Abstract
The relative fibroglandular tissue content in the breast, commonly referred to as breast density, has been shown to be the most significant risk factor for breast cancer after age. Currently, the most common approaches to quantify density are based on either semi-automated methods or visual assessment, both of which are highly subjective. This work presents a novel multi-class fuzzy c-means (FCM) algorithm for fully-automated identification and quantification of breast density, optimized for the imaging characteristics of digital mammography. The proposed algorithm involves adaptive FCM clustering based on an optimal number of clusters derived by the tissue properties of the specific mammogram, followed by generation of a final segmentation through cluster agglomeration using linear discriminant analysis. When evaluated on 80 bilateral screening digital mammograms, a strong correlation was observed between algorithm-estimated PD% and radiological ground-truth of r=0.83 (p<0.001) and an average Jaccard spatial similarity coefficient of 0.62. These results show promise for the clinical application of the algorithm in quantifying breast density in a repeatable manner.
Keywords: breast cancer, mammography, imaging biomarker, percent breast density, breast cancer risk estimation
1 Introduction
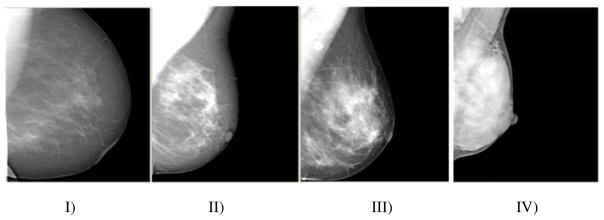
Beginning with the pioneering work of Wolfe [1], multiple studies have established that the relative amount of fibroglandular tissue seen within the breast, often referred to as breast density, is an image-derived biomarker that has been shown to be an independent risk factor for breast cancer, in fact the most significant after age [2]. Currently, the most commonly used methods to assess breast density rely either on visual assessment by radiologists in distinct categories [3] or through interactive, semiautomated image thresholding [4]. Both approaches are highly subjective with known limitations. Categorical methods, such as with the 4-class Breast Imaging Reporting and Data Systems (BIRADS) [3] system illustrated in figure 1, are associated with only moderate overall agreement, with poor concordance with respect to moderately dense breasts versus completely fatty or completely dense breasts [5]. Interactive image-thresholding methods, which require user interaction, are known to introduce reader-variability into density assessment [6].
Fig. 1.
Sample Digital Mammograms of BIRADS Categories I–IV in Increasing Order of Density. I) <25% fibroglandular content; II) fibroglandular content between 26–50%; III) fibroglandular content between 51–75%; IV): fibroglandular content >75%.
To address these limitations, automated methods have been proposed for extracting breast density information from digitized film mammography images. For example, Petroudi et al. have proposed a density classification scheme based on texture models [7] and Tagliafico et al have investigated adaptive thresholding techniques to identify the fibroglandular tissue regions of a mammogram [8]. However, the translation of these techniques into clinical practice has been limited due to the impracticality of incorporating mammographic film digitization into the clinical workflow solely for the purposes of estimating breast density.
As film mammography is rapidly being replaced by digital mammography, the opportunity arises to develop sophisticated fully-automated algorithms by quantifying breast density directly from the digital images. Digital mammograms capture richer gray-level intensity profiles compared to digitized film images, in which the digitization process also leads to different signal to noise ratio characteristics due to the inherent granularity of the film [9]. Here we propose a novel adaptive multi-cluster fuzzy c-means (FCM) segmentation algorithm for quantifying breast percent density (PD %) from digital mammography images. Our algorithm involves a series of steps; i) breast region and pectoral muscle segmentation; ii) adaptive histogram-based determination of the optimal number clusters for FCM segmentation; and iii) dense tissue cluster merging through a linear discriminant analysis (LDA) agglomeration classifier. The innovation of our algorithm lays in the adaptive nature of the FCM clustering segmentation, which determines the optimal number of clusters based on the breast tissue properties of the specific image, and the agglomeration classifier which combines imaging and patient characteristics to achieve optimal segmentation through cluster merging. We validate our algorithm by comparing to radiologist-provided ground truth of dense tissue on a set of 80 cases with bilateral digital screening mammograms (a total of 160 images) covering the full spectrum of breast densities seen in clinical practice. We compare our algorithm to the standard two-class FCM approach previously used for BIRADS density classification in digitized film mammograms [10].
2 Methods
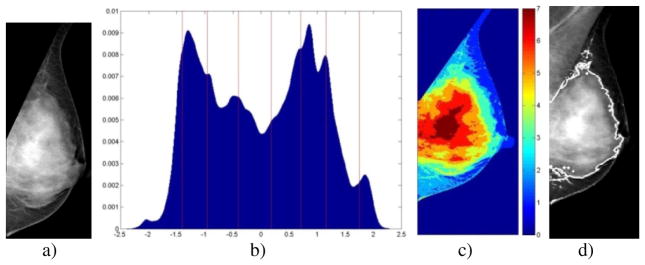
As a pre-processing step in the proposed algorithm a mask of the breast region is generated, here denoted as MB, using a previously validated algorithm [11], based on a combination of automated thresholding to identify the breast tissue versus air followed by identification of the pectoral muscle by the straight-line Hough transform (Figures 2a–b). To account for variable gray-level intensities between different mammographic images, the gray level values of the breast region are then normalized using z-score to a (μ, σ) of (0, 1).
Fig. 2.
Segmentation algorithm stages for a k=7 mammogram. a) Segmented breast region; b) Normalized breast-pixel intensity histogram with FCM cluster centroids (vertical lines); c) Pixel cluster-membership represented by shading; d) Final dense tissue segmentation.
2.1 Adaptive Fuzzy C-Means Clustering of Breast Fibroglandular Tissue
Once the breast area is identified, we perform an adaptive k-class fuzzy c-means clustering of the breast region gray-level intensities, where k is optimized for the given image based on the morphology of the normalized histogram of the corresponding image intensity-values. To determine the appropriate k, the grey-level histogram is convolved using a Gaussian kernel of an empirically determined window-size and standard deviation in order to both smooth out quantization noise while simultaneously enhancing concentrations of intensity values. The first and second derivatives of the smoothed histogram are then calculated, and the number of modes (defined as zero-crossings in the first derivative with negative second derivatives) are used to define k,
| (1) |
where H(gn) is the histogram of the normalized gray-levels values, gn, for those pixels found in the breast mask, MB. Once the appropriate number of clusters, k, is determined for a given image of the breast, we then perform k-class FCM clustering [12] on the gray-level values present in the breast region. FCM iteratively optimizes a weighted sum of square error function, which ultimately yields cluster centroids and a cluster-membership matrix for every intensity value in the breast mask. Once this is accomplished, every pixel is assigned to the cluster for which that pixel’s intensity value has the highest membership score. An example histogram for a k=7 case and the resultant FCM clustering can be seen in Figures 2b and 2c.
2.2 Cluster Agglomeration and Percent Density Calculation
To agglomerate the k-cluster output into a two-class segmentation of the dense versus fatty tissue, an LDA classifier is trained to determine the cluster cutoff for dense tissue. All clusters described by centroids of equal or higher intensity than the LDA cutoff are then combined (i.e., agglomerated) into a single segmented dense tissue region, MD.
Predictor variables of the LDA-classifier included image histogram statistics, which are often used to classify images into BIRADs categories [10]; image acquisition parameters and patient characteristics, which have been shown to correlate to PD% [13]; and parameters of the output of the FCM clustering. In order to reduce the dimensionality of the input predictor variables, stepwise feature selection is performed, where terms are systematically added to the LDA-model classifier based on the relative explanatory power of the terms as described by Draper et al. [14]. The final feature set included: the third central moment and 5th percentile of the histogram, kurtosis, mean and standard deviation of the un-normalized histogram, age, breast thickness, mammogram paddle compression-force, KVP, exposure, and parameters of a sigmoid-curve fit to horizontal and vertical extents of the resulting dense-tissue segmentation as a function increasing the cut-off cluster choice. An example of the agglomeration result can be seen in Figure 2d. From this final density mask, we calculate mammographic percent density, PD%, by computing
| (2) |
2.3 Dataset and Algorithm Evaluation
To validate our proposed algorithm, we identified 80 cases with bilateral MLO-view post-processed digital mammography images (PremiumView TM, GE Healthcare), which yielded a total of 160 images for analysis. All images were acquired using a standard screening protocol on a Senograph DS (GE Healthcare) full-field digital mammography (FFDM) system, with an isotropic 100μm resolution. For each image, a trained breast imaging radiologist provided a breast PD% estimation and dense tissue segmentation using a validated user-interactive image-thresholding tool for breast PD% estimation (Cumulus, Ver. 4.0, Univ. Toronto) [15].
For our experiments, the agglomeration LDA-classifier, including the feature selection stage, is trained using a leave-one-woman-out (2 mammograms) schema on the output of the unsurpervised FCM clustering to select the optimal cutoff point for dichotomizing the image into fatty and dense segmentations, in this case the lowest intensity cluster that is still a “dense” cluster, in order to maximize the agreement between computer-estimated and radiologist-defined PD%. To evaluate the accuracy of our algorithm in estimating breast PD%, we compute the Pearson product-moment correlation coefficient [16], r, between the algorithm-estimated PD% and the radiologist-provided PD%, considered here as ground-truth for our experiments. Spatial agreement between the algorithm-segmented and radiologist-segmented dense tissue regions is evaluated using the Jaccard index, J, [17] defined as
| (3) |
where RD and MD are the fibroglandular tissue segmentations generated by the radiologist and algorithm respectively. Finally, we also compare our algorithm with the standard two-class FCM segmentation that has been previously used for dense tissue segmentation in digitized film mammograms [10].
3 Results
For our experiments, image intensity histograms were constructed with a fine bin-width of 0.01 on the z-score normalized intensity histogram. With regards to cluster-count optimization, it was found that distribution of frequencies of k was approximately Gaussian, centered at k=6. We applied a bounding constraint on k to be between 2 (as there will always be an adipose and a fibroglandular tissue cluster) and 9 (for speed and memory considerations). Computation of k was not found to be particularly sensitive to changes in histogram construction or small peaks.
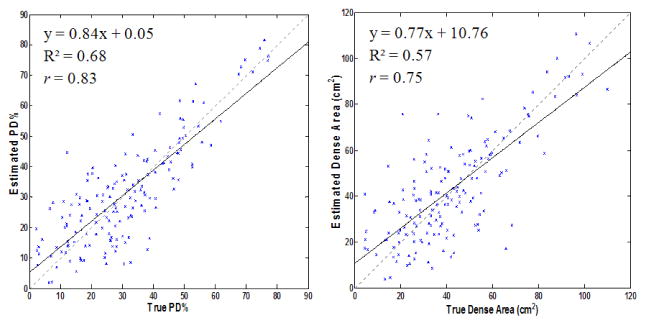
Feature selection during LDA-training was found to select the exact same feature set for all 80 iterations of the leave-one-woman-out training, indicating that the features used were robust to case variation and provide orthogonal information. When comparing radiologist-derived PD% to algorithm-estimated PD%, we were able to obtain a Pearson correlation of r=0.83. Correlation between estimated and true breast area calculations was found to be r=0.99, while the correlation between estimated and true absolute dense-tissue area was found to be r=0.75. Scatter-plots of the estimated vs. true PD% and dense area are provided in figure 3. Good spatial agreement between true and algorithm-derived dense area also found, with a corresponding distribution of Jaccard indices of J=0.62±0.22 for the 160 density segmentations. The 2-class FCM, previously used for segmenting dense tissue in film mammorgraphy, showed a low correlation of r=0.05 (p>0.1) between 2-class FCM estimated PD% and ground truth as well as lesser spatial agreement of J=0.57±0.24.
Fig. 3.
Scatter-plots of Algorithm-Estimated vs. Radiologist-Provided PD% (Left) and Dense-tissue Segmentation Area (Right). Regression-equations, R2, Pearson Correlations, the linear regression line (black) and the identity-lines (dashed-gray) are provided for reference.
4 Discussion
The proposed fully-automated algorithm was successful in identifying the fibroglandular tissue of the breast in digital mammographic images. Strong agreement with radiologist-provided ground truth was obtained, both in terms of the quantitative PD% estimate and the spatial agreement of the segmented dense tissue. Furthermore, the spatial agreement between automated-algorithm segmentations and radiologic ground-truth (J=0.62±0.22) was found to be similar to the human-observer inter-reader variability of 0.65±0.18 previously reported by Bakic et al. [15]. These findings indicate that the proposed algorithm can provide clinically relevant information from digital mammography for the assessment of breast density.
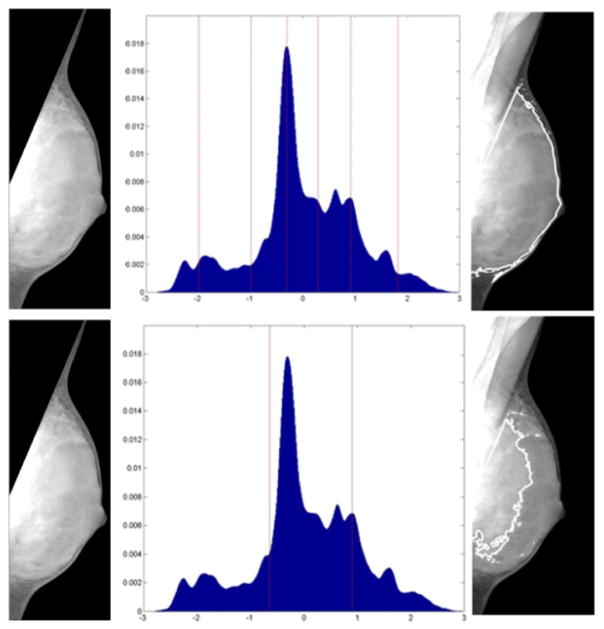
One surprising result was the relatively poor performance of the two-class fuzzy-c-means paradigm, previously used for density assessment in digitized film mammograms, in identifying the dense tissue region. Although BIRADS classification and not PD% quantification has often been the primary focus [10], this finding is in apparent contrast to findings by Torrent et al. who reported good visual agreement between FCM and expert markings [18]. Further investigation showed that one possible explanation for the poor performance of the 2-class FCM algorithm is the fact that majority gray-level intensity profiles of breast tissue as extracted from digital mammograms tend to be multi-modal, such as in the BIRADS-IV case illustrated in Figure 4. Given that appropriate selection of the number of clusters, k, is critical for proper clustering, the intensity profile of digital mammograms are complex enough that multi-class techniques, such as the one described in this work, may be required to appropriately analyze digital mammography images.
Fig. 4.
Comparison between 2-class (top) and adaptive k=6-class (bottom) FCM segmentation of a BIRADS-IV category breast. Left) Breast Mask; Center) Gray-level Histogram with marked FCM-centroids; Right) Final segmentation
As breast tissue seen mammographically is a 2D superimposition of different tissue types with different image properties, future studies should seek to expand density-based risk-stratification analysis beyond the dichotomous, fatty vs. dense tissue paradigm. Furthermore, as mammography is essentially limited by the effect of projection/tissue superimposition, volumetric analysis of fibroglandular tissue through emerging tomographic breast imaging modalities, such as breast tomosynthesis and magnetic resonance imaging, has been suggested as necessary to advance breast-cancer risk modeling [19] and the approach described in this work could become the foundation for fully-automated density quantification from three-dimensional images.
5 Conclusion
We have proposed and demonstrated the efficacy of a novel fully automated algorithm for fibroglandular tissue segmentation in digital mammography. We were able to obtain strong correlation between the output of the computerized algorithm and radiologist-provided ground truth. These results show promise for the potential clinical relevance and applicability of our method to quantify breast density in a repeatable and objective manner. This fully-automated method could accelerate the clinical translation of density-based cancer risk stratification and pave the way for new personalized screening and prevention strategies.
Acknowledgments
This work was supported by ACS Grant RSGHP-CPHPS-119586 and DOD Grant BC086591.The authors would also like to thank Dr. Martin Yaffe (University of Toronto) for providing Cumulus software tools as well as for useful discussions.
References
- 1.Wolfe JN. Breast patterns as an index of risk for developing breast cancer. Am J Roentgenol. 1976;126:1130–1137. doi: 10.2214/ajr.126.6.1130. [DOI] [PubMed] [Google Scholar]
- 2.Tice TA, Kerlikowske K. Screening and prevention of breast cancer in primary care. Prim Care. 2009;36:533–558. doi: 10.1016/j.pop.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 3.D’Orsi CJ, Bassett LW, Berg WA, Feig SA, Jackson VP, Kopans DB. Reston Am Col of Rad. 4. 2003. Breast imaging reporting and data system: ACR BIRADS-mammography. [Google Scholar]
- 4.Martin KE, Helvie MA, Zhou C, Roubidoux MA, Bailey JE, Paramagul C, Blane CE, Klein KA, Sonnad SS, Chan HP. Mammographic density measured with quantitative computeraided method: comparison with radiologists’ estimates and BIRADS categories. Radiology. 2006;240:656–665. doi: 10.1148/radiol.2402041947. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson BT, LoRusso AP, Smolkin M, Bovbjerg VE, Petroni GR, Harvey JA. Accuracy of assigned BIRADS breast density category definitions. Acad Radiol. 2006;13:1143–1149. doi: 10.1016/j.acra.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Yaffe MJ. Mammographic density. Measurement of mammographic density. Breast Cancer Res Treat. 2008;10:209–218. doi: 10.1186/bcr2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petroudi S, Kadir T, Brady M. Automatic classification of mammographic parenchymal patterns: A statistical approach. Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2003; 2003. pp. 798–801. [Google Scholar]
- 8.Tagliafico A, Tagliafico G, Tosto S, Chiesa F, Martinoli C, Derchi LE, Calabrese M. Mammographic density estimation: comparison among BIRADS categories, a semiautomated software and a fully automated one. Breast. 2009;18:35–40. doi: 10.1016/j.breast.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Yaffe M. PACS. Springer; Heidelberg: 2006. Digital mammography; pp. 363–371. [Google Scholar]
- 10.Oliver A, Freixenet J, Martí R, Pont J, Pérez E, Denton ERE, Zwiggelaar R. A novel breast tissue density classification methodology. IEEE Trans Inf Technol Biomed. 2008;12:55–65. doi: 10.1109/TITB.2007.903514. [DOI] [PubMed] [Google Scholar]
- 11.Karssemeijer N. Automated classification of parenchymal patterns in mammograms. Phys Med Biol. 1998;43:365–378. doi: 10.1088/0031-9155/43/2/011. [DOI] [PubMed] [Google Scholar]
- 12.Bezdek J. Pattern recognition with fuzzy objective function algorithms. Kluwer Academic Publishers; Norwell: 1981. [Google Scholar]
- 13.Lu LJW, Nishino TK, Khamapirad T, Grady JJ, Leonard MH, Brunder DG. Computing mammographic density from a multiple regression model constructed with image-acquisition parameters from a full-field digital mammographic unit. Phys Med Biol. 2007;52:4905–4921. doi: 10.1088/0031-9155/52/16/013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Draper NR, Smith H. Applied Regression Analysis. Wiley-Interscience; Hoboken: 1998. [Google Scholar]
- 15.Bakic PR, Carton AK, Kontos D, Zhang C, Troxel AB, Maidment ADA. Breast percent density: estimation on digital mammograms and central tomosynthesis projections. Radiology. 2009;252:40–49. doi: 10.1148/radiol.2521081621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson K. Mathematical contributions to the theory of evolution. III Regression, heredity and panmixia. Philos Trans Royal Soc London Ser A. 1896;187:253–318. [Google Scholar]
- 17.Jaccard P. The Distribution of Flora in the Alpine Zone. New Phytologist. 1912;11:37–50. [Google Scholar]
- 18.Torrent A, Bardera A, Oliver A, Freixenet J, Boada I, Feixes M, Martí R, Lladó X, Pont J, Pérez E, Pedraza S, Martí J. Breast Density Segmentation: A Comparison of Clustering and Region Based Techniques. In: Krupinski EA, editor. IWDM 2008. LNCS. Vol. 5116. Springer; Heidelberg: 2008. pp. 9–16. [Google Scholar]
- 19.Kopans DB. Basic physics and doubts about relationship between mammographically determined tissue density and breast cancer risk. Radiology. 2008;246:348–353. doi: 10.1148/radiol.2461070309. [DOI] [PubMed] [Google Scholar]