Summary
While several lung cancer susceptibility loci have been identified, much of lung cancer heritability remains unexplained. Here, 14,803 cases and 12,262 controls of European descent were genotyped on the OncoArray and combined with existing data for an aggregated GWAS analysis of lung cancer on 29,266 patients and 56,450 controls. We identified 18 susceptibility loci achieving genome wide significance, including 10 novel loci. The novel loci highlighted the striking heterogeneity in genetic susceptibility across lung cancer histological subtypes, with four loci associated with lung cancer overall and six with lung adenocarcinoma. Gene expression quantitative trait analysis (eQTL) in 1,425 normal lung tissues highlighted RNASET2, SECISBP2L and NRG1 as candidate genes. Other loci include genes such as a cholinergic nicotinic receptor, CHRNA2, and the telomere-related genes, OFBC1 and RTEL1. Further exploration of the target genes will continue to provide new insights into the etiology of lung cancer.
Lung cancer continues to be the leading cause of cancer mortality worldwide1. Although tobacco smoking is the main risk factor, the heritability of lung cancer has been estimated at 18%2. Genome-wide association studies (GWAS) have previously identified several lung cancer susceptibility loci including CHRNA3/5, TERT, HLA, BRCA2, CHEK23,4, but most of its heritability remains unexplained. With the goal of conducting a comprehensive characterization of common lung cancer genetic susceptibility loci, we undertook additional genotyping of lung cancer cases and controls using the OncoArray5 genotyping platform, which queried 517,482 SNPs chosen for fine mapping of susceptibility to common cancers as well as for de novo discovery (Supplementary Table 1, and Online Methods). All participants gave an informed consent, and each study obtained local ethics committee approval. After quality control filters (Online Methods), a total of 14,803 cases and 12,262 controls of European ancestry were retained and underwent imputation techniques to infer additional genotypes for genetic variants included in the 1000 Genomes Project data (Online Methods). Logistic regression was then used to assess the association between variants (n=10,439,017 SNPs) and lung cancer risk, as well as by predominant histological types and by smoking behavior (Online Methods). Fixed-effects models (Online Methods) were used to combine the OncoArray results with previously published lung cancer GWAS3,4,6, allowing for analysis of 29,266 patients and 56,450 controls of European descent (Table 1). There were no signs of genomic inflation overall or for any subtypes (Supplementary Figure 1) indicating little evidence for confounding by cryptic population structure (Online Methods). All findings with a P-value less than 1×10−5 are reported in Supplementary Table 2. As shown in Figure 1, the genetic architecture of lung cancer varies markedly among histological subtypes, with striking differences between lung adenocarcinoma and squamous cell carcinoma. Manhattan plots for small cell carcinoma (SCLC), ever and never smoking are displayed in Supplementary Figure 2. The array heritability estimates were comparable among histological subsets, but squamous cell carcinoma appeared to share more genetic architecture with small cell carcinoma (SCLC) than with adenocarcinoma (Supplementary Table 3).
Table 1.
Demographic characteristics of the participating studies after quality control filters
| Lung cancer patients
|
Controls
|
|||
|---|---|---|---|---|
| number | (%) | number | (%) | |
|
|
|
|
||
| OncoArray studies- passed QC | 14803 | (51) | 12262 | (22) |
| Published GWAS studiesa | 14463 | (49) | 44188 | (78) |
|
|
|
|
||
| Total | 29266 | 56450 | ||
|
|
|
|
||
| Age | ||||
| <=50 | 3112 | (12) | 6032 | (12) |
| >50 | 23025 | (88) | 44075 | (88) |
| Sex | ||||
| Male | 18208 | (62) | 27178 | (53) |
| Female | 11059 | (38) | 24069 | (47) |
| Smoking status | ||||
| Never | 2355 | (9) | 7504 | (31) |
| Ever | 23223 | (91) | 16964 | (69) |
| Former | 9037 | (35) | 8554 | (35) |
| Current | 13356 | (52) | 7477 | (31) |
| Histology c | ||||
| Adenocarcinoma | 11273 | (39) | 55483 b | |
| Squamous cell carcinoma | 7426 | (25) | 55627 b | |
| Small cell carcinoma | 2664 | (9) | 21444 b | |
Previous GWAS studies include IARC, MDACC, SLRI, ICR, Harvard, ATBC, CPSII, German and deCODE studies.
Number of non-cancer individuals included in the corresponding histology-specific analysis.
The remaining 27% includes other histological subsets, such as large cell carcinoma, non-small cell lung cancer, NOS, mixed histology, and unknown.
Figure 1. Manhattan plots of lung cancer risk overall and by histological subtypes.
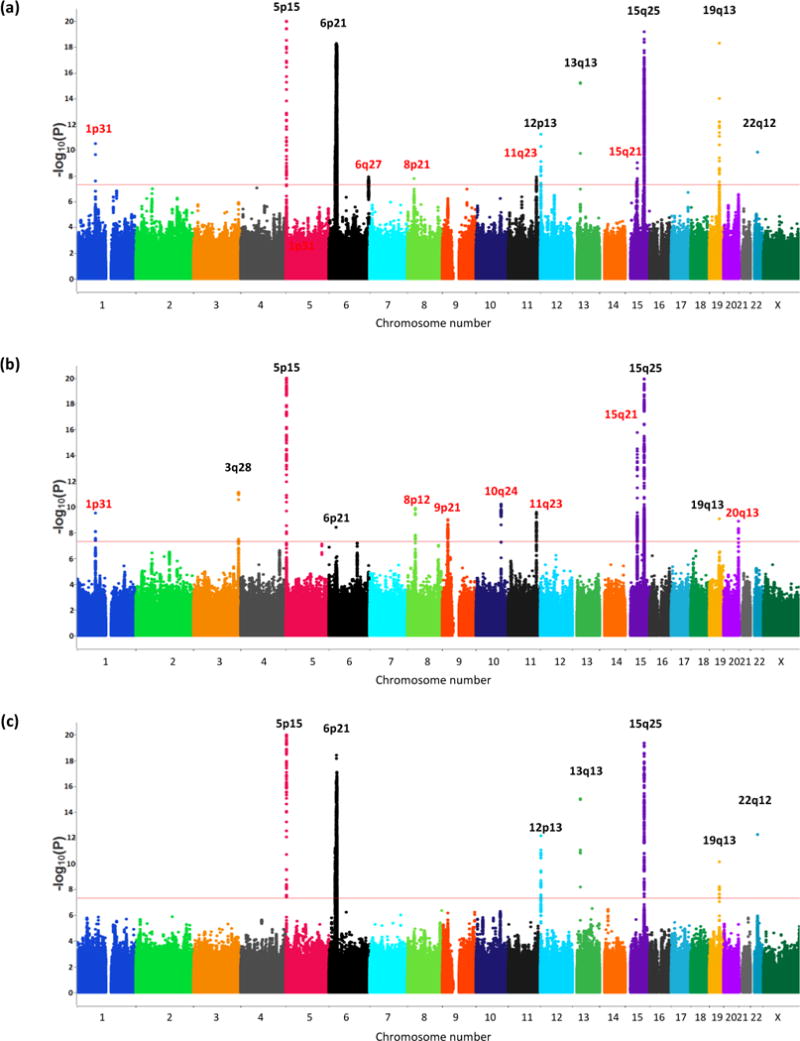
(a) lung cancer risk overall, 29,266 cases and 56,450 controls (b) adenocarcinoma, 11,273 cases and 55,483 controls (c) squamous cell carcinoma 7,426 cases and 55,627 controls. Each locus is annotated by their cytoband locations. The X‐axis represents chromosomal locations and the Y‐axis represents −log10(P‐value). Black denotes the previously known loci and Red denotes the new loci identified in this analysis
Table 2 presents summary results of all loci with sentinel variants (defined as the variant with the lowest P-value at each locus) that reached genome-wide significance (P-value < 5×10−8) for lung cancer overall and by histological subtypes. Sentinel variants stratified by new and previous genotyping and additional statistical significance assessed based on the number of effective tests, Approximate Bayes Factors, and Bayesian False Discovery Probability are presented in Supplementary Tables 4 and 5, respectively. Repeat genotyping of 12% of the OncoArray genotyped samples confirmed the fidelity of the genotyping or imputation for the risk loci, and showed excellent concordance of imputation for SNPs with MAF>0.05 (Online Methods, Supplementary note). Among the 18 loci that reached GWAS significance, 10 had not reached significance in a genome-wide scan (Figure 1). Of these, four novel loci were associated with lung cancer overall, and six with adenocarcinoma.
Table 2.
The association between sentinel variants representing each lung cancer locus and lung cancer risk.
| Strata | Locus* | rs number | Gene | Allelea | Imputed versus Oncoarray genotyped | Candidate Oncoarray | EAF | OR | 95%CI | P-value | CPD | FTND | FEV1 | FVC | FEV1/FVC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Customized panel | p- value | p- value | p- value | p- value | p- value | ||||||||||
| Lung | 1p31.1* | rs71658797 | FUBP1 | T_A | Oncoarray | No | 0.103 | 1.1 | 1.09–1.18 | 3.3E-11 | 0.056 | 0.334 | 0.445 | 0.898 | 0.334 |
| Lung | 6q27* | rs6920364 | RNASET2 | G_C | Imputed | eQTL | 0.456 | 1.1 | 1.05–1.10 | 1.3E-08 | 0.833 | 0.104 | 0.927 | 0.876 | 0.986 |
| Lung | 8p21.1* | rs11780471 | CHRNA2 | G_A | Imputed | Lung | 0.060 | 0.9 | 0.83–0.91 | 1.7E-08 | 0.646 | 0.403 | 6.9E-04 | 0.055 | 0.016 |
| Lung | 13q13.1 | rs11571833 | BRCA2 | A_T | Imputed | Lung | 0.011 | 1.6 | 1.43–1.80 | 6.1E-16 | 0.890 | 0.312 | 0.601 | 0.667 | 0.237 |
| Lung | 15q21.1* | rs66759488 | SEMA6D | G_A | imputed | Lung | 0.362 | 1.1 | 1.05–1.10 | 2.8E-08 | 0.266 | 0.888 | 0.739 | 0.200 | 0.202 |
| Lung | 15q25.1 | rs55781567 | CHRNA5 | C_G | Imputed | Lung | 0.367 | 1.3 | 1.27–1.33 | 3.1E-103 | 6.8E-38 | 9.7E-16 | 7.2E-03 | 0.020 | 0.144 |
| Lung | 19q13.2ˆ | rs56113850 | CYP2A6 | C_T | Oncoarray | Lung | 0.440 | 0.9 | 0.86–0.91 | 5.0E-19 | 8.1E-20 | 7.5E-04 | 0.822 | 0.826 | 0.319 |
| Adeno | 3q28 | rs13080835 | TP63 | G_T | Imputed | Lung | 0.493 | 0.9 | 0.87–0.92 | 7.5E-12 | 0.803 | 0.336 | 0.135 | 0.445 | 0.834 |
| Adeno | 5p15.33 | rs7705526 | TERT | C_A | Oncoarray | All | 0.342 | 1.3 | 1.21–1.29 | 3.8E-35 | 0.511 | 0.738 | 0.292 | 0.038 | 0.657 |
| Adeno | 8p12* | rs4236709 | NRG1 | A_G | Imputed | eQTL | 0.218 | 1.1 | 1.09–1.18 | 1.3E-10 | 0.991 | 0.957 | 0.503 | 0.151 | 0.403 |
| Adeno | 9p21.3* | rs885518 | CDNK2A | A_G | Imputed | Several | 0.101 | 1.2 | 1.11–1.23 | 9.96E-10 | 0.904 | 0.321 | 0.421 | 0.096 | 0.146 |
| Adeno | 10q24.3* | rs11591710 | OBFC1 | A_C | Imputed | Lung | 0.137 | 1.2 | 1.11–1.22 | 6.3E-11 | 0.500 | 0.152 | 0.027 | 0.019 | 0.533 |
| Adeno | 11q23.3* | rs1056562 | AMICA1 | C_T | Oncoarray | Breast | 0.473 | 1.1 | 1.07–1.14 | 2.8E-10 | 0.717 | 0.538 | 0.449 | 0.718 | 0.039 |
| Adeno | 15q21.1* | rs77468143 | SECISBP2L | T_G | Imputed | No | 0.253 | 0.9 | 0.83–0.89 | 1.7E-16 | 0.071 | 0.184 | 4.9E-03 | 0.440 | 1.4E-03 |
| Adeno | 20q13.33* | rs41309931 | RTEL1 | G_T | Imputed | Prost/ColR | 0.117 | 1.2 | 1.11–1.23 | 1.3E-09 | 0.146 | 0.939 | 0.964 | 0.657 | 0.284 |
| SQC | 6p21.33 | rs116822326 | MHC | A_G | Imputed | Lung | 0.155 | 1.3 | 1.19–1.32 | 3.8E-19 | 0.392 | 0.774 | 0.132 | 0.498 | 0.103 |
| SQC | 12p13.33 | rs7953330 | RAD52 | G_C | Oncoarray | Lung | 0.315 | 0.9 | 0.83–0.90 | 7.3E-13 | 0.800 | 0.463 | 0.019 | 3.3E-03 | 0.424 |
| SQC | 22q12.1 | rs17879961 | CHEK2 | A_G | Oncoarray | Lung | 0.005 | 0.4 | 0.32–0.52 | 5.7E-13 | 0.441 | 0.360 | 0.041 | 0.040 | 0.805 |
denote novel locus identified to GWAS significance by this study; a, reference_effect. Bolded p-values indicate significant associations with consistent direction as expected. Genome positions relative to GRCh37, EAF, effective allele frequency; OR, odds (log additive) ratio; 95%CI, 95% confidence interval. P-value, based on fixed-effect meta-analysis adjusted for age, sex and genetically derived ancestry; CPD, cigarette per day; FTND, Fagerstrӧm Test for Nicotine Dependence; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity.
Adeno, adenocarcinoma; SQC, squamous cell carcinoma.
marker had an acceptable, but not ideal concordance rate (see Supplementary Note)
To decipher the association between these 18 loci and lung cancer risk, we further investigated their association with gene expression level in normal lung tissues (n=1,425) (Supplementary Table 6, Supplementary Figure 3), genomic annotations (Supplementary Table 7), smoking propensity (cigarettes smoked per day (n=91,046) and Fagerström Test for Nicotine Dependence metrics (n=17,074)) (Table 2). Previous studies have shown shared risk for lung cancer and COPD through inflammation and ROS pathways7; therefore, we also assessed the association between sentinel SNPs and reduced lung capacity through spirometry measurements (forced expiratory volume in 1 second [FEV1], forced vital capacity [FVC], n =30,199) (Table 2 and Online Methods).
Variants at 4 novel loci (1p31.1, 6q27, 8p21, 15q21.1) were associated with lung cancer risk overall, with little evidence for heterogeneity among subtypes (Supplementary Figure 4). The 1p31.1 locus, recently identified in a pathway-based analysis of the TRICL data8, represented by rs71658797 (Odds Ratio [OR]=1.14, 95% Confidence Interval [CI] 1.09–1.18, P-value=3.25 × 10−11), is located near FUBP1/DNAJB4 (Supplementary Figure 4). At 6q27, rs6920364 was associated with lung cancer risk with an OR of 1.07 (95% CI 1.04–1.09, P-value=2.9×10−8) with little heterogeneity found by smoking status (Supplementary Figure 4). This locus is predicted to regulate RNASET2 (Supplementary Figure 5, Supplementary Table 7). We identified rs6920364 as a lung cis-eQTL for RNASET2, an extracellular ribonuclease, in all five cohorts tested (Supplementary Table 6), with increased lung cancer risk correlating with increased RNASET2 expression (Figure 2). Variants correlated with rs6920364 (r2>0.88) have been noted in GWAS of Crohn’s disease and inflammatory bowel disease9–13.
Figure 2. Scatter plots comparing variants across the 6q27, 15q21.1, 8p12 and 11q23.3 susceptibility loci and (Y-axis) their associated with lung cancer (or lung adenocarcinoma, as relevant) and (X-axis) the lung cis–eQTL (GTEx).
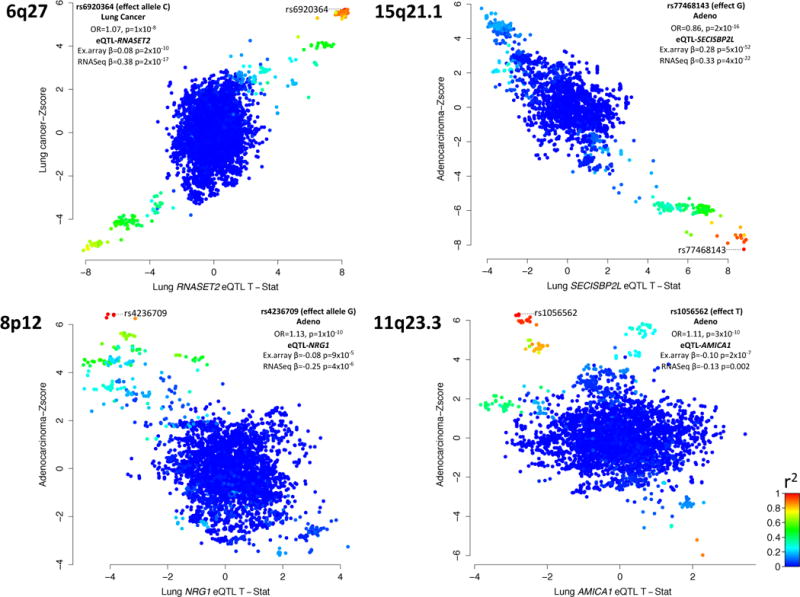
Each variant (dot) is colored relative the degree of linkage disequilibrium (r2) with sentinel lung cancer variant (marked) at that locus. Indented table, association between sentinel variant and lung cancer (or histological subtype) as well as the eQTL evidence in lung epithelium for the microarray (Laval, UBC, Groningen) and RNAseq (NCI and GTEx) cohorts. At 6q27, 15q21.1 and 8p12, the variants associated with lung cancer also tend to be those that that are lung cis-eQTL’s for RNASET2, SECISBP2L and NRG1, respectively. At 11q23.3, while the sentinel variant (rs1056562) is a lung cis-eQTL for AMICA1, additional variants are AMICA1 lung cis-eQTL’s, but not associated with lung adenocarcinoma and vice versa suggesting an alternate candidate gene may be responsible for this association or a pleiotropic effect at this locus.
The 8p21 locus has been suggested as a lung cancer susceptibility locus by pathway analysis14 and now confirmed at GWAS significance level. It is a complex locus represented by sentinel variant rs11780471 associated with lung cancer (OR=0.87, 95% CI 0.83–0.91, P-value=1.69×10−8) (Supplementary Figure 4), but this region contained additional uncorrelated variants (pairwise r2< 0.10) associated with lung cancer (Supplementary Table 8). Multivariate analysis was consistent with multiple susceptibility alleles at this locus (Supplementary Table 8). In contrast to lung tissue (Figure 3A, Supplementary Table 6, Supplementary Figure 3), we noted that the alleles associated with lung cancer tended to be associated with cerebellum expression of CHRNA2, a member of the cholinergic nicotinic receptor (Figure 3B). The CHRNA2 rs11780471 cis-eQTL effect in the brain was limited to the cerebellum (Figure 3C), a region not traditionally linked with addictive behavior, but where an emerging role is suggested15. We therefore investigated rs11780471 in the context of smoking behavior (Supplementary Methods). Unlike the well-described 15q25.1 (rs55781567) CHRNA5 locus (Table 2), rs11780471 was not associated with number of cigarettes smoked per day or the FTND metrics (Figure 3D). Nevertheless, lung cancer risk allele carriers of rs11780471 tended to be smokers and initiated smoking at earlier ages (Figure 3D), implying that this variant’s association with lung cancer could potentially be mediated via influencing aspects of smoking behavior. Another potentially relevant gene in this region is EPHX2, a xenobiotic metabolism gene. Although the sentinel variant is not an eQTL for EPHX2 in lung tissues, other associated variants in the region are (e.g. rs146729428, p-value of 1.77×10−7 (Supplementary Table 2) and 5 × 10−4 for lung cancer risk and eQTL, respectively). A potential synergistic role of both EPHX2 and CHRNA2 on lung cancer etiology cannot be excluded.
Figure 3. eQTL and smoking behavior analysis of the 8p21 lung cancer susceptibility locus. Upper panel.
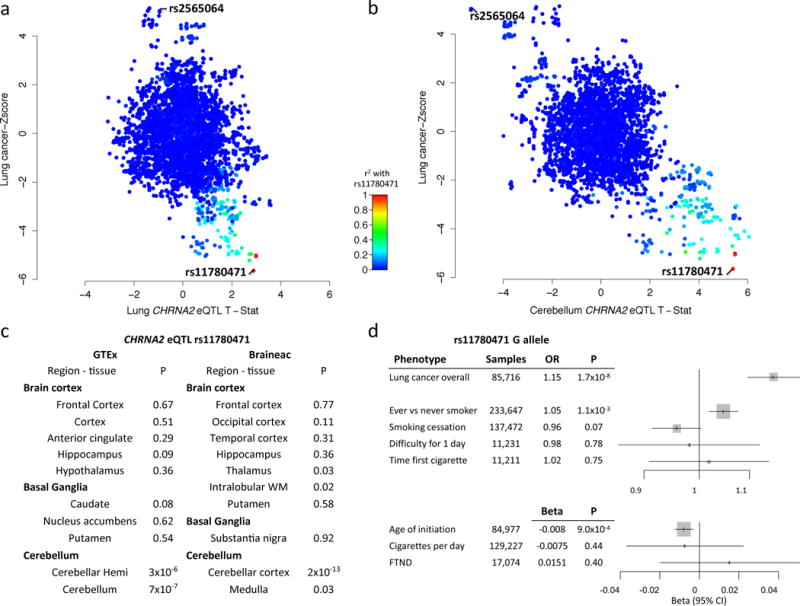
Scatter plots of variants across the 8p21 locus and their associated with lung cancer (Y-axis) and CHRNA2 eQTL (X-axis) in lung epithelial tissues (panel a) and CHRNA2 eQTL in brain cerebellum tissues (panel b). Panel C. eQTL association between rs11780471 across tissues from different parts of the brain from GTEx and Braineac consortia noting CHRNA2 cis- eQTL effect appears restricted to the brain cerebellum. Panel D. Association between rs11780471 and smoking phenotypes, noting evidence for association between smoking status (ever vs never) and age of initiation, with lung cancer risk allele carriers (G) more likely to be ever smokers and take up smoking earlier. Fagerström Test for Nicotine Dependence (FTND) index, error bars indicate the 95% confidence intervals.
The genetic locus at 15q21 (rs66759488) was shown to be associated with lung cancer (OR=1.07, 95% CI 1.04–1.10, p=2.83×10−8) overall and across lung cancer histologies (Supplementary Figure 4). Genomic annotation suggests that genetic variants correlated with rs66759488 may influence the SEMA6D gene (Supplementary Table 7), but there was no clear eQTL effect (Supplementary Table 6), and this variant did not appear to have a major influence on smoking propensity or lung function (Table 2).
For specific lung cancer histology subtypes, we identified 6 novel loci associated with lung adenocarcinoma (15q21, 8p12, 10q24, 20q13.33, 11q23.3 and 9p21.3) (Table 2). The locus at 15q21 (rs77468143, OR=0.86, 95% CI 0.82–0.89, p=1.15×10−16) is predicted to target SECISBP2L (Supplementary Figure 5), and expression analysis indicated rs77468143 to be a cis-eQTL for SECISBP2L in lung tissue in all eQTL cohorts tested (Supplementary Table 6). The genetic risk allele appears to correlate with decreased expression levels of SECISBP2L (Figure 2, Supplementary Figure 5), an observation that is consistent with SECISBP2L being down regulated in lung cancers16. rs77468143 was nominally associated with lung function (Table 2), potentially implicating inflammation of lung as part of the mechanism at this locus.
At 8p12, expression analysis indicated that the alleles associated with lung adenocarcinoma (represented by the sentinel variant rs4236709 (Table 2)), also appear to be a lung cis-eQTL for the NRG1 gene (Supplementary Table 6, Supplementary Figure 5). This region also contains putative regulatory regions (Supplementary Figure 5). Somatic translocations of NRG1 are infrequently observed in lung adenocarcinomas17. While somatic translocations at 8p12 generally take place in never smokers and are linked with ectopic activation of NRG1, rs4236709 was associated with lung cancer in both ever and never smokers (Supplementary Figure 4) and its genetic risk correlated with decreased NRG1 expression (Figure 2). Interestingly, 6q22.1 variants located near ROS1, another gene somatically translocated in lung adenocarcinoma and for which nearby germline variants were associated with never smoking lung adenocarcinoma in Asian women18, were associated with lung adenocarcinoma at borderline genome wide significance (rs9387479; OR=0.92, 95% CI 0.89–0.95, p=6.57×10−8) (Supplementary Table 2).
Three of the sentinel variants associated with lung adenocarcinoma are located near genes related to telomere regulation; rs7902587 (10q24) and rs41309931 (20q13.33) near OBFC1 and RTEL1, respectively, and rs2853677 near TERT as previously noted19,20. The variants at 10q24 associated with lung adenocarcinoma also appear to be associated with telomere length (Supplementary Figure 6). By contrast, and consistent with observations with 20q13.33 variants associated with glioma21, the variants associated with telomere length at 20q13.33 were not necessarily those associated with lung adenocarcinoma (Supplementary Figure 6). Nevertheless, more generally the variants associated by GWAS with longer telomere length22 appear linked with risk of lung adenocarcinoma23 and glioma21,24, a finding consistent with our expanded analysis here (Supplementary Figure 6).
We additionally identified a complex locus at 11q23.3. The sentinel variant rs1056562 (OR=1.11, 95% CI 1.07–1.14, p=2.7×10−10) is more prominently associated with lung adenocarcinoma (Supplementary Figure 4). rs1056562 was correlated with expression of two genes at this locus, AMICA1 and MPZL3 (Supplementary Table 6). However, there did not appear to be a consistent relationship between the alleles related with AMICA1 and MPZL3 gene expression and those with lung adenocarcinoma (Figure 2, Supplementary Table 9), suggesting that expression of these genes alone is unlikely to mediate this association.
At 9p21.3 we identified rs885518 that appeared to be associated with lung adenocarcinoma (OR=1.17, 95% CI 1.11–1.23, p=6.8×10−10). 9p21.3 is a region containing CDNK2A and variants associated with multiple cancer types, including lung cancer. Nevertheless, rs885518 is located approximately 200kb centromeric the previously described variants (Supplementary Figure 4) and shows little evidence for LD (all pairwise r2< 0.01) with rs1333040, a variant previously associated with lung squamous cell carcinoma3 and rs62560775, another variant suggested to be associated with lung adenocarcinoma25 that we confirm to genome significance here. Intriguingly, these variants appear to confer predominant associations with different lung cancer histologies suggesting that they are independent associations (Supplementary Figure 7).
Aside from the clear smoking-related effects on lung cancer risk through the CHRNA5 and CYP2A6 regions and association with CHRNA2 noted above, the rest of variants we have identified do not appear to clearly influence smoking behaviors (Table 2), implying that these associations are likely mediated by other mechanisms. Nevertheless, there is shared genetic architecture between smoking behavior and lung cancer risk, consistent with the notion that genetic variants do influence lung cancer risk also through behavioral mechanisms (Supplementary Figure 8).
In conclusion, the genetic susceptibility alleles we describe here explain approximately 12.3% of the familial relative risk previously reported in family cancer databases26,27, out of which 3.5% was accounted for by the novel loci. Our findings emphasize striking heterogeneity across histological subtypes of lung cancer. We expect that further exploration of the related target genes of these susceptibility loci, as well as validation and identification of new loci, will continue to provide insights into the etiology of lung cancer.
Online methods
This work is conducted based on the collaboration of Transdisciplinary Research of Cancer in Lung of the International Lung Cancer Consortium (TRICL-ILCCO) and the Lung Cancer Cohort Consortium (LC3). The participating studies are individually described in the Supplementary Note.
OncoArray genotyping
Genotyping was completed at the Center for Inherited Disease Research (CIDR), the Beijing Genome Institute, the HelmholtzCenter Munich (HMGU), Copenhagen University Hospital, and the University of Cambridge. Quality control steps follow the approach described previously for the OncoArray5 (Supplementary Note).
Genotype quality control
After removing the 1,193 expected duplicates, QC procedures for the 43,398 individuals are summarized in Supplementary Note Figure 1. Standard quality control procedures (detailed in the Supplementary Note) were used to exclude underperforming individuals (number of DNAs=1,708) and genotyping assays (judged by success rate, genotype distributions deviated from that expected by Hardy Weinberg equilibrium, number of variants=16,149). After filtering, there were 517,482 SNPs available for analysis.
Identity by Descent (IBD) was calculated between each pair of samples in the data using PLINK to detect unexpected duplicates and relatedness. Details are described in Supplementary Note. 340 unexpected duplicated samples (proportion IBD>0.95) and 940 individuals were removed as related samples with proportion IBD between 0.45 and 0.95. Of these, 721 of them were expected first degree relatives. In total, 0.56% of the total samples were removed as unexpected duplicates or relatives in the QC analysis. We additionally considered the potential that more distant familial relationships could have impacted the results. However, further restriction to proportion IBD > 0.2 identified 139 second degree relatives and excluding these had minimal impact on the association results (Supplementary Note Table 1).
Complete genotype data for X chromosomes were used to verify reported sex by using PLINK sex inference and a support vector machine procedure resulting in 306 non-concordant samples being removed (Supplementary Note).
We used the program FastPop (http://sourceforge.net/projects/fastpop/)28 was used to identify 5,406 individuals of non-European ancestry (Supplementary Note) resulting in a final association analysis including 14,803 lung cancer cases and 12,262 controls.
We confirmed the fidelity genotyping (directly and imputed) of the OncoArray platform by considering concordance of these genotypes relative to genotypes obtained from analogous genotyping platform (Supplementary Note).
Imputation analysis
A detailed description of the imputation procedures used by the OncoArray consortium and in this Lung Oncoarray project, has been described previously.5 Briefly, the reference Dataset was the 1000 Genomes Project (GP) Phase 3 (Haplotype release date October 2014). The forward alignment of SNPs genotyped on the Oncoarray was confirmed by blasting the sequences used for defining SNPs against the 1000 Genomes. Any ambiguous SNPs were subjected to a frequency comparison to 1000 Genomes variants. Allele frequencies were calculated from a large collection of control samples from Europeans (from 108,000 samples) and Asians (11,000 samples). A difference statistic is calculated by the formula: (|p1-p2|- 0.01)2/((p1+p2)(2-p1-p2)) where p1 and p2 are the frequencies our dataset and in the 1000 genomes respectively5. A cutoff of 0.008 in Europeans and 0.012 in Asians is needed to pass. SNPs where the frequency would match if the alleles were flipped were excluded from imputation, but not from the association analyses.5 AT/GC SNPs were not present in previously genotyped lower density arrays. Because all imputation was performed to the same standard all SNPs had the same orientation at the time of imputation. The OncoArray whole genome data were imputed in a two-stage procedure using SHAPEIT to derive phased genotypes, and IMPUTEv229 to perform imputation of the phased data. We included for imputation only the more common variant if more than one variant yielded a match at the same position. The detailed parameter settings are in the Supplementary Note.
Meta-analysis of lung cancer GWAS
FlashPCA30 was run for principal component analysis (PCA) to infer genetic ancestry by genotype. The regression model assumed an additive genetic model and included the first three eigenvalues from FlashPCA as covariates. For imputed data of smaller sample size, which was enrolled in our analysis later, we changed the method score to EM algorithm to accommodate smaller sample size.
We combined imputed genotypes from 14,803 cases and 12,262 controls from the OncoArray series with 14,436 cases and 44,188 controls samples undertaken by the previous lung cancer GWAS3,4,6, including studies of IARC, MDACC, SLRI, ICR, Harvard, NCI, Germany and deCODE as described previously3,4,6, and we ensured that there were no overlap between the ATBC, EAGLE and CARET studies included in both the previous GWAS and current OncoArray dataset by comparing the identity tags (IDs) of all study participants.
In addition to lung cancer, analyses by histological strata (adenocarcinoma, squamous cell carcinoma, small cell carcinoma (SCLC) and smoking status (Ever/Never) was assessed where data were available. Results from analyses defined by Ever and Never smoking strata did not identify any novel variants.
We conducted the fixed effects meta-analysis with the inverse variance weighting and random effects meta-analysis from the DerSimonian-Laird method31. All meta-analysis and calculations were performed using SAS version 9.4 (SAS Institute Inc., Cary, NC, USA). As the same referent panel was used for all studies, all SNPs showed the same forward alignment profiles. We excluded poorly imputed SNPs defined by imputation quality R2 < 0.3 or Info < 0.4 for each meta-analysis component and SNPs with a Minor allele frequency (MAF) >0.01 (except for CHEK2 rs17879961 and BRCA2 rs11571833 which we have validated extensively previously4. We generated the index of heterogeneity(I2) and P-value of Cochran’s Q statistic to assess heterogeneity in meta-analyses and considered only variants with little evidence for heterogeneity in effect between the studies (P-value of Cochran’s Q statistic >0.05). SNPs were retained for study provided the average imputation R-square was at least 0.4. For SNPs in the 0.4–0.8 range that reached genome wide significance results were evaluated for consistency with neighboring SNPs to assure a reliable inference. Due to the smaller sample size and fewer sites contributing in the strata of Never Smokers and SCLC, we additionally required variants to be present in each of the meta-analysis components to be retained for these 2 stratified analyses.
Conditional analysis was undertaken using SNPTEST where individual level data was available and GCTA32 packages for the previous lung cancer GWAS, with the LD estimates obtained from individuals of European origin for the later. Results were combined using fixed effects inverse variance weighted meta-analysis as described above33.
Assessing Statistical Significance
Genome wide statistical significance was considered at P-values of 5 × 10−8 or lower, but we also presented significance per alternative criteria following Bonferroni correction for the number of effective tests or Bayesian False Discovery Probability (BFDP) described below.
To evaluate the effective number of tests we used the Li and Ji (2005)33 method which performs an initial step of filtering out SNPs with MAF<0.01 (imputation is less reliable for these and power is also limited for most odds ratios). Among the 4,751,148 markers with that MAF there were 1,182,363 effective tests.
The BFDP combines significance level, study power, and cost of false discovery and non-discovery into consideration. The detailed procedures of this method are described in Wakefield, 200734. Essentially, the approximate Bayes Factor (ABF) which BFDP uses reflects how much the prior odds change in the light of the observed data (i.e. relative probability of the observed estimates under the null versus alternative hypothesis). Given the nature of GWA studies, we applied a flat prior for all variants at prior probability of 10−6 and 10−8 to demonstrate the range of BFDP.
Annotation of susceptibility loci
We combined multiple sources of in silico functional annotation from public databases to help identify potential functional SNPs and target genes, based on previous observations that cancer susceptibility alleles are enriched in cis-regulatory elements and alter transcriptional activity. The details are described in the Supplementary Note.
eQTL analysis of lung cancer sentinel variants
To investigate the association between the sentinel variants and mRNA expression, we used three different eQTL datasets: (i) Microarray eQTL study: The lung tissues for eQTL analyses were from patients who underwent lung surgery at three academic sites, Laval University, University of British Columbia (UBC), and University of Groningen. Whole-genome gene expression profiling in the lung was performed on a custom Affymetrix array (GPL10379). Microarray pre-processing and quality controls were described previously. Genotyping was carried on the Illumina Human 1M-Duo BeadChip array. Genotypes and gene expression levels were available for 409, 287 and 342 patients at Laval, UBC, and Groningen, respectively. (ii) NCI RNAseq eQTL study: RNA was extracted from lung tissue samples within the Environment and Genetics in Lung cancer Etiology (EAGLE) study. RNAseq was carried out on 90 lung tissue sampled from an area distant from the tumor (defined here as “non-malignant lung tissue”) to minimize the potential for local cancer field effects. Transcriptome sequencing of 90 non-tumor samples was performed on the Illumina HiSeq2000/2500 platform with 100-bp paired-end reads. Genotyping was undertaken using Illumina bead arrays as described previously. (iii) GTEx: eQTL summary statistics based on RNAseq analysis were obtained for eQTL summary statistics from the GTEx data portal http://www.gtexportal.org/home/35. This data included 278 individuals with data from lung tissue. Details of these three eQTL studies are included in the Supplementary Note.
The Microarray eQTL study was used as a discovery cohort. Probe sets located within 1 Mb up and downstream of lung cancer SNPs were considered for cis-eQTL analyses. We have also explored a 5 Mb interval for lung cancer-associated SNPs not acting as lung eQTL within the 1 Mb window. The top eQTL association for that sentinel variant (or if contained multiple eQTL’s with P-value<0.0005 each was considered), this particular eQTL was then chosen and assessed specifically in the independent NCI and GTEx RNAseq eQTL datasets. Statistical significance was defined the eQTL surpassed a locus specific Bonferroni correlation in the discovery cohort (P-value=0.05/number of probes at that locus) and subsequently there was evidence for replication of the eQTL effect with that variant and gene within the validation cohorts (NCI/GTEx RNAseq).
Lung cancer susceptibility variants in other phenotypes
We assessed associations between sentinel genetic variant associated with lung cancer and other phenotypes, including smoking behavior Fagerstrӧm Test for Nicotine Dependence, lung function and telomere length. Additional details of these analyses for other phenotypes are described in Supplementary Note. Briefly:
Smoking behaviors
The effects of lung cancer sentinel variants and smoking behavior were assessed based on the meta-analysis across 3 studies: ever-smoking controls with intensity information from the Oncoarray studies (N=8,120), deCODE (N=40,882) and UK Biobank (N=42,044). The association with nicotine dependence was evaluated based on Fagerstrӧm Test for Nicotine Dependence (FTND) data collected in 4 studies (n=17,074): deCODE Genetics, Environment and Genetics in Lung Cancer Etiology (EAGLE), Collaborative Genetic Study of Nicotine Dependence (COGEND), and Study of Addiction: Genetics and Environment (SAGE) and among current smokers in one other study [Chronic Obstructive Pulmonary Disease Gene (COPDGene). The study-specific SNP association results were combined using fixed effects, inverse variance-weighted meta-analysis with genomic control applied. Specifically, for the 8p21 variant rs11780471, we additionally considered other aspects of smoking behavior data from UKBiobank, deCODE and OncoArray controls. We additionally included summary statistics for the rs11780471 variants from the TAG consortium (described in detail in the Supplementary Note).
Lung function
The lung function in silico look up was conducted in SpiroMeta consortium, which included 38,199 European ancestry individuals. The genome-wide associations between genetic variants and forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC) and FEV1/FVC with 1000 Genomes Project (phase 1)-imputed genotypes in the GWAS with 38,199 individuals36.
Telomere Length (TL)
Sentinel genetic variants associated with telomere length were those described by Codd et al22. Telomere lengths in 6,766 individuals from the UK Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH) study controls using a real-time PCR methodology and genotyping as described in Pooley et al., 201337.
Genetic heritability and correlations
Genome-wide SNP heritability and correlation estimates were obtained using association summary statistics and linkage disequilibrium (LD) information through LD Score (LDSC) regression analyses38,39. These analyses were restricted to HapMap3 SNPs with minor allele frequency above 5% in European populations of 1000 Genomes. Association summary statistics used for these analyses were based on lung cancer histological/smoking types (lung cancer overall, adenocarcinoma, squamous cell, small cell, ever smokers and never smokers) and smoking behavior parameters (cigarettes per day (CPD), smoking status (ever vs never smokers), and smoking cessation (current vs former smokers) from TRICL-ILCCO OncoArray consortium and Tobacco And Genetics consortium (https://www.med.unc.edu/pgc/downloads)40.
Estimating the percentage of familial relative risks explained
The familial relative risk to a first degree relative accounted for by an individual variant (denoted as λi) is estimated based on relative risk per allele and allele frequency for that variant, using the method described in Hemminki et al41, and Bahcall42, under the assumption of log-additive effect. Assuming the effects of all susceptibility variants combined multiplicatively and not in linkage disequilibrium, the combined effect (λT) can then be expressed as the product of all λi. The proportion of the familial relative risk attributable to the totality of the susceptibility variants can then be computed as log(λT)/log(λP). For lung cancer, the λP is approximately 2.0 based on the family cancer databases26,27. The percentage reported is based on the 18 sentinel variants reported in Table 2. The multiple independent alleles in the same locus are not accounted for in this estimation.
Supplementary Material
Acknowledgments
Transdisciplinary Research for Cancer in Lung (TRICL) of the International Lung Cancer Consortium (ILCCO) was supported by (U19-CA148127 and CA148127S1). The ILCCO data harmonization is supported by Cancer Care Ontario Research Chair of Population Studies to R. H. and Lunenfeld-Tanenbaum Research Institute, Sinai Health System.
The TRICL-ILCCO OncoArray was supported by in-kind genotyping by the Centre for Inherited Disease Research (26820120008i-0-26800068-1).
IARC acknowledges and thanks V. Gaborieau, M. Foll, L. Fernandez-Cuesta, P. Chopard, T. Delhomme and A. Chabrier for their technical assistance in this project.
The authors would like to thank the staff at the Respiratory Health Network Tissue Bank of the FRQS for their valuable assistance with the lung eQTL dataset at Laval University. The lung eQTL study at Laval University was supported by the Fondation de l’Institut universitaire de cardiologie et de pneumologie de Québec, the Respiratory Health Network of the FRQS, the Canadian Institutes of Health Research (MOP – 123369). Y.B. holds a Canada Research Chair in Genomics of Heart and Lung Diseases.
Footnotes
Author contributions.
Drafted the Paper: JDM, RJH, CIA
Project Coordination: CIA, RJH, JDM, RJH, DCC, NEC, SC, PBr, MTL
Performed the Statistical Analysis: CIA, JDM, RJH, YH, XZ, RCT, XuJ, XX, YL, JBy, KAP, DCQ, MNT, YBr, DZh
eQTL analysis of candidate variants: JDM, YBo, RCT, MTL, BZ, LSu, MTL, ML
Genomic annotation of candidate variants: DCQ, GCT, JBe, RFT
Assessed impact of candidate variants on nicotine addiction JDM, TR, TET, GWR, KS, DBH, LJB, RJH, SPIRO, LK
Assessed impact of candidate variants on Telomere length: JDM, RJH, KAP, AD, LK
Assessed impact of candidate variants on lung function: MDTo, MSA, LVW, LK
Sample collection and development of the epidemiological studies RJH, TR, TET, GWR, DCC, NEC, MJ, GL, SEB, XW, LLM, DA, HBi, MCA, WSB, ATa, GR, MDT, JKF, LAK, PL, AH, SLa, MBS, ASA, HS, YCH, JMY, PAB, ACP, YY, ND, LSu, RZ, YB, NL, JSJ, AMe, WS, CAH, LRW, AF-S, GF-T, HFMH, JHK, JD, ZH, MPAD, MWM, HBr, JoM, OM, DCM, KO, ATr, RT, JAD, MPB, CC, GEG, AC, FT, PW, IB, H-EW, JuM, TRM, ARi, ARo, KG, MJ, FAS, M-ST, SMA, EBH, CB, IH, VJ, MK, JL, AMu, SO, TMO, GS, BS, DZa, PBa, VS, SHZ, EJD, LMB, W-PK, Y-TG, RSH, JMcL, VLS, PJ, ML, DCN, MO, WT, LSo, MSA, MDTe, MRS, NCG, SML, FG, EOJ, AK, CP, RJH, JDM, MLT
Genetic sharing analysis: RCT, SLi, XiJ, JDM, RJH
Data Availability
The datasets generated during the current study are available at the URL: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001273.v1.p1. Prior meta-analyses of genome-wide association studies contributing to this study are available at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000876.v1.p1.
The Oncoarray data deposited at dbGAP includes data excluded from the analyses presented in this paper to avoid overlap with prior studies. Readers interested in obtaining a copy of the original data can do so by completing a proposal request form that is located at http://oncoarray.dartmouth.edu)
Cluster plots of all SNPs on the Oncoarray are located at http://oncoarray.dartmouth.edu
URLs
OncoArray: http://epi.grants.cancer.gov/oncoarray/)
http://oncoarray.dartmouth.edu)
Fastpop http://sourceforge.net/projects/fastpop/)
PLINK: http://zzz.bwh.harvard.edu/plink/)
IMPUTE2: http://mathgen.stats.ox.ac.uk/impute/impute_v2.html)
SHAPEIN: https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html)
GTEx: http://www.gtexportal.org/home/)
BRAINEAC: http://braineac.org)
References
- 1.Ferlay J, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. International Agency for Research on Cancer; Lyon, France: 2013. [Google Scholar]
- 2.Mucci LA, et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. Jama. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Timofeeva MN, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–95. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46:736–41. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amos CI, et al. The OncoArray Consortium A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol Biomarkers Prev. 2017;26:126–135. doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Deciphering associations for lung cancer risk through imputation and analysis of 12,316 cases and 16,831 controls. Eur J Hum Genet. 2015;23:1723–8. doi: 10.1038/ejhg.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durham AL, Adcock IM. The relationship between COPD and lung cancer. Lung Cancer. 2015;90:121–7. doi: 10.1016/j.lungcan.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan H, et al. A Novel Genetic Variant in Long Non-coding RNA Gene NEXN-AS1 is Associated with Risk of Lung Cancer. Sci Rep. 2016;6:34234. doi: 10.1038/srep34234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGovern DP, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum Mol Genet. 2010;19:3468–76. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang SK, et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014;63:80–7. doi: 10.1136/gutjnl-2013-305193. [DOI] [PubMed] [Google Scholar]
- 14.Brenner DR, et al. Hierarchical modeling identifies novel lung cancer susceptibility variants in inflammation pathways among 10,140 cases and 11,012 controls. Hum Genet. 2013;132:579–89. doi: 10.1007/s00439-013-1270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moulton EA, Elman I, Becerra LR, Goldstein RZ, Borsook D. The cerebellum and addiction: insights gained from neuroimaging research. Addict Biol. 2014;19:317–31. doi: 10.1111/adb.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu CT, et al. The novel protein suppressed in lung cancer down-regulated in lung cancer tissues retards cell proliferation and inhibits the oncokinase Aurora-A. J Thorac Oncol. 2011;6:988–97. doi: 10.1097/JTO.0b013e318212692e. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Cuesta L, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov. 2014;4:415–22. doi: 10.1158/2159-8290.CD-13-0633. [DOI] [PubMed] [Google Scholar]
- 18.Lan Q, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet. 2012;44:1330–5. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truong T, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst. 2010;102:959–71. doi: 10.1093/jnci/djq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh KM, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–5. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Codd V, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45:422–7. 427e1–2. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C, et al. Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum Mol Genet. 2015;24:5356–66. doi: 10.1093/hmg/ddv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh KM, et al. Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget. 2015;6:42468–77. doi: 10.18632/oncotarget.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehringer G, et al. Cross-cancer genome-wide analysis of lung, ovary, breast, prostate and colorectal cancer reveals novel pleiotropic associations. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-15-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amundadottir LT, et al. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindelof B, Eklund G. Analysis of hereditary component of cancer by use of a familial index by site. Lancet. 2001;358:1696–1698. doi: 10.1016/S0140-6736(01)06721-6. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, et al. FastPop: a rapid principal component derived method to infer intercontinental ancestry using genetic data. BMC Bioinformatics. 2016;17:122. doi: 10.1186/s12859-016-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 30.Timofeeva MN, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–95. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software. 2010;36:1–48. [Google Scholar]
- 32.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–7. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 34.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–27. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soler Artigas M, et al. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat Comm. 2015;6:8658. doi: 10.1038/ncomms9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pooley KA, et al. Telomere length in prospective and retrospective cancer case-control studies. Cancer Res. 2010;70:3170–6. doi: 10.1158/0008-5472.CAN-09-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemminki K, Bermejo JL. Relationships between familial risks of cancer and the effects of heritable genes and their SNP variants. Mutat Res. 2005;592:6–17. doi: 10.1016/j.mrfmmm.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Bahcall OG. iCOGS collection provides a collaborative model. Foreword. Nature genetics. 2013;45:343. doi: 10.1038/ng.2592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


