromatin and antibody mixtures were further incubated with protein G agarose for 2–4 h at 4 °C. After washing, bound chromatins were eluted twice by elution buffer (0.5% SDS and 0.1 M NaHCO3) and reverse-crosslinked with 200 mM NaCl for at least 4 h at 65 °C. Proteins were digested by proteinase K and DNA was purified by AccuPrep Gel Purification Kit (K-3034, Bioneer). qPCR was performed to quantify the relative enrichment of promoter regions of Pea3 group genes in the immunoprecipitated DNA fragments.
For ETV5 ChIP experiment, CD4+ T cells were activated with plate bound anti-CD3 (145-2C11, BD) and anti-CD28 (37.51, BD) in the presence of rmIL-6. Sixteen hours after in vitro stimulation, 1 × 107 cells and 2 μg of anti-ETV5 antibody (sc-22807, Santa Cruz Biotechnology) were used for each ChIP experiment. To inhibit STAT3 activity, CD4+ T cells were activated with anti-CD3/CD28 in the presence of rmIL-6 for 12 h, and then Stattic (20 μM) was treated for 12 h. The primer sequences used are listed in Supplementary Data 2.
qRT-PCR
Total RNA was extracted using Trizol reagent (301-001, GeneAll), then 1–2 μg of the total RNA was subjected to cDNA synthesis using a GoScript Reverse Transcription System (A5000, Promega). Each gene expression level was normalized to Hprt levels and presented as relative to WT. The primers used in qRT-PCR analyses are listed in Supplementary Data 3.
RNA sequencing and data analysis
Spleens were dissected from 10-week-old Cicf/f and Cicf/fCd4-Cre mice. CD4+ T cells were pre-enriched using EasySep (19860 and 18001, StemCell), then further sorted on the basis of surface markers of naïve CD4+ T cells (CD4+CD44loCD62Lhi). Sorted naïve CD4+ T cells were activated in 24-well plates pre-coated with 2 μg ml−1 anti-CD3 (145-2C11, BD) and anti-CD28 (37.51, BD) for 3d. Total RNA was extracted from naïve and activated CD4+ T cells using ReliaPrep RNA Cell Miniprep Kit (Z6011, Promega). The library for mRNA sequencing was generated using a Illumnia TruSeq Preparation Kit (RS-122-2001) and sequenced on a NextSeq 500 sequencer (Illumnia). Tophat (v 2.0.10) was used to map sequencing reads to the mouse reference genome (mm9 RefSeq). Assembly of transcripts and identification of differentially expressed genes (DEGs, Fold change (log2)>1.5; P<0.05) were conducted by Cufflinks (v 2.1.1).
Statistical analysis
For statistical analysis, all experiments were performed more than three times independently. Statistical analyses were performed using the Student t-test (two-tailed, two-sample unequal variance). P<0.05 was considered significant. In bar graphs, bars indicate means and error bars indicate SEM. In box-and-whisker plots, boxes represent median with upper and lower quantiles and whiskers represent values of maximum and minimum.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information Files, or from the corresponding authors on reasonable request. The RNA-seq data were deposited in the Gene Expression Omnibus (NCBI) data repository under accession number GSE84125.
Additional information
How to cite this article: Park, S. et al. Capicua deficiency induces autoimmunity and promotes follicular helper T cell differentiation via derepression of ETV5. Nat. Commun. 8, 16037 doi: 10.1038/ncomms16037 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Dr Huda Y. Zoghbi for provision of the Cic floxed mice, Drs Alexander Y. Rudensky and Dipayan Rudra for provision of Foxp3-YFP-Cre mice, and Drs Yeonseok Chung, V. Narry Kim and Charles D. Surh for helpful discussions and comments on this study. This work was supported by Samsung Science and Technology Foundation under project number SSTF-BA1502-14 to Y.L. S.P., S.L., H.H., J.-S.L., Y.M.K. and G.Y.P. were supported by the BK21 Plus Program. S.-W.L. was supported by the grant from Cooperative Research Program for Agriculture Science and Technology Development under project number PJ01131603 (Rural Development Administration, Republic of Korea). S.-H.I. was supported by the grant from the Institute for Basic Science (IBS-R005-G1). D.H. was supported by the grant from the Institute for Basic Science (IBS-R013-G1).
Footnotes
The authors declare no competing financial interests.
Author contributions Y.L., S.-W.L., S.-H.I., S.P., S.L. and C.-G.L. designed the study. S.P., S.L., C.-G.L., H.H., J.-S.L., Y.M.K. and G.Y.P. performed the experiments. D.H. analysed RNA sequencing data. S.B.L., Y.S.C. and J.D.F. provided intellectual contributions. Y.L., S.-W.L. and S.P. wrote the paper. S.-H.I., Y.S.C. and J.D.F. edited the paper.
References
- Klein U. & Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat. Rev. Immunol. 8, 22–33 (2008). [DOI] [PubMed] [Google Scholar]
- Gatto D. & Brink R. The germinal center reaction. J. Allergy Clin. Immunol. 126, 898–907 (2010). [DOI] [PubMed] [Google Scholar]
- De Silva N. S. & Klein U. Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 15, 137–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C. G., Sanz I. & Cook M. C. Dysregulation of germinal centres in autoimmune disease. Nat. Rev. Immunol. 9, 845–857 (2009). [DOI] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson N. et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 62, 234–244 (2010). [DOI] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011). [DOI] [PubMed] [Google Scholar]
- Vinuesa C. G., Linterman M. A., Yu D. & MacLennan I. C. M. Follicular helper T cells. Annu. Rev. Immunol. 34, 335–368 (2016). [DOI] [PubMed] [Google Scholar]
- Kerfoot S. M. et al. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity 34, 947–960 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M. et al. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity 34, 961–972 (2011). [DOI] [PubMed] [Google Scholar]
- Johnston R. J. et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R. I. et al. Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D. et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468 (2009). [DOI] [PubMed] [Google Scholar]
- Bauquet A. T. et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10, 167–175 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu Y. et al. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-beta inhibits c-Maf-induced IL-21 production in CD4+ T cells. J. Leukoc. Biol. 87, 703–712 (2010). [DOI] [PubMed] [Google Scholar]
- Kroenke M. A. et al. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol. 188, 3734–3744 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez G., Shvartsman S. Y. & Paroush Z. The Capicua repressor--a general sensor of RTK signaling in development and disease. J. Cell Sci. 125, 1383–1391 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Y. C. et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 127, 1335–1347 (2006). [DOI] [PubMed] [Google Scholar]
- Fryer J. D. et al. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science 334, 690–693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.-C. et al. Disruption of the ATXN1-CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nat. Genet. 49, 527–536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayake K. et al. ERK/p90(RSK)/14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicúa. Biochem. J. 433, 515–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N. et al. miR-93/miR-106b/miR-375-CIC-CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget 6, 23533–23547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleize V. et al. CIC inactivating mutations identify aggressive subset of 1p19q codeleted gliomas. Ann. Neurol. 78, 355–374 (2015). [DOI] [PubMed] [Google Scholar]
- Kawamura-Saito M. et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 15, 2125–2137 (2006). [DOI] [PubMed] [Google Scholar]
- Kurpios N. A., Sabolic N. A., Shepherd T. G., Fidalgo G. M. & Hassell J. A. Function of PEA3 Ets transcription factors in mammary gland development and oncogenesis. J. Mammary Gland Biol. Neoplasia 8, 177–190 (2003). [DOI] [PubMed] [Google Scholar]
- Okimoto R. A. et al. Inactivation of Capicua drives cancer metastasis. Nat. Genet. 49, 87–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. et al. ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev. Cell 21, 746–757 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. et al. Deficiency of Capicua disrupts bile acid homeostasis. Sci. Rep. 5, 8272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-S. et al. A draft map of the human proteome. Nature 509, 575–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubberts E. The IL-23-IL-17 axis in inflammatory arthritis. Nat. Rev. Rheumatol. 11, 562 (2015). [DOI] [PubMed] [Google Scholar]
- Bour-Jordan H. & Bluestone J. A. Regulating the regulators: costimulatory signals control the homeostasis and function of regulatory T cells. Immunol. Rev. 229, 41–66 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. et al. Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells. Proc. Natl Acad. Sci. USA 106, 13439–13444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-J., Verbinnen B., Tang X., Lu L. & Cantor H. Inhibition of follicular T-helper cells by CD8(+) regulatory T cells is essential for self tolerance. Nature 467, 328–332 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.-H. & Chung Y. Regulatory T cells in B cell follicles. Immune Netw. 14, 227–236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh A. et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 16, 188–196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonelli M. et al. CD4+CD25−Foxp3+ T cells: a marker for lupus nephritis? Arthritis Res. Ther. 16, R104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutloff A. et al. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature 397, 263–266 (1999). [DOI] [PubMed] [Google Scholar]
- Akiba H. et al. The role of ICOS in the CXCR5+ follicular B helper T cell maintenance in vivo. J. Immunol. 175, 2340–2348 (2005). [DOI] [PubMed] [Google Scholar]
- Yu D. et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature 450, 299–303 (2007). [DOI] [PubMed] [Google Scholar]
- Lee S. K. et al. Interferon-γ excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity 37, 880–892 (2012). [DOI] [PubMed] [Google Scholar]
- Weinstein J. S. et al. TFH cells progressively differentiate to regulate the germinal center response. Nat. Immunol. 17, 1197–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C. G. et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435, 452–458 (2005). [DOI] [PubMed] [Google Scholar]
- Starr T. K., Jameson S. C. & Hogquist K. A. Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139–176 (2003). [DOI] [PubMed] [Google Scholar]
- Sohn S. J., Thompson J. & Winoto A. Apoptosis during negative selection of autoreactive thymocytes. Curr. Opin. Immunol. 19, 510–515 (2007). [DOI] [PubMed] [Google Scholar]
- Hwang S. et al. Reduced TCR signaling potential impairs negative selection but does not result in autoimmune disease. J. Exp. Med. 209, 1781–1795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper C. M. & Paul W. E. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science 236, 944–947 (1987). [DOI] [PubMed] [Google Scholar]
- Huang S. et al. Immune response in mice that lack the interferon-gamma receptor. Science 259, 1742–1745 (1993). [DOI] [PubMed] [Google Scholar]
- Chung Y. et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 17, 983–988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M. A. et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 17, 975–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage P. T., Francisco L. M., Carman C. V. & Sharpe A. H. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat. Immunol. 14, 152–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008). [DOI] [PubMed] [Google Scholar]
- Thornton A. M. et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184, 3433–3441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M. et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J. Exp. Med. 209, 1713–1722 S1-19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J. M. et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J. Exp. Med. 209, 1723–1742 S1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnden M. J., Allison J., Heath W. R. & Carbone F. R. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76, 34–40 (1998). [DOI] [PubMed] [Google Scholar]
- Lu K. T. et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity 35, 622–632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q. et al. Regulation of c-Maf and αA-crystallin in ocular lens by fibroblast growth factor signaling. J. Biol. Chem. 291, 3947–3958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-S. et al. JAK2-STAT3 blockade by AG490 suppresses autoimmune arthritis in mice via reciprocal regulation of regulatory T cells and Th17 cells. J. Immunol. 192, 4417–4424 (2014). [DOI] [PubMed] [Google Scholar]
- Schust J., Sperl B., Hollis A., Mayer T. U. & Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 13, 1235–1242 (2006). [DOI] [PubMed] [Google Scholar]
- Linterman M. A. et al. Follicular helper T cells are required for systemic autoimmunity. J. Exp. Med. 206, 561–576 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitari A. C. et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474, 403–406 (2011). [DOI] [PubMed] [Google Scholar]
- Suriben R. et al. β-cell insulin secretion requires the ubiquitin ligase COP1. Cell 163, 1457–1467 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. E3 ubiquitin ligase RFWD2 controls lung branching through protein-level regulation of ETV transcription factors. Proc. Natl Acad. Sci. USA 113, 7557–7562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Kato J.-Y., Nakamae I. & Yoneda-Kato N. COP1 targets C/EBPα for degradation and induces acute myeloid leukemia via Trib1. Blood 122, 1750–1760 (2013). [DOI] [PubMed] [Google Scholar]
- Pham D., Sehra S., Sun X. & Kaplan M. H. The transcription factor Etv5 controls TH17 cell development and allergic airway inflammation. J. Allergy Clin. Immunol. 134, 204–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jojic V. et al. Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 14, 633–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh B. et al. The ETS family transcription factors Etv5 and PU.1 function in parallel to promote Th9 cell development. J. Immunol. 197, 2465–2472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J. et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33, 314–325 (2003). [DOI] [PubMed] [Google Scholar]
- Lee P. P. et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001). [DOI] [PubMed] [Google Scholar]
- Rickert R. C., Roes J. & Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25, 1317–1318 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information Files, or from the corresponding authors on reasonable request. The RNA-seq data were deposited in the Gene Expression Omnibus (NCBI) data repository under accession number GSE84125.