Abstract
Introduction
It has been reported that APOA1 –75G/A polymorphism might be associated with susceptibility to coronary artery disease (CAD). Owing to mixed and inconclusive results, we conducted a meta-analysis to systematically summarize and clarify the association between APOA1-75G/A polymorphism and the risk of CAD.
Material and methods
A systematic search of studies on the association of single nucleotide polymorphisms (SNP) with susceptibility to CAD was conducted. A total of 9 case-control studies (1864 cases and 1196 controls) on the APOA1-75G/A polymorphism were included.
Results
We observed no statistically significant association between APOA1 –75G/A polymorphism and risk of CAD under the dominant genetic model (AA + AG vs. GG: OR = 1.03, 95% CI: 0.65–1.66), allelic contrast (A vs. G: OR = 0.88, 95% CI: 0.58–1.32), heterozygote model (AG vs. GG: OR = 1.24, 95% CI: 0.81–1.89) or homozygote model (AA vs. GG: OR = 0.52, 95% CI: 0.26–1.05). Significant heterogeneity between individual studies appears in all five models, but a strong association under the recessive genetic model (AA vs. AG + GG: OR = 0.51, 95% CI: 0.28–0.92). In the subgroup analysis by Hardy-Weinberg equilibrium (HWE; the presence or absence of HWE in controls), significantly decreased CAD risk and no significant heterogeneity were observed among controls consistent with HWE. Overall, the APOA1 A allele is one of the protective factors of CAD. A stronger association between APOA1-75G/A polymorphisms and CAD risk was present in the studies consistent with HWE.
Conclusions
The minor allele of the APOA1-75G/A polymorphism is a protective factor for CAD, especially in the studies consistent with HWE.
Keywords: apolipoprotein A1, odds ratio, gene polymorphism, confidence interval, coronary artery disease, Hardy-Weinberg equilibrium
Introduction
Coronary artery disease (CAD) is the leading cause of morbidity and mortality in developed and developing countries [1]. The precise mechanisms responsible for the incidence of CAD are still unknown, which is affected by interacting internal and external factors [2, 3]. The chief risk factors of CAD are hypertension, diabetes mellitus, abnormal serum cholesterol (low-density lipoprotein – LDL and high-density lipoprotein – HDL), cigarette smoking, high alcohol consumption, age, stress, family history of CAD and obesity [4–8]. Lipid metabolism disorder and genetic predisposition are major risk factors for CAD [9–11].
Lipoproteins are involved in the pathogenesis of atherosclerosis. Epidemiologic studies have established an inverse relationship between plasma levels of HDL cholesterol and the occurrence of CAD, while low-density lipoprotein has been established as an atherogenic factor [12–15]. There is more to HDL than just reverse cholesterol transport. Also quality (function) of HDL, not only quantity, may be very important [16, 17]. Apolipoprotein A-I (APOA1 gene, ApoA-I protein) is the major protein of HDL. It is a 243 amino acid long peptide, synthesized mainly in the liver and to some extent in the small intestine. ApoA-I has the ability to reverse cholesterol transport (RCT), to bind lipids, and to activate lecithin cholesterol acyltransferase (LCAT) to form mature HDL [18].
Genes influencing quantitative variation in plasma lipoproteins are being studied widely. The APOA1 gene is present along with APOC3 and APOA4 genes on chromosome 11 [19]. Variations in the APOA1-C3-A4 genes have been associated with dyslipidemia and CAD [20]. An association between the G to A substitution at –75 bp in the APOA1 gene and CAD has received most of the attention [21]. An Australian CAD study described a positive relationship between the A allele and severe forms of CAD [22]. However, studies on the Chinese population revealed that APOA1-75G/A polymorphism results in increased levels of apoA1 and HDL-C and is associated with a reduced risk of CAD [23]. Therefore, the relation between APOA1 G-75A polymorphisms and risk of CAD remains controversial. To elucidate this discrepancy, we performed a meta-analysis of all available case-control studies to explore the association between the APOA1 polymorphisms and risk of CAD.
Material and methods
Study selection
To identify all the articles that examined the association of APOA1 polymorphisms with coronary artery disease, we conducted a comprehensive search of PubMed, Embase, Web of Science, Cochrane library and CNKI (China National Knowledge Infrastructure) and EMBASE (the last search update was May 28, 2015). Search terms included apolipoprotein A-I or apolipoprotein AI or apolipoprotein A1 or APOAI or APOA-I or APO A1; gene polymorphism, or genetic mutation and myocardial infarct, myocardial infarction, coronary artery disease, coronary heart disease, myocardial ischemia, ischemic heart disease, ischemic cardiomyopathy, angina, angina pectoris, acute coronary syndrome, acute coronary syndrome (ACS), coronary calcification, coronary flow reserve, ischemic heart failure, heart failure. We also screened references of the retrieved articles and review articles by a hand search. Studies in this meta-analysis had to meet the following inclusion criteria: (1) evaluation of the association between APOA1-75G/A polymorphisms and CAD; (2) case-control study; (3) studies focusing on humans; (4) detailed genotype data could be acquired to calculate the odds ratios (ORs) and 95% confidence intervals (CIs). Exclusion criteria: (1) duplication of previous publications; (2) comment, review and editorial; (3) family-based studies of pedigrees; (4) study with no detailed genotype data. When there were multiple publications from the same population, only the largest study was included. Study selection was performed by two investigators independently, according to the inclusion and exclusion criteria by screening the title, abstract and full text. Any dispute was solved by discussion.
Data extraction
For each study that met our criteria, the following information was collected: first author, year of publication, country of origin, ethnicity, criteria of diagnosis, number of cases and controls, genotype distribution, genotyping methods and allele frequency, the criteria of CAD, Hardy-Weinberg equilibrium, number of cases and controls, and genotype frequency in cases and controls for APOA1-75G/A. All the searching work and data extraction work were conducted by two independent investigators (Xu and Sun). If dissent existed, they would recheck the original data of the included studies and have a discussion to reach a consensus. If the dissent still existed, the third investigator would be involved to adjudicate the disagreements (Zhou).
Quality assessment
The quality of the included studies was assessed by two authors separately according to the methodological quality assessment scale. In this scale, five items – representativeness of cases, source of controls, sample size, quality control of genotyping methods, and Hardy-Weinberg equilibrium (HWE) – were carefully checked. The quality score ranges from 0 to 10, and a high score means good quality of the study. Two investigators scored the studies independently and solved disagreement through discussion (Table I) [24–30].
Table I.
Characteristics of studies included in the meta-analysis
| Author | Year | Ethnicity | Source | Case/control (n) | Cases | Controls | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | GG | GA | AA | P-value for HWE | |||||
| Wang [22] | 1996 | Australian | HB | 462/182 | 306 | 133 | 23 | 125 | 55 | 2 | 0.129 |
| Reguero [24] | 1998 | Spanish | PB | 176/200 | 117 | 50 | 9 | 152 | 44 | 4 | 0.6983 |
| Shengli [25] | 2001 | Chinese | PB | 107/50 | 51 | 7 | 49 | 34 | 2 | 14 | < 0.01 |
| Zou [23] | 2003 | Chinese | HB | 92/45 | 78 | 8 | 6 | 26 | 12 | 7 | 0.0186 |
| Chhabra [26] | 2005 | Indian | HB | 164/36 | 102 | 51 | 11 | 29 | 7 | 0 | 0.5182 |
| Rai [27] | 2008 | Indian | PB | 140/100 | 45 | 44 | 51 | 51 | 39 | 10 | 0.5327 |
| Dawar [28] | 2010 | Indian | PB | 50/50 | 29 | 21 | 0 | 8 | 42 | 0 | < 0.01 |
| Ding [29] | 2012 | Chinese | PB | 229/254 | 94 | 95 | 40 | 108 | 124 | 22 | 0.1013 |
| Xuebiao [30] | 2014 | Chinese | HB | 444/279 | 250 | 165 | 29 | 121 | 137 | 21 | 0.0341 |
HWE – Hardy-Weinberg equilibrium.
Statistical analysis
The strength of association between APOA1 polymorphisms and CAD was measured by the odds ratio (OR) corresponding to a 95% confidence interval (CI) according to the method of Woolf [31]. Heterogeneity between studies was assessed by Cochran’s χ2-based Q statistic test [32]. Where the p-value for heterogeneity was less than 0.1, a random-effects model using the DerSimonian and Laird method [33] was used to pool the results; otherwise, a fixed-effects model using the Mantel-Haenszel method was adopted [34]. In order to better evaluate the extent of heterogeneity between studies, the I2 test was also used. This statistic yields results ranging from 0 to 100% (I2 = 0–25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2= 50–75%, large heterogeneity; I2 = 75–100%, extreme heterogeneity) [35].
For the APOA1 –75G>A promoter polymorphism, we investigated associations between the genetic variant and coronary artery disease risk in allelic contrast (A vs. G), homozygote comparison (AA vs. GG), heterozygote comparison (GA vs. GG), dominant (GA/AA vs. GG) and recessive (AA vs. GA/GG) models, respectively. The significance of the pooled OR was determined by the Z-test (p < 0.05 suggests a significant association). Subgroup analyses were also conducted to explore the effects of confounding factors: HWE (the presence or absence of HWE in controls). HWE was tested by the χ2 test at a significant level of p < 0.05 [36]. Publication bias was investigated by funnel plots [37] and by Egger’s linear regression test [38]. All analyses were performed using STATA version 12.0 (StataCorp LP, College Station, Texas).
Results
Study characteristics
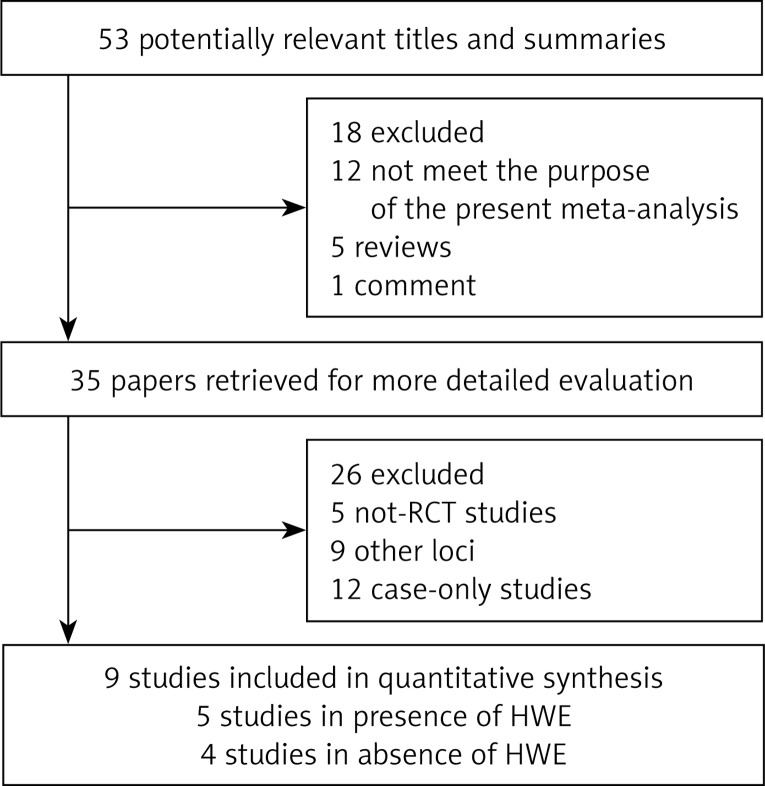
In total, 9 articles were identified according to inclusion and exclusion criteria. The detailed screening process is shown in Figure 1. For APOA1-75G/A polymorphisms, 9 studies involved a total of 1864 cases and 1196 controls. HWE of genotype distribution in the controls was tested in those studies and 5 studies were consistent with HWE [22, 24, 26, 27, 29]. Four studies were deviating from HWE [23, 25, 28, 30].
Figure 1.
Study selection process
Meta-analysis results
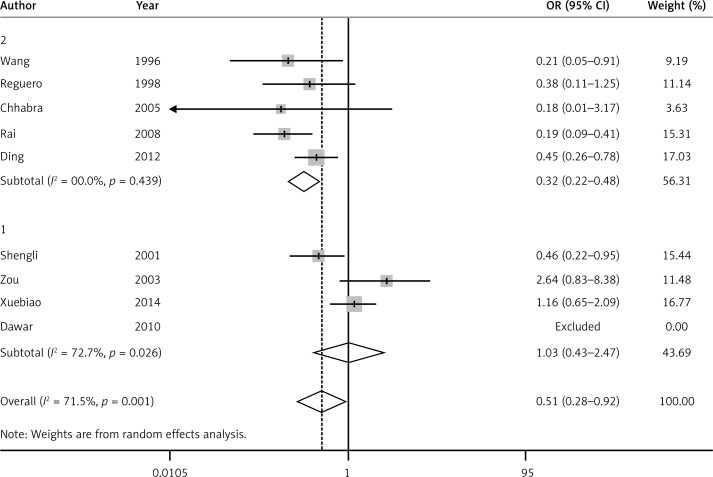
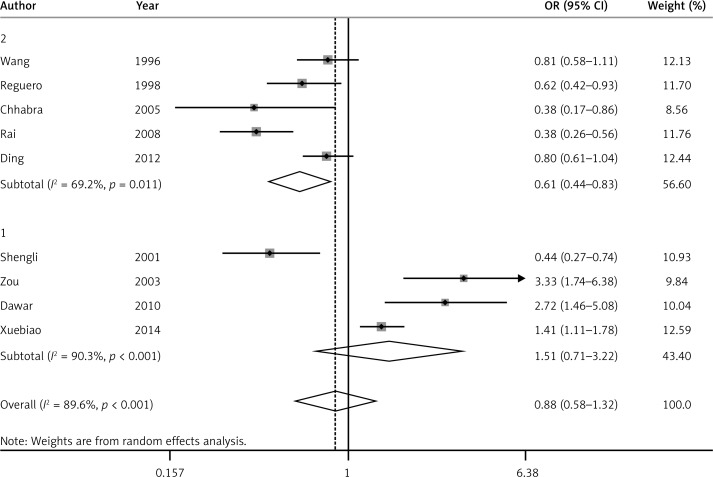
For the APOA1-75G/A polymorphism and its relationship to CAD, significant heterogeneity between individual studies appears obvious in all five models. Therefore, the random-effect model (DerSimonian and Laird) was applied in all five models. There was a statistically significant association between APOA1-75G/A polymorphism and CAD risk under the recessive model (OR = 0.51, 95% CI: 0.28–0.92, PH = 0.001) (Figure 2). Though there was no significant association between APOA1-75G/A polymorphism and CAD under the allele model (OR = 0.88, 95% CI: 0.55–1.32, PH < 0.01) (Figure 3), dominant model (OR = 1.03, 95% CI: 0.65–1.66, PH < 0.01), heterozygote model (OR = 1.24, 95% CI: 0.81–1.89, PH < 0.01) or homozygote model (OR = 0.52, 95% CI: 0.26–1.05, PH < 0.01), a trend of reduced risk could be drawn. Subgroup analysis was conducted according to HWE (the presence or absence of HWE in controls). In the presence of HWE in controls, significant heterogeneity was found under the allele model, so a random-effect model was used. A fixed-effect model was used in the other four genetic models. A statistically significant association was observed between APOA1-75G/A polymorphism and CAD risk under the recessive model (OR = 0.32, 95% CI: 0.22–0.48, PH = 0.439), dominant model (OR = 0.69, 95% CI: 0.51–0.94, PH = 0.078), allele model (OR = 0.61, 95% CI: 0.44–0.83, PH = 0.011) and homozygote model (OR = 0.30, 95% CI: 0.19–0.49, PH = 0.314), but there was no significant association between APOA1-75G/A polymorphism and CAD under the heterozygote model (OR = 0.88, 95% CI: 0.68–1.13, PH = 0.26).
Figure 2.
Forest plot of recessive model for overall comparison of APOA1-75G/A polymorphism and CAD (AA vs. AG + GG) (1 – controls deviating from HWE, 2 – controls consistent with HWE)
Figure 3.
Forest plot of allelic model for overall comparison of APOA1-75G/A polymorphism and CAD (A vs. G) (1 – controls deviating from HWE, 2 – controls consistent with HWE)
After restricting our analysis to the absence of HWE in controls, significant heterogeneity was found under all five models; therefore, the random-effect model was applied. We observed no significant association under any of the five models: recessive model (OR = 1.03, 95% CI: 0.43–2.47, PH = 0.026), dominant model (OR = 2.06, 95% CI: 0.77–5.54, PH < 0.01), allele model (OR = 1.51, 95% CI: 0.71–3.22, PH < 0.01), heterozygote model (OR = 2.49, 95% CI: 0.98–6.35, PH = 0.003) or homozygote model (OR = 1.23, 95% CI: 0.41–3.65, PH = 0.004) (Table II).
Table II.
Results of meta-analysis for APOA1-75G/A polymorphisms and CAD risks
| Study groups | Heterozygote codominant | Dominant model | Recessive model | ||||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | PH | OR (95% CI) | PH | OR (95% CI) | PH | ||
| Total (9) | 1.24 (0.81–1.89) | < 0.01 | 1.03 (0.65–1.66) | < 0.01 | 0.51 (0.28–0.92) | 0.001 | |
| HWE: | |||||||
| Yes (5) | 0.88 (0.68–1.13) | 0.26 | 0.69 (0.51–0.94) | 0.078 | 0.32 (0.22–0.48) | 0.439 | |
| No (4) | 2.49 (0.98–6.35) | 0.003 | 2.06 (0.77–5.54) | < 0.01 | 1.03 (0.43–2.47) | 0.026 | |
| Study groups | Allele contrast model | Homozygote codominant | |||||
| OR (95% CI) | PH | OR (95% CI) | PH | ||||
| Total (9) | 0.88 (0.58–1.32) | < 0.01 | 0.52 (0.26–1.05) | < 0.01 | |||
| HWE: | |||||||
| Yes (5) | 0.64 (0.44–0.83) | 0.011 | 0.30 (0.19–0.49) | 0.314 | |||
| No (4) | 1.51 (0.71–3.22) | < 0.01 | 1.23 (0.41–3.65) | 0.004 | |||
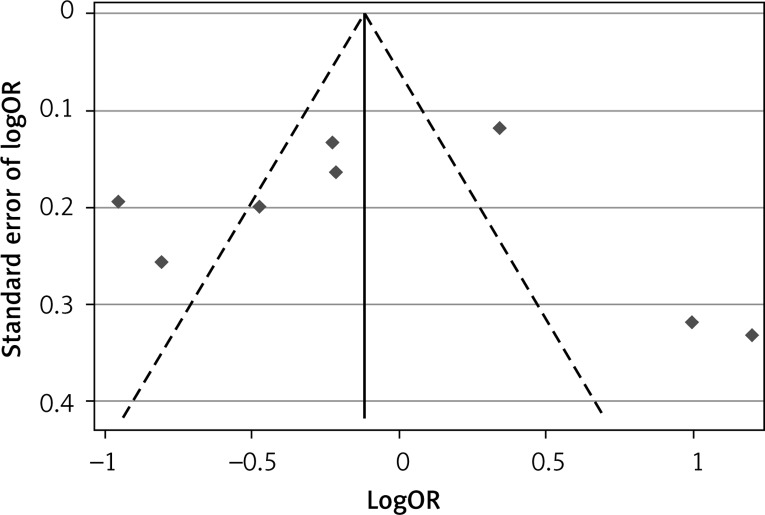
Publication bias
For the APOA1-75G/A polymorphisms, the shape of funnel plots showed no obvious asymmetry and the result of Egger’s test did not show statistical evidence for bias either (Figure 4).
Figure 4.
Funnel plot for publication bias test of the APOA1-75G/A polymorphism and CAD
Discussion
This is the first meta-analysis investigating the association between the ApoA1 polymorphisms and CAD. In this meta-analysis, 9 eligible studies including 1864 cases and 1196 controls were identified and analyzed. A positive relationship between APOA1-75G/A polymorphism and the risk of CAD was identified in the recessive model. Additionally, in the subgroup analysis for controls consistent with HWE, this association was significant under all the models except the heterozygote model. In conclusion, the minor allele of the APOA1-75G/A polymorphism is a protective factor for CAD, especially in the studies consistent with HWE.
Coronary artery disease is a complicated, polygenic disease driven by the coactions of various environmental and genetic factors, of which hereditary factors probably play a key role in disease development. ApoA1 is a component of HDL. High-density lipoprotein is a molecule that transports cholesterol and certain fats called phospholipids through the bloodstream from the body’s tissues to the liver. High-density lipoprotein is often referred to as “good cholesterol” because high levels of this substance reduce the chances of developing heart and blood vessel (cardiovascular) disease [39].
Up to now, several single-nucleotide polymorphisms (SNPs) have been identified in the APOA1 gene located on the long arm of chromosome 11 [40]. A common G-to-A transition located –75 base pairs (bp) upstream from the transcription start site of the APOA1 gene has been studied extensively [41]. The gene mutation might influence the individual susceptibility to CAD. The base changes from G to A at the –75 bp site increase circulating levels of ApoA1 and HDL-C, and the individuals with these changes are likely to have a lower risk of developing CAD [42]. Thus, there is a potential mechanism by which the minor allele of the APOA1-75G/A polymorphism is a protective factor for CAD.
Significant heterogeneity was found for the association between the ApoA1 polymorphisms and CAD in all the models. However, when stratified by status of HWE, the positive association still existed, but heterogeneity losses were found in controls consistent with HWE. Four studies deviating from the HWE have been included in the present meta-analysis; after omitting these studies, the pooled ORs became significant in four genetic models without evidence of heterogeneity. These disqualified studies may be potential sources of heterogeneity across studies for such differences.
The present study has some limitations. First, though we collected all eligible studies, the number of qualified studies was not large. Second, four studies did not conform to HWE. Third, although we designed our study to evaluate the effects of environmental modification such as smoking, alcohol intake, physical activities, and diet, few investigators have reported the effects of these environmental factors, and the definition of each stratum varied too much among studies. We failed to analyze modification of the effects of this polymorphism by environment factors.
In spite of the limitations, our meta-analysis has some key advantages. First, the results should be more reliable than those from a single study, as cases and controls were pooled from different studies and statistical power of the analysis was significantly increased. Second, no publication bias was found. Sensitive analyses conducted by deselecting studies one by one in chronological order revealed no significant changes or reversal of results, which suggested that the result of the present meta-analysis was stable and reliable.
In conclusion, this meta-analysis suggests that the minor allele of the APOA1-75G/A polymorphism is a protective factor for CAD, especially in the studies consistent with HWE. Primary studies of a large population are required to further evaluate gene-gene and gene-environment interaction effects of this polymorphism on CAD risk in different ethnicities.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81170174), Natural Scientific Fund of Jiangsu province (BK20161226) and Jiangsu Province’s Key Provincial Talents Program (RC2011111). The funders had no roles in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have declared that no competing interests exist.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Yusuf S, Hawken S, Ounpuu S, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–52. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 2.Zou JG, Ma YT, Xie X, et al. The association between CYP1A1 genetic polymorphisms and coronary artery disease in the Uygur and Han of China. Lipids Health Dis. 2014;13:145. doi: 10.1186/1476-511X-13-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaefer LE, Nechemias C. Endogenous hormones, lipid metabolism, and coronary artery disease. Prog Cardiovasc Dis. 1965;7:449–64. doi: 10.1016/s0033-0620(65)80038-x. [DOI] [PubMed] [Google Scholar]
- 4.van Hateren KJ, Bilo HJ. Hypertension control and cardiovascular outcomes among patients with diabetes and coronary artery disease. JAMA. 2010;304:1672. doi: 10.1001/jama.2010.1464. [DOI] [PubMed] [Google Scholar]
- 5.Dunn JP, Ipsen J, Elsom KO, Ohtani M. Risk factors in coronary artery disease, hypertension and diabetes. Am J Med Sci. 1970;259:309–22. doi: 10.1097/00000441-197005000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Turner RC, Millns H, Neil HA, et al. Risk factors for coronary artery disease in non-insulin dependent diabetes mellitus: United Kingdom Prospective Diabetes Study (UKPDS: 23) BMJ. 1998;316:823–8. doi: 10.1136/bmj.316.7134.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connelly PW, Stachenko S, MacLean DR, Petrasovits A, Little JA. The prevalence of hyperlipidemia in women and its association with use of oral contraceptives, sex hormone replacement therapy and nonlipid coronary artery disease risk factors. Canadian Heart Health Surveys Research Group. Can J Cardiol. 1999;15:419–27. [PubMed] [Google Scholar]
- 8.Li XL, Hong LF, Luo SH, et al. Impact of admission triglyceride for early outcome in diabetic patients with stable coronary artery disease. Lipids Health Dis. 2014;13:73. doi: 10.1186/1476-511X-13-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita H, Taguchi J, Kurihara H, et al. Genetic polymorphism of 5,10-methylenetetrahydrofolate reductase (MTHFR) as a risk factor for coronary artery disease. Circulation. 1997;95:2032–6. doi: 10.1161/01.cir.95.8.2032. [DOI] [PubMed] [Google Scholar]
- 10.Neutze J, Cormack B, Kerr B, Pepper L, Ramage C. Contribution of a risk factor clinic to lipid management in patients with coronary artery disease. N Z Med J. 1992;105:97–9. [PubMed] [Google Scholar]
- 11.Gruzdeva O, Uchasova E, Dyleva Y, et al. Multivessel coronary artery disease, free fatty acids, oxidized LDL and its antibody in myocardial infarction. Lipids Health Dis. 2014;13:111. doi: 10.1186/1476-511X-13-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon DJ, Probstfield JL, Garrison RJ, et al. High density lipoprotein cholesterol and cardiovascular disease: four prospective American studies. Circulation. 1999;79:8–15. doi: 10.1161/01.cir.79.1.8. [DOI] [PubMed] [Google Scholar]
- 13.Wilson PW, Abbott RD, Castelli WP. High density lipoprotein cholesterol and mortality: the Framingham Heart Study. Arteriosclerosis. 1998;8:737–41. doi: 10.1161/01.atv.8.6.737. [DOI] [PubMed] [Google Scholar]
- 14.Ballantyne CM, Hoogeven RC. Role of lipid and lipoprotein profiles in risk assessment and therapy. Am Heart J. 2003;146:227–33. doi: 10.1016/S0002-8703(02)94701-0. [DOI] [PubMed] [Google Scholar]
- 15.Elshazly MB, Quispe R, Michos ED, et al. Patient-level discordance in population percentiles of the total cholesterol to high-density lipoprotein cholesterol ratio in comparison with low-density lipoprotein cholesterol and non-high-density lipoprotein cholesterol: the Very Large Database of Lipids Study (VLDL-2B) Circulation. 2015;132:667–76. doi: 10.1161/CIRCULATIONAHA.115.016163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolic D, Rizzo M, Mikhailidis DP, Wong NC, Banach M. An evaluation of RVX-208 for the treatment of atherosclerosis. Expert Opin Investig Drugs. 2015;24:1389–98. doi: 10.1517/13543784.2015.1083010. [DOI] [PubMed] [Google Scholar]
- 17.Otocka-Kmiecik A, Mikhailidis DP, Nicholls SJ, Davidson M, Rysz J, Banach M. Dysfunctional HDL: a novel important diagnostic and therapeutic target in cardiovascular disease? Prog Lipid Res. 2012;51:314–24. doi: 10.1016/j.plipres.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Fielding CJ, Shore VG, Fielding PE. A protein co-factor of lecithin: cholesterol acyl transferase. Biochem Biophys Res Commun. 1972;46:1493–8. doi: 10.1016/0006-291x(72)90776-0. [DOI] [PubMed] [Google Scholar]
- 19.Karathanasis SK. Apolipoprotein multigene family: tandem organization of human apolipoprotein AI, CIII, and AIV genes. Proc Natl Acad Sci USA. 1985;82:6374–8. doi: 10.1073/pnas.82.19.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assmann G, von Eckardstein A, Funke H. High density lipoproteins, reverse transport of cholesterol, and coronary artery disease. Insights from mutations. Circulation. 1993;87(Suppl III):III28–34. [PubMed] [Google Scholar]
- 21.Groenendijk M, Cantor RM, de Bruin TW, Dallinga-Thie GM. The ApoAI-CIII-AIV gene cluster. Atherosclerosis. 2001;157:1–11. doi: 10.1016/s0021-9150(01)00539-1. [DOI] [PubMed] [Google Scholar]
- 22.Wang XL, Liu SX, McCredie RM, Wilcken DE. Polymorphisms at the 5’-end of the apolipoprotein AI gene and severity of coronary artery disease. J Clin Invest. 1996;98:372–7. doi: 10.1172/JCI118802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou Y, Hu D, Yang X, et al. Relationships among apolipoprotein A1 gene polymorphisms, lipid levels and coronary atherosclerosis disease. Chin Med J (Engl) 2003;116:665–8. [PubMed] [Google Scholar]
- 24.Reguero JR, Cubero GI, Batalla A, et al. Apolipoprotein A1 gene polymorphisms and risk of early coronary disease. Cardiology. 1998;90:231–5. doi: 10.1159/000006849. [DOI] [PubMed] [Google Scholar]
- 25.Shengli Y, Bingxian H, Xiufang H, Yangchun Z, Mingjun L. Apolipoprotein AI gene polymorphisms and coronary artery disease in the Chinese Xinjiang Uygur and population. South Chin J Cardiol. 2001;2:69–74. [Google Scholar]
- 26.Chhabra S, Narang R, Lakshmy R, et al. APOAl-75G to A substitution associated with severe forms of lower levels of HDL and apoA-I among northern Indians. Dis Markers. 2005;21:169–74. doi: 10.1155/2005/195078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rai TS, Khullar M, Sehrawat BS, et al. Synergistic effect between apolipoprotein E and apolipoprotein A1 gene polymorphisms in the risk for coronary artery disease. Mol Cell Biochem. 2008;313:139–46. doi: 10.1007/s11010-008-9751-3. [DOI] [PubMed] [Google Scholar]
- 28.Dawar R, Gurtoo A, Singh R. Apolipoprotein A1 gene polymorphism (G-75A and C+83T) in patients with myocardial infarction: a pilot study in a north Indian population. Am J Clin Pathol. 2010;134:249–55. doi: 10.1309/AJCPKPTXQ3QN1IFG. [DOI] [PubMed] [Google Scholar]
- 29.Ding Y, Zhu MA, Wang ZX, Zhu J, Feng JB, Li DS. Associations of polymorphisms in the apolipoprotein APOA1-C3-A5 gene cluster with acute coronary syndrome. J Biomed Biotechnol. 2012;2012:509420. doi: 10.1155/2012/509420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xuebiao W, Yaowang L, Wing Y, Danqing Y. Association between apolipoprotein A1-75 bp gene polymorphisms and risk for dyslipdemia and coronary artery disease. Chin J Cardiol. 2014;42:916–21. [PubMed] [Google Scholar]
- 31.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–14. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 33.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101. [PubMed] [Google Scholar]
- 35.Egger M, Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindley D. Statistical inference concerning Hardy-Weinberg equilibrium. Bayesian Stat. 1988;3:307–26. [Google Scholar]
- 37.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101. [PubMed] [Google Scholar]
- 38.Egger M, Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toth PP, Barylski M, Nikolic D, Rizzo M, Montalto G, Banach M. Should low high-density lipoprotein cholesterol (HDL-C) be treated? Best Pract Res Clin Endocrinol Metab. 2014;28:353–68. doi: 10.1016/j.beem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Karathanasis SK. Apolipoprotein multigene family: tandem organization of human apolipoprotein AI, CIII, and AIV genes. Proc Natl Acad Sci U S A. 1985;82:6374–8. doi: 10.1073/pnas.82.19.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saha N, Tay JS, Low PS, Humphries SE. Guanidine to adenine (G/A) substitution in the promoter region of the apolipoprotein AI gene is associated with elevated serum apolipoprotein AI levels in Chinese non-smokers. Genet Epidemiol. 1994;11:255–64. doi: 10.1002/gepi.1370110304. [DOI] [PubMed] [Google Scholar]
- 42.Colantonio LD, Bittner V, Reynolds K, et al. Association of serum lipids and coronary heart disease in contemporary observational studies. Circulation. 2016;133:256–64. doi: 10.1161/CIRCULATIONAHA.115.011646. [DOI] [PMC free article] [PubMed] [Google Scholar]