Abstract
Background
Structure profiling experiments provide single-nucleotide information on RNA structure. Recent advances in chemistry combined with application of high-throughput sequencing have enabled structure profiling at transcriptome scale and in living cells, creating unprecedented opportunities for RNA biology. Propelled by these experimental advances, massive data with ever-increasing diversity and complexity have been generated, which give rise to new challenges in interpreting and analyzing these data.
Results
We review current practices in analysis of structure profiling data with emphasis on comparative and integrative analysis as well as highlight emerging questions. Comparative analysis has revealed structural patterns across transcriptomes and has become an integral component of recent profiling studies. Additionally, profiling data can be integrated into traditional structure prediction algorithms to improve prediction accuracy.
Conclusions
To keep pace with experimental developments, methods to facilitate, enhance and refine such analyses are needed. Parallel advances in analysis methodology will complement profiling technologies and help them reach their full potential.
Keywords: RNA structure profiling, high-throughput sequencing, RNA secondary structure prediction, chemical structure probing, SHAPE-Seq
INTRODUCTION
RNAs are known to play essential roles in diverse cellular functions, extending well beyond the transfer of information from genes to proteins [1,2]. For example, small non-coding RNAs such as microRNAs and small interfering RNAs have regulatory roles in gene expression [3]. Long non-coding RNAs are also widely found in various regulatory roles at both transcriptional and post-transcriptional levels [4]. RNA function is closely linked with its ability to fold into and convert between specific complex structures. In fact, determining structure has become a crucial step in understanding RNA function [5]. Accurate and high-resolution structure models have been traditionally obtained using comparative sequence analysis or experimental techniques, such as X-ray crystallography and nuclear magnetic resonance (NMR) [6]. However, these methods require considerable manual labor and suffer technological limitations, which have precluded their use beyond a small scale [7]. Computational structure prediction from sequence information is a broadly applicable alternative that has been widely used [8,9], but reported structures often suffer from poor accuracy.
Structure profiling (SP), also known as structure probing or chemical probing, refers to a family of experiments that characterize RNA structure [10,11]. In these experiments, local structural characteristics are gleaned using structure-sensitive reagents that modify RNAs at nucleotide level. Well-studied reagents include dimethyl sulfate (DMS) [12], kethoxal [13], hydroxyl radicals [14], diethyl pyrocarbonate (DEPC) [15], CMCT [16], lead (II) [17,18], nucleases [19] and SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) [20]. Until very recently, limitations of probing reagents as well as sequencing and informatics challenges restricted SP to select few RNAs studied individually and primarily under in vitro conditions. The newest generation of SP experiments utilizes high-throughput sequencing techniques, which provide unprecedented multiplexing capacity in a cost-effective and automated manner. These advances have been used to study RNAs of varying lengths in vitro and in vivo, and more recently at transcriptome scale [21–42]. Despite shared principles, experiments differ in the information they extract and in the statistical properties of their measurements. Experimental protocols for SP and their biological applications have been reviewed previously; see, for example [11,43– 46].
Sequencing readouts from SP experiments are analyzed to extract structural parameters of interest for each nucleotide, in terms of its reactivity to the probing reagent. Nucleotide-level reactivity estimates are subsequently used to answer biological questions of interest, which may entail further analysis and interpretation. In this article, we focus on approaches to using reactivity data for comparative and integrative analysis — a central theme in recent studies. Comparative analysis of SP data has revealed structural patterns across different levels, ranging from low-resolution transcriptome level to high-resolution nucleotide level. Each level may warrant specialized analysis methods. Note that even at the same level, the ideal approach could possibly differ depending on the context and questions asked. We discuss three different contexts where technical, biological and systematic replicates of SP data are available. In addition to comparative analysis, we also review current progress in data-directed structure prediction, which is the most straightforward application of SP data in structural RNA biology. Unlike X-ray crystallography and NMR, in which RNA structure is explicitly modeled, SP does not directly reveal the pairing state of a nucleotide nor its pairing partner. However, it can complement structure prediction algorithms to enhance their performance [47,48].
This review is organized as follows. We begin with a discussion of shared principles of SP experiments and devote the bulk of the article to their data interpretation and analysis. We review current practices and principles in reactivity calculation. We then discuss recent approaches and emerging questions in comparative and integrative analysis, followed by discussion of quality control in large-scale SP datasets. Next, we review algorithms for secondary structure prediction and efforts to leverage SP data to improve their performance. Recent progress in public repositories, analysis tools and visualization platforms is also described.
OVERVIEW OF STRUCTURE PROFILING EXPERIMENTS
The general goal of an SP experiment is to obtain nucleotide-resolution structural characteristics of all RNAs in a sample [49]. Structural characteristics in the vicinity of a nucleotide are reflected in local stereochemical properties such as nucleotide dynamics, solvent accessibility and electrostatic environment [11,50]. In particular, pairing state of a nucleotide is known to be correlated with these stereochemical properties [51]. SP experiments utilize reagents that are sensitive to local stereochemistry [11]. These reagents react with nucleotides such that the reactivity to any particular nucleotide depends on its local stereochemistry, which in turn is affected by its pairing state. SP experiments aim to measure the sequence of reactivities corresponding to nucleotides of each transcript. High and low reactivities are indicative of unpaired and paired nucleotides, respectively [52]. Hence, it is understood that the sequence of nucleotide reactivities, henceforth called reactivity profile, is a representation of a transcript’s structure [53].
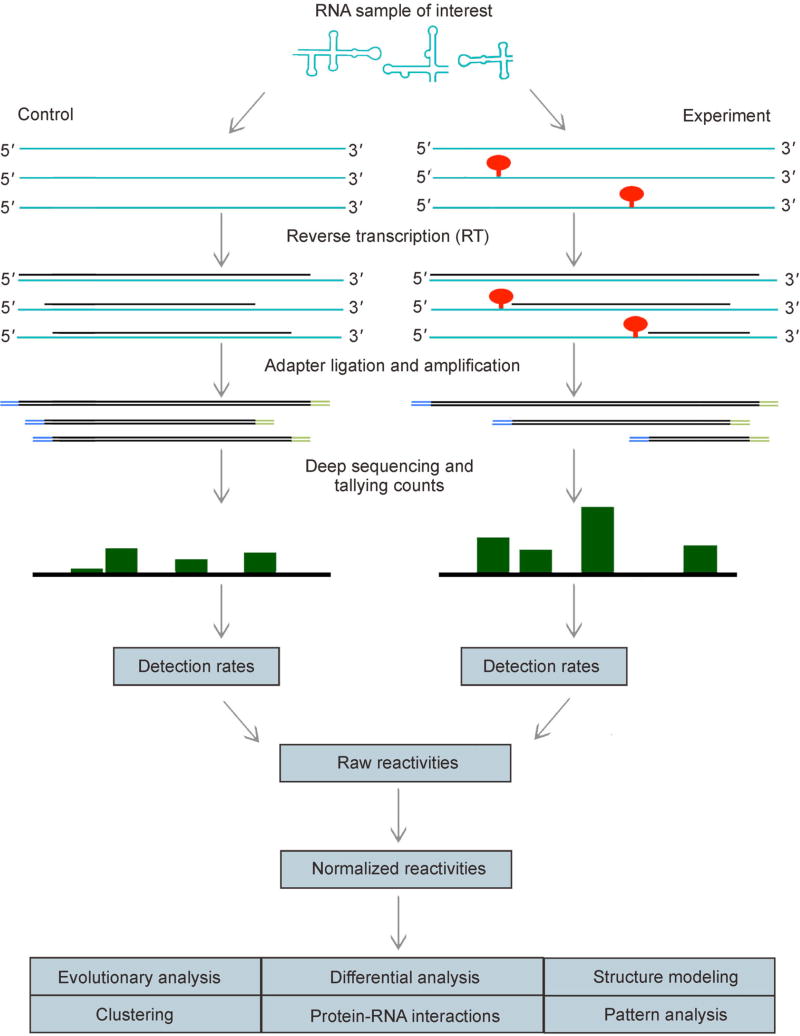
Most sequencing-based SP techniques share a common workflow (Figure 1) [43,44]. To start with, a sample of RNAs is allowed to react with a structure-sensitive reagent, resulting in chemical modifications of nucleotides. The degree of modification at each nucleotide is detected by reverse transcription (RT), which either stops or proceeds but introduces a mutation at modified nucleotides. The resulting cDNA library is sequenced and reads are mapped to target RNA sequences. Then, RT stops or mutations are counted for each nucleotide. To measure background noise in RT stops or mutations, parallel to the experiment, a control assay is similarly performed wherein the RNAs are not treated with reagents. This control assay also yields a stop or mutation count summary for each nucleotide. Counts from experiment and control assays are then combined to obtain reactivity profiles for all RNAs in the sample.
Figure 1. Overview of structure-profiling experiments.
RNA sample of interest (at the top) is probed with a structure-sensitive reagent, which introduces a modification (red pins) preferentially at unpaired nucleotides. Degree of modification is read via reverse transcription and sequencing. Next, the readouts are mapped to reference sequences and normalized reactivities are calculated from counts summary of mapped reads. Reactivity profiles of probed RNAs are used in diverse downstream applications, some of which are listed.
Despite these shared principles, measured reactivities are influenced by numerous intertwined factors that all impact the variability of readouts [54]. In fact, it has been found that single nucleotide variants can lead to substantially different reactivity profiles [55,56] and that identical sequences can have different reactivity profiles under different conditions [40,57,58]. Comparison of reactivity profiles reveals that quantitative differences persist even in the absence of structural differences between RNAs from one sample to another [54,59]. Listed below are factors that influence reactivity profiles.
Technical factors
Numerous technical factors add to variability in observed profiles. First, chemical reactions involved in SP occur in presence of limited quantities of reagents/transcripts. Concentrations of reagents are often controlled deliberately to limiting amounts to achieve desirable reaction kinetics [11]. In addition, many RNAs of interest are present in limited quantities [28]. As such, the reactions feature inherent stochasticity [54,60]. Secondly, these reactions are sensitive to stereochemistry and solvent conditions [11,61]. Nevertheless, they often occur in complex and dynamic solution environments. For example, RNAs often feature a dynamic ensemble of co-existing structures in vivo interacting with proteins and other biomolecules [62–64]. However, SP captures only the aggregate profile for all these structures, combining influences from intermolecular interactions [65]. In addition, cDNA library preparation involves numerous steps such as adapter ligation, reverse transcription and PCR, which also give rise to stochasticities. Finally, readouts from sequencing machines are also affected by stochasticities [54,66,67]. These factors collectively contribute to variance in reactivity profiles. In fact, they contribute to variance in any other parameter of interest that is estimated from data, e.g., Gini index of counts/reactivities [26,40,68]. Variance contribution of technical factors to any parameter of interest can be estimated by performing multiple replicates, called as technical replicates of experiment-control study, starting from biologically indistinguishable RNA samples. We refer to variance in estimates observed purely due to said technical factors as technical variation [69–71].
Biological factors
RNAs with significant structural diversity have been subjects of recent studies. For example, non-coding RNAs (ncRNA) are known to be highly structured, while mRNAs are thought to have a lesser degree of structure. Moreover, within an RNA, structure could significantly vary from one region to another. For example, mRNAs are believed to be less structured in protein coding regions than in untranslated regions [40]. Additionally, RNA structure is sensitive to factors such as solvent conditions, ligand and salt concentrations, temperature variations and interactions with proteins [61]. Should any of these factors differ between studies, detectable differences in the estimated reactivities may be observed. For example, reactivity profiles for the same transcript have been found to differ between in vitro and in vivo conditions [40,68,72]. We refer to variance of an estimate observed purely due to biological factors as biological variation. Additionally, it is to be noted that biological variation might be caused by differences in RNA-protein interactions besides structural differences [73]. Proteins can cover certain stretches of nucleotides, thereby influencing their reactivities to certain reagents. Two RNA samples known to have come from different biological sources are called biological replicates [69–71]. These contain information about biological differences between the samples.
Systematic factors
For biologically identical RNAs, reactivity measurements obtained in one experiment can differ from the profiles obtained through a different experimental protocol [53,74]. Technical replicates do not capture these variations, as they do not differ in protocol steps. Yet, such variations do not originate due to biological factors but rather can be attributed to discrepancies in key steps. For example, many current methods differ in choice of probing reagent. In fact, a variety of reagents are available, such as DMS, kethoxal, hydroxyl radical, 1M7, NMIA, NAI and NAI-N3, but each has its pros and cons [11,22,40,75]. These reagents differ in their stereochemical characteristics and reaction mechanisms. Consequently, reactivity profiles may reflect these differences. In addition, many reagents do not probe all nucleotides as well as feature biases that cause different reactivities, depending on nucleotide type, even in the absence of structural differences [11]. Besides choice of probing reagent, protocols often differ in priming method, modification detection approach (e.g., stop/mutation), ligation strategy, enrichment scheme, sequencing mode (single/paired-ended), and reactivity estimation method among others. These are a few noteworthy steps having equally plausible alternatives. Many of these steps contribute to biases, which interplay with other steps to result in miscellaneous effects on parameter estimates [54]. Nevertheless, biologically identical RNAs can be studied using different protocols to obtain detailed and comprehensive insights [74]. We refer to experiments involving SP of biologically indistinguishable samples using different protocols as systematic replicates and variances originating due to differences in protocols as systematic variation.
ESTIMATION OF STRUCTURAL PROFILE
As mentioned earlier, sequenced reads from both experiment and control assays are summarized as count of stops or mutations for each nucleotide. However, pernucleotide counts are not directly comparable because they can differ in magnitude due to a variety of factors. The number of reads mapped to a transcript, also known as its coverage, varies between transcripts due to dramatic differences in their relative abundances, which often range over five orders of magnitude [28,76]. Additionally, priming or ligation biases contribute to sequence-specific variations in counts within the same transcript [22,54,59,77,78]. Counts may also differ due to background noise in RT stops and mutations. In fact, for the same nucleotide between experiment and control, counts may not be comparable due to difference in sequencing depths. For these reasons, counts are processed into normalized reactivities, which are assumed to be comparable across transcripts and replicates.
Reactivity estimation methods differ between studies but share the following conceptual framework (see Figure 1). First, counts are adjusted to account for variations in coverage, yielding two detection rates per nucleotide — one for experiment and one for control. Second, comparison of detection rates yields an estimate of the degree of modification, or raw reactivity. Third, raw reactivities are normalized to ensure that values for all transcripts and replicates thereof span the same interval.
Detection rates
Detection rates are calculated to account for variations in coverage. Notably, variations in coverage exist at all levels. For example, substantial coverage differences have been noted between rRNAs and many mRNAs [28]. Significant differences in coverage also exist from one transcript to another within the same functional class. Additionally, within a transcript, coverage can be considered on regional basis (e.g., coverage of the 5′ untranslated region or the coding region, or 3′ end, etc.), sequence basis (e.g., more coverage in GC rich regions due to priming bias), or per-nucleotide basis. In general, coverage differences can be noticed at all levels of organization. Analysis methods in various studies differ in the level of detail at which they account for coverage variations. Many studies consider coverage variations between transcripts as significant while assuming uniformity of coverage within each transcript. Higher coverages for a transcript may be a result of its over-abundance in the sample or of priming biases among other factors. In such cases, counts corresponding to nucleotides of the transcript may be assumed to be proportionally higher. Hence, several studies adjust counts by their mean to account for coverage bias [28,30,31,35,36,55]. Additionally, Ding et al. [28] take the logarithm of counts to make count distribution symmetric. Others note that there could be local biases within the transcripts. For example, Rouskin et al. [26] adjust counts for each nucleotide by maximum counts in a local window. In fact, several studies [22,25,40,60,79,80] have accounted for nucleotide-level coverage variations. Through these adjustments, detection rates are estimated for both experiment and control.
Raw reactivities
Detections in control are attributed to noise in RT while detections in experiment arise from noise in RT as well as from modifications at nucleotides. Hence, it is expected that at any nucleotide, detection rate will be higher in experiment. One core assumption is that structure-sensitive modifications contribute additively to a background level of detection rates. Reactivities are therefore often calculated by subtracting detection rate in control from that in experiment [22,28,40,60,80]. Alternatively, reactivities have been estimated as odds ratio of experiment to control detection rates [35]. To control the range of reactivities, others take the logarithm of the odds ratio [30,31,36,55,81]. Occasionally, detection rates in experiment are found to be less than their counterparts in control. In such cases, a basal reactivity value of 0 (if subtracting detection rates) or 1 (if taking ratio) is assigned. This is done because the detection rate due to noise is often very low and if detection rates remain comparable or lower in the presence of modifications, it indicates negligible degree of modification.
Normalized reactivities
Profiles from different protocols could span disjoint intervals even for the same RNA. In fact, for different RNAs in the same experiment, profiles could span disjoint intervals because of biological variation. Raw reactivities are not considered comparable in absolute magnitude. Hence, all profiles are normalized such that the average reactivity of approximately 10% of the most reactive nucleotides is 1, excluding few unusually reactive nucleotides that are considered outliers [47]. Outliers can originate in datasets due to a variety of reasons, such as excessive degradation or over-modification at certain nucleotides, or over-representation of certain fragments due to various inherent biases in protocols. In fact, such hyper-reactive sites often appear in datasets [51,82].
Accordingly, most current approaches to normalization begin with identification of outliers in reactivity estimates [83]. This is done by either box plot analysis whereby reactivities greater than 1.5 times the interquartile range are deemed outliers [47,82], or by assuming that reactivities beyond a certain percentile are outliers [47]. Outliers are either ignored [47] in the process of calculating normalization constant or winsorized [21,26,36]. To estimate a normalizing constant, one approach is to take the mean of values greater than a certain percentile after removing outliers. For example, 2%–8% method assumes that the top 2% of reactivities are outliers and normalizes with mean of the next 8% of highest reactivities [47]. The winsorization approach aims to scale reactivities such that they range from 0 to 1 for all transcripts. Hence, after winsorization, the highest reactivity is chosen as the normalizing constant [21,26,36].
In the majority of analysis methods, the above workflow is preceded by conventional read alignment and counting routines. Recently, these pre-processing steps were integrated with reactivity estimation, such that counting and estimation are resolved simultaneously [79]. This is especially attractive in situations where multimapping reads (that is, reads which align to multiple sites in a transcriptome) abound, e.g., in studies of splicing isoforms. While common remedies discard such reads or allocate them uniformly among plausible alignment sites, Li et al. [79] expand on prior modeling and statistical inference work in RNA-Seq [84,85] and in SHAPE-Seq analyses [80] to address this issue. Another extension of the said statistical modeling work on SHAPE-Seq has been recently published by Selega et al. [81]. This method scores significance of modification levels from stop counts and nucleotide-level coverages under an assumption that modification states do not randomly switch, i.e., significantly reactive/unreactive nucleotides tend to appear in continuous stretches. The assumption is enforced using a Hidden Markov Model with transition probabilities based on empirically derived expected lengths of reactive and unreactive contiguous stretches in a training dataset.
COMPARATIVE ANALYSIS
Before the advent of high-throughput sequencing, probing was mostly applied to select highly structured ncRNAs under in vitro conditions. Recent advances have dramatically expanded the scope of SP and diverse RNAs can now be studied in biologically relevant conditions. In fact, applications of SP to numerous transcripts and transcriptomes have revealed novel insights [2,44]. Most such applications feature comparative analysis. Several recent examples of such analysis can be noted: i) Spitale et al. [40] compared mRNA profiles and identified conserved patterns around translation start sites. ii) Protein-RNA interactions were studied in viral RNA and mammalian ncRNAs and mRNAs by comparing reactivity profiles under different conditions [40,58,86,87], finding that interactions modulate reactivities significantly. iii) Comparison of mRNA coding regions revealed a three-nucleotide periodicity pattern in reactivities [28,30,40]. iv) Significant structural alterations have been identified in single-nucleotide variants [55,88]. v) Comparisons of entire transcriptomes at different temperatures identified structure-altering responses [26,89,90]. vi) Prevalence of specific noncanonical structure motifs has been found to differ between in vitro and in vivo conditions [68]. Interestingly, these studies involve comparisons at different levels such as structure at the level of regions within a transcript, at the transcript level, within functional classes, or at transcriptome level. In this section, we review recent approaches and emerging questions in addressing these challenges.
Notably, SP collects data at nucleotide level, but structural dynamics most often involve at least a few nucleotides or even entire functional domains. For example, many of the studies mentioned above seek signals that span protein-binding sites, codons and well-defined local structure motifs. Indeed, it is rare for a biological study to home in on isolated single-nucleotide reactivity changes. For this reason, comparative studies must also bridge between the resolution of measurements and that of sought-after effects. This is typically accomplished by integrating nucleotide information for scoping differential structural effects at various levels of lower resolution and/or by inspecting data-directed secondary structure predictions for detectable changes at that level [40,53,56,91].
Comparing technical replicates
Agreement between technical replicates indicates high quality of data. Technical replicates can be compared at the level of transcripts or at the level of nucleotides.
Transcript-level comparison
In high-throughput experiments or when profiling long transcripts, agreement between replicates of a transcript is commonly evaluated as Pearson correlation coefficient (PCC) for reactivity profiles. Transcripts with low PCC are filtered out for biological purposes, as their replicates do not agree. For each pair of profiles, PCC quantifies agreement in a single number that is invariant to scaling. However, PCC has its limitations as a measure of agreement [92–94].
First, PCC is sensitive to outliers [92]. PCC is based on the sample means of reactivities in the profiles that are being compared. Sample means are known to be sensitive to outliers, leading to similar sensitivity of PCC. Indeed, PCC is affected by both magnitude of outliers and the overall proportion of reactivities that is outlier. Hence, PCC is to be used with caution, especially for transcriptome-wide data, as outliers have indeed been routinely noted in experiments [47,59]. In our experience, we have found that a common practice in handling missing information often leads to outliers in reactivities. Specifically, while estimating reactivity profiles, poorly covered sites have a bias towards an apparent zero reactivity. This bias considerably adds to the proportion of outliers at the lower extreme of zero reactivity. However, most studies do not filter outliers while calculating PCC. Hence, PCC may be misleading in evaluating replicate agreement. Second, PCC does not quantify agreement at nucleotide level but rather summarizes it across a transcript. Third, PCC only evaluates correlation between two profiles and is unaffected by magnitude differences of nucleotide-level values. Nevertheless, to gauge significance of biological variation found in a study, it is important to first quantify technical variation. Since biological variation of interest is often manifested at nucleotide resolution, it is also desirable to quantify technical variation at that resolution.
Nucleotide-level comparison
At nucleotide level, replicates have been traditionally compared by taking mean and standard deviation of reactivities. In the absence of replicates, theoretical formulas and computational methods have been developed to evaluate technical variation at each nucleotide [22,59]. However, due to challenges in visualizing technical variation, most such nucleotide-level evaluations have been restricted to one or few transcripts. Recently, Choudhary et al. [59] proposed a method to quantify and visualize technical variation at nucleotide resolution for large-scale data, based on the classical signal-to-noise ratio (SNR) measure. For each nucleotide, SNR is defined as the ratio of sample mean to standard deviation of reactivities in all replicates. SNR is high when replicates are in strong quantitative agreement at a nucleotide and low otherwise. Nucleotide SNR values within a transcript could be visualized as box plot to glean overall agreement among multiple replicates from a single plot. Additionally, mean of SNR was proposed as a single-number or point summary for a transcript’s overall data quality. Mean SNR per transcript was found to correlate well with PCC and transcript coverage in diverse datasets.
Open questions
Nucleotide-resolution comparison of reactivities requires normalization strategies to render values in different replicates comparable. Clearly, the strategies described in the Section of “Estimation of Structural Profile” require optimizing two criteria: one for identifying outliers and another for selecting reactivities that will be used to estimate a normalizing constant. However, the proportion of outliers in a dataset could vary considerably depending on the length of transcripts involved, the protocol used and the experiment’s quality. Indeed, different labs and even same labs have made different choices of normalization steps when analyzing different datasets, although the general principle has been to eliminate outliers and scale reactivities such that they range approximately from 0 to 2 [39]. These strategies have been adopted based on experience with SP data before high-throughput technologies were introduced [47] or through validations with secondary structure prediction [82]. Hence, the field may benefit from a universal approach to normalization, which is assuring enough to dispense with the need for routine optimization of the normalization step. It is also worth noting that before SP became high-throughput, most of the RNAs that were chemically probed were highly structured rRNAs or short functional ncRNAs. Heuristic guidelines formulated based on such a specialized subset may not be ideally suited to all transcripts — in particular to long and less structurally constrained mRNAs. Furthermore, validation based on structure prediction itself involves parameter optimization and modeling assumptions, as reviewed in later sections. Given the recent advances in SP, methods of normalization warrant a revisit and possibly even generalization or standardization.
Comparing biological replicates
Comparison of reactivities from different biological replicates could potentially identify significant biological variation. If technical variation is high, statistically significant biological results might not be obtained from the data. To estimate significance of biological variation, it has to be examined in comparison with technical variation [69–71]. Recently, several studies have reported biological variation at all levels. At transcriptome level, differences in overall structural characteristics have been reported under different conditions and between different strains [26,40]. At transcript-to-transcript level, rRNAs have been described as being more structured than mRNAs. At a finer level, while differences in reactivities can be observed at nucleotide level, biological variation is commonly assumed to span a stretch of nucleotides [86]. In particular, within transcripts, biological variation has been described between regions, where significant differences in structure have been noted between UTRs and coding regions of mRNAs. Here, we review the methods used to measure biological variation.
Transcriptome-level comparison
Current normalization methods, as described in the Section of “Estimation of Structural Profile”, generally scale the reactivities such that they range from 0 to approximately 2 [39]. However, this does not ensure that reactivities within different transcripts are directly comparable. For example, although mRNAs are widely understood to be less structured than rRNAs [40], current normalization methods scale reactivities for both these classes of RNA such that they span a similar interval. Hence, comparing absolute values of reactivities on a transcriptome scale might be misleading. Differences in lengths of transcripts within the same functional class exacerbate the challenges in comparing profiles due to the need for reliable alignment. To facilitate nucleotide-level comparison of reactivities in case of differences in lengths, particularly for mRNAs, transcripts are often aligned by their start/stop codon, where arbitrary lengths (about 40–100 nt) are chosen upstream and downstream of the start/stop codon in all transcripts to be compared [28,30,36,40,89]. However, functional elements in UTRs differ in sequence and distance from the start/stop codon, thus presenting an additional challenge to direct comparisons.
Besides direct nucleotide-level comparison, another approach has been utilized, which is invariant to current normalization methods (due to properties as listed below) as well as applicable to transcripts of different lengths. At the transcriptome level, it has been found that RNAs are, in general, less structured in vivo than they are in vitro [40]. This conclusion was obtained by examining distributions of Gini indices for reactivity profiles. Gini index is a measure of inequality in a distribution [95]. It has two notable properties: i) It is a measure of inequality that is high if there is substantial gap in values across the nucleotides. Such high gaps (or inequalities) in distribution of counts and reactivities are expected in case of structured RNAs. Hence, Gini index can serve to quantitatively describe the overall degree of structure in a transcript. ii) It is invariant to scaling, i.e., Gini index does not change as long as the relative magnitudes of quantities remain the same. As current normalization methods essentially scale reactivity profiles linearly, scaling invariance is a significant merit of Gini index, as it obviates the need for optimizing normalization prior to conducting comparisons.
Transcript-level comparison
Structural similarities are often correlated with sequence and/or functional similarity [96]. Thus, in presence of known sequence and/or functional similarities, it may be reasonable to assume that reactivity profiles should span the same interval. Current normalization schemes do scale reactivity profiles such that they span the same interval from 0 to approximately 2 [39]. Hence, for cases with sequence and/or functional similarity, reactivity profiles have been compared by taking difference of normalized reactivities [23,40,58,86]. Additionally, based on models specific to the context, p-values can be calculated to characterize the significance of observed differences. Other approaches to establish statistical significance have also been used. For example, Smola et al. [86] used a modified version of a Z-factor test [97] instead of p-values to screen for sites with statistically significant differential reactivities. Z-factor is a screening coefficient that identifies nucleotides with biological variation substantially greater than technical variation. Recently, Choudhary et al. [59] have used a signal-to-noise ratio measure to quantify magnitudes of biological and technical variation. Besides these methods, comparability of profiles under conditions of sequence and/or functional similarity has been assumed when summarizing reactivity profiles for multiple RNAs via their mean. For example, mean of reactivities has been used to capture general characteristics of mRNA structure around the translation start site [26,28].
Regional comparison
Reactivity profiles often feature significant variations across the length of a transcript, indicating presence of structured and unstructured regions [28,40]. Several methods have been utilized to scan transcript regions for structural properties, which differ primarily in the structural characteristic they scan for. For example, Gini index has been applied to regions within a transcript [26,40] to identify those with high inequalities in counts/reactivities across nucleotides. Whereas Spitale et al. [40] applied Gini index to designated regions, such as UTRs and coding regions of mRNAs, Rouskin et al. [26] used it to scan rolling windows containing 50 probed nucleotides. Other studies scanned transcripts to identify regions with higher or lower reactivities. Reactivity level in a region can provide an idea about the number of base pairs in that region. To this end, the median of reactivities in a region has been used as a robust summary of regional structural characteristics [39,53,98]. Standard statistical tests such as Wilcoxon rank sum test have been used to evaluate statistical significance of differences between centers of reactivity distributions for two regions [36]. Additionally, Siegfried et al. [39] utilized Shannon entropy estimates to quantify a region’s structural properties. Entropy estimates were derived from base-pairing probabilities output by a data-directed ensemble-based secondary structure prediction algorithm (see the Section of “Secondary Structure Prediction”). Entropies are expected to be low in regions that either have well-defined structures or are predominantly single-stranded; they are expected to be high otherwise.
Open questions
Comparative analysis of SP data is in its nascent phase, and several issues are yet to be addressed. To date, the field has resorted to point summaries of structure (e.g., Gini index of counts). While statistical properties of a reactivity profile in one region/transcript have been compared with those of another, there is no consensus on the statistical property of reactivities that captures a desired structural property. Consequently, multiple metrics for quantifying regional structure have prevailed thus far. For example, measures of inequality and of non-uniformity in reactivities have both been used to characterize a high degree of structure or folding stability. At the transcriptome level, Gini index has been applied as a point summary of a transcript’s structure. However, there are several drawbacks to this index. One major issue is that it is highly influenced by outliers [99], which again underscores the importance of robust outlier detection. Another issue is that two transcripts could have vastly different reactivity profiles but the same Gini index, thus making it difficult to use it as a comparative feature. For example, consider two transcripts with the following compositions: (a) 50% of nucleotides with zero reactivity and 50% with equal and high reactivity (or more generally, 50% have high reactivity and 50% have low reactivity) and (b) 25% of nucleotides with reactivity 0.11 and 75% with reactivity 1 (or more generally, 75% have high reactivity and 25% have low reactivity). Despite their differences, both profiles result in a Gini index of 0.5.
Comparing systematic replicates
Reactivity profiles estimated from systematic replicates may provide more comprehensive insights into structure. For example, collecting and comparing information from multiple probing reagents has traditionally served as means of increasing confidence in structural inference from data [100]. Whereas such approach had been limited in applicability due to cost and labor constraints, as experiments have now become more accessible to the community, it appears to be gaining popularity [74,81,100–103]. To date, comparisons of systematic replicates have been mostly performed semi-quantitatively or via PCC [33,53]. While PCC only informs us of agreement of data, it is often desirable to merge data from systematic replicates. For example, data from systematic replicates could improve the accuracy of data-directed structure prediction if fused appropriately [103], such that correlations and systematic deviations are well characterized and accounted for. However, systematic replicates often derive from differing statistical distributions [53]. Therefore, besides scaling, systematic replicates might need more intricate normalization routines to ensure their comparable statistical properties. For this purpose, Wu et al. [104] used quantile normalization to transform reactivities in different datasets such that they follow the same distribution. Because the data throughput bottleneck has only recently been eliminated, much is yet to be done to address these emerging needs. Ensuring quantitative comparability and optimal integration of profiles from systematic replicates remains an open challenge.
SCREENING DATA FOR QUALITY
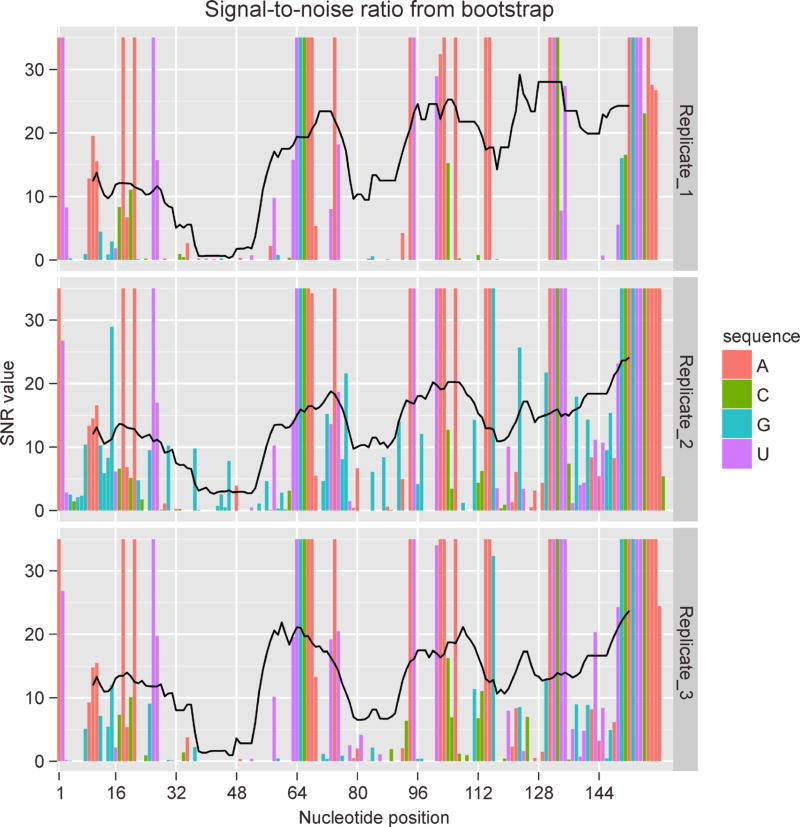
Since its days of inception, SP has moved towards large-scale transcriptome-wide and in vivo experiments. Despite significant advances, data quality remains non-uniform across the transcriptome. Data quality is primarily governed by coverage and by signal-over-background level [22,54,59]. Most studies filter out poor-quality data and draw biological insights from high-quality data subsets. Simple criteria based on a transcript’s coverage per unit length have been utilized to screen for high-quality components of a dataset. Several groups have considered coverage per unit length ⩾1 as acceptable criterion for quality [26,28,34,36], whereas others have opted for nucleotide-level coverage [22,39,40]. Several conditions have been used to optimize these criteria. For example, Smola et al. recommend nucleotide-level coverage above approximately 2,000 for high confidence in reactivity estimates [22]. This choice was guided by a desire to ensure high accuracy of data-directed structure prediction [39]. Spitale et al., on the other hand, optimized their criterion for high coverage such that transcripts meeting this criterion achieve high PCC between replicates [40]. Choudhary et al. [59] approached this from an experimental design perspective [54]. Building upon prior work on modeling SP experiments [60], they introduced a Coverage Quality Index (CQI), which quantifies the “goodness” of each nucleotide’s coverage. Given an acceptable level of variation in reactivities, a coverage level is computed for each nucleotide, which ensures (at a desired level of confidence, such as 95%) that variation is within admissible range. CQI is the ratio of the desired coverage of a nucleotide to its observed coverage. CQI < 1 is indicative of good quality while CQI > 1 is indicative of unacceptable quality. CQI calculations and other nucleotide-resolution quality measures, such as SNR, along with their visualizations from nucleotide to transcriptome level, are implemented in SEQualyzer — a quality assessment tool specialized to SP data (see Figure 2 for an example) [105]. Standardized methods for evaluating data quality as well as screening for high-quality components are essential to the maturation of this field.
Figure 2. Quality screening with SEQualyzer.
Bars represent per-nucleotide SNR and black lines represent rolling mean of per-nucleotide SNR for windows of 20 nt. SEQualyzer estimates SNR via bootstrap as described by Choudhary et al. [59]. Examination of quality profiles reveals that signal quality is good for entire RNA except a short region from nucleotides 35–53 where it is poor in all replicates. For illustration purpose, we used data for P4 – P6 domain of Tetrahymena group I intron ribozyme from Loughrey et al. [33].
SECONDARY STRUCTURE PREDICTION
Computational RNA structure prediction has been studied for several decades. Here, we focus on secondary structure prediction; readers are referred to [106] for a recent review on three-dimensional structure modeling. Typically, computational secondary structure prediction methods fall into three major categories: free energy minimization, ensemble-based prediction and comparative sequence analysis. It is worth noting that most existing methods do not allow pseudo-knots in predicted structures, as it will render the problem computationally intractable. Several solutions were developed, albeit with additional constraints on the type of considered pseudoknots [107–115].
Free energy minimization
The most widely used method for structure prediction from a single sequence aims to find the structure with minimum free energy (MFE). This method relies on the second law of thermodynamics, which states that the MFE structure is the most thermodynamically stable and the most prevalent in living cells. Free energy of a structure can be calculated based on a set of nearest-neighbor thermodynamic model (NNTM) parameters, which are obtained using optical melting experiments [116–118].
At the core of MFE prediction is a dynamic programming algorithm put forth in [119,120] and first proposed in [119,121] in the context of maximizing the number of predicted base pairs. It was subsequently extended by incorporating free energies of different structure motifs [122,123]. This algorithm has been implemented in popular software packages such as UNAFold [124], RNAstructure [125] and ViennaRNA [126]. For algorithmic details on various MFE prediction algorithms, readers are referred to the comprehensive reviews in [9,127–131].
While MFE predictions have been well studied and widely used, they often suffer from low prediction accuracies when utilizing sequence information alone, especially for long RNAs [132]. One possible reason is that the assumption that RNA folds into the MFE structure may not always hold [47]. On the other hand, RNA can interact with other biomolecules in the cell, stabilizing specific non-MFE conformations. In addition, the existing sets of NNTM parameters are neither perfect nor complete, although they have been improved over the years. The free energy of some structure motifs, such as multi-branch loops, are still not well understood and are thus obtained using simplified models [118].
In addition to the MFE structure, many programs have the option to also report a set of suboptimal structures. This is also a computational solution to the imperfect situation mentioned above. Such information is valuable for many downstream analysis applications. For example, one could generate energy dot plots from optimal and suboptimal structures, which could then be used to find frequent structure motifs [133].
Ensemble-based predictions
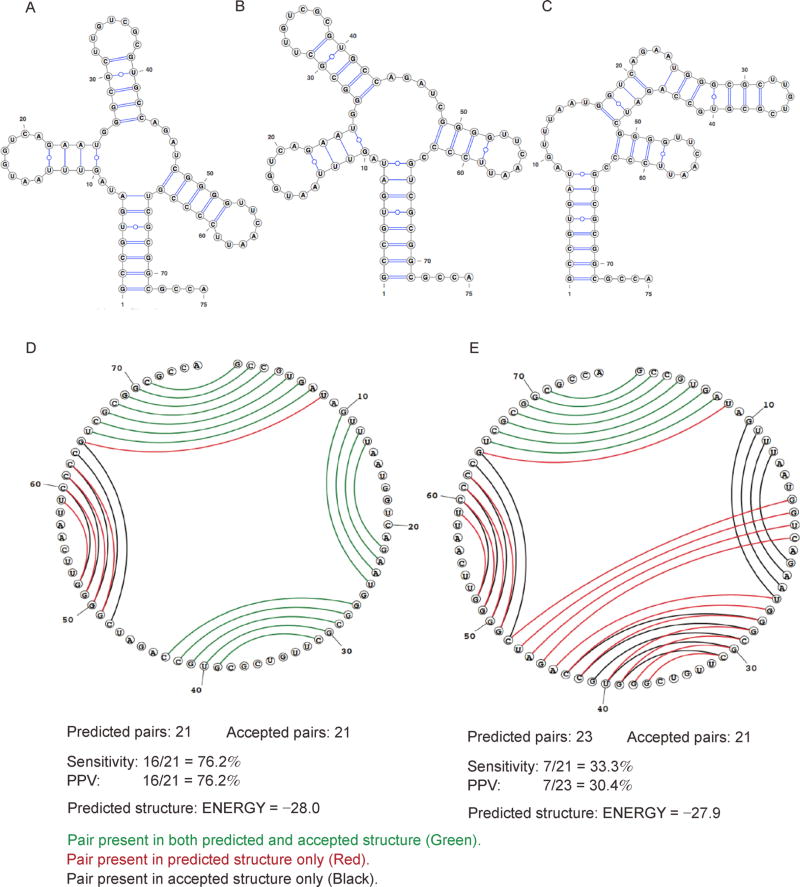
Prediction of suboptimal structures is complementary to the MFE structure. However, it is worth pointing out that suboptimal structures could be quite different than the MFE structure, even when the differences between their free energies are very small. Take the aspartic acid tRNA in yeast as an example (Figure 3). The energies of the predicted MFE structure and its closest suboptimal structure differ by 0.1 (−28 vs. −27.9), but their sensitivities differ quite a lot (76.2% vs. 33.3%); see the Subsection of “Performance Measures” for a formal definition of sensitivity. Furthermore, MFE predictions are highly sensitive in the sense that a minor change in NNTM parameters or experimental conditions might lead to a switch between the MFE and suboptimal structures; see, for example [135], for a discussion on ribosomal 30S subunit structure revealed in [136].
Figure 3. Comparison between MFE secondary structure and one of the suboptimal secondary structures for tRNA (asp), yeast.
(A) Reference (accepted) structure. (B) MFE structure. (C) Suboptimal structure. (D) Circular plot comparing the MFE structure in B to the reference structure in A. (E) Circular plot comparing the suboptimal structure in (C) to the reference structure in (A). Structures are predicted using the Fold program in RNAstructure package [125] with default parameters. Plots (A), (B) and (C) are prepared with VARNA [134]. Circular plots (D) and (E) are prepared with the CircleCompare program in RNAstructure. In (D) and (E), base pairs are indicated by lines. Pairs present in both the predicted and reference structures are in green; pairs which are present only in the predicted structure are in red; and pairs which are present only in the reference structure are in black.
A natural extension of suboptimal structures is to consider all possible structures. This can be accomplished by computing a partition function, which models the contribution of all structures weighted by their Boltzmann probabilities [62,137,138]. For a given sequence, the partition function, Q, can be calculated as
where ΔGk is the free energy of the k-th possible secondary structure, R is the gas constant and T is temperature. Furthermore, the probability of a base pair formed by nucleotides i and j can be calculated as
where the sum considers all structures that include base pair i-j.
Several algorithms that utilize the statistical nature of partition function calculations have been proposed for structure predictions. The Sfold program samples a user-specified number of structures from the Boltzmann ensemble. It then computes a centroid structure based on base-pair distances between structures [139]. Another type of approach predicts a secondary structure by maximizing the expected base-pair accuracy (MEA). Briefly, MEA seeks a structure that maximizes the sum (or weighted sum) of base-paired and single-stranded nucleotide probabilities. This objective function is inspired by an observation that base pairs with high pairing probabilities are more likely to be present in the known reference structure [137]. MEA was first proposed in CONTRA-fold, which learns a probabilistic model’s parameters from a set of known structures, based on conditional log-linear models [140]. Later, Lu et al. implemented another MEA approach that directly depends on base pairing probabilities derived from a partition function of the given sequence [141]. Related work that considers pseudo-expected accuracy is reported in [142].
It is most common for prediction algorithms to report a single optimal structure. However, some RNAs are known to have multiple functional structures in living cells. The function of these RNAs not only depends on these conformations but also on their ability to interconvert [143]. For example, riboswitches can adopt different structures upon binding a small molecule as a means of controlling gene expression [5,144]. In riboSNitches, single nucleotide polymorphisms (as analogous to binding of a small molecule in riboswitches) alter the structure of an RNA, which in turn regulates gene expression [88]. In such systems, analysis of structural ensembles would be a natural choice compared to MFE prediction.
Comparative sequence analysis
The structures of many RNAs, such as tRNAs and rRNAs, are usually highly conserved, despite possible discrepancies in their primary sequences [145]. Comparative sequence analysis aims to find a consensus structure from a set of homologous sequences [7,9,146]. This approach is highly accurate and has been widely used to study the structures of several RNAs, e.g., rRNAs [147]. Overall, three approaches currently exist to implement comparative analysis.
Align then fold aligns sequences first and then predicts the consensus structure [110,148,149]. Two of the widely used programs in this category are RNAalifold [150] and Pfold [151]. RNAalifold aims to find the minimum energy structure that is formed by a set of aligned sequences. It also supports the computation of partition function and the centroid structure, which is the structure with minimum base pair distance to other structures in the ensemble. Here, distance is defined based on base-pairing probabilities. Pfold uses a stochastic context-free grammar (SCFG) [152,153] to combine an evolutionary model of sequences with a probabilistic model for secondary structures.
Fold and align simultaneously aligns and folds input sequences [154–157]. This idea was first proposed by Sankoff [154] and utilizes a dynamic programming approach. The Sankoff algorithm has time complexity of O(n3m) for m sequences with maximum length n, and thus it is computationally expensive to apply to large inputs. By posing extra restrictions on the problem, several variations of the Sankoff algorithm with feasible complexity have been developed [156,158–160].
Fold then align predicts a structure from each input sequence, followed by alignment of structures. This method is particularly useful in scenarios where input sequences are not sufficiently conserved for direct alignment. Representatives of this method are reported in [161,162].
Although comparative sequence analysis is highly accurate, it has been successfully applied only to a limited number of RNAs with rich phylogenetic information available. This is because, analogous to many phylogenetic studies, high accuracy can only be achieved when input sequences are sufficiently divergent to contain enough co-variation information. At the same time, sequences need to be sufficiently similar in order to be aligned properly; otherwise it becomes infeasible to find a good consensus [47].
Performance measures
The accuracy of a predicted structure can be measured by comparing it to the known reference structure, where the latter is typically obtained through crystallography experiments or comparative sequence analysis [146]. Sensitivity and positive predictive value (PPV) are the two most commonly used metrics for this purpose. Sensitivity is the fraction of base pairs in the reference structure that are correctly predicted, while PPV is the fraction of correctly predicted base pairs in the predicted structure. Matthews correlation coefficient (MCC) is another widely used metric that combines sensitivity and PPV. Some studies approximate it by the geometric mean of sensitivity and PPV [146]. For partition-function-based predictions, one can measure the reliability of a prediction by calculating ensemble diversity and positional entropy, as proposed in [48].
When comparing different prediction algorithms, studies often use a benchmark dataset with multiple RNAs and compare their average performances. It is pointed out in [135] that this simple metric is not informative enough, as it is heavily biased by performances of short RNAs. To resolve this issue, this study proposed to use a “sequence-length-weighted average” (SLW-average) to replace the plain average. Intuitively, the SLW-average takes sequence length into consideration when averaging the performances of multiple RNAs.
DATA-DIRECTED SECONDARY STRUCTURE PREDICTION
In this section, we review data-directed prediction methods. While most methods seek a single optimal structure, they differ in their interpretation of SP data and/or in how they integrate it with computation.
Pseudoenergy-based approaches
The idea of converting SHAPE data into a pseudoenergy term was first proposed by Deigan et al. [82]. Serving as ad hoc energy modifications, pseudoenergies are incorporated into MFE predictions to find the structure that minimizes the sum of NNTM free energy and pseudoenergy. For a given reactivity α, its pseudoenergy is calculated using a linear-log formula, m log(1 + α) + b, where m and b are parameters determined from a training set of RNAs with known reference structures using grid search. Note that optimal values of m and b may differ quite noticeably between different data sets [33,163], as they depend on the statistical properties of the data as well as on its dynamic range. This method was first implemented in the RNAstructure package [125] and was recently included in the ViennaRNA package [48]. It is also integrated into a recent data analysis pipeline for transcriptome-wide SP experiments [164].
Deigan et al. ‘s approach has been widely used by the community and proved to significantly improve predictions for many RNAs [28,48,165,166]. For example, it has been included in RNAalifold program within the new version of the ViennaRNA package [48,167], which predicts the MFE structure and centroid structure given a set of aligned sequences. As another example, this approach is at the core of the experimental 3S technique for secondary structure determination of long non-coding RNAs [168]. 3S, also called shotgun SHAPE, is motivated by the observation that traditional thermodynamic-based prediction algorithms often have limited accuracy. It probes an entire RNA along with its shorter overlapping segments. By comparing reactivity profiles of short segments with that of the entire RNA, modular sub-domains are identified, whose structures are then predicted using Deigan et al. ‘s approach. However, it is worth mentioning that this linear-log model was not designed with biological assumptions in mind but rather in a data-driven manner [131,169]. Initially developed and optimized for SHAPE chemistry data, it is unknown how well this model fits other and newer types of SP data. In fact, Deng et al. showed, using mock-probe simulations, that Deigan et al. ‘s approach can give relatively poor performance when input data deviate from its assumed model [135]. To alleviate this problem and thereby provide broader applicability, several other methods have been developed. Most methods follow the “training and prediction” paradigm, where a model is first trained on SP data with known reference structures. The trained model is then used to direct structure prediction on new data. In an earlier work, pseudoenergies are derived from the log-likelihood ratio of a nucleotide being paired versus unpaired, given its reactivity [74]. Benchmarked on DMS data, this work uses two gamma distributions to model paired and unpaired likelihoods separately.
Motivated by the log-likelihood ratio approach in [74], the RME program converts reactivities into posterior probabilities before deriving pseudoenergies from them [104]. Pseudoenergies are then used to direct partition function calculation and to further obtain an MEA structure, in contrast to the MFE structure in [74,82]. Note that in RME, SP data are not only involved in the initial calculation of partition function but also in the post-calibration of base pairing probabilities, both in the form of posterior probabilities.
Interestingly, in [52], Eddy pointed out that Deigan et al. ‘s model actually signifies a base-pairing likelihood ratio. Furthermore, he proposed a principled and broadly applicable framework that directly derives from statistical modeling of SP data. Under the assumption that reactivities are only dependent on structural contexts (e.g., paired, unpaired, stacked, helix-end), the pseudoenergy of a reactivity for a given structural context can be derived from its likelihood. This framework has been implemented and extended in the RNAprob package for MFE prediction [135]. RNAprob investigates two different resolutions of structure context: a low resolution distinguishes between paired and unpaired nucleotides while a higher resolution further divides paired nucleotides into stacked and helix-end, resulting in three structure contexts. In RNAprob, pseudoenergies are applied once to each nucleotide, regardless of its structural context. In contrast, they are applied to every nearest-neighbor stack in [74,82,104]. Consequently, pseudoenergies are applied 0, 1 and 2 times for each unpaired, helix-end and stacked nucleotide, respectively. Note that RNAprob is implemented within the programming infrastructure of RNAstructure package [125], while providing enhanced applicability.
Similar to RNAprob, RNAsc includes pseudoenergies for all nucleotides, featuring two structure contexts (paired and unpaired) [170]. Unlike the aforementioned likelihood- and posterior-based pseudoenergy derivation, RNAsc first converts each reactivity i into pi, the probability of being unpaired. A pseudoenergy is then computed for each of the two structural contexts as β|xi – pi|, where β is a user-specified scaling factor and xi = 0 and 1 for unpaired and paired nucleotides, respectively.
RNApbfold extends the idea of pseudoenergy into perturbations in the context of the partition function, without explicitly converting SP data into ad hoc pseudoenergies [171]. Specifically, it aims to find a perturbation vector that minimizes the discrepancy between predictions and SP data. This perturbation vector applies only when SP data disagree with the thermodynamic model predictions.
Non-pseudoenergy-based approaches
While pseudoenergy-based approaches have attracted much attention in recent years, alternative data-directed prediction approaches have gained much progress. SeqFold adopts the “sample and select” strategy [172]. It first samples a set of structures from the entire structure ensemble of a given sequence, which are then clustered using Sfold [63]. Subsequently, one of the clusters is selected based on the distance of each sampled structure to the input structure profile, from which a consensus structure is further computed. The accuracy of this approach is largely determined by its ability to sample the “correct” structure. Since the number of possible structures is huge, there is no guarantee that the correct structure will be sampled. Ideas of sample and select were previously introduced in [65].
PPfold 3.0 extends the Pfold package [151] by combining phylogeny with SP data [173]. It uses i) a stochastic context-free grammars (SCFGs) to model structures; ii) a phylogeny model to compute the likelihood of input alignments; and iii) a probabilistic model to include SP data. In a more recent work, ProbFold combines SCFGs with probabilistic graphical models [174]. While SCFGs give prior knowledge over structures as in PPfold 3.0, the probabilistic graphical models account for sequence and SP data.
The above data-directed structure prediction methods all utilize SP data from a single experiment. The mutate-and-map (M2) strategy developed by the Das lab provides two-dimensional SP data [175]. For a sequence of length N, M2 performs N + 1 SP experiments: one for the wild type and others for each of the N point-mutated sequences. M2 is based on the assumption that mutation of a single nucleotide may result in local or global structural changes, which in turn result in reactivity changes. M2 data can be converted into Z-scores and then plugged into RNAstructure package as extra energy bonus for MFE structure prediction. Recently, M2 data have been used to predict multiple functional structures as well as their relative abundances in the REEFFIT algorithm [143].
Information content of SP data
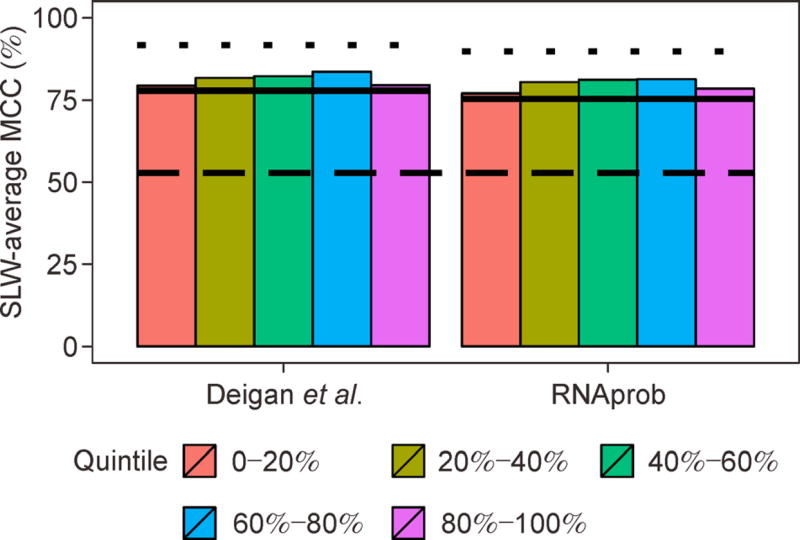
The addition of SP data to better predict RNA structure proved to be successful on a variety of RNAs. A natural question that arises is: do all reactivities contribute equally to drive structure prediction? This question was recently addressed in the context of SHAPE data [135]. Instead of evaluating the relative contribution (information content) of each single reactivity in a SHAPE profile, reactivities are divided into five equally populated subsets (a.k.a quintiles). The information content of each quintile is then quantified using a combination of leave-one-in and leave-one-out analyses. In the leave-one-in analysis, only a selected quintile is used to direct structure prediction, whereas in the leave-one-out analysis, all quintiles except for a selected one are used. Benchmarked on a set of 23 RNAs with known reference structures, this study showed that the top 20% reactivities are the major driving force in structure prediction, followed by the lowest 20%. In contrast, middle-range reactivities are less informative and have marginal contribution to improving prediction. Furthermore, the study showed, by a thought experiment, that middle-range reactivities are key to further improving predictions (Figure 4). Briefly, this experiment is done by inputting perfect information (0 and 1.6 for paired and unpaired nucleotides, respectively in [135]) to a selected quintile, while leaving reactivities in all other quintiles unchanged. Note that while it remains unknown if the conclusions reported above hold for other types of SP data, these analytical methods are readily applicable to any type of data.
Figure 4. Information content of SHAPE data.
Two data-directed structure prediction methods, Deigan et al.’s approach [82] and RNAprob [135], are tested on a set of 23 RNAs, as used in [135]. For RNAprob, the variant with two structure contexts and empirical decoder is used. Bars represent SLW-average MCC values of quintiles with perfect information. Upper dashed lines represent the performance with the entire struture profile set to perfect information. Solid lines indicate the performance with the original struture profile data and the bottom dashed line corresponds to the no-SHAPE control.
Understanding information content of SP data provides us with practical guidelines to data-directed predictions. For example, one may choose to be selective and use reactivities that are most informative while ignoring reactivities that are ambiguous. In addition, such insights facilitate new models with better discriminative power, which can potentially reduce the number of less informative reactivities and in turn improve structure prediction.
Open questions
Structure prediction has been greatly advanced by the rapid development of SP technologies. Studies have shown that data-directed predictions often lead to better performance. However, it is worth noting that the extent of improvement in prediction accuracy varies substantially among RNAs and appears to be sequence dependent. It sometimes can have minor or even negative effects on resulting predictions [135,176]. On the other hand, regardless of the availability of various strategies to incorporate SP data, to date, no method universally outperforms all others [135]. As such, further improvement is desired and can be possibly approached from the following angles: i) Pseudoenergy-based methods have solid performance in practice. We anticipate that performances may be improved with pseudoenergy derivation models that are more biologically and statistically meaningful. ii) As in [172–174], pseudoenergies are not the only way to integrate data and computation. Hence, it will be interesting to explore alternative strategies for modeling SP data. iii) The recent development of novel transcriptome-wide methods to probe RNA structures experimentally presents us with massive data of unprecedented complexity and diversity. These data, when judiciously combined, have the potential to lead to better performances. However, integrating information from multiple data sources within current algorithms is challenging due to their complex statistical dependencies. A first attempt in this direction is reported in [103]. Availability of new probabilistic methods, such as RNAprob and ProbFold, will certainly propel efforts in this direction.
SOFTWARE INFRASTRUCTURE
The rapid development of SP has generated massive amounts of diverse data. As for many other sequencing-based studies, tools for data sharing and analysis are two major needs. Here, we review recent progress towards addressing these needs.
Databases and visualization tools
Structure Surfer [177], RNAex [178] and FoldAtlas [179] are three recent tools for data sharing, which support experiments such as DMS-Seq [26], structure-Seq [28], icSHAPE [40], PARS [55] and ds/ssRNA-Seq [180]. In addition, they provide a set of useful inspection and visualization tools. Specifically, Structure Surfer allows to visually compare different data sets, while RNAex and FoldAtlas support visualization of predicted secondary structures. RNAex also supports annotated RNA editing, RNA modifications and SNP sites in predicted structures. A recent tool, SEQualyzer, specialized to SP data quality screening, is reported in [105].
Data preprocessing
Data analysis usually entails five major steps: i) Data cleaning removes adapters, PCR duplicates or other undesired sequences. ii) Read alignment maps reads to a reference set of transcripts. iii) Count summarization at nucleotide level. iv) Reactivity calculation. v) Data-directed secondary structure prediction. Steps ii), iii) and vi) are routinely featured in all platforms, while steps i) and v) are supported by a subset of tools.
Specialized analysis pipelines adjoin most recent SP protocols. Spats processes reads from SHAPE-Seq experiments [33], implementing a model-based maximum-likelihood estimation approach to calculate reactivities [60,80,181]. ShapeMapper and SuperFold are two distinct analysis pipelines for SHAPE-MaP experiments [39]. ShapeMapper converts raw sequencing reads into mutational profiles, which are then used as input to SuperFold for secondary structure prediction. They also facilitate de novo identification of well-defined and stable structure regions. Other specialized pipelines include Mod-Seeker [35], MAPseeker [38] and icSHAPE [40].
Tools designed with broader applicability in mind include StructureFold [164], RSF [182] and PROBer [79]. Deployed as part of the Galaxy platform [183], StructureFold supports conversion of reads into reactivities and structure prediction, each of which is available as a separate module. It implements a reactivity calculation method proposed in [28]. Another modular pipeline, RNA Structure Framework (RSF), supports similar functionality as well as data cleaning. Additionally, it offers flexibility in choosing from a number of reactivity calculation methods [26,28] and normalization strategies (2%–8%, 90% winsorizing and box plot). In contrast to the former two, PROBer is a closed-box solution that implements the statistical model-based approach of Li et al. (see the Section of “Estimation of Structural Profile”). PROBer is also unique in that it is applicable to a wider range of diverse experiments. In particular, it encompasses a number of recent non-SP techniques that share the following common workflow with SP: i) Chemical modification of nucleotides encodes a signal of interest. ii) The signal is detected via RT termination. iii) The cDNA products of RT are sequenced and mapped to estimate modification intensities per nucleotide. Examples of biological signals that can be studied under this framework include protein-RNA interactions [184,185], post-transcriptional RNA modifications [186–192] and sites of noncanonical RNA structure motifs such as G-quadruplexes [42]. Such unified view not only lends itself to shared analysis pipelines but also alludes to plausible commonalities in downstream comparative and integrative analysis challenges. Methods that approach these emerging challenges from a broader perspective may reach and serve a wider research community.
CONCLUSION
We reviewed current practices and emerging questions in comparative and integrative analysis of SP data. However, there are other emerging applications that we have not touched upon, which are timely as they directly leverage the new wealth of information. For example, SHAPE-based alignment has been recently shown to have comparable accuracy to traditional sequence-based alignment [167]. Alignment can be further improved when combining sequence information with SHAPE data. In addition, SP data-directed partition function can be used to calculate Shannon entropy, which in turn is useful in discovering well-defined RNA structures [39]. These and additional timely applications are described in a recent review [53]. Another exciting direction is the emergence of a new class of RNA structure experiments, which identify long-range and inter-molecular base-pairing interactions [193–198]. Integrating this type of information with SP data and with structure prediction algorithms is likely to pose newer challenges and spur dedicated methods development.
The advent of SP techniques has greatly expanded our capacity to understand structures of various RNAs and to deduce their functional roles. Propelled by these advances, we are standing in an era of large-scale data with increasing diversity and complexity, which in turn poses significant challenges in data interpretation and analysis. To maximize the potential of these datasets, there is a need to develop methods for accurate data interpretation, leveraging intrinsic statistical properties of an SP protocol. Additionally, there is a need to better suit methodology for comparative analysis to discover biological patterns of interest as well as methodology for characterizing SP information content to better utilize data within structure prediction algorithms.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grant (No. HG006860). We thank Chun Kit Kwok and Aviran lab members — Mirko Ledda, Sana Vaziri, Hua Li and Rob Gysel — for insightful comments during the preparation of this manuscript.
Footnotes
This article is dedicated to the Special Collection of Synthetic Biology, Aiming for Quantitative Control of Cellular Systems (Eds. Cheemeng Tan and Haiyan Liu).
COMPLIANCE WITH ETHICS GUIDELINES
Krishna Choudhary, Fei Deng and Sharon Aviran declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Strobel EJ, Watters KE, Loughrey D, Lucks JB. RNA systems biology: uniting functional discoveries and structural tools to understand global roles of RNAs. Curr. Opin. Biotechnol. 2016;39:182–191. doi: 10.1016/j.copbio.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Hashimi HM. Structural biology: aerial view of the HIV genome. Nature. 2009;460:696–698. doi: 10.1038/460696a. [DOI] [PubMed] [Google Scholar]
- 7.Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 8.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- 9.Mathews DH, Moss WN, Turner DH. Folding and finding RNA secondary structure. Cold Spring Harb. Perspect. Biol. 2010;2:a003665. doi: 10.1101/cshperspect.a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weeks KM. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 2010;20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brow DA, Noller HF. Protection of ribosomal RNA from kethoxal in polyribosomes: implication of specific sites in ribosome function. J. Mol. Biol. 1983;163:27–46. doi: 10.1016/0022-2836(83)90028-1. [DOI] [PubMed] [Google Scholar]
- 14.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr. Opin. Chem. Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Singer B. All oxygens in nucleic acids react with carcinogenic ethylating agents. Nature. 1976;264:333–339. doi: 10.1038/264333a0. [DOI] [PubMed] [Google Scholar]
- 16.Fritz JJ, Lewin A, Hauswirth W, Agarwal A, Grant M, Shaw L. Development of hammerhead ribozymes to modulate endogenous gene expression for functional studies. Methods. 2002;28:276–285. doi: 10.1016/s1046-2023(02)00233-5. [DOI] [PubMed] [Google Scholar]
- 17.Lindell M, Romby P, Wagner EGH. Lead(II) as a probe for investigating RNA structure in vivo. RNA. 2002;8:534–541. doi: 10.1017/s1355838201020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindell M, Brännvall M, Wagner EGH, Kirsebom LA. Lead(II) cleavage analysis of RNase P RNA in vivo. RNA. 2005;11:1348–1354. doi: 10.1261/rna.2590605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson KA, Merino EJ, Weeks KM. Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 21.Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods. 2017;14:75–82. doi: 10.1038/nmeth.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 2015;10:1643–1669. doi: 10.1038/nprot.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watters KE, Yu AM, Strobel EJ, Settle AH, Lucks JB. Characterizing RNA structures in vitro and in vivo with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Methods. 2016;103:34–48. doi: 10.1016/j.ymeth.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulsen LD, Kielpinski LJ, Salama SR, Krogh A, Vinther J. SHAPE Selection (SHAPES) enrich for RNA structure signal in SHAPE sequencing-based probing data. RNA. 2015;21:1042–1052. doi: 10.1261/rna.047068.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hector RD, Burlacu E, Aitken S, Le Bihan T, Tuijtel M, Zaplatina A, Cook AG, Granneman S. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 2014;42:12138–12154. doi: 10.1093/nar/gku815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. Determination of in vivo RNA structure in low-abundance transcripts. Nat. Commun. 2013;4:2971. doi: 10.1038/ncomms3971. [DOI] [PubMed] [Google Scholar]
- 28.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2013;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 29.Ding Y, Kwok CK, Tang Y, Bevilacqua PC, Assmann SM. Genome-wide profiling of in vivo RNA structure at single-nucleotide resolution using structure-seq. Nat. Protoc. 2015;10:1050–1066. doi: 10.1038/nprot.2015.064. [DOI] [PubMed] [Google Scholar]
- 30.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc. Natl. Acad. Sci. USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loughrey D, Watters KE, Settle AH, Lucks JB. SHAPE-Seq 2.0: systematic optimization and extension of high-throughput chemical probing of RNA secondary structure with next generation sequencing. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku909. 000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan Y, Qu K, Ouyang Z, Chang HY. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat. Protoc. 2013;8:849–869. doi: 10.1038/nprot.2013.045. [DOI] [PubMed] [Google Scholar]
- 35.Talkish J, May G, Lin Y, Woolford JL, Jr, McManus CJ. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA. 2014;20:713–720. doi: 10.1261/rna.042218.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Incarnato D, Neri F, Anselmi F, Oliviero S. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol. 2014;15:491. doi: 10.1186/s13059-014-0491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kielpinski LJ, Vinther J. Massive parallel-sequencing-based hydroxyl radical probing of RNA accessibility. Nucleic Acids Res. 2014;42:e70. doi: 10.1093/nar/gku167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seetin MG, Kladwang W, Bida JP, Das R. RNA Folding: Methods and Protocols. New York: Humana Press; 2014. Massively parallel RNA chemical mapping with a reduced bias MAP-seq protocol; pp. 95–117. [DOI] [PubMed] [Google Scholar]
- 39.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat. Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung J-W, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwok CK, Sahakyan AB, Balasubramanian S. Structural analysis using SHALiPE to reveal RNA G-quadruplex formation in human precursor microRNA. Angew. Chem. Int. Ed. Engl. 2016;55:8958–8961. doi: 10.1002/anie.201603562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwok CK, Marsico G, Sahakyan AB, Chambers VS, Balasubramanian S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016;13:841–844. doi: 10.1038/nmeth.3965. [DOI] [PubMed] [Google Scholar]
- 43.Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 2015;40:221–232. doi: 10.1016/j.tibs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Lu Z, Chang HY. Decoding the RNA structurome. Curr. Opin. Struct. Biol. 2016;36:142–148. doi: 10.1016/j.sbi.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwok CK. Dawn of the in vivo RNA structurome and interactome. Biochem. Soc. Trans. 2016;44:1395–1410. doi: 10.1042/BST20160075. [DOI] [PubMed] [Google Scholar]
- 46.Kubota M, Chan D, Spitale RC. RNA structure: merging chemistry and genomics for a holistic perspective. BioEssays. 2015;37:1129–1138. doi: 10.1002/bies.201300146. [DOI] [PubMed] [Google Scholar]
- 47.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lorenz R, Luntzer D, Hofacker IL, Stadler PF, Wolfinger MT. SHAPE directed RNA folding. Bioinformatics. 2015;32:145–147. doi: 10.1093/bioinformatics/btv523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 50.Lavery R, Pullman A. A new theoretical index of biochemical reactivity combining steric and electrostatic factors: an application to yeast tRNAPhe. Biophys. Chem. 1984;19:171–181. doi: 10.1016/0301-4622(84)85017-6. [DOI] [PubMed] [Google Scholar]
- 51.McGinnis JL, Dunkle JA, Cate JH, Weeks KM. The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eddy SR. Computational analysis of conserved RNA secondary structure in transcriptomes and genomes. Annu. Rev. Biophys. 2014;43:433–456. doi: 10.1146/annurev-biophys-051013-022950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kutchko KM, Laederach A. Transcending the prediction paradigm: novel applications of SHAPE to RNA function and evolution. WIREs RNA. 2016;8:e1374. doi: 10.1002/wrna.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aviran S, Pachter L. Rational experiment design for sequencing-based RNA structure mapping. RNA. 2014;20:1864–1877. doi: 10.1261/rna.043844.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ritz J, Martin JS, Laederach A. Evaluating our ability to predict the structural disruption of RNA by SNPs. BMC Genomics. 2012;13:S6. doi: 10.1186/1471-2164-13-S4-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watters KE, Abbott TR, Lucks JB. Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Res. 2016;44:e12. doi: 10.1093/nar/gkv879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bai Y, Tambe A, Zhou K, Doudna JA. RNA-guided assembly of Rev-RRE nuclear export complexes. eLife. 2014;3:e03656. doi: 10.7554/eLife.03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choudhary K, Shih NP, Deng F, Ledda M, Li B, Aviran S. Metrics for rapid quality control in RNA structure probing experiments. Bioinformatics. 2016;32:3575–3583. doi: 10.1093/bioinformatics/btw501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aviran S, Lucks JB, Pachter L. RNA structure characterization from chemical mapping experiments; the 49th Annual Allerton Conference on Communication, Control, and Computing; 2011. pp. 1743–1750. [Google Scholar]
- 61.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 63.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rogers E, Heitsch C. New insights from cluster analysis methods for RNA secondary structure prediction. Wiley Interdiscip. Rev. RNA. 2016;7:278–294. doi: 10.1002/wrna.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quarrier S, Martin JS, Davis-Neulander L, Beauregard A, Laederach A. Evaluation of the information content of RNA structure mapping data for secondary structure prediction. RNA. 2010;16:1108–1117. doi: 10.1261/rna.1988510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo JU, Bartel DP. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016;353:aaf5371. doi: 10.1126/science.aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anders S, Huber W. Differential expression of RNA-Seq data at the gene level-the DESeq package. Heidelberg: European Molecular Biology Laboratory; 2012. [Google Scholar]
- 71.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leamy KA, Assmann SM, Mathews DH, Bevilacqua PC. Bridging the gap between in vitro and in vivo RNA folding. Q. Rev. Biophys. 2016;49:e10. doi: 10.1017/S003358351600007X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu X, Wu Y, Lu ZJ, Yip KY. Analysis of sequencing data for probing RNA secondary structures and protein- RNA binding in studying posttranscriptional regulations. Brief. Bioinform. 2015;17:1032–1043. doi: 10.1093/bib/bbv106. [DOI] [PubMed] [Google Scholar]
- 74.Cordero P, Kladwang W, VanLang CC, Das R. Quantitative dimethyl sulfate mapping for automated RNA secondary structure inference. Biochemistry. 2012;51:7037–7039. doi: 10.1021/bi3008802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee B, Flynn RA, Kadina A, Guo JK, Kool ET, Chang HY. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA. 2016 doi: 10.1261/rna.058784.116. rna.058784.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 77.Sorefan K, Pais H, Hall AE, Kozomara A, Griffiths-Jones S, Moulton V, Dalmay T. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence. 2012;3:4. doi: 10.1186/1758-907X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li B, Tambe A, Aviran S, Pachter L. Prober: a general toolkit for analyzing sequencing-based ‘toeprinting’ assays. bioRxiv. 2016:063107. doi: 10.1016/j.cels.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aviran S, Trapnell C, Lucks JB, Mortimer SA, Luo S, Schroth GP, Doudna JA, Arkin AP, Pachter L. Modeling and automation of sequencing-based characterization of RNA structure. Proc. Natl. Acad. Sci. USA. 2011;108:11069–11074. doi: 10.1073/pnas.1106541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Selega A, Sirocchi C, Iosub I, Granneman S, Sanguinetti G. Robust statistical modeling improves sensitivity of high-throughput RNA structure probing experiments. Nat. Methods. 2017;14:83–89. doi: 10.1038/nmeth.4068. [DOI] [PubMed] [Google Scholar]
- 82.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sloma MF, Mathews DH. Improving RNA secondary structure prediction with structure mapping data. Methods Enzymol. 2015;553:91–114. doi: 10.1016/bs.mie.2014.10.053. [DOI] [PubMed] [Google Scholar]
- 84.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unan-notated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smola MJ, Calabrese JM, Weeks KM. Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry. 2015;54:6867–6875. doi: 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. USA. 2016;113:10322–10327. doi: 10.1073/pnas.1600008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Solem AC, Halvorsen M, Ramos SB, Laederach A. The potential of the riboSNitch in personalized medicine. Wiley Interdiscip. Rev. RNA. 2015;6:517–532. doi: 10.1002/wrna.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wan Y, Qu K, Ouyang Z, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY. Genome-wide measurement of RNA folding energies. Mol. Cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Righetti F, Nuss AM, Twittenhoff C, Beele S, Urban K, Will S, Bernhart SH, Stadler PF, Dersch P, Narberhaus F. Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA. 2016;113:7237–7242. doi: 10.1073/pnas.1523004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Corley M, Solem A, Qu K, Chang HY, Laederach A. Detecting riboSNitches with RNA folding algorithms: a genome-wide benchmark. Nucleic Acids Res. 2015;43:1859–1868. doi: 10.1093/nar/gkv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Abdullah MB. On a robust correlation coefficient. Statistician. 1990;39:455–460. [Google Scholar]
- 93.Goodwin LD, Leech NL. Understanding correlation: factors that affect the size of r. J. Exp. Educ. 2006;74:249–266. [Google Scholar]
- 94.Müller R, Büttner P. A critical discussion of intraclass correlation coefficients. Stat. Med. 1994;13:2465–2476. doi: 10.1002/sim.4780132310. [DOI] [PubMed] [Google Scholar]
- 95.Gastwirth JL. The estimation of the Lorenz curve and Gini index. Rev. Econ. Stat. 1972;54:306–316. [Google Scholar]
- 96.Eddy SR, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang J-H, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 98.Pollom E, Dang KK, Potter EL, Gorelick RJ, Burch CL, Weeks KM, Swanstrom R. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog. 2013;9:e1003294. doi: 10.1371/journal.ppat.1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cowell FA, Victoria-Feser M-P. Robustness properties of inequality measures. Econometrica. 1996;64:77–101. [Google Scholar]
- 100.Liang R, Kierzek E, Kierzek R, Turner DH. Comparisons between chemical mapping and binding to isoenergetic oligonucleotide microarrays reveal unexpected patterns of binding to the Bacillus subtilis RNase P RNA specificity domain. Biochemistry. 2010;49:8155–8168. doi: 10.1021/bi100286n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hawkes EJ, Hennelly SP, Novikova IV, Irwin JA, Dean C, Sanbonmatsu KY. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Reports. 2016;16:3087–3096. doi: 10.1016/j.celrep.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xue Z, Hennelly S, Doyle B, Gulati AA, Novikova IV, Sanbonmatsu KY, Boyer LA. A G-rich motif in the lncRNA braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol. Cell. 2016;64:37–50. doi: 10.1016/j.molcel.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rice GM, Leonard CW, Weeks KM. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA. 2014;20:846–854. doi: 10.1261/rna.043323.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu Y, Shi B, Ding X, Liu T, Hu X, Yip KY, Yang ZR, Mathews DH, Lu ZJ. Improved prediction of RNA secondary structure by integrating the free energy model with restraints derived from experimental probing data. Nucleic acids res. 2015;43:7247–7259. doi: 10.1093/nar/gkv706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choudhary K, Ruan L, Deng F, Shih N, Aviran S. SEQualyzer: interactive tool for quality control and exploratory analysis of high-throughput RNA structural profiling data. Bioinformatics. 2016:btw627. doi: 10.1093/bioinformatics/btw627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rother K, Rother M, Skiba P, Bujnicki JM. RNA Sequence, Structure, and Function: Computational and Bioinformatic Methods. New York: Humana Press; 2014. Automated modeling of RNA 3D structure; pp. 395–415. [DOI] [PubMed] [Google Scholar]
- 107.Tabaska JE, Cary RB, Gabow HN, Stormo GD. An RNA folding method capable of identifying pseudoknots and base triples. Bioinformatics. 1998;14:691–699. doi: 10.1093/bioinformatics/14.8.691. [DOI] [PubMed] [Google Scholar]
- 108.Rivas E, Eddy SR. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol. 1999;285:2053–2068. doi: 10.1006/jmbi.1998.2436. [DOI] [PubMed] [Google Scholar]
- 109.Lyngsø RB, Pedersen CN. RNA pseudoknot prediction in energy-based models. J. Comput. Biol. 2000;7:409–427. doi: 10.1089/106652700750050862. [DOI] [PubMed] [Google Scholar]
- 110.Ruan J, Stormo GD, Zhang W. An iterated loop matching approach to the prediction of RNA secondary structures with pseudoknots. Bioinformatics. 2004;20:58–66. doi: 10.1093/bioinformatics/btg373. [DOI] [PubMed] [Google Scholar]
- 111.Reeder J, Giegerich R. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics. 2004;5:104. doi: 10.1186/1471-2105-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ren J, Rastegari B, Condon A, Hoos HH. HotKnots: heuristic prediction of RNA secondary structures including pseudo-knots. RNA. 2005;11:1494–1504. doi: 10.1261/rna.7284905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cao S, Chen S-J. Predicting RNA pseudoknot folding thermodynamics. Nucleic Acids Res. 2006;34:2634–2652. doi: 10.1093/nar/gkl346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reeder J, Steffen P, Giegerich R. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res. 2007;35:W320–W324. doi: 10.1093/nar/gkm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sato K, Kato Y, Hamada M, Akutsu T, Asai K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011;27:85–i93. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Andronescu M, Condon A, Turner DH, Mathews DH. RNA Sequence, Structure, and Function: Computational and Bioinformatic Methods. New York: Humana Press; 2014. The determination of RNA folding nearest neighbor parameters; pp. 45–70. [DOI] [PubMed] [Google Scholar]
- 117.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded Nearest-Neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 118.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 119.Nussinov R, Pieczenik G, Griggs JR, Kleitman DJ. Algorithms for loop matchings. SIAM J. Appl. Math. 1978;35:68–82. [Google Scholar]
- 120.Waterman MS, Smith TF. RNA secondary structure: a complete mathematical analysis. Math. Biosci. 1978;42:257–266. [Google Scholar]
- 121.Nussinov R, Jacobson AB. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc. Natl. Acad. Sci. USA. 1980;77:6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zuker M, Sankoff D. RNA secondary structures and their prediction. Bull. Math. Biol. 1984;46:591–621. [Google Scholar]
- 124.Markham NR, Zuker M. Bioinformatics: Structure, Function and Applications. New York: Humana Press; 2008. UNAFold; pp. 3–31. [Google Scholar]
- 125.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA package 2.0. Algorithms. Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Eddy SR. How do RNA folding algorithms work? Nat. Biotechnol. 2004;22:1457–1458. doi: 10.1038/nbt1104-1457. [DOI] [PubMed] [Google Scholar]
- 128.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 129.Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Bridging the gap in RNA structure prediction. Curr. Opin. Struct. Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 130.Bai Y, Dai X, Harrison A, Johnston C, Chen M. Toward a next-generation atlas of RNA secondary structure. Brief. Bioinform. 2016;17:63–77. doi: 10.1093/bib/bbv026. [DOI] [PubMed] [Google Scholar]
- 131.Ge P, Zhang S. Computational analysis of RNA structures with chemical probing data. Methods. 2015;79–80:60–66. doi: 10.1016/j.ymeth.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Doshi KJ, Cannone JJ, Cobaugh CW, Gutell RR. Evaluation of the suitability of free-energy minimization using nearest-neighbor energy parameters for RNA secondary structure prediction. BMC Bioinformatics. 2004;5:105. doi: 10.1186/1471-2105-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 134.Darty K, Denise A, Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Deng F, Ledda M, Vaziri S, Aviran S. Data-directed RNA secondary structure prediction using probabilistic modeling. RNA. 2016;22:1109–1119. doi: 10.1261/rna.055756.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McGinnis JL, Liu Q, Lavender CA, Devaraj A, McClory SP, Fredrick K, Weeks KM. In-cell SHAPE reveals that free 30S ribosome subunits are in the inactive state. Proc. Natl. Acad. Sci. USA. 2015;112:2425–2430. doi: 10.1073/pnas.1411514112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bernhart SH, Hofacker IL, Stadler PF. Local RNA base pairing probabilities in large sequences. Bioinformatics. 2006;22:614–615. doi: 10.1093/bioinformatics/btk014. [DOI] [PubMed] [Google Scholar]
- 139.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Do CB, Woods DA, Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006;22:e90–e98. doi: 10.1093/bioinformatics/btl246. [DOI] [PubMed] [Google Scholar]
- 141.Lu ZJ, Gloor JW, Mathews DH. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA. 2009;15:1805–1813. doi: 10.1261/rna.1643609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hamada M, Sato K, Asai K. Prediction of RNA secondary structure by maximizing pseudo-expected accuracy. BMC Bioinformatics. 2010;11:586. doi: 10.1186/1471-2105-11-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cordero P, Das R. Rich RNA structure landscapes revealed by mutate-and-map analysis. PLoS Comput. Biol. 2015;11:e1004473. doi: 10.1371/journal.pcbi.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012;4:a003566. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Parsch J, Braverman JM, Stephan W. Comparative sequence analysis and patterns of covariation in RNA secondary structures. Genetics. 2000;154:909–921. doi: 10.1093/genetics/154.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gardner PP, Giegerich R. A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics. 2004;5:140. doi: 10.1186/1471-2105-5-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rupert L, Stefan G, Gerhard S. ConStruct: a tool for thermodynamic controlled prediction of conserved secondary structure. Nucleic Acids Res. 1999;27:4208–4217. doi: 10.1093/nar/27.21.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 150.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sakakibara Y, Brown M, Hughey R, Mian IS, Sjölander K, Underwood RC, Haussler D. Stochastic context-free grammars for tRNA modeling. Nucleic Acids Res. 1994;22:5112–5120. doi: 10.1093/nar/22.23.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Knudsen B, Hein J. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics. 1999;15:446–454. doi: 10.1093/bioinformatics/15.6.446. [DOI] [PubMed] [Google Scholar]
- 154.Sankoff D. Simultaneous solution of the RNA folding, alignment and protosequence problems. SIAM J. Appl. Math. 1985;45:810–825. [Google Scholar]
- 155.Havgaard JH, Lyngsø RB, Stormo GD, Gorodkin J. Pairwise local structural alignment of RNA sequences with sequence similarity less than 40% Bioinformatics. 2005;21:1815–1824. doi: 10.1093/bioinformatics/bti279. [DOI] [PubMed] [Google Scholar]
- 156.Mathews DH, Turner DH. Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 2002;317:191–203. doi: 10.1006/jmbi.2001.5351. [DOI] [PubMed] [Google Scholar]
- 157.Harmanci AO, Sharma G, Mathews DH. Efficient pairwise RNA structure prediction using probabilistic alignment constraints in Dynalign. BMC Bioinformatics. 2007;8:130. doi: 10.1186/1471-2105-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gorodkin J, Heyer LJ, Stormo GD. Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res. 1997;25:3724–3732. doi: 10.1093/nar/25.18.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Perriquet O, Touzet H, Dauchet M. Finding the common structure shared by two homologous RNAs. Bioinformatics. 2003;19:108–116. doi: 10.1093/bioinformatics/19.1.108. [DOI] [PubMed] [Google Scholar]
- 160.Hofacker IL, Bernhart SH, Stadler PF. Alignment of RNA base pairing probability matrices. Bioinformatics. 2004;20:2222–2227. doi: 10.1093/bioinformatics/bth229. [DOI] [PubMed] [Google Scholar]
- 161.Hochsmann M, Toller T, Giegerich R, Kurtz S. Local similarity in RNA secondary structures; Proceedings of the IEEE Bioinformatics Conference; 2003. pp. 159–168. [PubMed] [Google Scholar]
- 162.Siebert S, Backofen R. MARNA: a server for multiple alignment of RNAs; Proceedings of the German Conference on Bioinformatics; 2003. pp. 135–140. [Google Scholar]
- 163.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. USA. 2013;110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tang Y, Bouvier E, Kwok CK, Ding Y, Nekrutenko A, Bevilacqua PC, Assmann SM. StructureFold: genome-wide RNA secondary structure mapping and reconstruction in vivo. Bioinformatics. 2015;31:2668–2675. doi: 10.1093/bioinformatics/btv213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Montaseri S, Ganjtabesh M, Zare-Mirakabad F. Evolutionary algorithm for RNA secondary structure prediction based on simulated SHAPE data. PLoS One. 2016;11:e0166965. doi: 10.1371/journal.pone.0166965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lavender CA, Lorenz R, Zhang G, Tamayo R, Hofacker IL, Weeks KM. Model-free RNA sequence and structure alignment informed by SHAPE probing reveals a conserved alternate secondary structure for 16S rRNA. PLoS Comput. Biol. 2015;11:e1004126. doi: 10.1371/journal.pcbi.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Novikova IV, Dharap A, Hennelly SP, Sanbonmatsu KY. 3S: shotgun secondary structure determination of long non-coding RNAs. Methods. 2013;63:170–177. doi: 10.1016/j.ymeth.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 169.Lorenz R, Wolfinger MT, Tanzer A, Hofacker IL. Predicting RNA secondary structures from sequence and probing data. Methods. 2016;103:86–98. doi: 10.1016/j.ymeth.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 170.Zarringhalam K, Meyer MM, Dotu I, Chuang JH, Clote P. Integrating chemical footprinting data into RNA secondary structure prediction. PLoS One. 2012;7:e45160. doi: 10.1371/journal.pone.0045160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Washietl S, Hofacker IL, Stadler PF, Kellis M. RNA folding with soft constraints: reconciliation of probing data and thermodynamic secondary structure prediction. Nucleic Acids Res. 2012;40:4261–4272. doi: 10.1093/nar/gks009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ouyang Z, Snyder MP, Chang HY. SeqFold: genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data. Genome Res. 2013;23:377–387. doi: 10.1101/gr.138545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sükösd Z, Knudsen B, Kjems J, Pedersen CN. PPfold 3.0: fast RNA secondary structure prediction using phylogeny and auxiliary data. Bioinformatics. 2012;28:2691–2692. doi: 10.1093/bioinformatics/bts488. [DOI] [PubMed] [Google Scholar]
- 174.Sahoo S, Świtnicki MP, Pedersen JS. ProbFold: a probabilistic method for integration of probing data in RNA secondary structure prediction. Bioinformatics. 2016;32:2626–2635. doi: 10.1093/bioinformatics/btw175. [DOI] [PubMed] [Google Scholar]
- 175.Kladwang W, VanLang CC, Cordero P, Das R. A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat. Chem. 2011;3:954–962. doi: 10.1038/nchem.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sükösd Z, Swenson MS, Kjems J, Heitsch CE. Evaluating the accuracy of SHAPE-directed RNA secondary structure predictions. Nucleic Acids Res. 2013;41:2807–2816. doi: 10.1093/nar/gks1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Berkowitz ND, Silverman IM, Childress DM, Kazan H, Wang L-S, Gregory BD. A comprehensive database of high-throughput sequencing-based RNA secondary structure probing data (Structure Surfer) BMC Bioinformatics. 2016;17:215. doi: 10.1186/s12859-016-1071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wu Y, Qu R, Huang Y, Shi B, Liu M, Li Y, Lu ZJ. RNAex: an RNA secondary structure prediction server enhanced by high-throughput structure-probing data. Nucleic Acids Res. 2016;44:W294–W301. doi: 10.1093/nar/gkw362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Norris M, Cheema J, Kwok CK, Hartley M, Morris RJ, Aviran S, Ding Y. FoldAtlas: a repository for genome-wide RNA structure probing data. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw611. DOI: 10.1093/bioinformatics/btw611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li F, Zheng Q, Vandivier LE, Willmann MR, Chen Y, Gregory BD. Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell. 2012;24:4346–4359. doi: 10.1105/tpc.112.104232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mortimer SA, Trapnell C, Aviran S, Pachter L, Lucks JB. SHAPE-Seq: high-throughput RNA structure analysis. Curr Protoc Chem Biol. 2012;4:275–297. doi: 10.1002/9780470559277.ch120019. [DOI] [PubMed] [Google Scholar]
- 182.Incarnato D, Neri F, Anselmi F, Oliviero S. RNA structure framework: automated transcriptome-wide reconstruction of RNA secondary structures from highthroughput structure probing data. Bioinformatics. 2015;32:459–461. doi: 10.1093/bioinformatics/btv571. [DOI] [PubMed] [Google Scholar]
- 183.Goecks J, Nekrutenko A, Taylor J. The Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat. Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 188.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013;9:e1003602. doi: 10.1371/journal.pgen.1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:43–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Incarnato D, Anselmi F, Morandi E, Neri F, Maldotti M, Rapelli S, Parlato C, Basile G, Oliviero S. High-throughput single-base resolution mapping of RNA 2′-O-methylated residues. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw810. doi: 10.1093/nar/gkw810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. USA. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ramani V, Qiu R, Shendure J. High-throughput determination of RNA structure by proximity ligation. Nat. Biotechnol. 2015;33:980–984. doi: 10.1038/nbt.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D’Ambrogio A, Luscombe NM, Ule J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519:491–494. doi: 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sharma E, Sterne-Weiler T, O’Hanlon D, Blencowe BJ. Global mapping of human RNA-RNA interactions. Mol. Cell. 2016;62:618–626. doi: 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 197.Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016;165:1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, Boon K-L, Tapsin S, Chan Y-S, Tan C-P, Sim AY, et al. in vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol. Cell. 2016;62:603–617. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]