Abstract
The mycelial fungus Sordaria macrospora was first used as experimental system for meiotic recombination. This review shows that it provides also a powerful cytological system for dissecting chromosome dynamics in wild-type and mutant meioses. Fundamental cytogenetic findings include: (1) The identification of presynaptic alignment as a key step in pairing of homologous chromosomes. (2) The discovery that biochemical complexes that mediate recombination at the DNA level concomitantly mediate pairing of homologs. (3) This pairing process involves not only resolution but also avoidance of chromosomal entanglements and the resolution system includes dissolution of constraining DNA recombination interactions, achieved by a unique role of Mlh1. (4) Discovery that the central components of the synaptonemal complex directly mediate the re-localization of the recombination proteins from on-axis to in-between homologue axis positions. (5) Identification of putative STUbL protein Hei10 as a structure-based signal transduction molecule that coordinates progression and differentiation of recombinational interactions at multiple stages. (6) Discovery that a single interference process mediates both nucleation of the SC and designation of crossover sites, thereby ensuring even spacing of both features. (7) Discovery of local modulation of sister-chromatid cohesion at sites of crossover recombination.
Keywords: Sordaria, pairing, synaptonemal complex, meiotic recombination
1. Introduction
Recent interesting results concerning the formation of DNA double-strand breaks (DSBs) and their processing into crossing over (CO) or non-crossover (NCO) products are the extension of an over 80 year long progression of studies on meiotic recombination in which fungi like Neurospora crassa, Sordaria macrospora and Sordaria fimicola played a central and often pioneering role. The fantastic advantage afforded by the recovery of all four meiotic products within a tube-like cell (ascus), in a linear order has allowed several founding concepts of inheritance.
First, utilization of ascospore colour-mutants provides easy detection of pre- and post-segregation of allelic pairs, thus allowing to directly “read” if, yes or no, a CO has occurred between the spore-colour gene and its centromere (Fig.1A). Moreover, meiosis is followed by a post-meiotic mitosis, again without spindle overlap, giving a linear series of eight haploid nuclei. Each of these nuclei represents therefore the genetic character of one of the eight DNA strands of the four chromosomes produced by meiosis (e.g. [1,2]). This is how, using two linked markers of N. crassa, Lindegren [3] showed definitively in 1933 that CO occurs at the “the four-strand stage” and involves only two of the four chromatids: one from each of the homologous chromosomes, with the centromeres segregating at the first meiotic division. Also, detailed analyses of linkage data of multifactor crosses revealed that positive CO interference, first discovered by Sturtevant [4] and Muller [5] in analyzing the ratio of observed to expected double COs in Drosophila, is a general feature of recombination [6].
Figure 1.
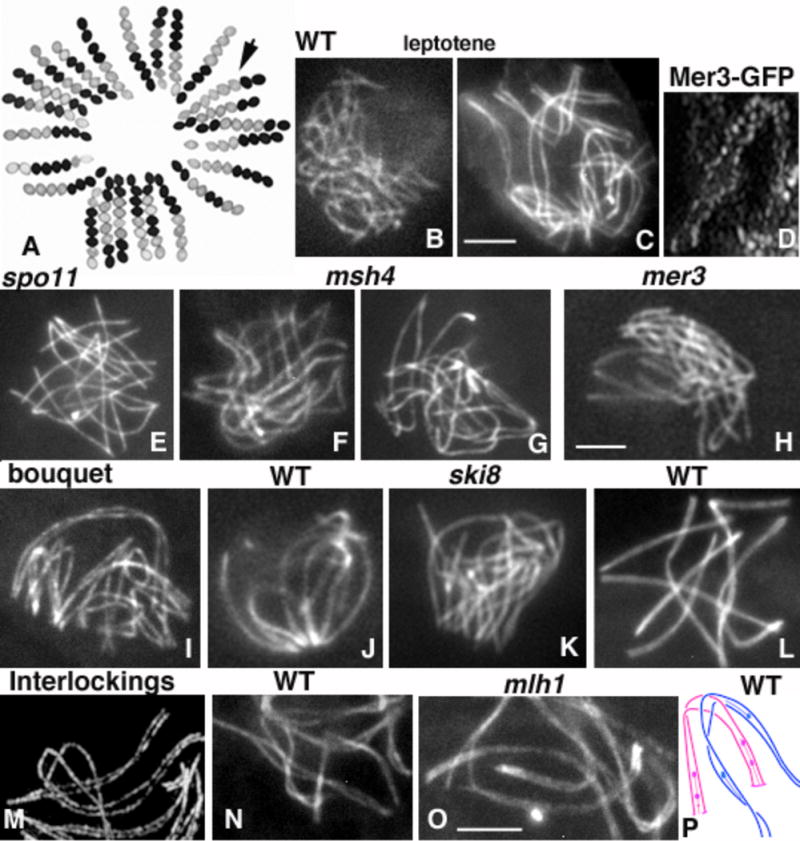
A. Pre- and Post-Segregations from a cross between a white-spore-mutant and a wild-type (WT) strain (black ascospores). Arrow points to a 2:6 conversion ascus. B–O: Chromosome axes are stained by Spo76/Pds5-GFP. B and C: WT nuclei at early (B) and late (C) leptotene. D. Mer3-GFP foci at matching sites along co-aligned axes. E. Asynaptic spo11D nucleus. F and G: msh4D nuclei at early (F) and late (G) stages of pairing. H. mer3D nucleus. I–K. WT bouquet at early zygotene (I) and pachytene (J); (K) bouquet of asynaptic ski8D. L. Bouquet release at pachytene. M–O. Interlocking during WT leptotene (M) and zygotene (N) nuclei, and (O) in mlh1D pachytene; arrows point to the interlocking sites. P. Cartoon of an interlocking between two bivalents (blue and pink), which have already developed SCs stretches (lines) plus RNs (balls). Bars = 2microns.
Second, in 1934, Heinrich Zickler [7] (no relation to the author) showed that aside from the usual 2:2 parental color-marker segregation in heterozygous crosses of Bombardia lunata, a small percentage of asci contained 2:6 and 6:2 black and white ascospores (see equivalent segregation in Sordaria Fig.1A, arrow). He named this phenomenon “conversion” (now usually called “gene conversion”). The existence of such events was “rediscovered” in budding yeast by Lindegren [8] in 1953 and definitively demonstrated in N. crassa by Mitchell [1] in 1955, with the use of linked markers on either side of the studied locus. When 2:6 segregation was observed at the pyridoxine locus, the linked markers segregated with normal 2:2 ratios, indicating that the aberrant segregation occurred at the pyridoxin mutant site and not as a consequence of aberrant chromosome segregation. Molecular analysis of recombination has confirmed that in 1:3 segregations DSBs interact with their homologous partner but without accompanying exchange of their flanking regions resulting in non-Medelian segregation of the studied marker (review in [9–11]).
Third, the discovery of S. fimicola asci with postmeiotic segregation, in which both parental alleles appear in mitotic segregants from a single chromatid, provided direct evidence for the presence of heteroduplex DNA at a site. (i) 5:3 and 3:5 segregations were first described by Olive in 1959 [12]. (ii) Kitani et al. [13] reported the existence of “aberrant 4:4” segregations, which arise through a reciprocal exchange between the two subunits of different parental chromatids. (iii) Leblon [14] showed that there is a strong correlation between the mutagenic origin of the mutations and the type of non-mendelian segregation patterns, since understood to reflect biases of the mismatch repair process with respect to the nature of the mismatch.
Fourth, using white spore-color mutants of the Ascomycete Ascobolus immersus (with non-ordered 8-spored asci), Lissouba and Rizet showed [15], that intragenic frequencies of gene conversion decline from one end of a gene to the other. Moreover, in heteroallelic crosses, the converted allele was always on the same side of the gene, relative to the other allele, suggesting that recombinational events had preferential starting and ending sites [16]. Confirmation of “polarity” was obtained from studies of several other genes, and in other fungi, substantiating the concept that gene conversion reflects the non-reciprocal transfer of information from one parental DNA duplex to the other parental duplex during meiosis (reviewed in [10,17]). Polarity across genes is also consistent with the presence of initiating hotspots of recombination in flanking regions (e.g. [17]), now confirmed by high-resolution mapping of recombination events through direct molecular analyses of DNA sequences (review in [18,19]).
Fifth, the frequency of various types of asci with aborted non-colored ascospores and the order of those aborted ascospores in the asci revealed both several types of chromosome rearrangements (translocations, duplications, etc) and how far the rearrangement breakpoint is from the centromere (e.g. [2,20–22]). Translocations were also used to map genetically all linkage groups, and to assign them to the respective chromosomes [23].
The S. macrospora cell cycle affords also several advantages for the study of meiosis per se (see Supplemental materials and Supplemental Fig. 1). Being homothallic the fungus is able to enter the sexual cycle from a single ascospore. Self-fertility is interesting because it allows the direct recovery of recessive and dominant mutants after mutagenesis as well as to directly obtain haploid (and then homozygous diploid) versions of molecularly transformed strains. Also, all perithecia contain over 100 eight-spored asci/meiocytes and 100% viable ascospores.
Sordariales provide also a powerful system for visualizing and dissecting chromosome dynamics in wild type and mutants. The small chromosomal size of most fungi was long considered as a handicap for the study of meiosis. However, Barbara McClintock was right when she said in her 1961 conference “You may think they (prophase chromosomes of N. crassa) are small but when you look at them, they get bigger and bigger and bigger” (cited in [24]). And in fact, thanks to new cytological tools, mycelial fungi, and especially S. macrospora (with larger asci and nuclei than N. crassa; see also Supplemental Materials for detailed comparison) provide powerful systems for cytological analyses of the early steps of homologous pairing for at least four reasons.
First, in both wild-type and mutant meioses, prophase stages can be identified independently of chromosomal morphology on the basis of nuclear volume and ascus size, which increases progressively from 10 μm after karyogamy to 150 μm at the end of prophase. Also, clear changes in overall chromatin conformation (by DAPI) allow easy sub-staging of pachytene nuclei [25,26]. Second, contrary to most eukaryotes, the two sets of homologous chromosomes are in separated nuclei before entering meiosis [27,28]. Thus, as their first contacts occur only after karyogamy, homologous juxtaposition cannot involve contributions resulting from pre-existing pairing (review in [29,30]). Also, premeiotic replication occurs just before karyogamy, which thus defines a clear “start” for the study of meiotic stages (review in [31]). Third, chromosome axes are developed by early leptotene, making it possible to follow axis status and spatial relationships among different chromosomes over early periods, especially because the seven chromosomes can be easily identified by length differences (e.g. [25,27,32–35]). Finally, as in higher plants, meiosis usually proceeds to its end even when severe recombination or SC defects are present, rather than arresting as in yeast and mammals, thus facilitating complete multi-stage analysis of mutant defects.
This chapter focuses on topics that have been of central importance to our laboratory and surveys our recent progress in delineating the links between meiotic recombination and pairing. Trying to understand the coordination between chromosome pairing, synapsis and recombination in Sordaria allowed us to provide many unexpected insights into these aspects of meiosis. It allowed also to uncover complex networks of interactions among axis components and recombination plus synaptonemal complex proteins.
2. The four steps of chromosome co-alignment require DSBs and the MusS homolog Msh4
One of the most fascinating features of meiosis is the fact that homologous chromosomes are able to identify one another and come together in the rather “crowded” early leptotene nucleus (Fig. 1B). What have we learned thus far from Sordaria analyses concerning the juxtaposition of homologous chromosomes (prior to installation of the synaptonemal complex) during early prophase?
Pioneering three-dimensional EM reconstructions from serial sections of karyogamy, leptotene, zygotene, pachytene and diplotene nuclei [27] and recent analyses using fluorescent-tagged proteins and/or high resolution microscopy have revealed that homologous chromosomes come together in four distinct steps. (i) Just after karyogamy, homologues are far apart with no evidence of any specific relationship (Fig. 1B) and their ends/ axial elements (AEs) are already attached to the nuclear envelope [27]. (ii) By mid-leptotene most homologous chromosomes have moved into rough long-range co-alignment with a tendency for telomeres to co-align first. Although there is a general tendency for shorter chromosomes to align first, there is no individual chromosome rule for the progression of pairing from one nucleus to the other. (iii) At end of leptotene, all homologous pairs are co-aligned along their entire length at a distance of ~ 400nm (Fig. 1C). Similar complete co-alignment prior to SC formation (synapsis) was also observed in N. crassa, Coprinus and higher plants [36–39] and also in budding yeast in absence of Zip1, the synaptonemal complex central-component [40]. (iv) Homologs then become aligned at ~200nm separation and stay perfectly aligned at this distance along their entire length through late leptotene. At stages (iii) and (iv), homolog axes are connected by bridges which appear to include both DNA, as suggested by DAPI-stained bridges (Figure 2G) and recombination proteins as suggested by two-by-two association of Mer3 foci in Sordaria (Fig. 1D), by RPA bridges in human spermatocytes [41] and by bridges with associated nodules in spread SCs of Allium [39].
Figure 2.
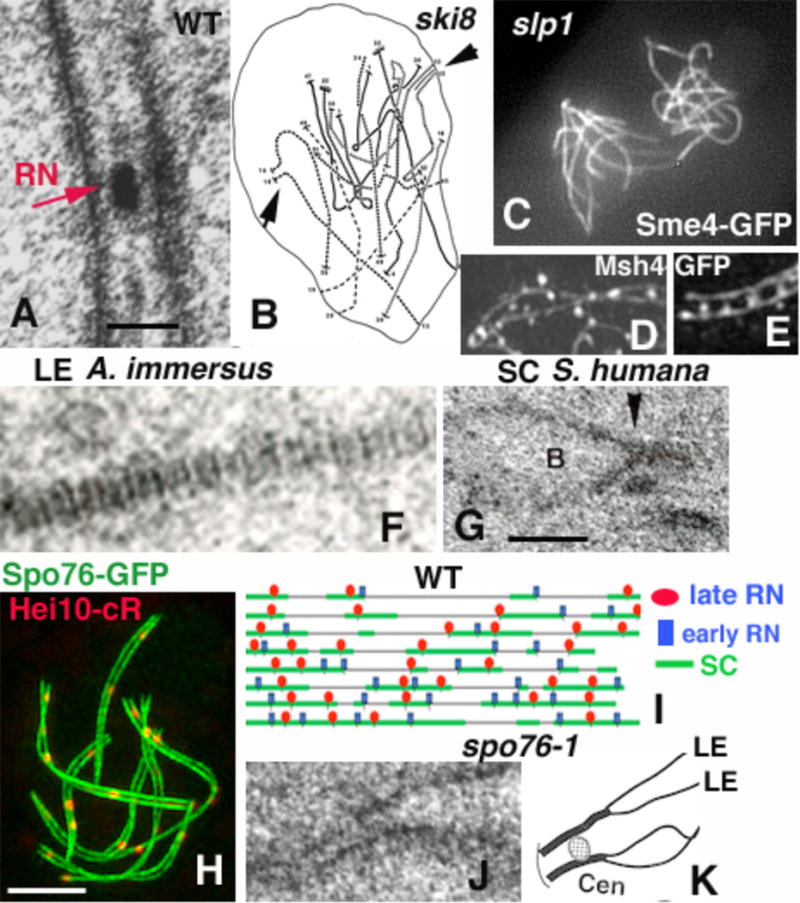
A. Sordaria SC with RN (red arrow). B. Reconstruction from serial sections of a ski8D nucleus; arrows point to two SC initiation sites. C. All chromosomes from the slp1D twin haploid-nuclei load Sme4 all along their lengths. D and E: Msh4-GFP foci. (D) Between homologous axes in WT pachytene and (E) on axes in co-aligned sme4D axes (axes are stained by Spo76-GFP). F. Stripped AE of A. immersus. G. SC of S. humana with a buldge (B) on the upper LE initiating at the RN site (arrowhead). H. WT pachytene nucleus double-stained by Spo76-GFP and Hei10-cherryRed in high-resolution microscopy. I. Eight mid-zygotene chromosomes 1 (from serial section reconstructions) with localization of late RNs (red), early RNs (blue) and SC initiation sites (green). J. LE of spo76-1 with widely open sister-chromatid LEs (arrows). K. Cartoon of a spo76-1 SC (LE = lateral element; Cen = centromere). Bar = 2 microns; 100nm for A and G.
The mechanism(s) by which homologous chromosomes identify one another and achieve co-alignment remain(s) an unsolved problem. However, in Sordaria, as in most organisms, recombination as initiated by DSBs, is the central mediator of co-alignment (review in [19]). In the absence of DSBs, i.e. in Sordaria spo11 null, spo11Tyrosin and ski8 null mutants, homologues do not align or do so only rarely (Fig. 1E; [32,33]). Co-alignment defects in these cases are not attributable to absence of the corresponding proteins, but to the absence of DSBs per se, because exogenous DSBs induced by Gamma irradiation can restore co-alignment in 20% of the meioses. However, the extent of alignment is variable, suggesting that programmed DSBs mediated by Spo11/Ski8 occur with tightly regulated positioning of DSB sites [32,33].
Pairing is also dependent upon, and mediated via, early post-DSB stages of DNA recombination. At the DNA level, one end of a DSB interacts with the homologous position on a homolog partner duplex to form a nascent D-loop [11]; however, concomitantly, unknown events cause whole homologous chromosomes axes to come together in space. In Sordaria, involvement of recombination proteins is indicated by the fact that the mismatch repair MutS-homolog Msh4, which binds to branched DNAs and stabilizes early recombination structures [42], is involved in specifying distance between co-aligned axes. When Msh4 is absent, homologues co-align but at a distance of 600–800nm, rather than the 400nm seen in wild type (Fig. 1F, compare with 1C) suggesting that Msh4 participates directly in the determination of the inter-axis distance.
Moreover, analysis of Sordaria nuclei lacking Msh4 provided two new insights into the pairing process. First, in wild type, pairing is highly coordinated throughout each nucleus (Fig. 1C); in contrast, msh4D nuclei show a mixture of completely co-aligned pairs and others, which are either only aligned in short regions (often at telomere regions) or still completely separated (Fig. 1G). Such mixture points to per-chromosome and/or per-region asynchrony of co-alignment-related events in the mutant. Second, while msh4D mutant defects are visible at early leptotene, the biochemically-defined role for Msh4 occurs at zygotene [11]: perhaps Msh4 acts during pairing to stabilize nascent D-loops. Third, Msh4 foci appear only at early zygotene on synapsing regions [35]. Thus, Msh4 plays a central role in homologous juxtaposition and co-alignment at one stage preceding both its supposed enzymatic role and its cytologically visible localization [35]. Meiotic helicase Mer3 protein is also involved (Fig. 1H), as discussed below.
3. Chromosomes co-align before bouquet formation
Co-alignment is accompanied by telomere-led chromosome movements, which culminate in the well-conserved meiotic configuration called the bouquet. An important prerequisite for these movements is the attachment of all chromosome ends to the nuclear envelope (NE). Attachment occurs at early leptotene (as revealed by serial sections analyzed in EM; [27]) and in yeasts requires the SUN protein Mps3, Ndj1/Taz1 and Csm4, which link telomeres to movement-mediating components of the cytoskeleton [43,44]. In wild-type Sordaria meiosis, chromosomes first co-align at a distance of 400-200nm before entering into the bouquet (Fig. 1C). Bouquet forms progressively first into sub-clusters in a loose bouquet configuration (Fig. 1I). It is only at late zygotene-early pachytene that all chromosome ends are clustered in a small area of the NE (Fig. 1J). This timing implies that telomere-led movements and the bouquet are not required for homolog co-alignment. This conclusion is directly confirmed by treating young Sordaria asci with the cytoskeletal inhibitors nocadazole or cytochalasin D, which probe respectively the roles of microtubules and actin microfilaments. In either treatment, asci remain blocked with homologs perfectly co-aligned, but with chromosome ends distributed throughout the nuclear volume (like in Fig. 1C; DZ, unpublished). Thus the bouquet plays no, or only a minor role in pairing. While first described in Sordaria [25], similar uncoupling of pairing and bouquet formation was further observed in several other organisms (review in [25,44,45]). Also, in budding yeast, high levels of chromosome movements occur only at zygotene/pachytene, thus again after pairing [46,47]. Interestingly, also, while co-alignment is mediated by DSBs, bouquet formation and thus chromosome movement during leptotene are both independent of DSBs formation and the downstream process of recombination: in Sordaria, bouquet forms in the absence of Spo11, Ski8 (Fig. 1K), Mer3, Msh4 and Mlh1 [33, 35].
Chromosome movements are required at a second step of prophase: the release from the bouquet configuration at mid-pachytene. As during its formation, bouquet release is progressive before final dispersal of all chromosome ends (Fig. 1L). Release is also mediated by cytoskeleton components: when asci are treated with cytoskeletal inhibitors at a stage when they are at the zygotene-pachytene stage, meiosis stops progression in the bouquet configuration. Also, while not necessary for formation of the bouquet, Spo11, Ski8, Mer3, Msh4 and Mlh1 are required for timely bouquet resolution, which is highly delayed in their absence in Sordaria [33, 35] and in budding yeast, worm and mouse [48, 49].
Thus, although the existence of a clear temporal link between bouquet formation and homolog juxtaposition during pairing, the bouquet is not the main mediator of that juxtaposition. Bouquet formation and pairing could be in fact concomitant components of a general nuclear reorganization (starting by release of the Rabl centromere clustering at meiosis onset) rather than being linked by a simple cause-and-effect relationship (review in [25,50]). Nevertheless, exit from the bouquet could help to destabilize ectopic recombination interactions and essentially help to resolve chromosome interlocks (see below). Plus, the fact that exit, and not entry into the bouquet is under the control of the recombination process could indicate that the bouquet works as a checkpoint sensor to evaluate the status of the chromosome and/or the CO recombination process before allowing completion of synapsis.
4. Preventing and resolving interlocks: involvement of recombination proteins Mer3, and Mlh1
Mer3 and Mlh1 are direct participants in the DNA events of recombination. Mer3 is a meiosis-specific 3′–5′ helicase that stimulates Rad51-mediated DNA heteroduplex extension and stabilizes recombinational interactions [51,52]. The mismatch repair protein Mhl1 is implicated at a later stage, in finalization of CO recombinational interactions during pachytene [11,53]. In Sordaria, in correspondence with their times of action during pairing and recombination, foci of Mer3 (and Msh4, above) are observed prior to and during SC formation whereas, in many studied organisms, foci of Mlh1 are not visible until after completion of synapsis [35,54–57]. Moreover, in Sordaria, by the time homologue axes are co-aligned at ~200nm, Mer3 foci are exclusively on axes and they mostly occur at matching sites (Fig. 1D). This configuration, and the correlated fact that the total number of all Mer3 foci is double that of Rad51 foci at this stage, suggest that Mer3 foci mark both DSB ends, one associated with each axis [35].
Absence of Mer3 confers no defect in DSB formation but produces strikingly aberrant co-alignment, in two respects. First, in contrast to wild-type meiosis where all chromosomes in each nucleus exhibit synchronous pairing configurations (Fig. 1C), in mer3D nuclei, chromosomes exhibit either absence of co-alignment or different states of partial co-alignment, implying both asynchrony and inefficiency. Second, at what should be early pachytene by ascus size, this defective co-alignment yields strikingly “interwoven” chromosomes (Fig. 1H). This phenotype points to the existence of a process by which wild-type chromosomes avoid, or at least minimize, such configurations, with Mer3 required for that process. Interwoven chromosomes is the outcome one might expect if, in the mutant situation, long-range DNA connections between homologues were made all throughout the genome prior to onset of co-alignment which, when it does occur, simply locks in the already present meshwork of interactions. In accord with this interpretation, the first discernible defect of mer3D meiosis is a long delay in the onset of co-alignment. Thus, the further implication of the mer3D phenotype is that, during wild-type meiosis, establishment of a long-range DNA/partner connection at one position is followed rapidly by axis juxtaposition at that position. This would not only draw the two involved regions into a regular arrangement, but would increase the likelihood of juxtaposition at nearby positions, thus resulting in fast propagation of co-alignment along the involved pair of chromosomes. The role of Mer3 could thus be to ensure that establishment of a long-distance contact is rapidly followed by initiation of spatial axis juxtaposition [35].
Analysis of Sordaria mlh1D mutant enabled to show that aside being a central participant in recombination, Mlh1 is also required for interlock resolution. Initiation of SC formation at two or more sites along a pair of homologues creates a potential risk that any chromosome or bivalent located between the points of association will be entrapped. Occurrence of such interlocked chromosomes, as well as the existence of mechanisms that resolve such entrapments, are manifested by significant levels of interlockings at zygotene, and their absence at pachytene (review in [29,30,58]). In wild-type Sordaria, interlocked chromosomes are already observable at late leptotene (Fig. 1M), implying that they occur as part of the pairing process, and 20% of the zygotene nuclei show at least one entanglement (Fig. 1N). However, at pachytene, no interlocks are visible and the presence of 100% 8-spored-asci in all perithecia demonstrates that they are all resolved.
In the absence of Mlh1, although onset of co-alignment is delayed by 12h for unknown reasons, chromosomes do then co-align and ultimately fully synapse. However, strikingly, 67% of the pachytene nuclei exhibit at least one or two bivalents with a non-synapsed region, of variable length. Moreover, 70% of these non-synapsed regions correspond to interlockings, in which the entrapped chromosome (either a single chromosome or a synapsed pair of homologs) is still present in the open region (Fig. 1O). The other 30% likely represent cases of “left-behind regions” that did not re-synapse after resolution [35].
The mechanism of interlock resolution is unknown, but two models are commonly proposed. Relying on the fact that transient breakages of axes are often seen at the sites of interlockings (Fig. 1O), one model suggests DNA topoisomerase II to catalyse the passage of the trapped chromosomes (review in [58]). The other model suggests a role of the bouquet to allow sliding of the entrapped chromosomes [25,59,60]. However, most entrapped homologues show both synaptonemal complex (SC) and late recombination nodules (RNs, see below) along part of their lengths (Fig. 1P), both of which must be eliminated before resolution. SC formation being a highly dynamic process that is readily amenable to reversal [30] destabilization between the interlocking site and the end(s) of the entrapping homolog pair plus following SC re-polymerisation will likely not prevent resolution of interlocks. In contrast, removal of constraining recombinational connections (e.g. RNs) requires a specific molecular process devoted to that task. Interestingly, Mlh1 is known to mediate specific rejection of CO-fated recombination intermediates during meiosis when the involved DNA molecules contain heteroduplex mismatches [53]. Resolution of interlocks will finally be constrained only by mature CO products, rather than by NCOs or inter-sister events. Thus, perhaps, Mlh1 acts at zygotene, in advance of CO finalization, to abort CO-fated interactions, which might then undergo repair of sister chromatids.
5. The Synaptonemal complex: formation and role in the maintenance and/or turnover of the recombination proteins
Following co-alignment, Sordaria homolog axes become more tightly associated, at 100nm separation, by installation of the synaptonemal complexes. SCs appear first at chromosome ends and later interstitially, excluding a zipper-like mechanism (examples in [22,61]). As in other organisms, the SC consists of two dense lateral elements (LEs), separated by a less dense central region which exhibits “electron-dense” structures called “recombination nodules” (RNs), on the basis of their correlation with the number of COs/chiasmata (Fig. 2A); e.g. review in [29,62,63]). SCs are shed progressively from the chromosomes at end of pachytene when homologues start to separate.
Sordaria SCs form progressively between homologues during zygotene. The specific pattern of SC installation is dependent on both the number of DSBs and Msh4. In hypomorphic ski8 mutants of Sordaria (as in budding yeast spo11 mutants), only partial SC initiation is observed (Fig. 2B; [32,64,65]), suggesting that nucleation of SC formation at a single or few sites is not sufficient to permit spreading of SC all along the homologues. Furthermore, SC appears to be nucleated initially at sites of recombination interactions that eventually mature into COs: in wild-type Sordaria the number of early interstitial SC initiation sites corresponds well to the number of COs and RNs and are decreased in two mutants with decreased COs [22]. Interestingly also, Msh4 is not only required to establish correct co-alignment distances (above), but it is also required for timely and correct per-chromosome synchronous SC formation [35].
Strikingly, SC can form between sister-chromatids in haploid meiosis (Fig. 2C). In the absence of the Sordaria SUN-like protein Slp1, karyogamy does not occur but meiosis proceeds efficiently in the two haploid twin nuclei by the same timing as in diploid wild-type nuclei. Recombination proteins Rad51, Mer3, Msh4 and Hei10 as well as the SC proteins Zip4 and Sme4 are loaded synchronously in both nuclei but between sister chromatids instead of between homologues (Fig. 2C; [28]). Thus, absence of karyogamy converts the entire recombination/synapsis program from an inter-homolog process to an inter-sister process.
Sordaria studies showed furthermore that SC central-region components mediate the re-localization of the recombination proteins from on-axis to in-between homologue axis. In wild-type Sordaria, recombination proteins like Rad51, Mer3 (Fig. 1D) and Msh4 are first located on chromosome axes, but after SC nucleation, they are delocalized from axes within the SC central region (Fig. 2D; [35,66]; see [67] for mouse). This is clearly illustrated by the Sordaria Sme4 protein (a transverse filament protein with Zip1-like structure and function). Sme4 is not required for co-alignment but is essential specifically for synapsis. When Sme4 is absent, Rad51, Mer3 foci and Msh4 foci form normally on axis but remain in this on-axis configuration through pachytene by ascus size (Fig. 2E; [66]). These observations show that, while DSB-initiated recombinational interactions directly mediate both homology searching and co-alignment, the SC is in turn required, through its central components, for the maintenance and/or turnover of the recombination proteins required for maturation of the DSBs into COs (e.g. [26,35,66, 68]). Furthermore, this shift, concomitantly with SC formation, indicates an intimate association of those early recombination proteins with SC central-region components [66]. Interestingly also, we Identified the putative STUbL protein Hei10 as a structure-based signal transduction molecule that coordinates progression and differentiation of recombinational interactions from zygotene through diplotene [26].
Finally, it is worth noting that while most organisms show LEs without obvious substructures, fungi from the Discomycete group of Ascomycetes like Neotiella rutilans, Ascobolus immersus and Ascobolus stercorarius exhibit a characteristic banded pattern with species-specific repeating units of alternating thick and thin bands (Fig. 2F; [69,70]). Interestingly, the spacing of the bands is constant at about 25 per micron of LE and the same periodicity is also observed when the mammal LE component SCP3 is expressed in somatic cells [71]. Furthermore, striking transitory changes in LE morphology are observed along Sordaria humana SCs [72]. LEs are tubular and form bulges (of variable lengths) at zygotene at the junctions between synapsed and unsynapsed regions and at early pachytene in regions surrounding late recombination nodules (Fig. 2G). Bulges are gone at late pachytene. As S. humana is homothallic, bulges cannot result from homolog length adjustment. Their role, if any, and why this fungus has developed such unusual LEs remain unknown.
6. Crossover interference and nucleation of the synaptonemal complex
Analysis of Sordaria Mer3, Msh4 and Hei10 localization, in parallel with analysis of the RNs seen in EM, provided further insights into the nature of those proteins for CO interference and SC formation. First, Mer3 and Msh4 foci are dramatically evenly spaced along chromosomes, for Mer3 in matched pairs along co-aligned homologs at leptotene (Fig. 1D), for Msh4 in synapsed regions at zygotene and in the SC central region at early pachytene (Fig. 2D). Even distribution of RPA and Msh4 foci is also seen in mouse at late zygotene [41,73]. A tendency for even spacing can be explained by the operation of “interference”, in which occurrence of a determining event at one position decreases the occurrence of another such determining event nearby. Interference among COs (or their cytological correlates chiasmata or MLH1/Hei10 foci) is well documented (e.g. review in [74,75]. Even spacing of Mer3 and Msh4 foci imply that interference operates at earlier stage(s), in Sordaria and in mouse [41,73] and, therefore, is not just a feature of COs. Such effects, which apply to all recombinational interactions, can be related to the fact that COs interfere not only with each other but also with adjacent NCOs [76] and to the fact that a DSB at one position along a budding-yeast chromosome disfavors occurrence of another DSB nearby [77].
Concomitant analysis of E3 ligase Hei10 foci localization (Fig. 2H) and localization of RNs plus the SC initiation sites (as analyzed from EM serial sections; Fig. 2I) showed that the process of interference includes also nucleation of SC formation [61]. A hierarchical process is defined in which early SC nucleation sites occur at recombination sites that also undergo designation for future maturation into COs, followed by later SC initiations at sites of other recombination interactions. Such a process makes biological sense because it ensures that SC nucleation sites are evenly spaced and, concomitantly, that CO-designated interactions may be especially embedded within the SC. These conclusions derive from the following findings:
(i) Late RNs, that arise from the array of total recombination interactions (defined by Mer3 and Msh4 foci), appear at zygotene just at/after the first SC nucleation sites (Fig. 2I) and at pachytene their number is similar to the number of Hei10 foci (which mark the CO sites; Fig. 2H) and chiasmata. Further, COs assayed genetically, RNs, and Hei10 foci exhibit similar CO interference [22,26,61]. Taken together these findings imply that: (ii) CO-designation and CO interference occur no later than zygotene, and concomitantly with SC formation. (iii) EM-defined RNs include both CO-correlated late RNs and so-called early RNs (Fig. 2I), which apparently correspond to recombinational interactions that are fated to mature as NCOs [62]. Detailed analysis of the relationship between zygotene SC segments with and without associated RNs on 3D EM reconstructions (e.g. Fig. 2I) showed that SC is always initiated at sites of recombination interactions, with a 1:1 relationship between SC nucleation sites and late and early RNs. This analysis also provided two more insights: (a) SC initially forms asymmetrically, away from its nucleation site and (b) SC stretches with late RNs are longer than stretches associated with early RNs, suggesting a qualitative difference and, specifically, that SCs are nucleated at late RN sites prior to nucleation of SCs at early RN sites. (iv) Simulation of the experimental data by the beam-film model for CO interference predicts that there is a single interference-mediated patterning process in Sordaria [61]. Finally, late RNs (and thus, likely, CO designation) occurring at zygotene is also observed in tomato and mammals [78,79], indication that the Sordaria findings are likely universal.
7. Local modulation of sister-chromatid cohesion at sites of CO recombination
Analysis of Sordaria mutants of the Rec8 kleisin and the cohesin-associated Spo76/Pds5 revealed several interesting features concerning the relationships among sister cohesion, SC formation and the recombination process.
First, although wild-type sister chromatids appear tightly conjoined in a single Les (Fig. 2A), mutants of both Rec8 and Spo76/Pds5, revealed that in certain conditions, separated sister chromatids can each build an AE/LE (Fig. 2J [34,80]). Similarly, separation of the sister-chromatid AEs is also observed in mouse rec8 null mutants with SC locally built between the two sister LEs [81]. Moreover, presence of SC components along all haploid slp1D chromosomes (Fig. 2C) revealed that each sister-chromatid can indeed serve as an LEs for SC formation [28].
Second, analysis of Sordaria rec8D mutant showed that recombination at the DNA level is accompanied by, and/or nucleates, changes at the axis level at the corresponding positions. (a) While Spo76-GFP staining occurs as continuous lines along all chromosome axes in wild type, in rec8D, these lines exhibit gaps from zygotene through pachytene in which the underlying chromatin is diffuse. Moreover, Msh4 foci localize to those gaps, which suggests a strong correlation between the positions of CO-fated sites and the positions of the gaps. (b) These gaps are dependent on recombination: they are absent in the rec8D spo11D double mutant. (c) Similarly, in spo76-1, a non-null mutant of SPO76 (SPO76/PDS5 is an essential gene in Sordaria), axes are single at leptotene but show partial widely open sister-chromatids from zygotene on (Fig. 2K). In contrast, axes remain single in the spo11D spo76-1 double mutant [33,80], thus providing another indication that repair of DSBs destabilizes intersister connections and axis continuity at the leptotene/zygotene transition. (d) Local separation of the two sister chromatids (indicated by double AEs) specifically at the sites of COs (marked by late RNs) is also seen in wild-type pachytene nuclei analyzed in EM, again indicating local destabilization of chromosome axes specifically at sites of CO interactions ([34]). (e) While Rec8 and Spo76/Pds5 play distinct roles in sister cohesion and axial structure, the double mutant rec8D spo76-1 confers dramatically synergistic effects on chromosome organization at all stages. As Rec8 loads normally in spo76-1 and Spo76 loads correctly in rec8D, these defects do not reflect interdependence of loading. The different patterns of axis destabilization observed in rec8D and spo76-1 mutants following recombination initiation (respectively local loss of cohesion at CO sites and dramatic loss of sister cohesion plus broken AEs) suggest that Rec8 may be devoted primarily to maintenance of axis status locally at CO sites while Spo76/Pds5 may be devoted primarily to maintenance of axis status between these sites [34]. This likely allows maintenance of structural integrity along the chromosomes despite the need for local loss of sister cohesion at sites of DSB repair. (f) Finally, as unlike Rec8, Spo76/Pds5 plays important roles during mitosis [82] the synergistic effects seen in the double mutant could reflect a functional collaboration between meiosis-specific and general cohesion components. Interestingly also, double staining of Spo76-GFP with MPM2 (which marks Topoisomerase II, thus the axes per se) shows that Spo76 is located above MPM2, leading to the idea that axis components comprise a “super-axial meshwork” [80,83].
A more general idea emerged from the finding that the transition from pachytene to the diffuse stage is accompanied by a global tendency for loss of sister connections that is constrained both by Rec8 and Spo76/Pds5. This transition period is the equivalent of the human female prophase period called the “dictyate stage”, at which female meiosis arrests from the time of prophase (which occurs in utero) to the time of ovulation, sometimes decades later. This finding thus provided an interesting potential explanation for the well known “maternal age effect” of human and mouse female meiosis. More specifically, we proposed that abnormal or weaker than normal levels of Rec8, without the ability to renew the protein, conjugated with time-dependent decay, could well contribute to the age-related observed aneuploidies [33]. These ideas have since been confirmed by several observations that there is none or very little turnover of Rec8 during the growing phase of mouse oocytes (thus including the dictyate stage; e.g. [84]).
8. Concluding remarks
We hope that the contribution of Sordaria in the comprehension of the links between meiotic pairing and recombination underlines once more that a wealth of information can be provided by the use of other organisms than the handful of major model organisms. Since an important feature of meiosis is that most steps of recombination occur in recombination complexes that are associated with chromosome axes, future studies with Sordaria will be dedicated to the understanding of the relationships among the different components of this interaction and their roles in the different steps of the recombination process. The power of this system for cytological analysis, added to new high-resolution microscopy tools, should also provide new hints regarding both bouquet and interlocking formation plus resolution.
Supplementary Material
Acknowledgments
We are extremely grateful to Nancy Kleckner for constructive discussions and comments on the manuscript. This review is dedicated to the memory of David D. Perkins who contributed in many ways to advancements of filamentous fungi in science and who always stimulated with steady interest our work. This work, D.Z. and E.E. were supported by grants from the Centre National de la Recherche Scientifique (Unite Mixte de Recherche 8621, now I2BC) and by a sub-contract collaboration with N. Kleckner supported by NIH RO1-GM044794.
References
- 1.Mitchell MB. Aberrant recombination of pyridoxine mutants of Neurospora. Proc Natl Acad Sci USA. 1955;41:216–220. doi: 10.1073/pnas.41.4.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perkins DD. The manifestation of chromosome rearrangements in unordered asci of Neurospora. Genetics. 1974;77:459–489. doi: 10.1093/genetics/77.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindegren CC. The genetics of Neurospora — III. Pure bred stocks and crossing-over in N. crassa Bull Torrey bot Club. 1933;60:133–154. [Google Scholar]
- 4.Sturtevant AH. The behavior of the chromosomes as studied through linkage. Z indukt Abstamm-u VererbLehre. 1915;13:234–287. [Google Scholar]
- 5.Müller HJ. The mechanism of crossing-over. American Naturalist. 1916;50:193–221. [Google Scholar]
- 6.Perkins DD. Crossing-over and interference in a multiply marked chromosome arm of Neurospora. Genetics. 1962;47:1253–1274. doi: 10.1093/genetics/47.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zickler H. Genetische Untersuchungen an einer heterothallischen Askomyzeten (Bombardia lunata nov spec.) Planta. 1934;22:573–613. [Google Scholar]
- 8.Lindegren CC. Gene conversion in Saccharomyces. J Genet. 1953;51:625–637. [Google Scholar]
- 9.Whitehouse HLK. In: Genetic Recombination: Understanding the mechanisms. J Wiley & sons, editor. Wiley-Interscience Publication; Chichester New York Brisbane Toronto Singapore: 1982. [Google Scholar]
- 10.Rossignol J-L, Nicolas A, Hamza H, Kalogeropoulos A. Recombination and gene conversion. In: Low B, editor. Ascobolus. The Recombination of Genetic Material, Academic Press; New York: 1988. pp. 23–72. [Google Scholar]
- 11.Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. 2015;7:361–395. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olive LS. Aberrant tetrads in Sordaria fimicola. Proc Nat Acad Sci USA. 1959;45:727–732. doi: 10.1073/pnas.45.5.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitani Y, Olive LS, El-Ani AS. Genetics of Sordaria fimicola. V. Aberrant segregation at the g locus. Am J Bot. 1962;49:697–706. [Google Scholar]
- 14.Leblon G. Mechanism of gene conversion in Ascobolus immersus. I. Existence of a correlation between the origin of the mutants induced by different mutagens and their conversion spectrum. Mol Gen Genet. 1972;115:36–48. [Google Scholar]
- 15.Lissouba P, Rizet G. Sur l’existence d’une unité génétique polarisée ne subissant que des échanges non réciproques. C R Acad Sci Paris. 1960;250:3408–3410. [PubMed] [Google Scholar]
- 16.Lissouba P, Mousseau J, Rizet G, Rossignol J-L. Fine structure of genes in the ascomycete Ascobolus immersus. Adv Genet. 1962;11:343–380. [Google Scholar]
- 17.Nicolas A, Petes TD. Polarity of meiotic recombination in fungi: contrasting views. Experientia. 1994;50:242–252. doi: 10.1007/BF01924007. [DOI] [PubMed] [Google Scholar]
- 18.Lichten M, de Massy B. The impressionistic landscape of meiotic recombination. Cell. 2011;147:267–270. doi: 10.1016/j.cell.2011.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Massy B. Initiation of Meiotic Recombination: How and Where? Conservation and Specificities Among Eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 20.Perkins DD. Chromosome rearrangements in Neurospora and other filamentous fungi. Adv Genet. 1997;36:239–398. doi: 10.1016/s0065-2660(08)60311-9. [DOI] [PubMed] [Google Scholar]
- 21.Leblon G, Zickler D, Le Bilcot S. Most U.V. induced reciprocal translocations in Sordaria macrospora occur in or near the centromere regions. Genetics. 1986;112:183–204. doi: 10.1093/genetics/112.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zickler D, Moreau PJ, Huynh AD, Slezec AM. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics. 1992;132:135–148. doi: 10.1093/genetics/132.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zickler D, Leblon G, Haedens V, Collard A, Thuriaux P. Linkage group-chromosome correlations in Sordaria macrospora: chromosome identification by three dimensional reconstruction of their synaptonemal complex. Current Genetics. 1984;8:57–67. doi: 10.1007/BF00405433. [DOI] [PubMed] [Google Scholar]
- 24.Perkins DD. Neurospora chromosomes. In: Federoff N, Botstein D, editors. The dynamic genome: B. McClintock’s ideas in the century of genetics. Cold Spring Harbor Laboratory Press; 1992. pp. 33–44. [Google Scholar]
- 25.Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- 26.De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, Zickler D. E3 ligase Hei10: a multi-faceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev. 2014;28:1111–1123. doi: 10.1101/gad.240408.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zickler D. Development of the synaptonemal complex and the “recombination nodules” during meiotic prophase in the seven bivalents of the fungus Sordaria macrospora Auersw. Chromosoma. 1977;61:289–316. doi: 10.1007/BF00288615. [DOI] [PubMed] [Google Scholar]
- 28.Vasnier C, de Muyt A, Zhang L, Tesse S, Kleckner NE, Zickler D, Espagne E. Absence of SUN-domain protein Slp1 blocks karyogamy and switches meiotic recombination and synapsis from homologs to sister chromatids. Proc Natl Acad Sci USA. 2014;111:E4015–4023. doi: 10.1073/pnas.1415758111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 30.Zickler D, Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol. 2015;7:397–422. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zickler D. Meiosis in mycelial fungi. In: Kües & Fischer, editor. The Mycota I, Growth, differenciation and sexuality. Springer-Verlag; Berlin Heidelberg: 2009. pp. 407–424. [Google Scholar]
- 32.Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci USA. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storlazzi A, Tessé S, Gargano S, James F, Kleckner N, Zickler D. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 2003;17:2675–2687. doi: 10.1101/gad.275203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storlazzi A, Tesse S, Ruprich-Robert G, Gargano S, Pöggeler S, Kleckner N, Zickler D. Coupling meiotic chromosome axis integrity to recombination. Genes Dev. 2008;22:796–809. doi: 10.1101/gad.459308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell. 2010;141:94–106. doi: 10.1016/j.cell.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bojko M. Two kinds of “recombination nodules” in Neurospora crassa. Genome. 1989;32:309–317. doi: 10.1139/g89-446. [DOI] [PubMed] [Google Scholar]
- 37.Pukkila PJ, Lu BC. Silver staining of meiotic chromosomes in the fungus, Coprinus cinereus. Chromosoma. 1985;91:108–112. doi: 10.1007/BF00294053. [DOI] [PubMed] [Google Scholar]
- 38.Loidl J. The initiation of meiotic chromosome pairing: The cytological view. Genome. 1990;33:759–778. doi: 10.1139/g90-115. [DOI] [PubMed] [Google Scholar]
- 39.Albini SM, Jones GH. Synaptonemal complex-associated centromeres and recombination nodules in plant meiocytes prepared by an improved surface-spreading technique. Exp Cell Res. 1984;155:588–592. doi: 10.1016/0014-4827(84)90219-2. [DOI] [PubMed] [Google Scholar]
- 40.Fung JC, Rockmill B, Odell M, Roeder GS. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell. 2004;116:795–802. doi: 10.1016/s0092-8674(04)00249-1. [DOI] [PubMed] [Google Scholar]
- 41.Oliver-Bonet M, Campillo M, Turek PJ, Ko E, Martin RH. Analysis of replication protein A (RPA) in human spermatogenesis. Mol Hum Reprod. 2007;13:837–844. doi: 10.1093/molehr/gam076. [DOI] [PubMed] [Google Scholar]
- 42.Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4–hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 43.Trelles-Sticken E, Dresser ME, Scherthan H. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J Cell Biol. 2000;151:95–106. doi: 10.1083/jcb.151.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CY, Conrad MN, Dresser ME. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 2012;8:e1002730. doi: 10.1371/journal.pgen.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rog O, Dernburg AF. Chromosome pairing and synapsis during Caenorhabditis elegans meiosis. Curr Opin Cell Biol. 2013;25:349–356. doi: 10.1016/j.ceb.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scherthan H. Factors directing telomere dynamics in synaptic meiosis. Biochem Soc Trans. 2006;34:550–553. doi: 10.1042/BST0340550. [DOI] [PubMed] [Google Scholar]
- 49.MacQueen AJ, Colaiácovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 51.Nakagawa T, Kolodner RD. Saccharomyces cerevisiae Mer3 is a DNA helicase involved in meiotic crossing over. Mol Cell Biol. 2002;22:3281–3291. doi: 10.1128/MCB.22.10.3281-3291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC. Saccharomyces cerevisiae Mer3 helicase stimulates 3′–5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell. 2004;117:47–56. doi: 10.1016/s0092-8674(04)00294-6. [DOI] [PubMed] [Google Scholar]
- 53.Hunter N. Molecular recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Springer; New York: 2007. pp. 381–442. (Topics in Current Genetics). [Google Scholar]
- 54.Oliver-Bonet M, Turek PJ, Sun F, Ko E, Martin RH. Temporal progression of recombination in human males. Mol Hum Reprod. 2005;7:517–522. doi: 10.1093/molehr/gah193. [DOI] [PubMed] [Google Scholar]
- 55.de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci USA. 2006;103:9607–9612. doi: 10.1073/pnas.0600418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J Cell Sci. 2007;120:1017–1027. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- 57.Franklin FC, Higgins JD, Sanchez-Moran E, Armstrong SJ, Osman KE, et al. Control of meiotic recombination in Arabidopsis: role of the MutL and MutS homologues. Biochem Soc Trans. 2006;34:542–544. doi: 10.1042/BST0340542. [DOI] [PubMed] [Google Scholar]
- 58.von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- 59.Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harbor Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- 60.Wang CR, Carlton PM, Golubovskaya IN, Cande WZ. Interlock formation and coiling of meiotic chromosome axes during synapsis. Genetics. 2009;183:905–915. doi: 10.1534/genetics.109.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, Espagne E, de Muyt A, Zickler D, Kleckner NE. Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc Natl Acad Sci USA. 2014;111:5059–68. doi: 10.1073/pnas.1416411111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carpenter ATC. Gene conversion, recombination nodules, and the initiation of meiotic synapsis. Bioessays. 1987;6:232–236. doi: 10.1002/bies.950060510. [DOI] [PubMed] [Google Scholar]
- 63.Anderson LK, Lohmiller LD, Tang X, Hammond DB, Javernick L, Shearer L, et al. Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proc Natl Acad Sci USA. 2014;111:13415–20. doi: 10.1073/pnas.1406846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henderson KA, Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc Natl Acad Sci USA. 2004;101:4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rockmill B, Lefrançois P, Voelkel-Meiman K, Oke A, Roeder GS, Fung JC. High throughput sequencing reveals alterations in the recombination signatures with diminishing Spo11 activity. PLoS Genet. 2013;9:e1003932. doi: 10.1371/journal.pgen.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Espagne E, Vasnier C, Storlazzi A, Kleckner NE, Silar P, Zickler D, Malagnac F. Sme4 coiled-coil protein mediates synaptonemal complex assembly, recombinosome relocalization, and spindle pole body morphogenesis. Proc Natl Acad Sci USA. 2011;108:10614–10619. doi: 10.1073/pnas.1107272108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J Cell Sci. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- 68.Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 69.von Wettstein D. The synaptinemal complex and four-strand crossing over. Proc Natl Acad Sci USA. 1971;68:851–855. doi: 10.1073/pnas.68.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zickler D. Fine structure of chromosome pairing in ten Ascomycetes: meiotic and premeiotic (mitotic) synaptonemal complexes. Chromosoma. 1973;40:401–416. doi: 10.1007/BF00399431. [DOI] [PubMed] [Google Scholar]
- 71.Yuan I, Pelttari J, Brundell E, Bjorkroth B, Zhao J, et al. The synaptonemal complex protein SCP3 can form multistranded, cross-striated fibers in vivo. J Cell Biol. 1998;142:331–339. doi: 10.1083/jcb.142.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zickler D, Sage J. Synaptonemal complexes with modified lateral elements in Sordaria humana: development of and relationship to the “recombination nodules”. Chromosoma. 1981;84:305–318. [Google Scholar]
- 73.de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci USA. 2006;103:9607–9612. doi: 10.1073/pnas.0600418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mézard C, Vignard J, Drouaud J, Mercier R. The road to crossovers: plants have their say. Trends Genet. 2007;23:91–99. doi: 10.1016/j.tig.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 75.Wang S, Zickler D, Kleckner N, Zhang L. Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle. 2015;14:305–314. doi: 10.4161/15384101.2014.991185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu TC, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lhuissier FG, Offenberg HH, Wittich PE, Vischer NO, Heyting C. The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell. 2007;19:862–876. doi: 10.1105/tpc.106.049106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bojko M. Human meiosis. IX. Crossing over and chiasma formation in oocytes. Carlsberg Res Commun. 1985;50:43–72. [Google Scholar]
- 80.van Heemst D, James F, Pöggeler S, Berteaux-Lecellier V, Zickler D. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell. 1999;98:261–271. doi: 10.1016/s0092-8674(00)81020-x. [DOI] [PubMed] [Google Scholar]
- 81.Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 82.van Heemst D, Kafer E, John T, Heyting C, van Aalderen M, Zickler D. BimD/SPO76 is at the interface of cell cycle progression, chromosome morphogenesis, and recombination. Proc Natl Acad Sci USA. 2001;98:6267–6272. doi: 10.1073/pnas.081088498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kleckner N, Zhang L, Weiner B, Zickler D. Meiotic chromosome dynamics. In: Rippe K, editor. Genome Organization and Function in the Mammalian Cell Nucleus. Wiley-VCH; Weinheim, Germany: 2011. pp. 487–534. [Google Scholar]
- 84.Tachibana-Konwalski K, Godwin J, van der Weyden L, Champion L, Kudo NR, Adams DJ, Nasmyth K. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev. 2010;24:2505–16. doi: 10.1101/gad.605910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


