Abstract
Although most modern dog breeds are less than 200 years old, the symbiosis between man and dog is ancient. Since prehistoric times, repeated selection events have transformed the wolf into man’s guardians, laborers, athletes, and companions. The rapid transformation from pack predator to loyal companion is a feat that is arguably unique among domesticated animals. How this transformation came to pass remained a biological mystery until recently: Within the past decade, the deployment of genomic approaches to study population structure, detect signatures of selection, and identify genetic variants that underlie canine phenotypes is ushering into focus novel biological mechanisms that make dogs remarkable. Ironically, the very practices responsible for breed formation also spurned morbidity; today, many diseases are correlated with breed identity. In this review, we discuss man’s best friend in the context of a genetic model to understand paradigms of heritable phenotypes, both desirable and disadvantageous.
Keywords: morphology, skull, body size, breed standard, disease, genomics
INTRODUCTION
Today, nearly 500 breeds of dogs are recognized worldwide (Fogle 2000, Wilcox & Walkowicz 1995), each with its own unique constellation of physical and behavioral traits (Parker et al. 2010; Wayne&Ostrander 1999, 2007). In the United States alone, it is estimated that 37% of households own a dog and that 47% of these animals are reported to be breed dogs (http://www.americanhumane.org/assets/pdfs/pets-fact-sheet.pdf). At its most recent low point, in 2010, the American Kennel Club (AKC) still reported more than half a million new registrants (http://www.akc.org/). Thus, breed dogs are not only diverse but also plentiful (see review Parker et al. 2010).
In the dog world, beauty lies in the eye of the breeder. A trait that is desirable in one breed could just as easily be considered grounds for disqualification in another. And whereas we call the physical differences between canines traits or characteristics, equivalent conditions in humans would often be classified as pathologies. Moreover, canine traits, although judged and measured in adults, are developmental outcomes. In this review, we attempt to highlight findings from the dog genetics community and relate them to developmental paradigms, particularly those that regulate growth, morphology, and their diseases.
THE PEDIGREE DOG: AN ONGOING EXPERIMENT
The Making of an Unconventional Genetic Model
For good reason, pedigree dogs will never supplant the laboratory mouse as the poster child for basic research. However, it can be argued that dogs’ special status is what makes them so intriguing to geneticists in the first place. Unlike cows, swine, and other livestock, dogs, as a species, are uncoupled from the global economic pressures to maintain uniformity in size, shape, and productivity. Rather, because dogs complemented our lifestyles so well, they radiated to fill a variety of purposes in modern societies, including hunting, tracking, herding, guiding, guarding, drafting, and providing companionship. As a result, they vary enormously in size, shape, disease susceptibility, and behavior (Figure 1) (Parker et al. 2010, Shearin & Ostrander 2010b).
Figure 1.
More than 180 breeds exist in the United States, 215 are recognized by the Kennel Club in the United Kingdom, and approximately 490 are noted worldwide. The closest living ancestor to the dog is the gray wolf. Shown here are (a) a gray wolf and (b–i ) some interesting breed examples, including the (b) Pembroke Welsh Corgi, (c) Boxer, (d ) French Bulldog, (e) Papillon, ( f ) Miniature Dachshund, ( g) Borzoi, (h) Greyhound, and (i ) Mastiff. Photo credit: Mary Bloom courtesy of American Kennel Club (panels a, b, d–i ) and courtesy of Wikipedia (panel c).
Consequences of Artificial Selection and Standardization
Despite their obvious limitations and the understandable public disdain for using dogs in experimental research, there is great potential to extract data from the pet population to propel our understanding of morphology and disease susceptibility. Studies of the dog’s genetic code have advanced humankind’s fundamental understandings of genomic architecture and the genetic basis of heritable phenotypes that are pertinent to dogs and humans alike. Nearly unanimously, the advances that have been and continue to be made began not in the laboratory but rather at blood draws hosted at local dog shows, from saliva swaps collected in the homes of pet owners, and from samples collected at veterinary clinics that treat family pets.
Viewed from the lens of a geneticist, each dog breed is effectively a subpopulation or strain of the same species. This characterization depends on morphological, mitochondrial DNA, and nuclear DNA analyses, all of which generally support the idea that modern dogs are all descendants of the gray wolf, Canis familiaris (Larson et al. 2012, Savolainen et al. 2002, Vilà et al. 1997, vonHoldt et al. 2010).
Recent studies based on mitochondrial sequencing data from ancient dog fossils suggest that dogs were domesticated between 11,000 and 32,000 years ago (Freeman et al. 2014, Thalmann et al. 2013). Further, there were probably several domestication events taking place in multiple places that led to domesticated lineages and selection of various gene families (Freedman et al. 2014, Thalmann et al. 2013, vonHoldt et al. 2010). This may account, in part, for the extraordinary variation observed in modern dog breeds. Studies show that multiple bottlenecks underlie modern dogs, including a major one during domestication and another that occurred during breed formation (Larson et al. 2012, Lindblad-Toh et al. 2005, (Stern et al. 2013, Thalmann et al. 2013, Wang et al. 2013).
When fanciers formed clubs like the United Kingdom’s Kennel Club and the AKC, stud books closed. Thereafter, membership to these prestigious clubs required that registrants be sired from registered parents of the same breed. Today, numerous breed clubs exist worldwide, each maintaining the breed and pedigree information of their canine constituents. These clubs also maintain strict criteria (breed standards) that specify the ideal physical and behavioral conformation of their members. At club-sponsored conformation events, animals are judged for their adherence to these criteria. Not surprisingly, winners of such events are highly sought after as sires and dams, ensuring the propagation of ideal traits for each breed’s future generations. Thus, dog breeds are genetically isolated from one another, and selective breeding continues to maintain standardization within each breed such that morphological traits remain uniform for individuals of the same breed.
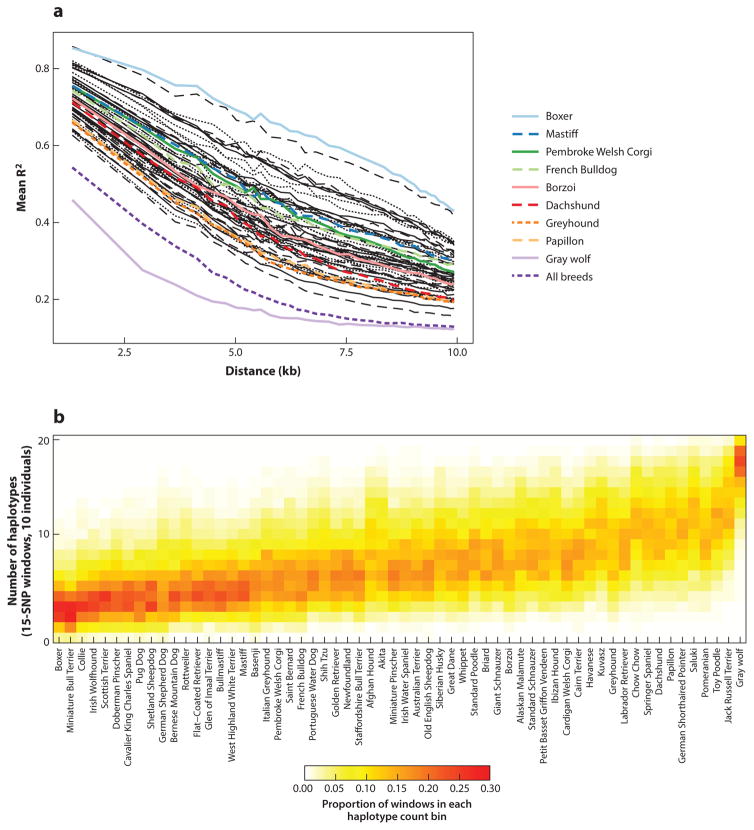
Other phenomena added to pedigree dogs’ genetic isolation. For example, both world wars greatly reduced the effective population sizes of some European and Asian breeds, particularly those of large and giant sizes (Grandjean 2000). Breeders themselves accelerated the genetic isolation of pedigree dogs through such practices as line breeding and widespread insemination using popular sires. As a result, intrabreed genetic diversity is markedly reduced, as evidenced by extensive linkage disequilibrium (LD), reduced haplotype diversity, and broad runs of homozygosity (ROH) (Figure 2) (Boyko et al. 2010, Lindblad-Toh et al. 2005, Stern et al. 2013, Sutter et al. 2004).
Figure 2.
The dog’s morphological diversity belies its genetic diversity. (a) Linkage disequilibrium (LD) by breed. Each line represents the average R2 of markers within a single breed or between groups of ten breeds (dashed lines). For simplification, only breeds highlighted in Figure 1 are assigned colored lines. Considered individually, the LD of each breed of dog is more extensive than that of wolves. Considered collectively, the LD of breed dogs approaches that of gray wolves. (b) Intrabreed haplotype diversity is greatly reduced relative to the gray wolf. Panels a and b were adapted with permission from Boyko et al. (2010).
Canine Breed Variation
The reduction in intrabreed genetic heterogeneity, breed standardization, readily available pedigree information, and in many settings routine access to veterinary diagnostics provide a strong foundation from which genetic mapping studies can be designed to identify the genetic basis of heritable traits and diseases. Again, viewed through the lens of a geneticist, pedigree dogs are unique because breeders take advantage of spontaneous mutations rather than depending on traditional methods of mutagenesis (e.g., X rays, chemicals, retroviral insertion). Indeed, it is the naturally occurring genetic variation that breeders, over the course of centuries, selected for and against in breed dogs. This distinction is important because the types of genetic variation that cause canine phenotypes are not biased by the type or genomic location of the derived genetic change, as is commonly the case with mutagenesis. In fact, the emerging data from genetic mapping studies indicate that breed-defining traits among dogs are driven by an ever-growing array of polymorphisms, including single-nucleotide polymorphisms (SNPs), copy number variation (CNV), insertions/deletions (indels), and retrogene and short interspersed nucleotide element (SINE) insertions (Clark et al. 2006; Karlsson et al. 2007; Karyadi et al. 2013; Parker et al. 2009, 2010; Salmon-Hillbertz et al. 2007).
Another major difference between traditional mutagenesis screens using laboratory animals and the screens conducted by dog breeders is that in the latter, breed standards were arrived at incrementally through selective breeding of highly admixed animals. Two profound consequences result from this design. First, one or more loci can contribute to any particular trait. Second, by virtue of the fact that traits had to be recognized by breeders for selection to occur in the first place, the effect size of the polymorphisms underlying a trait must be substantial.
GENETIC APPROACHES TO MAPPING CANINE TRAITS AND DISEASES
Canine genetic and genomic studies stand apart from most other animal studies in that their designs rely heavily on public participation. By necessity, canine geneticists rely on dog owners not only to donate biological samples from their pets but also to provide pedigrees, phenotypic information, and health status. The first canine studies aimed at mapping traits and diseases used extended pedigree information to conduct linkage analysis (Acland et al. 1998, Jónasdóttir et al. 2000, Yuzbasiyan-Gurkan et al. 1997). In the early days of canine genetics, microsatellites were the markers of choice used in mapping (Mellersh et al. 1994; Ostrander et al. 1992, 1995). Later, markers discovered from sequencing the Boxer and Standard Poodle genomes, along with select regional sequencing of other breed dogs and wild canids, enabled commercially available SNP chips, facilitating the transition from familial pedigree-based studies to population-based, interbreed studies (Boyko et al. 2010, Kirkness et al. 2003, Lindblad-Toh et al. 2005, Vaysse et al. 2011).
With the availability of SNP chips, genome-wide association studies (GWAS) became the tool of choice for finding loci of interest. By most accounts, the use of GWAS to locate heritable traits has been wildly successful in dogs, identifying loci for eye diseases, skin disorders, cancer, deafness, neurological disorders, heart disease, and a host of other disorders (Ahonen et al. 2013, Grall et al. 2012, Karyadi et al. 2013, Olsson et al. 2011, Tengvall et al. 2013, Yokoyama et al. 2012), as well as morphological traits (Boyko et al. 2010, Vaysse et al. 2011), including body size, skull shape, leg length, and fur type and color (Cadieu et al. 2009, Clark et al. 2006, Dreger et al. 2013, Drögemüller et al. 2008, Karlsson et al. 2007, Parker et al. 2009, Rimbault et al. 2013, Schoenebeck et al. 2012). Loci of large effect, a fraction of which are described below, were rapidly identified in population-based approaches that compared SNP allele frequencies with a myriad of phenotypic traits. Regional allele frequencies, ROH, and LD are now routinely leveraged to identify selective sweeps that occur as a result of artificial selection. Particular examples include both disease loci, such as finding a locus for a skin phenotype and periodic fevers in the Chinese Shar-Pei (Olsson et al. 2011), and morphological traits, such as brachycephalic skulls (Bannasch et al. 2010), short legs (Parker et al. 2009), or variation in body size (Sutter et al. 2007).
In dogs, identifying a quantitative trait locus (QTL) or association is relatively easy, whereas sifting through a critical interval to identify a causative polymorphism is often difficult, especially when such mutations reside outside the open reading frame of a candidate gene. Extensive LD within breed dogs facilitates coarse mapping of traits to a genomic region. However, it works against fine-mapping efforts, as many marker alleles cosegregate with the causal variant. Fortunately, as the costs of conducting whole-genome sequencing (WGS) decrease, platform performance improves, and the tools needed to reliably call variants are refined, fine mapping to identify the causal genetic underpinnings of traits and diseases is becoming less laborious (Frischknecht et al. 2013, Karlsson et al. 2013, Owczarek-Lipska et al. 2013a, Schoenebeck et al. 2012).
GENETICS OF CANINE DISEASES INFORM HUMAN CONDITIONS
When considering dog diseases that have been informative for parallel human conditions (Ostrander 2012), several examples come to mind and have been reviewed previously (Karlsson & Lindblad-Toh 2008, Ostrander 2012, Shearin & Ostrander 2010a), most notably cancer (Cadieu & Ostrander 2007, Dobson 2013, Parker & Ostrander 2014, Ranieri et al. 2013), neurological diseases (Ostrander & Beale 2012), and cardiac diseases (Parker et al. 2006). In the interest of space, it is not possible to consider all of these, but we consider three instances where particular progress has been made in the past several months.
Osteosarcoma
When scientists first began to consider the dog as a system to better understand disorders that are problematic in humans, cancer, in particular osteosarcoma, was one of the first diseases proposed. Like many cancers, there is a strong breed disposition to osteosarcoma (Dobson 2013, Dobson et al. 2002). Long-limbed breeds, such as Scottish Deerhounds and Irish Wolfhounds, are at particular risk, as are Rottweilers and Greyhounds.
In humans, the mortality rate for osteosarcoma approaches 40% (Ta et al. 2009), but GWAS on humans have not been particularly fruitful (Savage et al. 2013). The argument has been made that dogs are good for finding single gene traits but are not as advantageous for disentangling complex traits. It was therefore exciting to see a group of collaborating investigators tackle the question of osteosarcoma loci in Greyhounds, Rottweilers, and Irish Wolfhounds (Karlsson et al. 2013). The investigators genotyped more than 500 dogs, of which approximately 300 were affected, with a chip that includes 170,000 SNPs. They identified more than 30 loci believed to contribute to risk. Surprisingly, risk loci largely did not overlap between breeds, despite the recent relatedness of breeds (Parker et al. 2004, vonHoldt et al. 2010). However, a locus on canine chromosome 11 (CFA11) was fixed in Rottweilers and Irish Wolfhounds.
The CFA11 locus is interesting, as it spans a 15-kb noncoding region near the cyclin-dependent kinase inhibitor 2 (CDKN2) genes CKN2A (encodes p16INK4a and p14ARF) and CKN2B (encodes p15INK4b), as well as some noncoding sequences. Both CDKN2A and CDKN2B are well-known tumor suppressors and cell-cycle regulators. It is also notable that the same region was defined previously as being important in susceptibility to canine histiocytic carcinoma in Bernese Mountain Dogs (Shearin et al. 2012). The authors of the osteosarcoma paper, however, defined a risk allele that they show is also correlated with osteosarcoma in Leonbergers and Great Pyrenees (Karlsson et al. 2013).
In evaluating the other loci, the authors used GRAIL (Raychaudhuri et al. 2009), a statistical method that mines published abstracts for relationships corresponding to specified genomic locations (e.g., regions of association), to identify numerous gene connections relating to the words bone, differentiation, and development. Among the candidate genes that were statistically significant were bmp binding endothelial regulator and von Willebrand factor C domain containing 2, both of which have been shown to be “negative regulators of osteoblast differentiation” (Karlsson et al. 2013, p. 7). Both now become prime candidates as osteosarcoma-causing genes (Karlsson et al. 2013, Koike et al. 2007), as do engrailed homeobox 1, a gene whose product regulates osteoblast differentiation (Deckelbaum et al. 2006), and delta-like 3, which encodes a Notch ligand previously shown to be important in human skeletal growth (Bulman et al. 2000).
At this point, it is hard to know which of the 33 loci will bear fruit and which will be false positives, but the provocative candidate genes identified thus far suggest that the dog is indeed a viable model for osteosarcoma, and at a minimum, it is sure to yield data that are important in managing both human and animal osteosarcoma.
Heart Disease
Heart disease is observed in many dog breeds (Parker et al. 2006), from dilated cardiomyopathy in the Doberman Pinscher (Steudemann et al. 2013) to subaortic stenosis in Newfoundlands (Reist-Marti et al. 2012), Golden Retrievers (Stern et al. 2012), and Dogue de Bordeaux dogs (Ohad et al. 2013). Congestive heart failure is observed in many breeds (Isono et al. 2012), as are cardiomyopathy (Mausberg et al. 2011, Meurs et al. 2012, Owczarek-Lipska et al. 2013b), ventricular arrhythmia ( Jesty et al. 2013), and mitral valve disease (Fox 2012, French et al. 2012). For some disorders, such as dilated cardiomyopathy in the Irish Wolfhound, many loci are involved (Philipp et al. 2012). Other disorders seem to be single-gene traits (for review, see Parker et al. 2006).
In terms of genetics, one of the most interesting stories to date is that of familial cardiomyopathy in the Doberman Pinscher (Meurs et al. 2012). The disease is characterized by congestive heart failure and sudden cardiac death. A GWAS in 2012 identified an area of significance on canine chormosome 14. This was followed by DNA sequence analysis, which identified a deletion in pyruvate dehydrogenase kinase 4 (PDK4) as the causative mutations.
The GWAS in this case used just 66 cases and 66 controls. While 54 of 66 affected dogs carried the mutation, only 26 unaffected dogs had it, providing strong statistical evidence that the mutation had been found (P < 0.0001). In addition, none of 100 dogs of 11 other breeds carried a PDK4 mutation, providing good supportive evidence. The mutation was a 16-bp deletion at the donor splice site of intron 10 in PDK4, whose encoded protein regulates energy metabolism by phosphorylating pyruvate dehydrogenase. mRNA levels were associated with 14-fold reduction in Doberman Pinscher carriers compared with what was observed in normal dogs of other breeds. However, a subsequent European study of Doberman Pinschers with dilated cardiomyopathy did not replicate the findings, reporting essentially the same mutation rate (16%) among cases and controls (Owczarek-Lipska et al. 2013b). It remains to be seen how this story plays out. At a minimum it suggests that additional genes remain to be found (Owczarek-Lipska et al. 2013b). Indeed, a GWAS by this group reports a major locus for the disease in Dobermans on chromosome 5 (Mausberg et al. 2011).
Another interesting story has been that of arrhythmogenic right ventricular cardiomyopathy, which is associated with a mutation in the striatin gene in Boxer dogs (Meurs et al. 2010). The disease appeared to be autosomal dominant with incomplete and age-dependent penetrance (Meurs et al. 2010). A GWAS identified a region CFA17 in the Boxer as being highly associated. Evaluation of the underlying gene striatin, a calmodulin binding protein (STRN), revealed an 8-bp deletion in the 3′ untranslated region (Meurs et al. 2010). Both homozygotes and heterozygotes were identified, but the dogs that were homozygous for the deletion had a more severe form of the disease. A subsequent study identified the second clinical form of myocardial disease seen in Boxers, dilated cardiomyopathy, as being caused by a deletion in the same gene in at least some families (Meurs et al. 2013).
Interestingly, the encoded protein is believed to serve as a scaffold that functions in a calcium-dependent manner in both signaling and trafficking. These authors made the novel observation that STRN protein colocalizes with the desmosomal proteins plakophilin-2, plakoglobin, and desmoplakin. All are proteins that are involved in the human forms of ventricular cardiomyopathy. STRN now becomes a superb candidate gene for unexplained familial and sporadic forms of the human disease.
Rare Diseases
Sometimes the very traits selected by breeders inadvertently cause disease. One example of this is the case of hereditary periodic fever syndrome, a group of diseases characterized by recurrent episodes of inflammation followed by fever. Olsson et al. (2011) initially used a GWAS to identify a locus for Shar-Pei fever. Upon close examination, they identified two highly associated duplications, both unique to the Shar-Pei, that were upstream of the hyaluronan synthase 2 (HAS2) gene. HAS2 is integral for making hyaluronan, a major component of skin (Laurent & Fraser 1992). Interestingly, previous work suggested that Shar-Pei dogs have an excess of hyaluronan in their skin (Zanna et al. 2009) and that excess hyaluronan is associated with abnormal skin phenotypes in both dogs and man (Ramsden et al. 2000). In the Shar-Pei, the excess hyaluronan linked to skin wrinkling may occur by cis-acting misregulation of HAS or its negative regulator, HAS2 antisense RNA1, in a manner that is dose dependent in terms of copy number (Olsson et al. 2011). Interestingly, an imbalance between high molecular weight and fragmented forms of hyaluronan can trigger the immune system, producing fever and inflammation, even though no infectious particle is present (Puré & Assoian 2009).
The Shar-Pei periodic fever study is important for several reasons. First, it advances our knowledge of a canine disease that is important to a breed community. Second, it introduces both a type of mutation and a gene family as being potentially important in undiagnosed human disorders. More recently, a CNV was described whose dosage not only increases blackness of hair color but also inadvertently increases susceptibility to squamous cell carcinoma of the toes (Karyadi et al. 2013). Studies such as these demonstrate that strong selection to enhance appearances can be pushed too far, resulting in a disease that is common for a breed. So far, the disease examples published for dogs have been rare in humans. It seems almost certain that additional studies of seemingly rare traits that are common in dogs will contribute to our understanding of human and canine disease.
FINE MAPPING USING NEXT-GENERATION SEQUENCING APPLICATIONS.
Two approaches to fine mapping are becoming more commonly employed, both with the intent of producing a near-comprehensive list of genetic variants, which in turn are filtered and subsequently validated through expanded genotyping. First, a select number of study participants with phenotypic extremes can be used for whole-genome sequencing (WGS). Although this approach produces far more data than just the critical interval, the benefit is that with enough depth of sequencing coverage (~30-fold coverage or more), SNPs, indels, and CNVs can be called. Furthermore, WGS data can be used for de novo assembly, with the goal of closing gaps within the current dog genome assembly. The drawback is that despite technological improvements, WGS remains expensive, limiting the number of animals that can be used. In the second approach, often referred to as targeted resequencing, the critical interval is initially enriched. Libraries created from the enrichment are then barcoded for multiplexing and sequenced using a next-generation sequencing platform, thus reducing the costs. The disadvantage of targeted resequencing is that coverage is only as good as the enrichment—DNA corresponding to gaps in the assembly or repetitive elements or with poor thermodynamic profiles will not be enriched. Also, CNV callers and de novo assemblers cannot use targeted resequencing data.
WHOLE-GENOME SEQUENCING IN DOGS
The previous papers describe a common and very successful method for identifying mutations. In each, the mutations described were found after resequencing superb candidate genes in a locus identified through GWAS. However, as new technologies are developed and perfected, there is a rapidly growing trend in canine genomics and other model organisms to use large-scale sequencing platforms to combine these steps. Indeed, canine genetics is moving toward WGS of small numbers of cases and controls, thus bypassing the GWAS step to try and find genes associated with particular diseases (see sidebar, Fine Mapping Using Next-Generation Sequencing Applications).
WGS is considerably more expensive, not just in actual sequencing but in its analysis and data storage. With rigorous variant calling and effective filtering, the millions of SNPs produced by WGS can be reduced significantly, hopefully to a point where the number of candidate mutations to consider is manageable. The more dogs that are sequenced at >30-fold depth and whose data are made public, the better off the entire community will be because the variants derived from these endeavors can serve as a filter to distinguish benign polymorphisms from those that are potentially pertinent to one’s own study population.
BODY SIZE AS A BREED-DEFINING TRAIT
No other morphological trait is more striking than the size differences between breed dogs. At their extremes, dogs can differ nearly fiftyfold in weight. For example, the AKC breed standard specifies that Chihuahuas should weigh between 4 and 6 lb, approximately fifty times smaller than the English Mastiff, a breed that can exceed 200 lb (Am. Kennel Club 1998). Altogether, breed dogs form a continuum of body sizes that suit their historic roles. Companion dogs are small, perfect for holding on the lap. Working breeds, whose strength is required for drafting or guarding property, are among the tallest and heaviest of all breeds. Dogs from herding and hunting breeds were selected to run long distances; these breeds tend to be medium sized.
GENETIC MAPPING OF BODY SIZE LOCI
In dogs, size can be approximated using a variety of measurements, including breed average weight, height at the withers, or skull landmarks. To date, at least nine loci are associated with canine size determination (Boyko et al. 2010, Chase et al. 2002, Hoopes et al. 2012, Rimbault et al. 2013, Sutter et al. 2007, Vaysse et al. 2011). At each QTL, promising candidate genes were identified, including insulin-like growth factor 1 (IGF1) (Sutter et al. 2007), insulin-like growth factor 1 receptor (IGF1R) (Hoopes et al. 2012), high mobility group AT-hook 2 (HMGA2), SMAD family member 2 (SMAD2), growth hormone receptor (GHR), stanniocalcin 2 (STC2), ligand dependent nuclear receptor corepressor-like (LCORL), cyclin-dependent kinase 4 (CDK4), and two regions on canine chromosome X (CFA X), one of which is in close proximity to glypican 3 (GPC3) and glypican 4 (GPC4) ( J.J. Schoenebeck, M. Rimbault & E.A. Ostrander, unpublished data) (see Supplemental Table 1, follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org). Seven of the nine genes cluster based on several biological themes that support growth, notably positive regulation of cellular proliferation and metabolism, phosphorylation, regulation of cell death, and extracellular matrix composition.
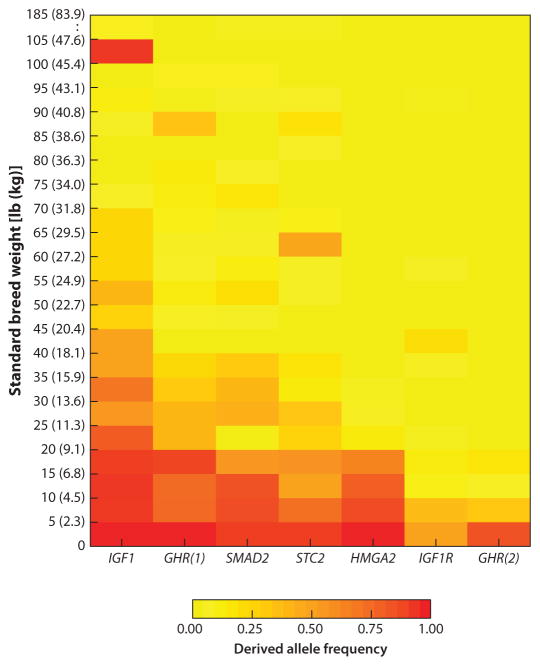
Fine mapping of QTL associated with GHR, HMGA2, IGF1, IGF1R, SMAD2, and STC2 indicates that the causal variants at these loci must be derived because the variant is absent in gray wolves and golden jackals (a species of wild canid evolutionarily basal to wolves). Moreover, extensive LD, ROH, and associated haplotypes are restricted to dog breeds that are smaller in size than wolves. These data indicate that mutations that limited dog size were most likely artificially selected following domestication. Toy breeds like the Chihuahua typically carry derived alleles at all six genes, giant breeds like the Great Dane typically carry none of the derived variants, and breeds smaller than 90 lb (41 kg) can carry any combination of the six derived variants (Figure 3). Finally, the combinatorial effects of the six loci are strongly predictive of overall animal size for dog breeds <90 lb, with conservative estimates indicating that 53% of size variation can be explained by these six loci alone (Rimbault et al. 2013).
Figure 3.
Derived alleles that are putatively causal (or tightly linked to causal variants) are carried at higher frequencies among small and toy breeds. Standard breed weights are listed on the y-axis as pounds and kilograms. One exception to this trend is the Rottweiler. Despite weighing ~100–105 lb, Rottweilers appear to carry the derived IGF1 variant, suggesting either that the causal mutation occurred on an ancestral haplotype carried by this breed or that an epistatic genetic interaction occurs to mute the derived IGF1 variant’s effects on size. Figure adapted from Rimbault et al. (2013).
The genetic mechanisms responsible for canine gigantism remain murky. Both loci on CFA X are associated with breeds of large and giant sizes, although not exclusively so (see Skull and Axial Skeleton section below). Similarly, large- and giant-breed dogs display the signatures of selective sweeps, including extensive LD and ROH (Rimbault et al. 2013). Fine mapping of these loci is thwarted by reduced genetic diversity and the extensive LD that are typical of the X chromosome (Sandor et al. 2006). Moving forward to identifying mechanisms of causality will likely necessitate the use of WGS approaches to comprehensively interrogate these regions.
A salient theme has emerged from these efforts, as well as those to map human stature and livestock carcass traits and the work carried out in animal models: The genes that regulate size are functionally conserved across species (see Supplemental Table 2). Compared with humans, in which hundreds of variants appear to explain a modest amount of human height variation, far fewer variants have been found in dogs; however, these variants are expected to be of large effect size (Chase et al. 2002). It is worth noting that this comparison is deceiving. Human GWAS have typically looked for genetic variation within populations (e.g., Europeans, African Americans). In dogs, GWAS primarily focus on differentiation between populations (breeds). Moreover, canine studies remain vastly underpowered compared with human studies, limiting the detection of rare variants that may not be fixed within breeds. Moving forward, it would be interesting to map size variants within individual breeds like the Portuguese Water Dog, whose breed standard does not strictly define size.
Growth Hormone and Insulin Signaling as Related to Body Size
Across species, growth hormone (GH), IGF1, and insulin endocrine signaling drive postnatal growth (Supplemental Table 2) (Wang et al. 1999). It is widely accepted that GH is released into circulation from the hypothalamus. IGF1 stimulates cell division via binding to its tyrosine kinase receptor, IGF1R, which is ubiquitously expressed. In response to GH, the liver and other tissues secrete IGF1. IGF1 also feeds back on the thymus to limit GH production (Tannenbaum et al. 1983).GH and insulin(-like) signaling is highly conserved across the animal kingdom. Mouse knockouts clearly indicate that GH, IGF1, and insulin are needed to achieve normal size (Mathews et al. 1988, Powell-Braxton et al. 1993).
In dogs, variants that restrain the GH-IGF1 signaling axis were selected repeatedly to control for animal size. Although the most significant signal of association comes from the IGF1 locus (Boyko et al. 2010), the causative mutation driving artificial selection at the IGF1 locus remains unknown. GHR appears to have been targeted twice in succession: Two nonsynonymous SNPs were identified, and one nonsynonymous mutation occurred in complete LD with the other. As one might expect, breed average weights of dogs that carry both GHR mutations are smaller than breed averages of dogs with just one GHR mutation (Rimbault et al. 2013). Finally, among the tiniest of dog breeds, a nonsynonymous mutation in IGF1R was identified (Hoopes et al. 2012). Additional evidence that the insulin-signaling pathways were manipulated in dogs comes by way of selective sweep mapping: Vaysse et al. (2011) report one such sweep associated with size that is in proximity to CDK4, a gene whose product is necessary for β-islet cell–mediated insulin production (Rane et al. 1999).
HMGA2 and Body Size
HMGA2 encodes a small chromatin-associated protein that binds other nuclear proteins to modulate chromatin conformation and transcription, the latter role through its involvement at enhancer sites (Thanos&Maniatis 1995). Multiple genetic studies indicate that HMGA2 is a key modulator of size. HMGA2 modulates human stature (Supplemental Table 2) (Lettre et al. 2008; Ligon et al. 2005; Menten et al. 2007; Sanna et al. 2008; Weedon et al. 2007, 2008), as well as size in various domesticated animals, including mice, cattle, chickens, horses, pigs, and dogs (Supplemental Table 2) (Boyko et al. 2010, Carty et al. 2011, Chung et al. 2013, Gudbjartsson et al. 2008, Jones et al. 2008, Makvandi-Nejad et al. 2012, N’Diaye et al. 2011, Pryce et al. 2011, Rimbault et al. 2013, Song et al. 2011, Wilkinson et al. 2013). In mice, disruption of Hmga2 renders mice 60% smaller than their wild-type littermates (Benson & Chada 1994, Zhou et al. 1995). Conversely, overexpression of a truncated form causes gigantism, obesity, and lipomas (Battista et al. 1999). In dogs, the causal mutation(s) at the HMGA2 locus remains unknown.
SMAD2 and Body Size
Among breed dogs, the most strongly associated variants at the SMAD2 locus are two neighboring deletions that are in perfect LD and located approximately 15 kb downstream of the gene (Rimbault et al. 2013). The deletions, which are found mostly among small and toy breeds, remove a CpG island. Syntenic comparisons and in silico analyses indicate that they also remove clusters of predicted transcription factor binding sites, possibly indicating an enhancer (Rimbault et al. 2013). A potential clue about SMAD2’s role in size determination comes from gene expression profiling: In response to Smad2-mediated TGF-β signaling, Hmga2 expression is significantly up- or downregulated (Murakami et al. 2010, Valcourt et al. 2005). By extrapolation, these data raise the hypothesis that SMAD2 and HMGA2 operate within the same signal transduction pathway to modulate canine size.
STC2 and Body Size
STC2 and its paralog, STC1, are evolutionarily conserved, secreted peptide hormones (Chang & Reddel 1998, Ishibashi et al. 1998, Jellinek et al. 2000) that promote cell survival during endoplasmic reticulum stress (Ito et al. 2004, Law & Wong 2010) and are implicated in disease processes (Bouras et al. 2002, Chang et al. 2003, Kita et al. 2011, Law & Wong 2010). Mouse data indicate that STC2 regulates postnatal growth. Stc2−/− mice are approximately 10–15% heavier than their wild-type littermates, a finding attributed to organomegaly (Chang et al. 2008). The converse appears to be true of the mouse overexpression model. In these transgenics, STC2 retarded skeletal development and organ size (Gagliardi et al. 2005). Interestingly, both studies concluded that STC2’s effects were independent of GH/IGF1 signaling.
Despite extensive fine mapping, the causal genetic variation underlying the canine size QTL spanning STC2 remains unknown (Rimbault et al. 2013). No compelling genetic variants were identified in the open reading frame of the gene, suggesting that the causal genetic variant affects STC2 transcription in cis or, alternatively, that a structural variation may be at play. Based on the mouse models and the observation that the derived variant is found among small and toy dogs, the causal genetic variant likely increases mRNA expression or STC2 protein activity.
LEG LENGTH
A great deal remains to be learned about the genes that control the final length of the dog’s leg, from the very short Chihuahua to the long-legged hounds, such as the Borzoi, Irish Wolfhound, and Scottish Deerhound (Figure 1). Experiments done to date focused on the approximately 20 breeds with chondrodysplasia, such as the Basset Hound, Corgi breeds, and Dachshund. These dogs have much shorter legs than their axial skeleton size would suggest, indicating appendicular growth defects. For some breeds, disproportional dwarfism is part of their standard, as breeders believed that their low profile was advantageous for hunting small game or avoiding the kick of disgruntled livestock.
A multibreed GWAS identified an insert on CFA18 as being highly associated with breed-specific chondrodysplasia. A selective sweep of 24 kb narrowed the region (Parker et al. 2009). The causal variant identified was a retrogene encoding fibroblast growth factor 4 (FGF4), suggesting that the phenotype arises from FGF4 overexpression.
The fact that the chondrodysplastic breeds were developed in many different countries at different times hints that they do not share obvious common ancestry (Am. Kennel Club 1998). However, haplotype analysis shows that the mutation evolved only once. Sequence data suggest that the mutation arose from wolves in Europe and the Middle East, providing supportive evidence that these geographic locations played host to the development of the modern dog, as was suggested by both fossil evidence (Germonpré et al. 2009, Sablin & Khlopachev 2002) and recent mitochondrial data (Thalmann et al. 2013).
SKULL AND AXIAL SKELETON
Variation in the Axial Skeleton
Although size differences account for the most morphological variation among breed dogs, skull shapes are arguably their most identifiable feature. Nearly everyone recognizes a Pug for its flattened face; the Greyhound for its long, tapered snout; and the Bull Terrier for its “Roman nose.” The dog’s posterior was also subjected to man’s intervention: Breeds like the Pembroke Welsh Corgi and the Australian Shepherd are frequently born with shortened or no tails, as are many of the bull-type breeds (Am. Kennel Club 1998).
Skull Shape
Within the past ten years, the genetics of canine skull shape have begun to emerge. In 2004, Fondon & Garner (2004) evaluated coding tandem repeats of developmental genes, including RUNX2, and showed strong correlation and predictive modeling to demonstrate that the collective effects of coding repeat length could drive craniofacial features, such as face length and angle. Subsequent mapping efforts have continued to build on the idea that breed dog skull shapes are driven by complex genetics (Bannasch et al. 2010, Boyko et al. 2010, Quilez et al. 2011, Schoenebeck et al. 2012, Vaysse et al. 2011). As a consequence, numerous loci are now implicated as determinants of canine craniofacial shape, primarily as they pertain to canine brachycephaly (CBa) and dolichocephaly (CDo).
CBa is defined by the rostrocaudal flattening of the viscero- and neurocranium and is usually accompanied by dorsalization of the rostrum. CBa is common to breeds with pushed-in faces, such as the Bulldog, Shih Tzu, and Boxer. In contrast, CDo breeds tend to have long, narrower rostra and neurocrania; their rostra tilt ventrally, such as in the Borzoi, Collies, and Greyhounds.
Both CBa and CDo are of interest because of their resemblance to human craniofacial anomalies, namely craniosynostoses. Craniosynostosis occurs with premature loss of suture patency, leading to malformations in skull shape. Craniosynostosis occurs in ~1 in 2,500 children (Boulet et al. 2008, Lajeunie et al. 1995). Approximately 80% of craniosynostosis cases are nonsyndromic, and estimates suggest that only 28% of cases can be assigned a genetic diagnosis ( Justice et al. 2012, Morriss-Kay & Wilkie 2005). Human studies paint a picture in which bone morphogenetic protein, FGF, and Hedgehog signaling; their downstream transcription factors; and cilia function feature prominently in regulating suture biology. For more details, see Jezela-Stanek & Krajewska-Walasek (2013).
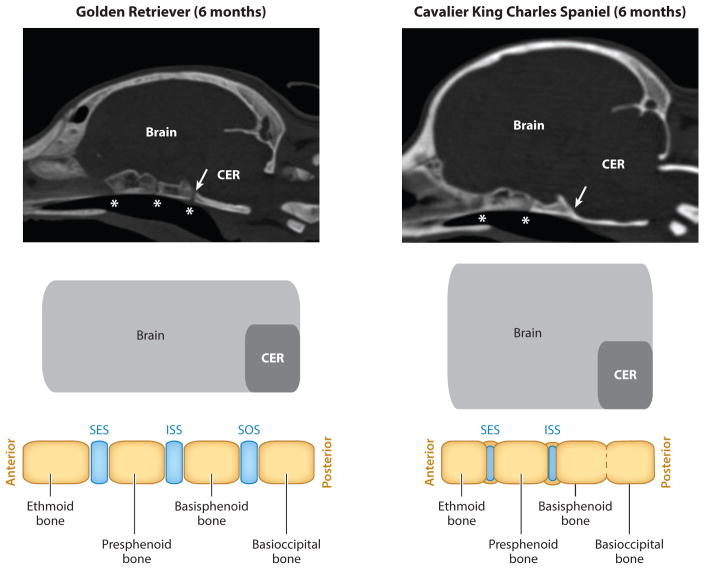
In CBa and CDo dogs, deregulation of suture patency plays a less obvious role in the skull morphology of these extreme head shapes. For CBa, evidence suggests that the morphogenetic changes associated with this conformation are related to the cartilaginous growth zones located at the cranial base, called synchondroses. For years, morphologists noted that the skull base was disproportionately shorter among CBa and chondrodysplastic breeds (Lups 1974, Stockard 1941). More recently, magnetic resonance imaging and computed tomography scans were used to monitor the ossification of cranial base synchondroses. Although the scope of their study was limited, the authors reported that the sphenooccipital synchondrosis closes significantly earlier among CBa breed dogs (Figures 4 and 5) (Schmidt et al. 2013). The CBa findings are interesting in the context of human pathology: For unclear reasons, craniosynostosis is often correlated with precocious closure of cranial base synchodroses (Tannenbaum et al. 1983). Although the mechanisms of cross talk between cranial sutures and synchodroses remain unclear, it could be that in humans, as in CBa breeds, certain types of craniosynostosis could precipitate from morphogenetic defects originating in the cranial base.
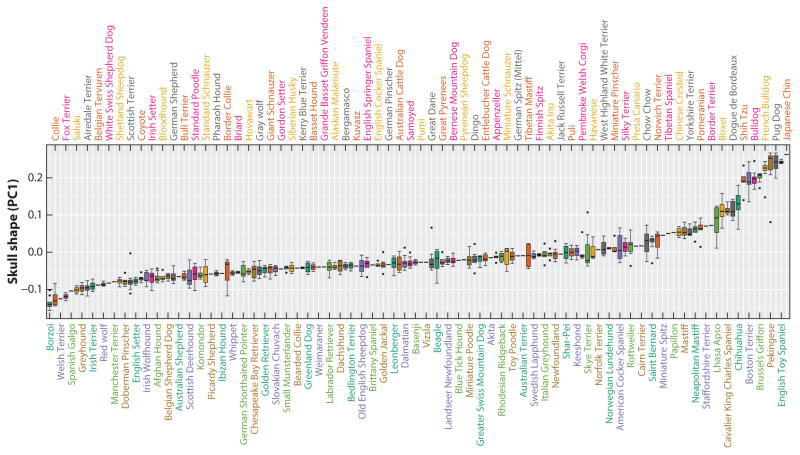
Figure 4.
Canine brachycephaly and its connection to the cranial base. Boxplots of skull PC1 breed averages illustrate the continuum of shape that exists between brachycephalic and dolichocephalic breeds. Figure adapted from Schoenebeck et al. (2012) to include additional dog breeds.
Figure 5.
Computed tomography scans of a six-month-old Golden Retriever and Cavalier King Charles Spaniel (a CBa breed). Asterisks indicate the position of the synchondroses. In this example, the spheno-occipital synchondrosis of the Cavalier King Charles Spaniel is ossified (arrow). Accompanying schematics illustrate the changes that occur at the cranial base between bones ( yellow) and the cartilaginous synchondroses (blue). Abbreviations: CER, cerebellum; ISS, intersphenoid synchondrosis; SES, spheno-ethmoidal synchondrosis; SOS, spheno-occipital synchondrosis. Scanned images provided courtesy of Dr. Martin Schmidt.
Empowered by GWAS approaches, attempts to map genetic drivers of canine skull shape have begun to yield successes. The strongest signal associated with canine head shape is driven by dogs with CBa. The signal occurs on CFA1 (Bannasch et al. 2010, Boyko et al. 2010, Schoenebeck et al. 2012). Bannasch et al. (2010) described a haplotype that spans thrombospondin 2 (THSB2), a gene whose product is found in the bone matrix (Young 2003). Mouse studies indicate that together THSB2 and its paralog, THSB1, are required for craniofacial morphogenesis: The anteroposterior axis of Thsb1−/− Thsb2−/− mice crania is shorter and its mediolateral axis wider than wild type and single knockouts of either gene alone (Nishiwaki et al. 2006). Morphologically, the mouse data appear consistent with CBa. However, the causal mutation driving the signal on CFA1 of dogs remains unknown, and as the authors acknowledge, the causal genetic variant may promote CBa through mechanisms that affect genes outside the critical interval defined by haplotype analysis. Also, it is unclear whether the CFA1 QTL promotes CBa among large and giant breeds of dog, like the Bullmastiff and Dogue de Bordeaux.
Another QTL associated with CBa is located on CFA32. Fine mapping through haplotype comparisons and WGS variant filtering led to the discovery of a Phe→Leu missense mutation in the signaling domain of bone morphogenetic protein 3 (BMP3) (Schoenebeck et al. 2012). The mutation is intriguing for many reasons. First, nearly all members of the TGF-β superfamily bear either a tyrosine or a phenylalanine at this position, indicating a high degree of conservation throughout vertebrate evolution. Moreover, the mutation is located close to where receptor-ligand interactions occur (Allendorph et al. 2007). In zebrafish, knockdown of Bmp3 leads to a hypoplasia phenotype of the head cartilages (Schoenebeck et al. 2012); however, craniofacial defects were not seen in Bmp3−/− mice. Rather, null mice show increased long bone trabeculation, which led to the conclusion that BMP3 restricts bone density (Daluiski et al. 2001). An intriguing observation from mice is Bmp3 RNA expression in the cranial base synchondroses during postnatal development (Kettunen et al. 2006). Additional work is needed to understand the differences between BMP3 function in mice and dogs, including introduction of a comparable Phe→Leu mutation into transgenic mice, as well as biochemistry to determine whether the mutation adversely affects receptor binding.
In addition to QTL on CFA1 and CFA32, CBa is also highly associated with regions on CFA5, 30, and X. Interestingly, the CFA X QTL coincides with the same region that is also associated with canine gigantism, leading to speculation that the genetics are one and the same for these two traits. Although quite broad, the critical interval at this region is within proximity to GPC3 and GPC4 (M. Rimbault and J.J. Schoenebeck, unpublished data). In humans, disruption of GPC3 function is known to cause Simpson-Golabi-Behmel syndrome, diagnostic features of which include overgrowth and coarse facial features (Cottereau et al. 2013).
Tail Length
Tail length is yet another trait that breeders have modified in certain dog breeds. Breeds with naturally short tails (so-called bobtails) occur when animals are born without some or all caudal vertebrae. Two modes of inheritance are observed. In some dog breeds, like the Pembroke Welsh Corgi, the inheritance of a bobtail is dominant and embryonic lethal in its homozygous state. Based on the inheritance pattern and phenotypic similarities to murine T−/−, Haworth et al. (2001) sequenced and discovered an Ile→Met mutation in canine T that segregates with the tail phenotype. Based on gel-shift assays, the authors concluded that the mutation disrupts DNA binding of the transcription factor to its targets. Among other breeds, short tail length was shown to occur independent of mutations in T. Many of these breeds are also CBa type, suggesting that perhaps one of the genetic drivers of CBa is pleiotropic, affecting development of the caudal vertebrae as well as craniofacial structures (Hytönen et al. 2009).
SKIN AND PELAGE
The skin and pelage of the dog are composed of several attributes often regulated by breed standards, for example, the skin’s color or the length, curl, and growth pattern of the fur. Leegwater et al. (2007) first identified the white spotting locus in the Boxer dog by using a genome-wide linkage scan with 1,500 SNPs. In a subsequent GWAS, the locus was mapped to a region containing the pigment-related gene microphthalmia-associated transcription factor (Karlsson et al. 2007). Since then, numerous other studies described additional coat color characteristics, all of which are beyond the scope of this discussion. Note, however, that coat color variation in dogs, in at least two cases, is due to the insertion of a SINE element. Merle is due to an insertion into premelanosome protein (Clark et al. 2006), and black-and-tan saddle color is due to an insertion into the agouti signaling protein (Dreger & Schmutz 2011).
Aside from color, growth pattern, fur length, and curl are attributes that contribute enormously to the appearance of the dog. These three traits were mapped in a large GWAS of more than 900 dogs (Cadieu et al. 2009). The investigators found that the R-spondin 2 gene (RSPO2) was responsible for the pattern of a mustache and eyebrows and the wire hair coat that are characteristic of breeds like the Giant Schnauzer. The critical mutation was a 167-bp insertion in the 3′ untranslated region of the gene that may affect message stability. The FGF5 gene was found to be responsible, at least in part, for fur length; here, a missense change Cys95Phe in exon one was implicated at the critical mutation. This was not surprising, as previous studies had identified FGF5 as being responsible for the fluffy coat of the Pembroke Welsh Corgi, an atypical pattern (Housley & Venta 2006), as well as pelage anomalies in other species (Sundberg et al. 1997). Given the variation in length observed across breeds, additional genes are certain to play a role. Fur curl, such as that common to Poodles, was controlled by mutations in keratin 71 (KRT71). Mutations in this gene have been described in curly-coated mice (Runkel et al. 2006).
When the variations at RSPO2, FGF5, and KRT71 were considered together, these three genes explained approximately 80% of the variation that is observed in canine coat length, growth pattern, and curl. Striking differences between breeds, such as the tight coat of the Labrador, the curly coat of the Poodle, and the long coat of the Old English Sheepdog, could all be explained by variants in just these three genes (Cadieu et al. 2009). This was important, as it established a pattern observed in subsequent papers. There appears, for many traits, to be a primary set of genes responsible for most of the variation observed in the species. Secondary genetic modifiers likely exist that overlay more subtle layers of physical variation; however, these are likely to occur at lower allele frequencies within and across breeds.
The dog has also proven useful for studies of canine skin cancers, particularly melanoma. Simpson has argued that the sporadic melanoma in the dog is a good preclinical model for human melanoma: Because canine melanomas, like those in humans, rarely occur in sun-exposed areas, there may also exist a set of subtypes in dog that are similar to those in human, and both canine and human melanomas harbor AKT and MAPK activation mutations (Simpson et al. 2014). Epidemiologic analyses demonstrate that Poodles are at a particularly high risk of developing oral melanoma, whereas, for instance, Schnauzers mostly develop cutaneous melanoma. Breed-specific differences occur, indicating heritable, lineage-specific modifiers. Understanding these genetics is sure to inform the human disease.
Supplementary Material
SUMMARY POINTS.
Approximately 500 breeds of dogs worldwide are recognized in part by morphological criteria outlined in their breed standard, leading to gross intrabreed uniformity for traits such as size and skull shape.
Historical events, such as domestication, the use of popular sires, bottlenecks, and artificial selection of traits, have each left genetic signatures in the pedigree dog genome.
Extensive linkage disequilibrium that is commonly observed within breeds is reduced with the use of interbreed comparisons, thus aiding fine mapping.
Body size in dogs is controlled by a small number of genes of major effect. This may help us better understand genetic regulation of human height and livestock productivity.
The genetic underpinnings and mechanisms of canine brachycephaly could lead to new insights into human craniofacial anomalies that originate from morphogenetic defects of the skull base and sutures.
Dogs and humans are afflicted by etiologically similar diseases; the genetics of these diseases are often highly related.
Dogs serve as excellent models for understanding comparable complex human conditions, including cancer, heart disease, and autoimmune disorders.
FUTURE POINTS.
A comprehensive understanding of allelic contributions to traits will require exploration of intrabreed morphological and genetic variation.
The roles of indels, copy number variation, and methylation deserve further consideration, as they are expected to contribute to traits and diseases.
Although the Boxer serves as the current reference sequence for the dog, many more domestic and wild canids must be sequenced at least 30-fold coverage to build a comprehensive understanding of the dog’s genetic variation.
The utility of canine models of disease depends heavily on the degree to which diseases are accurately phenotyped in dogs.
Pathology, histology, and physical manifestations of disease all play a key role in advancing canine models of human disease.
In many organisms, GWAS are augmented by WGS and may ultimately be replaced by directly sequencing study subjects.
As the costs of sequencing decrease, read lengths are extended, and genotype calling improves, the use of SNP chip genotyping will be supplanted by low pass WGS, thus enabling a more comprehensive view of genetic variation within study populations.
Acknowledgments
We thank the many dog owners who have generously shared samples and information with us. We gratefully acknowledge the Roslin Institute and Intramural Program of the National Human Genome Research Institute for continued support. We also thank Dr. Maud Rimbault, Alexandra Byers, and Erica Chapman for helping to generate figures and tables, as well as Dr. Martin Schmidt for providing unpublished computed tomography images.
Abbreviation
- Breed
a named population of dogs with a set of highly specified traits (e.g., body size, coat type, leg length)
- Breed standard
a set of morphological measures set by a fancier club that define the critical characteristics of a dog breed
- Linkage disequilibrium (LD)
nonrandom association of alleles at two or more loci on the same DNA strand; measured as a correlation between markers
- Haplotype
alleles on a single strand of DNA that are coinherited owing to physical proximity or linkage disequilibrium
- Genetic variation
differences in the DNA sequence used to distinguish individuals and populations; these differences can be benign or causal
- Single-nucleotide polymorphism (SNP)
a single base position within the genome that has one or more alleles
- Admixture
genetic intermixing of two or more different populations
- Genome-wide association study (GWAS)
comparison of allele frequencies with the goal of identifying trait- or disease-causing loci
- Allele
one of two or more variants located at a specific position on a chromosome
- Selective sweep
a signature of natural or artificial selection frequently detected by runs of homozygosity, linkage disequilibrium, or genetic differentiation
- Quantitative trait loci (QTL)
regions of the genome that contribute to a complex trait or disease; inheritance is non-Mendelian
- Whole-genome sequencing (WGS)
often termed next-generation sequencing or massively parallel sequencing to distinguish technologies from low-output Sanger sequencing
- Targeted resequencing
enrichment and sequencing of specific regions of DNA; used for fine mapping for the purposes of causal variant discovery
- Derived allele
the allele that is found in the evolved population
- Craniosynostosis
premature fusion of developing bones of the skull; in humans, presentation can be uni- or bilateral, syndromic or nonsyndromic
- Cranial synchondroses
cartilage growth plates located along the midline of the skull base
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Acland GM, Ray K, Mellersh CS, Gu W, Langston AA, et al. Linkage analysis and comparative mapping of canine progressive rod-cone degeneration ( prcd ) establishes potential locus homology with retinitis pigmentosa (RP17) in humans. Proc Natl Acad Sci USA. 1998;96:3048–53. doi: 10.1073/pnas.95.6.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahonen SJ, Pietilä E, Mellersh CS, Tiira K, Hansen L, et al. Genome-wide association study identifies a novel canine glaucoma locus. PLOS ONE. 2013;8:e70903. doi: 10.1371/journal.pone.0070903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorph GP, Isaacs MJ, Kawakami Y, Izpisua Belmonte JC, Choe S. BMP-3 and BMP-6 structures illuminate the nature of binding specificity with receptors. Biochemistry. 2007;46:12238–47. doi: 10.1021/bi700907k. [DOI] [PubMed] [Google Scholar]
- Am. Kennel Club. The Complete Dog Book. New York: Howell Book House; 1998. p. 790. [Google Scholar]
- Bannasch D, Young A, Myers J, Truvé K, Dickinson P, et al. Localization of canine brachycephaly using an across breed mapping approach. PLOS ONE. 2010;10:e9632. doi: 10.1371/journal.pone.0009632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battista S, Fidanza V, Fedele M, Klein-Szanto AJ, Outwater E, et al. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res. 1999;59:4793–97. [PubMed] [Google Scholar]
- Benson KF, Chada K. Mini-mouse: phenotypic characterization of a transgenic insertional mutant allelic to pygmy. Genet Res. 1994;64:27–33. doi: 10.1017/s0016672300032511. [DOI] [PubMed] [Google Scholar]
- Boulet SL, Rasmussen SA, Honein MA. A population-based study of craniosynostosis in metropolitan Atlanta, 1989–2003. Am J Med Genet A. 2008;146A:984–91. doi: 10.1002/ajmg.a.32208. [DOI] [PubMed] [Google Scholar]
- Bouras T, Southey MC, Chang AC, Reddel RR, Willhite D, et al. Stanniocalcin 2 is an estrogen-responsive gene coexpressed with the estrogen receptor in human breast cancer. Cancer Res. 2002;62:1289–95. [PubMed] [Google Scholar]
- Boyko A, Quignon P, Li L, Schoenebeck J, Degenhardt J, et al. A simple genetic architecture underlies morphological variation in dogs. PLOS Biol. 2010;8:e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulman MP, Kusumi K, Frayling TM, McKeown C, Garrett C, et al. Mutations in the human Delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat Genet. 2000;24:438–41. doi: 10.1038/74307. [DOI] [PubMed] [Google Scholar]
- Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, et al. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326:150–53. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieu E, Ostrander EA. Canine genetics offers new mechanisms for the study of human cancer. Cancer Epidemiol Biomark Prev. 2007;16:2181–83. doi: 10.1158/1055-9965.EPI-07-2667. [DOI] [PubMed] [Google Scholar]
- Carty CL, Johnson NA, Hutter CM, Reiner AP, Peters U, et al. Genome-wide association study of body height in African Americans: the Women’s Health Initiative SNP Health Association Resource (SHARe) Hum Mol Genet. 2011;21:711–20. doi: 10.1093/hmg/ddr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AC, Hook J, Lemckert FA, McDonald MM, Nguyen MA, et al. The murine stanniocalcin 2 gene is a negative regulator of postnatal growth. Endocrinology. 2008;149:2403–10. doi: 10.1210/en.2007-1219. [DOI] [PubMed] [Google Scholar]
- Chang AC, Jellinek DA, Reddel RR. Mammalian stanniocalcins and cancer. Endocr Relat Cancer. 2003;10:359–73. doi: 10.1677/erc.0.0100359. [DOI] [PubMed] [Google Scholar]
- Chang AC, Reddel RR. Identification of a second stanniocalcin cDNA in mouse and human: stanniocalcin 2. Molecular and cellular endocrinology. Mol Cell Endocrinol. 1998;141:95–99. doi: 10.1016/s0303-7207(98)00097-5. [DOI] [PubMed] [Google Scholar]
- Chase K, Carrier DR, Adler FR, Jarvik T, Ostrander EA, et al. Genetic basis for systems of skeletal quantitative traits: principal component analysis of the canid skeleton. Proc Natl Acad Sci USA. 2002;99:9930–35. doi: 10.1073/pnas.152333099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Zhang X, Colins B, Howard K, Simpson S, et al. Disruption of the high mobility group at-hook 2 (HMGA2) gene in swine reduces postnatal growth. Reprod Fertil Dev. 2013;26:117. [Google Scholar]
- Clark LA, Wahl JM, Rees CA, Murphy KE. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc Natl Acad Sci USA. 2006;103:1376–81. doi: 10.1073/pnas.0506940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottereau E, Mortemousque I, Moizard MP, Bürglen L, Lacombe D, et al. Phenotypic spectrum of Simpson-Golabi-Behmel syndrome in a series of 42 cases with a mutation in GPC3 and review of the literature. Am J Med Genet. 2013;163C:92–105. doi: 10.1002/ajmg.c.31360. [DOI] [PubMed] [Google Scholar]
- Daluiski A, Engstrand T, Bahamonde ME, Gamer LW, Agius E, et al. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat Genet. 2001;27:84–88. doi: 10.1038/83810. [DOI] [PubMed] [Google Scholar]
- Deckelbaum RA, Majithia A, Booker T, Henderson JE, Loomis CA. The homeoprotein engrailed 1 has pleiotropic functions in calvarial intramembranous bone formation and remodeling. Development. 2006;133:63–74. doi: 10.1242/dev.02171. [DOI] [PubMed] [Google Scholar]
- Dobson JM. Breed-predispositions to cancer in pedigree dogs. ISRN Vet Sci. 2013;2013:941275. doi: 10.1155/2013/941275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson JM, Samuel S, Milstein H, Rogers K, Wood JL. Canine neoplasia in the UK: estimates of incidence rates from a population of insured dogs. J Small Anim Pract. 2002;43:240–46. doi: 10.1111/j.1748-5827.2002.tb00066.x. [DOI] [PubMed] [Google Scholar]
- Dreger DL, Parker HG, Ostrander EA, Schmutz SM. Identification of a mutation that is associated with the saddle tan and black-and-tan phenotypes in Basset Hounds and Pembroke Welsh Corgis. J Hered. 2013;104:339–406. doi: 10.1093/jhered/est012. [DOI] [PubMed] [Google Scholar]
- Dreger DL, Schmutz SM. A SINE insertion causes the black-and-tan and saddle tan phenotypes in domestic dogs. J Hered. 2011;1:S11–18. doi: 10.1093/jhered/esr042. [DOI] [PubMed] [Google Scholar]
- Drögemüller C, Karlsson EK, Hytönen MK, Perloski M, Dolf G, et al. A mutation in hairless dogs implicates FOXI3 in ectodermal development. Science. 2008;321:1462. doi: 10.1126/science.1162525. [DOI] [PubMed] [Google Scholar]
- Fogle B. The New Encyclopedia of the Dog. New York: Dorling Kindersley; 2000. p. 416. [Google Scholar]
- Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101:18058–63. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PR. Pathology of myxomatous mitral valve disease in the dog. J Vet Cardiol. 2012;14:103–26. doi: 10.1016/j.jvc.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Freedman AH, Gronau I, Schweizer RM, Ortega-Del Vecchyo D, Han E, et al. Genome sequencing highlights the dynamic early history of dogs. PLOS Genet. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AT, Ogden R, Eland C, Hemani G, Pong-Wong R, et al. Genome-wide analysis of mitral valve disease in Cavalier King Charles Spaniels. Vet J. 2012;193:283–86. doi: 10.1016/j.tvjl.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Frischknecht M, Niehof-Oellers H, Jagannathan V, Owczarek-Lipska M, Drögemüller C, et al. A COL11A2 mutation in Labrador retrievers with mild disproportionate dwarfism. PLOS ONE. 2013;8:e60149. doi: 10.1371/journal.pone.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi AD, Kuo EYW, Raulic S, Wagner GF, DiMattia GE. Human stanniocalcin-2 exhibits potent growth-suppressive properties in transgenic mice independently of growth hormone and IGFs. Am J Physiol Endocrinol Metab. 2005;288:E92–E105. doi: 10.1152/ajpendo.00268.2004. [DOI] [PubMed] [Google Scholar]
- Germonpré M, Sablin MV, Stevens RE, Hedges RE, Hofreiter M, et al. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. J Archaeol Sci. 2009;36:473–90. [Google Scholar]
- Grall A, Guaguère E, Planchais S, Grond S, Bourrat E, et al. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat Genet. 2012;44:140–47. doi: 10.1038/ng.1056. [DOI] [PubMed] [Google Scholar]
- Grandjean D. The Dog Encyclopedia. Paris: R. Canin; 2000. [Google Scholar]
- Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–15. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- Haworth KE, Putt W, Cattanach B, Breen M, Binns M, et al. Canine homolog of the T-box transcription factor T; failure of the protein to bind to its DNA target leads to a short-tail phenotype. Mamm Genome. 2001;12:212–18. doi: 10.1007/s003350010253. [DOI] [PubMed] [Google Scholar]
- Hoopes BC, Rimbault M, Liebers D, Ostrander EA, Sutter NB. The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm Genome. 2012;23:780–90. doi: 10.1007/s00335-012-9417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housley DJ, Venta PJ. The long and short of it: evidence that FGF5 is a major determinant of canine ‘hair’-itability. Anim Genet. 2006;37:309–15. doi: 10.1111/j.1365-2052.2006.01448.x. [DOI] [PubMed] [Google Scholar]
- Hytönen MK, Grall A, Hédan B, Dréano S, Seguin SJ, et al. Ancestral T-box mutation is present in many, but not all, short-tailed dog breeds. J Hered. 2009;100:236–40. doi: 10.1093/jhered/esn085. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Miyamoto K, Taketani Y, Morita K, Takeda E, et al. Molecular cloning of a second human stanniocalcin homologue (STC2) Biochem Biophys Res Commun. 1998;250:252–58. doi: 10.1006/bbrc.1998.9300. [DOI] [PubMed] [Google Scholar]
- Isono T, Matsumoto T, Wada A, Suzaki M, Chano T. A global transcriptome analysis of a dog model of congestive heart failure with the human genome as a reference. J Card Fail. 2012;18:872–78. doi: 10.1016/j.cardfail.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Ito D, Walker JR, Thompson CS, Moroz I, Lin W, et al. Characterization of stanniocalcin 2, a novel target of the mammalian unfolded protein response with cytoprotective properties. Mol Cell Biol. 2004;24:9456–69. doi: 10.1128/MCB.24.21.9456-9469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinek DA, Chang AC, Larsen MR, Wang X, Robinson PJ, Reddel RR. Stanniocalcin 1 and 2 are secreted as phosphoproteins from human fibrosarcoma cells. Biochem J. 2000;350:453–61. [PMC free article] [PubMed] [Google Scholar]
- Jesty SA, Jung SW, Cordeiro JM, Gunn TM, Di Diego JM, et al. Cardiomyocyte calcium cycling in a naturally occurring German shepherd dog model of inherited ventricular arrhythmia and sudden cardiac death. J Vet Cardiol. 2013;15:5–14. doi: 10.1016/j.jvc.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezela-Stanek A, Krajewska-Walasek M. Genetic causes of syndromic craniosynostoses. Eur J Paediatr Neurol. 2013;17:221–24. doi: 10.1016/j.ejpn.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Jónasdóttir T, Mellersh CS, Moe L, Heggebo R, Gamlem H, et al. Genetic mapping of a naturally occurring hereditary renal cancer syndrome in dogs. Proc Natl Acad Sci USA. 2000;97:4132–37. doi: 10.1073/pnas.070053397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Chase K, Martin A, Davern P, Ostrander EA, Lark KG. Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics. 2008;179:1033–44. doi: 10.1534/genetics.108.087866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice CM, Yagnik G, Kim Y, Peter I, Jabs EW, et al. A genome-wide association study identifies susceptibility loci for nonsyndromic sagittal craniosynostosis near BMP2 and within BBS9. Nat Genet. 2012;44:1360–64. doi: 10.1038/ng.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NHC, Zody MC, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–28. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Lindblad-Toh K. Leader of the pack: gene mapping in dogs and other model organisms. Nat Rev Genet. 2008;9:713–24. doi: 10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Sigurdsson S, Ivansson E, Thomas R, Elvers I, et al. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 2013;14:R132. doi: 10.1186/gb-2013-14-12-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karyadi DM, Karlins E, Decker B, vonHoldt BM, Carpintero-Ramirez G, et al. A copy number variant at the KITLG locus likely confers risk for canine squamous cell carcinoma of the digit. PLOS Genet. 2013;9:e1003409. doi: 10.1371/journal.pgen.1003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen P, Nie X, Kvinnsland IH, Luukko K. Histological development and dynamic expression of Bmp2-6 mRNAs in the embryonic and postnatal mouse cranial base. Anat Rec Part A. 2006;288:1250–58. doi: 10.1002/ar.a.20402. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898–903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Kita Y, Mimori K, Iwatsuki M, Yokobori T, Leta K, et al. STC2: a predictive marker for lymph node metastasis in esophageal squamous-cell carcinoma. Ann Surg Oncol. 2011;18:261–72. doi: 10.1245/s10434-010-1271-1. [DOI] [PubMed] [Google Scholar]
- Koike N, Kassai Y, Kouta Y, Miwa H, Konishi M, Itoh N. Brorin, a novel secreted bone morphogenetic protein antagonist, promotes neurogenesis in mouse neural precursor cells. J Biol Chem. 2007;282:15843–50. doi: 10.1074/jbc.M701570200. [DOI] [PubMed] [Google Scholar]
- Lajeunie E, Le Merrer M, Bonaïti-Pellie C, Marchac D, Renier D. Genetic study of nonsyndromic coronal craniosynostosis. Am J Med Genet. 1995;55:500–4. doi: 10.1002/ajmg.1320550422. [DOI] [PubMed] [Google Scholar]
- Larson G, Karlsson EK, Perri A, Webster MT, HoS Y, et al. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc Natl Acad Sci USA. 2012;109:8878–83. doi: 10.1073/pnas.1203005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent TC, Fraser JRE. Hyaluronan. FASEB J. 1992;7:2397–404. [PubMed] [Google Scholar]
- Law AYS, Wong CKC. Stanniocalcin-2 is a HIF-1 target gene that promotes cell proliferation in hypoxia. Exp Cell Res. 2010;316:466–76. doi: 10.1016/j.yexcr.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Leegwater PA, van Hagen MA, van Oost BA. Localization of white spotting locus in Boxer dogs on CFA20 by genome-wide linkage analysis with 1500 SNPs. J Hered. 2007;98:549–52. doi: 10.1093/jhered/esm022. [DOI] [PubMed] [Google Scholar]
- Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–91. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon AH, Moore SDP, Parisi MA, Mealiffe ME, Harris DJ, et al. Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas. Am J Hum Genet. 2005;76:340–48. doi: 10.1086/427565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lups P. Biometric analysis of the skull base of domestic dogs. Zool Anz. 1974;192:383–413. (in German) [Google Scholar]
- Makvandi-Nejad S, Hoffman GE, Allen JJ, Chu E, Gu E, et al. Four loci explain 83% of size variation in the horse. PLOS ONE. 2012;7:e39929. doi: 10.1371/journal.pone.0039929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews LS, Hammer RE, Brinster RL, Palmiter RD. Expression of insulin-like growth factor I in transgenic mice with elevated levels of growth hormone is correlated with growth. Endocrinology. 1988;123:433–37. doi: 10.1210/endo-123-1-433. [DOI] [PubMed] [Google Scholar]
- Mausberg TB, Wess G, Simak J, Keller L, Drögemüller M, et al. A locus on chromosome 5 is associated with dilated cardiomyopathy in Doberman Pinschers. PLOS ONE. 2011;6:e20042. doi: 10.1371/journal.pone.0020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh C, Holmes N, Binns M, Sampson J. Dinucleotide repeat polymorphisms at four canine loci (LEI 003, LEI 007, LEI 008 and LEI 015) Anim Genet. 1994;25:125–26. doi: 10.1111/j.1365-2052.1994.tb00098.x. [DOI] [PubMed] [Google Scholar]
- Menten B, Buysse K, Zahir F, Hellemans J, Hamilton SJ, et al. Osteopoikilosis, short stature and mental retardation as key features of a new microdeletion syndrome on 12q14. J Med Genet. 2007;44:264–68. doi: 10.1136/jmg.2006.047860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs KM, Lahmers S, Keene BW, White SN, Oyama MA, et al. A splice site mutation in a gene encoding for PDK4, a mitochondrial protein, is associated with the development of dilated cardiomyopathy in the Doberman pinscher. Hum Genet. 2012;131:1319–25. doi: 10.1007/s00439-012-1158-2. [DOI] [PubMed] [Google Scholar]
- Meurs KM, Mauceli E, Lahmers S, Acland GM, White SN, Lindblad-Toh K. Genome-wide association identifies a deletion in the 3′ untranslated region of striatin in a canine model of arrhythmogenic right ventricular cardiomyopathy. Hum Genet. 2010;128:315–24. doi: 10.1007/s00439-010-0855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs KM, Stern JA, Sisson DD, Kittleson MD, Cunningham SM, et al. Association of dilated cardiomyopathy with the striatin mutation genotype in Boxer dogs. J Vet Intern Med. 2013;27:1437–40. doi: 10.1111/jvim.12163. [DOI] [PubMed] [Google Scholar]
- Morriss-Kay GM, Wilkie AO. Growth of the normal skull vault and its alteration in craniosynostosis: insights from human genetics and experimental studies. J Anat. 2005;207:637–53. doi: 10.1111/j.1469-7580.2005.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Suzuki M, Nishino Y, Funaba M. Regulatory expression of genes related to metastasis by TGF-β and activin A in B16 murine melanoma cells. Mol Biol Rep. 2010;37:1279–86. doi: 10.1007/s11033-009-9502-x. [DOI] [PubMed] [Google Scholar]
- N’Diaye A, Chen GK, Palmer CD, Ge B, Tayo B, et al. Identification, replication, and fine-mapping of loci associated with adult height in individuals of African ancestry. PLOS Genet. 2011;7:e1002298. doi: 10.1371/journal.pgen.1002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T, Yamaguchi T, Zhao C, Amano H, Hankenson KD, et al. Reduced expression of thrombospondins and craniofacial dysmorphism in mice overexpressing Fra1. J Bone Min Res. 2006;21:596–604. doi: 10.1359/jbmr.051216. [DOI] [PubMed] [Google Scholar]
- Ohad DG, Avrahami A, Waner T, David L. The occurrence and suspected mode of inheritance of congenital subaortic stenosis and tricuspid valve dysplasia in Dogue de Bordeaux dogs. Vet J. 2013;197:351–57. doi: 10.1016/j.tvjl.2013.01.012. [DOI] [PubMed] [Google Scholar]
- Olsson M, Meadows JRS, Truvé K, Rosengren Pielberg G, Puppo F, et al. A novel unstable duplication upstream of HAS2 predisposes to a breed-defining skin phenotype and a periodic fever syndrome in Chinese Shar-Pei dogs. PLOS Genet. 2011;7:e1001332. doi: 10.1371/journal.pgen.1001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA. Franklin H. Epstein Lecture. Both ends of the leash—the human links to good dogs with bad genes. N Engl J Med. 2012;367:636–46. doi: 10.1056/NEJMra1204453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Beale HC. Leading the way: finding genes for neurologic disease in dogs using genome-wide mRNA sequencing. BMC Genet. 2012;13:56. doi: 10.1186/1471-2156-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Jong PM, Rine J, Duyk G. Construction of small-insert genomic DNA libraries highly enriched for microsatellite repeat sequences. Proc Natl Acad Sci USA. 1992;89:3419–23. doi: 10.1073/pnas.89.8.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Mapa FA, Yee M, Rine J. One hundred and one simple sequence repeat-based markers for the canine genome. Mamm Genome. 1995;6:192–95. doi: 10.1007/BF00293011. [DOI] [PubMed] [Google Scholar]
- Owczarek-Lipska M, Jagannathan V, Drögemüller C, Lutz S, Glanemann B, et al. Aframeshift mutation in the cubilin gene (CUBN) in border collies with Imerslund-Gräsbeck Syndrome (selective cobalamin malabsorption) PLOS ONE. 2013a;8:e61144. doi: 10.1371/journal.pone.0061144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owczarek-Lipska M, Mausberg TB, Stephenson H, Dukes-McEwan J, Wess G, Leeb T. A 16-bp deletion in the canine PDK4 gene is not associated with dilated cardiomyopathy in a European cohort of Doberman Pinschers. Anim Genet. 2013b;44:239. doi: 10.1111/j.1365-2052.2012.02396.x. [DOI] [PubMed] [Google Scholar]
- Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–64. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Parker HG, Meurs KM, Ostrander EA. Finding cardiovascular disease genes in the dog. J Vet Cardiol. 2006;8:115–27. doi: 10.1016/j.jvc.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Ostrander EA. Cancer. Hiding in plain view—an ancient dog in the modern world. Science. 2014;343:376–78. doi: 10.1126/science.1248812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Shearin AL, Ostrander EA. Man’s best friend becomes biology’s best in show: genome analyses in the domestic dog. Annu Rev Genet. 2010;44:309–36. doi: 10.1146/annurev-genet-102808-115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–98. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp U, Vollmar A, Häggstrom J, Thomas A, Distl O. Multiple loci are associated with dilated cardiomyopathy in Irish wolfhounds. PLOS ONE. 2012;7:e36691. doi: 10.1371/journal.pone.0036691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell-Braxton L, Hollingshead P, Warburton C, Dowd M, Pitts-Meek S, et al. IGF-I is required for normal embryonic growth in mice. Genes Dev. 1993;7:2609–17. doi: 10.1101/gad.7.12b.2609. [DOI] [PubMed] [Google Scholar]
- Pryce JE, Hayes BJ, Bolormaa S, Goddard ME. Polymorphic regions affecting human height also control stature in cattle. Genetics. 2011;187:981–84. doi: 10.1534/genetics.110.123943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puré E, Assoian RK. Rheostatic signaling by CD44 and hyaluronan. Cell Signal. 2009;21:651–55. doi: 10.1016/j.cellsig.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilez J, Short AD, Martínez V, Kennedy LJ, Ollier W, et al. A selective sweep of>8 Mb on chromosome 26 in the Boxer genome. BMC Genomics. 2011;12:339. doi: 10.1186/1471-2164-12-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden CA, Bankier A, Brown TJ, Cowen PS, Frost GI, et al. A new disorder of hyaluronan metabolism associated with generalized folding and thickening of the skin. J Pediatr. 2000;136:62–68. doi: 10.1016/s0022-3476(00)90051-9. [DOI] [PubMed] [Google Scholar]
- Rane SG, Dubus P, Mettus RV, Galbreath EJ, Boden G, et al. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in β-islet cell hyperplasia. Nat Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- Ranieri G, Gadaleta CD, Patruno R, Zizzo N, Daidone MG, et al. A model of study for human cancer: spontaneous occurring tumors in dogs. Biological features and translation for new anticancer therapies. Crit Rev Oncol Hematol. 2013;88:187–97. doi: 10.1016/j.critrevonc.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLOS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reist-Marti SB, Dolf G, Leeb T, Kottmann S, Kietzmann S, et al. Genetic evidence of subaortic stenosis in the Newfoundland dog. Vet Rec. 2012;170:597. doi: 10.1136/vr.100019. [DOI] [PubMed] [Google Scholar]
- Rimbault M, Beale HC, Schoenebeck JJ, Hoopes BC, Allen JJ, et al. Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 2013;23:1985–95. doi: 10.1101/gr.157339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel F, Klaften M, Koch K, Böhnert V, Büssow H, et al. Morphologic and molecular characterization of two novel Krt71 (Krt2-6g) mutations: Krt71rco12 and Krt71rco13. Mamm Genome. 2006;17:1172–82. doi: 10.1007/s00335-006-0084-9. [DOI] [PubMed] [Google Scholar]
- Sablin MV, Khlopachev GA. The earliest Ice Age dogs: evidence from Eliseevichi I. Curr Anthropol. 2002;43:795–99. [Google Scholar]
- Salmon-Hillbertz NHC, Isaksson M, Karlsson EK, Hellmén E, Pielberg GR, et al. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat Genet. 2007;39:1318–20. doi: 10.1038/ng.2007.4. [DOI] [PubMed] [Google Scholar]
- Sandor C, Farnir F, Hansoul S, Coppieters W, Meuwissen T, Georges M. Linkage disequilibrium on the bovine X chromosome: characterization and use in quantitative trait locus mapping. Genetics. 2006;173:1777–86. doi: 10.1534/genetics.106.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA, Mirabello L, Wang Z, Gastier-Foster JM, Gorlick R, et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet. 2013;45:799–803. doi: 10.1038/ng.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–13. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, Volk H, Klingler M, Failing K, Kramer M, Ondreka N. Comparison of closure times for cranial base synchondroses in mesaticephalic, brachycephalic, and Cavalier King Charles Spaniel dogs. Vet Radiol Ultrasound. 2013;54:497–503. doi: 10.1111/vru.12072. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Hutchinson SA, Byers A, Beale HC, Carrington B, et al. Variation of BMP3 contributes to dog breed skull diversity. PLOS Genet. 2012;8:e1002849. doi: 10.1371/journal.pgen.1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearin A, Hedan B, Cadieu E, Erich SA, Schmidt EV, et al. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol Biomark Prev. 2012;21:1019–27. doi: 10.1158/1055-9965.EPI-12-0190-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearin AL, Ostrander EA. Leading the way: canine models of genomics and disease. Dis Model Mech. 2010a;3:27–34. doi: 10.1242/dmm.004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearin AL, Ostrander EA. Canine morphology: hunting for genes and tracking mutations. PLOS Biol. 2010b;8:e1000310. doi: 10.1371/journal.pbio.1000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RM, Bastian BC, Michael HT, Webster JD, Prasad ML, et al. Sporadic naturally occurring melanoma in dogs as a preclinical model for human melanoma. Pigment Cell Melanoma Res. 2014;27:37–47. doi: 10.1111/pcmr.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Gu X, Feng C, Wang Y, Gao Y, et al. Evaluation of SNPs in the chicken HMGA2 gene as markers for body weight gain. Anim Genet. 2011;42:333–36. doi: 10.1111/j.1365-2052.2010.02141.x. [DOI] [PubMed] [Google Scholar]
- Stern JA, Meurs KM, Nelson OL, Lahmers SM, Lehmkuhl LB. Familial subvalvular aortic stenosis in Golden Retrievers: inheritance and echocardiographic findings. J Small Anim Pract. 2012;53:213–16. doi: 10.1111/j.1748-5827.2011.01187.x. [DOI] [PubMed] [Google Scholar]
- Stern JA, White SN, Meurs KM. Extent of linkage disequilibrium in large-breed dogs: chromosomal and breed variation. Mamm Genome. 2013;24:409–15. doi: 10.1007/s00335-013-9474-y. [DOI] [PubMed] [Google Scholar]
- Steudemann C, Bauersachs S, Weber K, Wess G. Detection and comparison of microRNA expression in the serum of Doberman Pinschers with dilated cardiomyopathy and healthy controls. BMC Vet Res. 2013;9:12. doi: 10.1186/1746-6148-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockard CR. The Genetic and Endocrinic Basis for Differences in Form and Behavior. Philadelphia: Wistar Inst. Anat. Biol; 1941. p. 775. [Google Scholar]
- Sundberg JP, Rourk MH, Boggess D, Hogan ME, Sundberg BA, Bertolino AP. Angora mouse mutation: altered hair cycle, follicular dystrophy, phenotypic maintenance of skin grafts, and changes in keratin expression. Vet Pathol. 1997;34:171–79. doi: 10.1177/030098589703400301. [DOI] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–15. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Eberle MA, Parker HG, Pullar BJ, Kirkness EF, et al. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–96. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta HT, Dass CR, Choong PF, Dunstan DE. Osteosarcoma treatment: state of the art. Cancer Metastasis Rev. 2009;28:247–63. doi: 10.1007/s10555-009-9186-7. [DOI] [PubMed] [Google Scholar]
- Tannenbaum GS, Guyda HJ, Posner BI. Insulin-like growth factors: a role in growth hormone negative feedback and body weight regulation via brain. Science. 1983;220:77–79. doi: 10.1126/science.6338593. [DOI] [PubMed] [Google Scholar]
- Tengvall K, Kierczak M, Bergvall K, Olsson M, Frankowiack M, et al. Genome-wide analysis in German shepherd dogs reveals association of a locus on CFA 27 with atopic dermatitis. PLOS Genet. 2013;9:e1003475. doi: 10.1371/journal.pgen.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann O, Shapiro B, Cui P, Schuenemann VJ, Sawyer SK, et al. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science. 2013;342:871–74. doi: 10.1126/science.1243650. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-β and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Rosengren Pielberg G, et al. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLOS Genet. 2011;7:e1002316. doi: 10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilà C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, et al. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–89. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-D, Zhai W, Yang H-C, Fan R-X, Cao X, et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 2013;4:1860. doi: 10.1038/ncomms2814. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou J, Powell-Braxton L, Bondy C. Effects of Igf1 gene deletion on postnatal growth patterns. Endocrinology. 1999;140:3391–94. doi: 10.1210/endo.140.7.7045. [DOI] [PubMed] [Google Scholar]
- Wayne RK, Ostrander EA. Origin, genetic diversity, and genome structure of the domestic dog. Bio Essays. 1999;21:247–57. doi: 10.1002/(SICI)1521-1878(199903)21:3<247::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Wayne RK, Ostrander EA. Lessons learned from the dog genome. Trends Genet. 2007;23:557–67. doi: 10.1016/j.tig.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Weedon M, Lango H, Lindgren CM, Wallace C, Evans DM, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon M, Lettre G, Freathy RM, Lindgren CM, Voight BF, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–50. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox B, Walkowicz C. Atlas of Dog Breeds of the World. Neptune City, NJ: T.F.H. Publ; 1995. [Google Scholar]
- Wilkinson S, Lu ZH, Megens HJ, Archibald AL, Haley C, et al. Signatures of diversifying selection in European pig breeds. PLOS Genet. 2013;9:e10003453. doi: 10.1371/journal.pgen.1003453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama JS, Lam ET, Ruhe AL, Erdmanm CA, Robertson KR, Webb AA. Variation in genes related to cochlear biology is strongly associated with adult-onset deafness in border collies. PLOS Genet. 2012;8:e1002898. doi: 10.1371/journal.pgen.1002898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MF. Bone matrix proteins: their function, regulation, and relationship to osteoporosis. Osteoporos Int. 2003;14(Suppl 3):S35–42. doi: 10.1007/s00198-002-1342-7. [DOI] [PubMed] [Google Scholar]
- Yuzbasiyan-Gurkan V, Blanton SH, Cao V, Ferguson P, Li J, et al. Linkage of a microsatellite marker to the canine copper toxicosis locus in Bedlington terriers. Am J Vet Res. 1997;58:23–27. [PubMed] [Google Scholar]
- Zanna G, Docampo MJ, Fondevila D, Bardagi M, Bassols A, Ferrer L. Hereditary cutaneous mucinosis in shar pei dogs is associated with increased hyaluronan synthase-2 mRNA transcription by cultured dermal fibroblasts. Vet Dermatol. 2009;20:377–82. doi: 10.1111/j.1365-3164.2009.00799.x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:725–26. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.